Significance
Seeds are the main human food source and germination characteristics are critical in modern agriculture. For plants, seeds allow propagation over long distances and time periods and provide protection of progeny from hostile environments. For germination and seedling establishment the embryo relies fully on its stored energy resources. We report a sharp transition in mitochondrial metabolic activity and thiol redox status that coincides with seed rehydration. Hundreds of Cys-based redox switches are operated, providing early and direct metabolic regulation, long before gene expression-based control is established to underpin hormonal checkpoints. Seeds lacking proteins of the mitochondrial thiol redox machinery use their resources less efficiently, suggesting that the early reset of thiol redox status orchestrates metabolic efficiency for successful germination.
Keywords: redox regulation, in vivo biosensing, redox proteomics, mitochondria, seed germination
Abstract
Seeds preserve a far developed plant embryo in a quiescent state. Seed metabolism relies on stored resources and is reactivated to drive germination when the external conditions are favorable. Since the switchover from quiescence to reactivation provides a remarkable case of a cell physiological transition we investigated the earliest events in energy and redox metabolism of Arabidopsis seeds at imbibition. By developing fluorescent protein biosensing in intact seeds, we observed ATP accumulation and oxygen uptake within minutes, indicating rapid activation of mitochondrial respiration, which coincided with a sharp transition from an oxidizing to a more reducing thiol redox environment in the mitochondrial matrix. To identify individual operational protein thiol switches, we captured the fast release of metabolic quiescence in organello and devised quantitative iodoacetyl tandem mass tag (iodoTMT)-based thiol redox proteomics. The redox state across all Cys peptides was shifted toward reduction from 27.1% down to 13.0% oxidized thiol. A large number of Cys peptides (412) were redox switched, representing central pathways of mitochondrial energy metabolism, including the respiratory chain and each enzymatic step of the tricarboxylic acid (TCA) cycle. Active site Cys peptides of glutathione reductase 2, NADPH-thioredoxin reductase a/b, and thioredoxin-o1 showed the strongest responses. Germination of seeds lacking those redox proteins was associated with markedly enhanced respiration and deregulated TCA cycle dynamics suggesting decreased resource efficiency of energy metabolism. Germination in aged seeds was strongly impaired. We identify a global operation of thiol redox switches that is required for optimal usage of energy stores by the mitochondria to drive efficient germination.
Dry orthodox seeds can preserve their ability to germinate for years, and even millennia in extreme cases (1), which is linked to their minimal metabolic activity when desiccated. At seed imbibition, stored metabolites and enzymes become accessible to reboot metabolic activity in the presence of oxygen. In oilseeds, like those of Arabidopsis, the breakdown of triacylglycerols in peroxisomes then progressively provides citrate and succinate as substrate for mitochondrial metabolism (2, 3), which delivers metabolic intermediates, reductant and ATP essential for biosynthesis and cell expansion. Translation of stored transcripts and de novo transcription require the successful reestablishment of energy physiology, and only commence in the range of several hours after imbibition (4, 5). This raises the problem that hormonal control relying on genetic programs can be executed only at a stage when physiological, thermodynamic, and organizational conditions have already been reestablished. Before that, more direct regulatory principles have to apply to first ensure the orderly establishment of cellular metabolism and physiology.
Posttranslational protein modifications have been implicated as a critical regulatory mechanism of metabolic function in the early phases of seed germination and Cys-based redox regulation provides a particularly good candidate mechanism (6, 7). In metabolically reactivated seeds, the reduction of the thiol redox machineries entirely depends on NADPH derived from primary metabolism (8). NADPH provides electrons either to the glutathione/glutaredoxin (GRX)-based redox machineries via glutathione reductases (GRs) or to the different thioredoxins (TRXs) via NADPH-thioredoxin reductases (NTRs) (9).
Already in 1943, reduced glutathione (GSH) could be detected in seed extracts 4 h after imbibition, indicating the early establishment of reducing activity, since GSH amounts were much lower in extracts from dry seeds (10). More recently, TRX-dependent reduction of the primary storage proteins in response to imbibition was observed in seeds from different species (11–13), with an impact on the storage protein solubility and accessibility (14). Since TRX-mediated reduction during imbibition was also observed for nonstorage proteins of different cellular localization and function (13), the reduction of TRX-linked proteins during seed rehydration is likely to be a global and conserved phenomenon for orthodox seeds. However, specific proteins, such as protein disulfide isomerases and several glutathione S-transferases, were found to be oxidized during imbibition (13), which probably reflects the reestablishment of the cell compartment-specific physiological redox environments, i.e., oxidizing in the endoplasmic reticulum. The direct mechanistic connection of Cys redox switching with metabolism raises the hypothesis of Cys redox switching as a fundamental strategy to provide rapid control when hormonal and genetic programs are not yet fully active. However, testing the plausibility of this hypothesis requires precise information about timing, identity, and physiological operation of redox-switched thiol proteins in seeds during metabolic reactivation, which has hitherto been missing.
Here, we focus on the earliest events during seed imbibition before hormonal and genetic programs start dominating germination control. Making use of in vivo monitoring of ATP, thiol redox, and oxygen dynamics in combination with metabolite analysis, we define the kinetics of reactivation of mitochondrial energy and redox metabolism. Finding that energy and redox rebooting are linked, we dissect the significance of the redox transition in the mitochondria. We capture the transition by establishing a controlled in organello model and use iodoacetyl tandem mass tag (iodoTMT)-based redox proteomics to identify and quantify individual target cysteines that are operated as thiol switches in situ. Using genetic ablation of the proteins most strongly affected we pinpoint the physiological role of the redox transition in maintaining resource efficiency and germination vigor identifying thiol redox regulation as a rapid and direct control strategy.
Results
Mitochondrial Energy Metabolism of Seeds Starts Rapidly at Imbibition.
To assess the cellular energy dynamics during early germination, we made use of Arabidopsis seeds expressing the MgATP2−-specific Förster resonance energy transfer (FRET)-sensor ATeam 1.03 nD/nA (ATeam) in the cytosol (15). The embryo dominates the sensor signal from the intact seed and the fluorescence intensity was sufficient to be detected through the seed coat (Fig. 1A). Emission spectra showed the characteristic sensor maxima, and the sensor spectrum could be reliably separated from the autofluorescence through subtraction of the spectra from seeds without the sensor, both in the dry and the imbibed state (Fig. 1A and SI Appendix, Fig. S1A). To monitor cytosolic MgATP2− dynamics live during seed imbibition, intact seeds were rehydrated through online water injection in a multiwell plate reader (Fig. 1B). A rapid increase of FRET directly at imbibition suggested an immediate onset of MgATP2− accumulation in the cytosol (Fig. 1C). The FRET response moved toward a plateau, which was reached at about 70 min (half-maximal sensor response T1/2 after 34 ± 2 min), suggesting either stable cytosolic MgATP2− concentration or reaching sensor saturation. We observed similar sensor dynamics also by confocal imaging of embryos (SI Appendix, Fig. S1B), which could be isolated in an intact state from 10 min after imbibition onwards. Quantification of adenosine phosphate concentrations in total seed extracts validated a rapid reestablishment of the energy charge (EC) of the adenosine phosphates, which was largely completed after 60 min (Fig. 1D). AMP and ADP levels were dominant in extracts from dry seeds and were decreased by imbibition, while ATP was dominant in imbibed seeds. The EC increased from 0.2 in dry seeds to 0.6 after 1 h of imbibition and remained constant afterward (4 h). As the EC is typically ≥0.9 in fully active cells, the EC is likely to increase further beyond the 4-h time point. A lower apparent EC may alternatively be explained by tissue gradients within the seeds. To assess the source of ATP production and the shift in EC, we next assessed oxygen uptake by the seeds as a measure of mitochondrial respiration. Oxygen consumption was established immediately from the start of the measurement after addition of water, whereas the addition of the respiratory inhibitor cyanide decreased the oxygen consumption (Fig. 1E). We next hypothesized that the fast transition from quiescence to metabolic reactivation may give rise to transient changes of metabolite pools, as a result of fluxes that have not yet reached steady state. Analysis of a set of organic acids associated with mitochondrial energy metabolism (Fig. 1F) revealed high malate content in dry seeds, whereas pyruvate, 2-oxoglutarate, succinate, fumarate, and lactate were present at lower concentrations. Isocitrate/citrate was below the detection limit, but accumulated to malate levels within 1 h of imbibition and dropped again beyond detection limit at 4 h of imbibition. Succinate and lactate accumulated gradually, possibly reflecting partial hypoxia in central embryo tissues. Pyruvate, 2-oxoglutarate, and fumarate pools remained stable. The pronounced isocitrate/citrate transient indicates sequential activity changes within the glyoxylate cycle and the TCA cycle during the first hours of seed imbibition, as mediated by citrate synthase, aconitase, isocitrate dehydrogenase, or ATP-citrate lyase. Free amino acid (AA) pools were unchanged between dry and imbibed seeds at 1 and 4 h (SI Appendix, Fig. S2), suggesting balanced fluxes or no major degradation of storage proteins at this early stage of germination.
Fig. 1.
Restart of respiratory energy metabolism in Arabidopsis seeds at imbibition. (A and B) Experimental setup for plate reader-based detection of genetically encoded probes in seeds at imbibition exemplified for the MgATP2− FRET-sensor ATeam. (A) Representative brightfield and fluorescence (GFP-filter) images of dry seeds, either with or without expression of cytosolic ATeam (cyt-ATeam). Emission spectra of the seeds are plotted below; dashed magenta line for wildtype (Col-0) seeds, dashed green line for cyt-ATeam seeds, and green line for cyt-ATeam seeds corrected for autofluorescence of Col-0 seeds (n = 3 to 4 seed batches). (B and C) Monitoring cytosolic MgATP2− levels in intact seeds through online imbibition and detection of the cyt-ATeam FRET ratio. 1: dry seed, 2: addition of water, 3: imbibition. The arrows indicate addition of water and T1/2 equals the time point of half-maximal sensor response (n = 6 seed batches; mean normalized to last value before injection of water ± SD and corrected for Col-0 autofluorescence). (D) Total ATP, ADP, and AMP concentrations in dry seeds and after 1 and 4 h of imbibition (n = 4; mean normalized to seed dry weight + SD; 2-way ANOVA with Tukey’s multiple comparisons test with a: P > 0.05, b: P < 0.05, and e: P < 0.0001, significant differences between 1 and 4 h imbibition are indicated by a second character). Corresponding EC ([ATP] + 1/2 [ADP])/([ATP] + [ADP] + [AMP]) is shown on secondary axis (mean + SD). (E) Oxygen consumption of imbibed seeds measured with MitoXpress Xtra (fluorescence lifetime of the probe is physically quenched by oxygen). Increase of fluorescence lifetime (Δ FLT) recorded for seeds at control conditions (white circles) or supplemented with 500 µM potassium cyanide (KCN) (gray circles; n = 6 seed batches; mean normalized to amount of dry seeds ± SD). Inset shows Δ FLT rate in the timeframe of 60 to 90 min (2-sided Student’s t test with ***P < 0.001) as indicated by the green and magenta line. (F) Concentration of organic acids analyzed in dry seeds and after 1 and 4 h of imbibition (n = 4; mean normalized to seed dry weight + SD; 2-way ANOVA with Tukey’s multiple comparisons test with a: P > 0.05, b: P < 0.05, c: P < 0.01, d: P < 0.001, and e: P < 0.0001, significant differences between 1 and 4 h imbibition are indicated by a second character). Pyruvate (pyr), (iso-)citrate ((iso-)cit), 2-oxoglutarate (2OG), succinate (succ), fumarate (fum), malate (mal), and lactate (lac). (G) Working model of the restart of the respiratory energy metabolism of dry seeds early during imbibition. Width of arrow lines indicate flux rates. OXPHOS, oxidative phosphorylation.
Taken together these data support the idea that mitochondrial respiration is started rapidly with imbibition and before gene expression-based control can underpin hormonal signaling (Fig. 1G). Although the velocity of the activation suggests that the restart is largely driven by physical rehydration of metabolites and other cell constituents, as well as oxygen supply, early rebooting of mitochondrial energy metabolism makes it a likely prerequisite for early metabolic regulation and germination control.
An Early Kick-Start of the Thiol Redox Machinery.
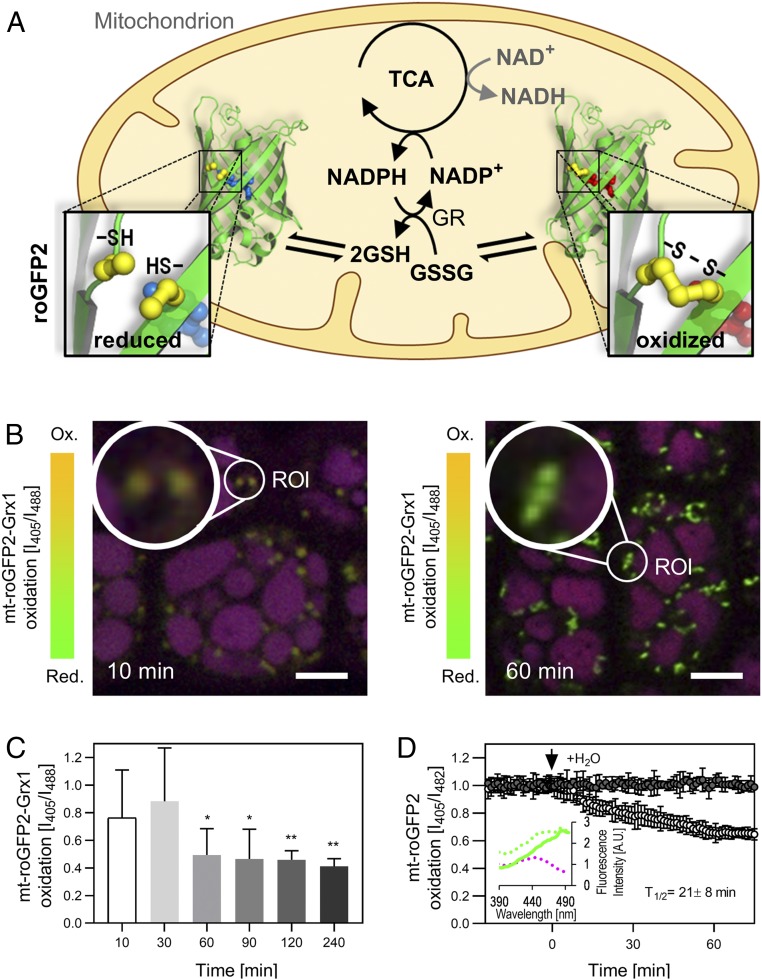
We reasoned that direct regulation through posttranslational protein modifications provides a particularly plausible regulatory framework to ensure orderly progression of early germination, before hormonal and transcriptional circuits take control after several hours (16, 17). Focusing on cysteine-linked redox switches, which can be operated rapidly as linked to metabolic electron fluxes, we employed the mitochondrial matrix-targeted genetically encoded fluorescent biosensor roGFP2-Grx1 that responds to the glutathione redox potential (EGSH) (18, 19), to explore the dynamics of thiol redox status specifically in the mitochondrial matrix, i.e., the site of action of most respiratory enzymes (Fig. 2A). Our rationale was that EGSH reduction in the mitochondrion indicates local NADPH provision within the mitochondrial matrix, which is required to reduce glutathione disulfide (GSSG) through glutathione reductase 2 (GR2) in the matrix (20) (Fig. 2A).
Fig. 2.
Activation of the mitochondrial thiol redox machinery in Arabidopsis seeds at imbibition. (A) Model of glutathione and roGFP reduction via mitochondrial metabolism. TCA cycle-derived NADPH, but not NADH, acts as electron donor to the matrix thiol redox machineries, e.g., to reduce glutathione disulfide (GSSG) to glutathione (GSH) by glutathione reductase (GR), and to the thioredoxin-system (not depicted). Redox state of the matrix glutathione pool can be monitored by roGFP2 sensors. (B and C) Change in matrix glutathione redox state in imbibed seeds lacking seed coat and endosperm over time analyzed by confocal microscopy monitoring the 405/488-nm excitation ratio of roGFP2-Grx1. (B) Representative confocal images showing embryo epidermal cells imbibed for 10 and 60 min. ROIs with mitochondria are highlighted and magnified. Overlay of the 405-nm and the 488-nm channels in red and green, protein storage vacuoles appear in purple, due to an autofluorescence channel displayed in blue. (Scale bars, 10 µm.) (C) Changes in matrix roGFP2-Grx1 oxidation during seed imbibition analyzed by confocal imaging (n = 400 to 700 individual mitochondria per time point in 8 to 12 individual embryos; mean + SD; 2-sided Student’s t test with *P < 0.05 and **P < 0.01). (D) Mitochondrial roGFP2 dynamics at imbibition of intact seeds monitored by plate reader-based fluorimetry (Fig. 1 A and B). The arrow indicates addition of water and T1/2 equals the time point of half-maximal sensor response (n = 6 seed batches; mean normalized to last value before injection of water ± SD and corrected for Col-0 autofluorescence). Inserted panel shows excitation spectra of dry seeds, dashed magenta line for Col-0 seeds, dashed green line for mt-roGFP2 seeds, and green line for mt-roGFP2 seeds corrected for autofluorescence of Col-0 seeds (n = 3 to 4 seed batches).
Similar to imaging of MgATP2−, we used ratiometric confocal imaging to measure the dynamics of matrix EGSH starting from 10 min after imbibition when intact embryos could be isolated. For separation of the sensor signal from the autofluorescence of the protein storage vacuoles, we analyzed several hundred individual mitochondria in several embryos by a region-of-interest (ROI) analysis (21). Embryos isolated at 10 min showed high 405/488-nm excitation ratios indicating high probe oxidation, which decreased markedly after 60 min, indicating a pronounced transition toward probe reduction (Fig. 2 B and C). To control for potential effects of embryo isolation, we adjusted the plate reader-based analysis protocol (Fig. 1B) for roGFP2 to assess the probe response in intact seeds. Despite a low signal-to-noise ratio due to a high proportion of autofluorescence in the mitochondrial sensor line, the characteristic excitation spectrum of roGFP2 was reliably recorded (Fig. 2D and SI Appendix, Fig. S3A) to monitor the redox dynamics in mitochondria of intact seeds. A rapid sensor reduction was detectable at imbibition within the first minutes that reached a plateau within 60 min (half-maximal sensor response T1/2 after 21 ± 8 min), indicating rapid establishment of a highly reduced glutathione pool (Fig. 2D) as typically found in the mitochondria of vegetative plant tissues (19). Rapid EGSH reduction was also observed using a cytosolic sensor, although with distinct, biphasic kinetics (SI Appendix, Fig. S3B). The sensor reduction dynamics in both subcellular compartments coherently indicate that the thiol redox machineries are restarted very early at imbibition.
Rapid and simultaneous reestablishment of subcellular energy and redox physiology through mitochondrial activity may be a characteristic of germinating, orthodox seeds (Figs. 1 and 2), and a prerequisite to drive and regulate subsequent steps of germination. Specifically, activation of the thiol redox machinery in the mitochondrial matrix provides a mechanism to reset the landscape of operational protein thiol switches to control mitochondrial function.
Rebooting the Thiol Redox Machinery in Purified Mitochondria.
To reveal which specific protein thiol switches are operated within the functional mitochondria we developed a strategy based on quantitative redox proteomics. Maintaining meaningful thermodynamic, kinetic, and spatial constraints for the operation of thiol switches was a key priority, since kinetic control delivers specificity in thiol redox regulation (22), and cannot be straightforwardly studied in vitro (23). Since whole seed extracts are dominated by storage proteins, leaving mitochondrial proteins strongly underrepresented in proteome analyses (24), we optimized a model system to capture the fast release of metabolic quiescence that we observed in seeds using purified seedling mitochondria. Respiratory metabolism is quiescent in dry seeds due to lack of free water for substrate solubilization and enzyme activity; consequently, no NADPH is generated in the matrix and thiols cannot be effectively maintained in a reduced state, as indicated by the oxidized roGFP2 sensor (Fig. 2). Analogously, isolated plant mitochondria are metabolically quiescent due to respiratory substrate depletion and cannot generate NADPH (Fig. 3A).
Fig. 3.
Rebooting the thiol redox machinery of isolated mitochondria to define the proteomic landscape of mitochondrial proteinaceous Cys redox switches. (A) Schematic model of quiescent mitochondria and their metabolic reactivation by substrate addition. Substrates are metabolized reducing NAD+ and NADP+. Both NADH and NADPH can be oxidized by the electron transport chain (ETC), but only NADPH can act as reductant for the matrix thiol redox machineries. Matrix-targeted roGFP2-Grx1 acts as artificial target of the glutathione redox machinery. (B) Representative roGFP2-Grx1 reduction kinetics of quiescent mitochondria, either supplemented with 10 mM 2OG or 10 mM citrate (n = 4 technical replicates, mean ± SD; dashed line indicates addition of substrates and red box highlights the oxidation state after 25 min). The mt-roGFP2-Grx1 redox state calculated for each metabolic condition after 25 min on the Right; for calibration of the sensor, mitochondria were incubated with 5 mM dipyridyldisulfide (DPS, oxidant) or 20 mM DTT (DTT, reductant) for 20 min (2-sided Student’s t test with **P < 0.01 and ****P < 0.0001). Inset shows state II oxygen consumption of isolated mitochondria supplemented either with 2OG or citrate (n = 5, mean + SD). (C and D) Abundance profiles of the redox states of Cys peptides in quiescent and respiring mitochondria. Isolated mitochondria supplemented with citrate (C) or 2OG (D) and incubated for 25 min before differential thiol labeling with iodoTMTs. Redox states of individual peptides quantified for isolated mitochondria with and without respiratory substrate (blue and red bars, respectively). The distribution of cysteine peptide oxidation levels is shown by the proportion of the total number of peptides in each 2% quantile of percentage oxidation (n = 3, mean + SD). In Insets, percentage of Cys oxidation without substrate addition is plotted against percentage of Cys oxidation after substrate addition for individual Cys peptides. Cys peptides with significant change in their Cys-oxidation state are displayed in blue (t test corrected for multiple comparisons by Benjamini, Krieger, and Yekutieli with <2% false discovery rate [FDR]). (E) The 5 most enriched GO terms of the “cellular component” and “biological process” categories of a PANTHER-overrepresentation analysis of all significantly redox-switched Cys peptides. (F) All identified Cys peptides of the individual subunits of the mitochondrial complex I mapped on the schematic structure according to Braun et al. (32) (Left); all Cys peptides identified for enzymes of the TCA cycle and closely associated (Right). Blue: significantly redox-switched Cys peptide; red: not significantly redox-switched Cys peptide. (G) Redox shift analysis through immunodetection of aconitase proteins (ACO) in seed protein extracts alkylated with methylpolyethylene glycolmaleimide reagent (MM-PEG24). Protein extracts from dry seeds (dry), seeds imbibed for 1 h (1 h), and dry seeds and treated with Tris(2-carboxyethyl)phosphine (TCEP) for reduction (n = 3 biological replicates, see SI Appendix, Fig. S8A). Note that the 3 Arabidopsis ACO proteins are localized in the mitochondrial matrix and the cytosol. (H) Relative activities of alternative oxidase (AOX, Left panels, see also SI Appendix, Fig. S8B), NAD-malic enzyme (NAD-ME), and aconitase (ACO) in isolated mitochondria assayed at different redox potentials adjusted by a DTT/dithiane buffer. RoGFP2 to validate the redox conditions in the assay media (Lower panels). Calculated redox potentials are adjusted for assay pH. AOX: 7.5, NAD-ME: 6.7, ACO: 7.8 (n = 3 biological replicates, mean + SD, 1-way ANOVA with Dunnett’s multiple comparisons test, *P < 0.05, ***P < 0.001, and ****P < 0.0001).
To recreate the rapid redox transition as observed in mitochondria of intact seeds we supplied purified mitochondria, expressing mitochondrial roGFP2-Grx1, with metabolic substrates. Different matrix dehydrogenases couple substrate oxidation to the reduction of NAD+ and NADP+ (25). Yet, only NADPH can act as an electron donor to the thiol redox machinery (Fig. 3A). Citrate feeding triggered a pronounced reduction of the sensor (Fig. 3 B, Left). After 25 min, the sensor signal reached a plateau at a highly reduced state. The rapid and pronounced sensor response indicates active and sustained NADPH generation in the matrix, with the NADP-dependent isoform of isocitrate dehydrogenase (ICDH) as a likely source. As a control substrate, we used 2-oxoglutarate (2OG), for which there is no obvious NADPH-producing enzyme downstream (25). 2OG triggered only a weak reduction of the sensor. Mitochondrial oxygen consumption rates triggered by the 2 substrates were comparable, indicating that both substrates were taken up into the matrix and metabolized at similar rates (Fig. 3 B, Inset Right). Taken together the data suggest that 2OG oxidation predominantly produced NADH, while citrate oxidation caused significant rates of NADPH production in addition. The data further suggest that the redox states of NAD and NADP pools are separated in the matrix, consistent with the lack of any mitochondrial transhydrogenase protein homolog in higher plant genomes.
The Landscape of Operational Cysteinyl Thiol Redox Switches of the Mitochondrial Matrix.
Next, we aimed to identify those mitochondrial thiol switches that are operated in situ during the transition from quiescence to activity of respiratory matrix metabolism. We developed and optimized fast differential Cys labeling in intact mitochondria using thiol-specific iodoTMTs. Differential iodoTMT-based redox proteomics allows the quantification of the degree of oxidation (% oxidation) of individual proteinaceous cysteinyl thiols by tandem mass spectrometry (MS/MS). Absolute quantitation of the Cys redox landscape at Cys-peptide resolution was not explored by previous redox proteomic approaches in plants. Previous approaches typically pinpointed redox changes at the protein level and/or rather focused on the relative percentage of thiol redox switching (Δ% oxidation) as induced by a treatment. Δ% oxidation can also be determined as a difference between the absolute % oxidation of the 2 metabolic states. However, the iodoTMT approach as applied here does not resolve the chemical identity of Cys modification, which will include several different types of oxidative Cys-peptide modifications, such as disulfides, sulfenylation, and nitrosylation, for which complementary methods exist (e.g., ref. 26). Potential changes in protein abundance between individual replicates are accounted for by internal normalization of the differential labeling approach. Protein abundance remained stable (SI Appendix, Fig. S4). The comparison between quiescent and citrate-respiring mitochondria (sampled after 25 min) allowed for the quantitation of 741 Cys peptides, mapping to 425 different proteins (734 Cys peptides mapping to 443 proteins for the control using 2OG supplementation) (Dataset S1). Activating mitochondrial metabolism with citrate resulted in a global reductive shift from 27.1% Cys oxidation on average under quiescence to only 13.0% Cys oxidation (Δ14.1%; Fig. 3C). By contrast, inducing respiration with 2OG did not change Cys oxidation significantly (27.1% under quiescence to 27.0% with 2OG; Δ0.1%), and the characteristic distribution of the redox states of Cys peptides was fully preserved (Fig. 3D). The individual Cys peptides responded highly differentially to citrate respiration, with 412 peptides mapping to 245 different proteins undergoing a significant reductive shift, while no peptide underwent oxidation (Fig. 3 C, Inset and SI Appendix, Fig. S5D). By contrast not a single Cys peptide was significantly shifted in its degree of oxidation in response to 2OG respiration (Fig. 3 D, Inset and SI Appendix, Fig. S6D). Those effects were highly reproducible as evidenced by very similar patterns across the independent replicates (SI Appendix, Figs. S5 A and B and S6 A and B). The pronounced reductive shift that we had observed fluorimetrically under citrate respiration using matrix roGFP2-Grx1 (Fig. 3B) was also observed in the proteomic approach, where a roGFP2 Cys peptide was detected and showed a reductive shift of Δ30.4% (Dataset S1).
The individual thiol-switched Cys peptides differed strongly in their degree of redox shifting (Dataset S1). The Cys peptides showing the most pronounced reductive shifts mapped to the plastidial/mitochondrial GR2 (Δ61.7%) (20), the cytosolic/mitochondrial NADPH-dependent thioredoxin reductases a and b (NTR a/b; Δ49.1%) (27, 28) and mitochondrial TRX-o1 (Δ48.1%) (29). All 3 peptides included the catalytic cysteines of their proteins, mirroring their role as active thiol redox enzymes, consistent with previous findings of particularly reactive catalytic cysteines (30, 31). Since 2 Cys residues were present in all 3 peptides, the quantified redox shift represents an average and may be even larger for 1 of the 2 cysteinyl thiols. However, over 80% of Cys peptides contained a single Cys, making the quantification unambiguous for that specific site (SI Appendix, Fig. S7A).
To investigate if, in addition to the thiol redox machineries, specific biological functions were preferentially associated with those thiol switches that were operational in situ, we used a gene ontology (GO) enrichment analysis (http://geneontology.org). Despite bias through the input of enriched mitochondrial fractions, and known weaknesses in the classification and annotation of proteins, the analysis has proven useful to highlight overrepresented pathways (31). Central components and processes of the mitochondrial respiratory metabolism were enriched, including those of the mitochondrial respiratory chain and the TCA cycle (Fig. 3E), suggesting central respiratory metabolism harbors hotspots of operational thiol switching. The enrichment of “chloroplast stroma” proteins can be accounted for by a combination of plastidic contamination (SI Appendix, Fig. S7B) and by proteins assigned as stromal that are actually also mitochondrial. At least 20 out of the 46 redox switched proteins in this category are also mitochondrial matrix proteins. An example is GR2, which is annotated as plastidic, but was demonstrated to be dual targeted, also to the mitochondria (20). Further functions represented by several different proteins with active thiol redox switches included iron–sulfur metabolism, amino acid metabolism, and mitochondrial RNA processing (PPR proteins) (Dataset S1).
As an independent way to identify hotspots of operational thiol switching in the intact mitochondrion we looked for proteins for which more than 1 thiol-switched Cys peptide was identified. A total of 94 proteins contained 2 or more Cys peptides that were significantly redox switched. The 75-kDa subunit (AT5G37510) of the peripheral arm of complex I contained even 10 functional thiol redox switches (Fig. 3F). Since complex I was also pinpointed by the GO term analysis (Fig. 3E), we further assessed the locations of the redox-switched Cys within the complex. The relevant proteins were all part of the peripheral arm (21 Cys peptides switched, 3 nonswitched) and the plant-specific carbonic anhydrase domain (6 Cys peptides switched, 1 nonswitched), which may be partly explained by accessibility to soluble redox enzymes. In contrast, all 7 identified Cys peptides of the membrane arm were exposed to the intermembrane space (IMS) (32) and were not redox switched, which validates the experimental system in which citrate feeding provides NADPH in the matrix, but not the IMS.
Recent work demonstrated Trx-mediated redox regulation of 2 TCA cycle enzymes, succinate dehydrogenase and fumarase (29). Our dataset validates the presence of operational thiol switches on both players and pinpoints operational Cys switches. It further expands the current picture by the identification of at least 1 active thiol switch for each enzymatic step of the TCA cycle (Fig. 3F), as a mechanistic basis for comprehensive redox control of this hub of central metabolism. The E1 subunit of the oxoglutarate dehydrogenase complex (OGDC) was identified with 7 redox-switched Cys peptides (AT5G65750, 3 nonunique peptides also map to the isoform AT3G55410). Fourteen Cys peptides from different pyruvate dehydrogenase complex (PDC) subunits were redox shifted. For the succinate dehydrogenase complex (SDH) 6 out of 9 identified Cys peptides were redox switched, while only 3 out of 8 Cys peptides identified from aconitase (ACO) 1 and 3 were redox switched. ACO showed a reproducible reductive shift also in seeds within 1 h of imbibition (Fig. 3G and SI Appendix, Fig. S8A), providing independent validation of the isolated mitochondria model.
The proteomic compendium of active mitochondrial thiol switches that we have generated demonstrates that a large number of proteinaceous Cys thiols cannot only be reduced in vitro (33–37), but are operated in the intact mitochondrion linked to metabolic activity. All shifts were reductive, reflecting the influx of electrons into the matrix thiol redox systems, and quantitative, since most peptides were maintained partially reduced in the quiescent metabolic state. It is likely that the operation of a thiol switch in situ is typically not an all-or-nothing process, although even a partial redox shift of an individual Cys on mammalian uncoupling protein 1 (Cys253) by less than 10% has been shown to be able to trigger dramatic metabolic changes and induce thermogenesis (38). Yet, more conceivably during early seed imbibition the concerted operation of many switches on many different enzymes at once may have a considerable impact on the fluxes through mitochondrial energy metabolism. Indeed, several plant respiratory enzymes have been found to be Cys redox-dependent in vitro (23). To investigate whether redox modulation can be confirmed and extrapolated also to other TCA cycle enzymes, we performed activity assays of mitochondrial enzymes in the presence of different redox potentials (as set by the dithiothreitol [DTT]/dithiane buffer and confirmed using recombinant roGFP2 protein) (Fig. 3H and SI Appendix, Fig. S8C). The redox-dependence of alternative oxidase (AOX) activity served as positive control (Fig. 3H and SI Appendix, Fig. S8B). Interestingly, we observed quantitative redox-dependence also of NAD-malic enzyme (NAD-ME) and ACO (Fig. 3H). 2-oxoglutarate dehydrogenase (OGDH), NAD-isocitrate dehydrogenase (IDH), and ICDH showed only marginal responses (SI Appendix, Fig. S8C), indicating that the functional impact of Cys redox switch operation is enzyme-specific.
Mutants of the Mitochondrial Thiol Redox Machinery Are Impaired in Germination.
To assess the physiological significance of the rapid reduction of the mitochondrial thiol redox machinery during early germination, we aimed to understand its impact on seed energy metabolism and germination characteristics. Since the most strongly thiol-switched peptides belonged to the catalytic sites of GR2, NTR a/b and TRX-o1, we selected Arabidopsis mutant lines lacking those proteins (20, 28, 29). In addition, those proteins operate upstream in potential redox regulation cascades, enabling an integrated picture of thiol switching on several target enzymes at once, even though a degree of compensation between the thiol redox systems needs to be anticipated, based on previous observations (20, 28, 39).
Strikingly, oxygen consumption rates early after imbibition (here measured between 60 and 90 min) were strongly increased in the seeds of all 3 mutants (Fig. 4A), indicating increased respiration rates with the potential of boosting oxidative phosphorylation and ATP synthesis rates. Yet, there was no consistent impact on the different adenosine phosphate pools. Only ntr a/b showed a change in EC after 1 h of imbibition, which was even lowered, however. The adenylate pools of the other seeds recovered as in Col-0 (Fig. 4B). Increased respiration rates coinciding with unchanged adenylate charge clearly point to less efficient ATP generation or higher consumption. To test the hypothesis of deregulated mitochondrial enzymatic activity as a common cause of inefficient respiration in the mutants, we assessed the free organic acid pools of the seeds of all lines. In dry seeds the different pools were unchanged; only malate content differed between the lines, albeit without a consistent pattern (Fig. 4 C, i), indicating a similar metabolic situation at the start of imbibition. As observed in Col-0 (Fig. 1F), the organic acid pools responded markedly to imbibition, and strikingly a consistently lowered (iso-)citrate content was observed for all mutants as compared to Col-0 at the 1-h time point (Fig. 4 C, ii). Since (iso-)citrate accumulation occurs as a transient (Fig. 1F), the modification of its amplitude and/or timing shows that the flux into and/or out of the (iso-)citrate pool is affected in the mutants and provides evidence for deregulated respiratory carbon metabolism in the thiol redox mutants during early germination. Of the 19 free amino acid pools, 16 did not differ between the lines and remained stable at imbibition (SI Appendix, Fig. S8). As an exception, Glu, Asn, and Pro were slightly increased in dry ntr a/b seeds and accumulated in gr2 seeds at imbibition, indicating that deregulation of amino acid metabolism was limited to specific pathways, some of which are located in the mitochondria (40).
Fig. 4.
The effect of impairing the mitochondrial thiol redox machinery on energy metabolism and germination. Synchronized seeds of Col-0, gr2, trx-o1, and ntr a/b lines were analyzed for their performance during germination. (A) Oxygen consumption of imbibed seeds measured with MitoXpress Xtra. (i) Oxygen uptake as indicated by increase in fluorescence lifetime (Δ FLT) over time (n = 5 seed batches; mean normalized to amount of dry seeds + SD). (ii) Δ FLT rate in the 60- to 90-min time window (2-sided Student’s t test with **P < 0.01 and ***P < 0.001 if compared with Col-0). (B) Total ATP, ADP, and AMP concentration (i) in dry seeds and (ii) after 1 h of imbibition (n = 4; mean normalized to seed dry weight + SD; 2-way ANOVA with Dunnett’s multiple comparisons test with b: P < 0.05, c: P < 0.01, d: P < 0.001, e: P < 0.0001 if compared with Col-0). Corresponding EC ([ATP] + 1/2 [ADP])/([ATP] + [ADP] + [AMP]) is shown on secondary axis (mean + SD). (C) Concentration of organic acids analyzed (i) in dry seeds and (ii) after 1 h of imbibition (n = 4; mean normalized to seed dry weight + SD; 2-way ANOVA with Dunnett’s multiple comparisons test with b: P < 0.05, c: P < 0.01, d: P < 0.001 and e: P < 0.0001 if compared with Col-0). Pyruvate (pyr), (iso-)citrate [(iso-)cit], 2-oxoglutarate (2OG), succinate (succ), fumarate (fum), malate (mal), and lactate (lac). (D) Germination over 5 d (i) for freshly harvested seeds and (ii) after a CDT by storage at 37 °C and 75% relative humidity for 19 d to mimic seed aging. The statistical analysis of germination rates of gr2, trx-o1, and ntr a/b as compared to Col-0 seeds is shown in SI Appendix, Table S1.
To investigate the potential impact of respiratory deregulation on seed germination, we analyzed germination vigor in freshly harvested seeds over 5 d after imbibition. All lines showed efficient germination with 100% germination rate within 5 d. Only trx-o1 showed temporarily lowered germination efficiency after 2 d (Fig. 4 D, i and SI Appendix, Table S1). The potential defect in trx-o1 was consistent with a previous observation (29), where seeds showed much lower germination vigor overall, however. We hypothesized that, in fresh seeds, efficient carbon metabolism may not be limiting and investigated the germination characteristics of seeds that were exposed to a controlled deterioration treatment (CDT). CDT at 37 °C, 75% relative humidity for 19 d decreased germination vigor for all lines, and only 60% of Col-0 seeds germinated within 5 d (Fig. 4 D, ii). Yet, all 3 redox mutants showed much stronger defects in germination ability and this effect was observed consistently across different aging regimes (SI Appendix, Fig. S9 and Table S2). Interestingly, the trx-o1 line was consistently more severely impaired than the gr2 and ntra/b lines (Fig. 4 and SI Appendix, Fig. S9), indicating a primary role of TRX-o1 that is least efficiently backed up for by the other redox systems. Taken together the different results demonstrate the importance of mitochondrial thiol redox switching in the efficient use of stored resources by respiratory metabolism in germinating seeds.
Discussion
MgATP2− synthesis and oxygen consumption start almost instantaneously upon rehydration, illustrating how rapidly the transition from seed quiescence to metabolic activity occurs (Fig. 1 C–F). The rapid onset of respiration requires structurally intact, functional mitochondria in the desiccated embryo cells, as observed after 10 min of imbibition (Fig. 2B). Those observations are in contrast to the previous postulate that a maturation phase is required for respiratory incompetent protomitochondria before respiration can commence (41), but in agreement with recent observations in pea and Arabidopsis seeds (42, 43). Work in lettuce seed extracts has pinpointed the early establishment of adenylate charge, but the in vivo dynamics of the response could not be resolved (44). Oxidative phosphorylation sets the balance between the adenylate pools together with adenylate kinase (42), explaining the rapid conversion of AMP to ATP (Figs. 1D and 4B). While oxidative phosphorylation appears to dominate within the first hour, the accumulation of lactate between 1 and 4 h suggests a contribution by fermentation to ATP synthesis (Fig. 1F). That makes the rapid establishment of respiratory energy homeostasis a likely prerequisite for other early germination events, such as translation or de novo transcription, which underpin major hormonal checkpoints.
RoGFP-based in vivo sensing suggests that reestablishment of the thiol redox status of the mitochondrial matrix and the cytosol is intimately linked to the reestablishment of energy metabolism (Figs. 1 and 2). Low EC coincides with oxidized glutathione status, which reflects the absence of sufficient metabolic flux to provide phosphorylation potential and reductant in the form of NADPH. The rapid reactivation of the thiol redox machinery is consistent with the observation of strongly increased GSH levels in seed extracts 4 h after imbibition (10). Further, our data demonstrate that an active thiol redox machinery is not only a side effect of metabolic reactivation, but plays an important regulatory role in germination (Fig. 4). Mutants of 3 different genes (gr2, ntr a/b, and trx-o1), all of which are part of the mitochondrial thiol redox machinery, consistently showed increased respiratory rates in the presence of unaltered adenylate dynamics, suggestive of lower efficiency of energy conservation from respiring stored carbon resources. Specific deregulation of (iso-)citrate is consistent with this interpretation since major citrate and succinate flux is expected from peroxisomal triacylglycerol degradation in Arabidopsis seeds (2). Based on our data we propose a model, in which the dampened (iso-)citrate transient at the 1-h time point is caused by derepressed efflux from the (iso-)citrate pool (Figs. 1F and 4C), leading to increased respiration rates (Fig. 4A) and carbon loss in the mutants. While fresh seeds can tolerate inefficient mitochondrial metabolism up to a degree, aging further compromises seed mitochondria (45) leading to drastically decreased germination vigor (Fig. 4D). Our observation that every enzymatic step of the TCA cycle features at least 1 operational thiol switch (Fig. 3F) makes it likely that several TCA cycle proteins contribute to the overall activity pattern of the pathway, which is further strengthend by redox-dependent activity changes of NAD-malic enzyme and aconitase (Fig. 3G). Interestingly, TRX-o1–mediated reduction repressed the activity of succinate dehydrogenase and fumarase (29), and recently, increased respiration rates in green leaves of trx-o1 lines were linked to deregulation of respiratory carbon metabolism (46). Also photorespiratory activity was altered in the absence of TRX-o1 (47).
For 2 mutants of the TRX-based redox machinery we observed comparable germination defects. Our model provides a plausible explanation for the data, but it is unlikely to be comprehensive, due to the central role of mitochondrial respiration and carbon metabolism in the cell, which means that defects are likely to trigger further pleiotropic rearrangements. The start from a joint quiescent situation (dry seeds), may minimize such effects, as supported by the measured metabolite profiles (Fig. 4), but differences already set during seed filling and/or maturation cannot be ruled out. For instance, a wrinkled seed phenotype was reported for the ntr a/b mutant, which was shown to be due to the role of NTR a/b in the cytosol and not in the mitochondria, however (28).
Several candidate target proteins of the plant mitochondrial TRX and GRX machineries have been identified by in vitro trapping approaches using protein extracts (33–35, 37). Those approaches have been instrumental in identifying thiol proteins that can react with TRX and GRX active site thiols in principle. However, they cannot account for microcompartmentalization, kinetic competition, and thermodynamic conditions, which are key constraints governing thiol redox regulation in vivo (23). In situ trapping approaches, as performed in mitochondria of human embryonic kidney cells (48) or the cytosol of cultured Arabidopsis cells (49) circumvent several limitations. However, they often rely on nonendogenous baits (TRXs or GRXs from other cell compartments or species, mutated YAP1 from yeast), which introduce bias since the selectivity between TRX-type proteins and their targets can differ profoundly based on electrostatic surface properties (22). A study combining in vitro TRX trapping and in vivo monobromobimane labeling identified a total of 114 TRX targets from germinating Medicago seeds, including a small number of abundant mitochondrial proteins, such as HSP70, β-subunit of the mitochondrial ATP synthase, pyruvate decarboxylase, succinate dehydrogenase, succinic semialdehyde dehydrogenase, and peroxiredoxin IIF (13).
Here we establish respiring seedling mitochondria as a model to detect and quantify those Cys-thiol switches that are operated in response to an in situ redox transition that mimics the reactivation occurring during early seed germination (Fig. 3A). The application of this model to germination was validated by the redox responses of mt-roGFP2-Grx1 (Fig. 3B), and aconitase (Fig. 3G and SI Appendix, Fig. S8A). There are also obvious limits to the model. For instance, mitochondria could not be isolated from dry Arabidopsis seeds. Due to active mitochondrial biogenesis in the later stages of germination their quantitative protein composition is likely to differ. At the qualitative level, however, a large majority of the mitochondrial proteins with operational thiol switches are present in the Arabidopsis seed proteome (SI Appendix, Fig. S10A) (24). Further, there are several additional influences on mitochondrial physiology within the seed cells, such as different flux rates, availability of a mixture of respiratory substrates, rehydration of endogenous proteins and active restructuring of membranes, which may have secondary impacts on the redox dynamics. Yet, it is reasonable to anticipate that the endogenous generation of NADPH dominates redox dynamics of proteinaceous Cys in both systems. The reduction of target proteins is, in both cases, achieved by the endogenous thiol redox machinery, ensuring target-specificity. In the isolated mitochondria, the reduction of target proteins is specific to the matrix, where the full redox cascade can operate, as exemplified for respiratory complex I, which showed thiol switching on matrix-exposed subunits, but not on the IMS exposed subunits (Fig. 3F). Indeed, the comparison of the consensus subcellular localization of all proteins identified from the mitochondrial fractions vs. those that showed thiol switching revealed enrichment for mitochondrial proteins (SI Appendix, Fig. S7B) (50). Individual cases of reduction of nonmatrix Cys may be due to redox interaction between proteins during the short time window of the blocking step when mitochondria are ruptured. In contrast, simultaneous activation of the redox systems occurs in different subcellular locations, such as the IMS or the cytosol, by separate machineries (SI Appendix, Fig. S3B)
It is a strength of the differential iodoTMT-based approach that the individual cysteine thiol to undergo switching can be pinpointed and quantified in their degree of oxidation in most cases, whereas many other techniques only report relative changes. This provides the opportunity to compare the 2 approaches of fluorescent protein-based redox sensing and redox proteomics in the same system: in citrate-activated mitochondria roGFP2 underwent a reductive shift by Δ43.1% as quantified by fluorimetry, compared to Δ30.4% as quantified by MS/MS (only 1 of the 2 Cys peptides involved in the roGFP disulfide was identified), indicating a qualitative and close to quantitative agreement between the 2 approaches.
Thiol switching can impact on the activity, stability, conformation, and localization of a protein, as well as its interaction with other proteins. Although activity of a switch, as demonstrated here, is necessary for those changes, it is not sufficient for a protein to be redox regulated (23). Several of the proteins we identify have been studied in vitro, through Cys mutagenesis and with a focus on the impact of Cys redox status on enzymatic activity. Fumarase and succinate dehydrogenase activity were inhibited by TRX-mediated reduction (29). Also for mitochondrial citrate synthase redox regulation was observed (51), while no change of activity was found for malate dehydrogenase 1 (52). Our study now provides the missing evidence for the operation of those thiol switches in situ and reveals activity changes to physiological redox changes also for NAD-ME and ACO (Fig. 3H). The well-characterized thiol switch of the AOX dimer was previously shown to respond to specific respiratory substrates supplied to isolated tobacco mitochondria, including citrate, but not others, such as 2OG (53). Those observations elegantly match our findings at the proteome level and by in situ redox sensing. They further validate our interpretation that endogenous reduction of NADPH, but not NADH, by matrix metabolism leads to a restart of the endogenous thiol redox systems in mitochondria. The thiol-switched Cys peptide of the AOX1a, however, was not identified in our dataset, probably due to the cutoff criterion for the proteomic data evaluation (≥7 AA; the predicted Cys peptide is 5 AA) (SI Appendix, Fig. S10B). While there is no prior evidence for Cys-based redox control of other respiratory chain components in plants, our observation of the respiratory chain as a hotspot of operational thiol switching is mirrored by thiol switches that regulate the activities of mitochondrial complex I, complex V in mammalian systems (recently reviewed in ref. 23).
A growing number of thiol redox proteomes have been generated for various organisms under resting and stress conditions, but none captures the transition between a quiescent and an active state of metabolism (38, 54–60). Global oxidation of 27.1% in the inactive plant mitochondria is higher than degrees of oxidation found previously in various organisms (10 to 20%), while metabolic activation established 13.1% oxidation, which matches observations in other metabolically active systems. A noteworthy exception is a redox proteome of the heads and thoraces from fasting Drosophila, which showed 30.5% oxidation overall (shifted from 19.6% in the nonfasting control), i.e., a similar value as the quiescent plant mitochondria (59). Overexpression of catalase to scavenge H2O2 as a potential oxidant did not decrease the degree of oxidation in the fasting flies, indicating a dominant effect of the reductant supply through metabolism. Fasting Drosophila and quiescent plant mitochondria may have in common that metabolic flux is insufficient to provide enough NADPH to operate the thiol redox machinery effectively. By providing substrate to mitochondrial metabolism, the degree of Cys oxidation is effectively lowered, reaching comparable levels to the in vivo values of metabolically active organisms.
We conclude that redox regulation contributes to efficient metabolism during early seed germination. To investigate if this regulatory strategy is conserved as a principle of metabolic regulation it will be interesting to look at other systems that show similarly pronounced metabolic transitions in the future, for instance, the metabolic activation of an egg cell after fertilization. That would make Cys-based redox regulation of chloroplast metabolism, which is the founding example and particularly well studied (61), a specific case of a more fundamental principle of regulation during metabolic transitions.
Materials and Methods
Plant Material and Growth.
Synchronized plants were used for seed production. Nondormant and not-stratified seeds of Arabidopsis thaliana (L.) Heynh. Columbia-0 were used for all assays. Cytosolic ATeam 1.03 nD/nA (15) and mitochondrial roGFP2(-Grx1) (18, 19) lines were used for in vivo sensing and redox proteomics. A detailed description of the plant lines and growth conditions is provided in SI Appendix, Materials and Methods.
Fluorimetry and Confocal Imaging.
Fluorimetry and confocal imaging of fluorescent biosensors was performed as described (15, 62). For confocal imaging of isolated Arabidopsis embryos, seeds were imbibed on wet filter paper before the embryo was isolated using tweezers. For fluorimetry, intact seeds were rehydrated in 384-well microplates during the measurement by automated water injection in a CLARIOstar plate reader (BMG LABTECH GmbH) at 25 °C. Oxygen consumption of intact seeds was measured with MitoXpress Xtra fluorescent dye (Agilent) as described previously (63).
Differential iodoTMT Labeling and Protein Mass Spectrometry.
A detailed description is provided in SI Appendix, Materials and Methods.
Data Availability.
All plant lines are available upon request. Mass spectrometry proteomics data are available via the JPOST repository: https://repository.jpostdb.org/preview/11304555215d0a5e5be8428.
Supplementary Material
Acknowledgments
We thank Jürgen Eirich and Paulina Pieloch (Münster) for support with proteomic data handling and Western blotting, and the Deutsche Forschungsgemeinschaft (DFG) for financial support through the Emmy-Noether program (SCHW1719/ 1-1), the priority program SPP1710 “Dynamics of thiol-based redox switches in cellular physiology” (SCHW1719/ 7-1, ME1567/9-1/2, LI 984/3-1/2), the infrastructure grant INST 211/744-1 FUGG and the project grants (SCHW1719/5-1, FI1655/3-1) as part of the package PAK918.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Mass spectrometry proteomics data are available via the JPOST repository: https://repository.jpostdb.org/preview/11304555215d0a5e5be8428, access code: 7446.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910501117/-/DCSupplemental.
References
- 1.Sallon S., et al. , Germination, genetics, and growth of an ancient date seed. Science 320, 1464 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Pracharoenwattana I., Cornah J. E., Smith S. M., Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17, 2037–2048 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eastmond P. J., Graham I. A., Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6, 72–78 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Galland M., et al. , Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol. Cell. Proteomics 13, 252–268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Née G., Xiang Y., Soppe W. J. J., The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14 (2017). [DOI] [PubMed] [Google Scholar]
- 6.El-Maarouf-Bouteau H., Bailly C., Oxidative signaling in seed germination and dormancy. Plant Signal. Behav. 3, 175–182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arc E., et al. , Reboot the system thanks to protein post-translational modifications and proteome diversity: How quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics 11, 1606–1618 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Buchanan B. B., The path to thioredoxin and redox regulation beyond chloroplasts. Plant Cell Physiol. 58, 1826–1832 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Meyer Y., Belin C., Delorme-Hinoux V., Reichheld J. P., Riondet C., Thioredoxin and glutaredoxin systems in plants: Molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 17, 1124–1160 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Hopkins F. G., Morgan E. J., Appearance of glutathione during the early stages of the germination of seeds. Nature 152, 288–290 (1943). [Google Scholar]
- 11.Kobrehel K., et al. , Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol. 99, 919–924 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yano H., Wong J. H., Cho M. J., Buchanan B. B., Redox changes accompanying the degradation of seed storage proteins in germinating rice. Plant Cell Physiol. 42, 879–883 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Alkhalfioui F., et al. , Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol. 144, 1559–1579 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong J. H., et al. , Thioredoxin reduction alters the solubility of proteins of wheat starchy endosperm: An early event in cereal germination. Plant Cell Physiol. 45, 407–415 (2004). [DOI] [PubMed] [Google Scholar]
- 15.De Col V., et al. , ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 6, e26770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E., Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697–709 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Shu K., Liu X. D., Xie Q., He Z. H., Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Albrecht S. C., et al. , Redesign of genetically encoded biosensors for monitoring mitochondrial redox status in a broad range of model eukaryotes. J. Biomol. Screen. 19, 379–386 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Schwarzländer M., et al. , Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 231, 299–316 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Marty L., et al. , Arabidopsis glutathione reductase 2 is indispensable in plastids, while mitochondrial glutathione is safeguarded by additional reduction and transport systems. New Phytol. 224, 1569–1584 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Fricker M. D., Quantitative redox imaging software. Antioxid. Redox Signal. 24, 752–762 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Berndt C., Schwenn J. D., Lillig C. H., The specificity of thioredoxins and glutaredoxins is determined by electrostatic and geometric complementarity. Chem. Sci. 6, 7049–7058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nietzel T., Mostertz J., Hochgräfe F., Schwarzländer M., Redox regulation of mitochondrial proteins and proteomes by cysteine thiol switches. Mitochondrion 33, 72–83 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Xiang Y., et al. , Sequence polymorphisms at the REDUCED DORMANCY5 pseudophosphatase underlie natural variation in Arabidopsis dormancy. Plant Physiol. 171, 2659–2670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Møller I. M., Rasmusson A. G., The role of NADP in the mitochondrial matrix. Trends Plant Sci. 3, 21–27 (1998). [Google Scholar]
- 26.Huang J., et al. , Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. U.S.A. 116, 21256–21261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichheld J. P., Meyer E., Khafif M., Bonnard G., Meyer Y., AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 579, 337–342 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Reichheld J. P., et al. , Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19, 1851–1865 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daloso D. M., et al. , Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, E1392–E1400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weerapana E., et al. , Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bak D. W., Pizzagalli M. D., Weerapana E., Identifying functional cysteine residues in the mitochondria. ACS Chem. Biol. 12, 947–957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braun H. P., et al. , The life of plant mitochondrial complex I. Mitochondrion 19, 295–313 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Balmer Y., et al. , Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 101, 2642–2647 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouhier N., et al. , Identification of plant glutaredoxin targets. Antioxid. Redox Signal. 7, 919–929 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Yoshida K., Noguchi K., Motohashi K., Hisabori T., Systematic exploration of thioredoxin target proteins in plant mitochondria. Plant Cell Physiol. 54, 875–892 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Winger A. M., Taylor N. L., Heazlewood J. L., Day D. A., Millar A. H., Identification of intra- and intermolecular disulphide bonding in the plant mitochondrial proteome by diagonal gel electrophoresis. Proteomics 7, 4158–4170 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Martí M. C., et al. , Mitochondrial and nuclear localization of a novel pea thioredoxin: Identification of its mitochondrial target proteins. Plant Physiol. 150, 646–657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chouchani E. T., et al. , Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature 532, 112–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marty L., et al. , The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 9109–9114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hildebrandt T. M., Nunes Nesi A., Araújo W. L., Braun H. P., Amino acid catabolism in plants. Mol. Plant 8, 1563–1579 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Law S. R., et al. , Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol. 158, 1610–1627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raveneau M. P., Benamar A., Macherel D., Water content, adenylate kinase, and mitochondria drive adenylate balance in dehydrating and imbibing seeds. J. Exp. Bot. 68, 3501–3512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paszkiewicz G., Gualberto J. M., Benamar A., Macherel D., Logan D. C., Arabidopsis seed mitochondria are bioenergetically active immediately upon imbibition and specialize via biogenesis in preparation for autotrophic growth. Plant Cell 29, 109–128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hourmant A., Pradet A., Oxidative-phosphorylation in germinating lettuce seeds (Lactuca sativa) during the first hours of imbibition. Plant Physiol. 68, 631–635 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benamar A., Tallon C., Macherel D., Membrane integrity and oxidative properties of mitochondria isolated from imbibing pea seeds after priming or accelerated ageing. Seed Sci. Res. 13, 35–45 (2003). [Google Scholar]
- 46.Florez-Sarasa I., et al. , The lack of mitochondrial thioredoxin TRXo1 affects in vivo alternative oxidase activity and carbon metabolism under different light conditions. Plant Cell Physiol. 60, 2369–2381 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Reinholdt O., et al. , Redox-regulation of photorespiration through mitochondrial thioredoxin o1. Plant Physiol. 181, 442–457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelhard J., et al. , In situ kinetic trapping reveals a fingerprint of reversible protein thiol oxidation in the mitochondrial matrix. Free Radic. Biol. Med. 50, 1234–1241 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Waszczak C., et al. , Sulfenome mining in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 111, 11545–11550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hooper C. M., et al. , SUBAcon: A consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30, 3356–3364 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Schmidtmann E., et al. , Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol. Plant 7, 156–169 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Yoshida K., Hisabori T., Adenine nucleotide-dependent and redox-independent control of mitochondrial malate dehydrogenase activity in Arabidopsis thaliana. Biochim. Biophys. Acta 1857, 810–818 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Vanlerberghe G. C., Day D. A., Wiskich J. T., Vanlerberghe A. E., McIntosh L., Alternative oxidase activity in tobacco leaf mitochondria (Dependence on tricarboxylic-acid cycle-mediated redox regulation and pyruvate activation). Plant Physiol. 109, 353–361 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leichert L. I., et al. , Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 8197–8202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brandes N., Reichmann D., Tienson H., Leichert L. I., Jakob U., Using quantitative redox proteomics to dissect the yeast redoxome. J. Biol. Chem. 286, 41893–41903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brandes N., et al. , Time line of redox events in aging postmitotic cells. eLife 2, e00306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Araki K., et al. , Redox sensitivities of global cellular cysteine residues under reductive and oxidative stress. J. Proteome Res. 15, 2548–2559 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Knoefler D., et al. , Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol. Cell 47, 767–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menger K. E., et al. , Fasting, but not aging, dramatically alters the redox status of cysteine residues on proteins in Drosophila melanogaster. Cell Rep. 11, 1856–1865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Topf U., et al. , Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 9, 324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buchanan B. B., The path to thioredoxin and redox regulation in chloroplasts. Annu. Rev. Plant Biol. 67, 1–24 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Nietzel T., et al. , The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H2O2 and thiol redox integration and elucidates intracellular H2O2 dynamics during elicitor-induced oxidative burst in Arabidopsis. New Phytol. 221, 1649–1664 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Sechet J., et al. , The ABA-deficiency suppressor locus HAS2 encodes the PPR protein LOI1/MEF11 involved in mitochondrial RNA editing. Mol. Plant 8, 644–656 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plant lines are available upon request. Mass spectrometry proteomics data are available via the JPOST repository: https://repository.jpostdb.org/preview/11304555215d0a5e5be8428.