Significance
Circadian clocks are present in most cells of the body and act in concert to coordinate our daily physiology and behavior with the environment. Circadian misalignment between body clocks and the environment has been implicated in various pathologies. Here, we show that circadian misalignment can also occur between clocks of different peripheral tissues following exposure to hypoxia and that it stems from the differential response of core clock components to hypoxia. Intertissue circadian misalignment also occurs upon intermittent hypoxia, a model for obstructive sleep apnea (OSA). Our findings highlight the potential role of internal circadian misalignment in the pathophysiology of OSA and potentially other hypoxia-related diseases.
Keywords: circadian clock, metabolism, hypoxia, PER2::LUC mice, obstructive sleep apnea
Abstract
The occurrence and sequelae of disorders that lead to hypoxic spells such as asthma, chronic obstructive pulmonary disease, and obstructive sleep apnea (OSA) exhibit daily variance. This prompted us to examine the interaction between the hypoxic response and the circadian clock in vivo. We found that the global transcriptional response to acute hypoxia is tissue-specific and time-of-day–dependent. In particular, clock components differentially responded at the transcriptional and posttranscriptional level, and these responses depended on an intact circadian clock. Importantly, exposure to hypoxia phase-shifted clocks in a tissue-dependent manner led to intertissue circadian clock misalignment. This differential response relied on the intrinsic properties of each tissue and could be recapitulated ex vivo. Notably, circadian misalignment was also elicited by intermittent hypoxia, a widely used model for OSA. Given that phase coherence between circadian clocks is considered favorable, we propose that hypoxia leads to circadian misalignment, contributing to the pathophysiology of OSA and potentially other diseases that involve hypoxia.
Hypoxia plays a critical role in various pathologies, including ischemic myocardial infarction, cerebrovascular accidents, asthma, chronic obstructive pulmonary disease, and obstructive sleep apnea (OSA) (1). Intriguingly, the occurrence as well as the severity of their sequelae often exhibit daily variance (2, 3), hinting at a potential involvement of the circadian clock in these hypoxia-related disorders.
The mammalian circadian clock consists of a master clock in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus and a multitude of cellular oscillators throughout the body. These molecular oscillators rely on negative transcription-translation feedback loops of several genes, so-called core-clock components and their products (e.g., Arntl [also known as Bmal1], Clock, Per1,2, Cry1,2, Nr1d1,2 [also known as Rev-erbα,β], and Rorα,β,γ) (4, 5). Their concerted action drives the daily rhythms of myriad metabolic, physiologic, and behavioral processes (6–8). Despite their autonomous and self-sustained nature, circadian clocks need to be adjusted to the external time as well as synchronized with each other to be effective. This is achieved through various external and internal timing cues, known as zeitgebers, which convey the time to all clocks (6).
Recent studies point toward an interaction between circadian clocks and the hypoxia signaling pathway. Under physiological conditions, circadian clocks drive daily oscillations in oxygen levels, which in turn can synchronize clocks through HIF1α (9, 10). Concurrently, clock components physically interact with and modulate HIF1α levels and activity (11–15). However, to date, the day-time–dependent effects of hypoxia on different tissues and their circadian control have not been directly tested in vivo. To this end, we exposed mice to hypoxic conditions and asked 1) whether the transcriptional response to hypoxia in different tissues is time-of-day–dependent and clock-controlled; and 2) what the effect of hypoxia is on clocks of different tissues. We observed the global transcriptional response to hypoxia to be highly tissue-specific, time-of-day–dependent, and circadian-clock–controlled. Moreover, several core clock genes responded to hypoxia in a tissue-specific manner and consequently elicited intertissue circadian clock misalignment. Importantly, this phenomenon also occurred upon exposure to intermittent hypoxia, a model for OSA, supporting its potential role in the disease’s pathophysiology.
Results
The Transcriptional Response to Hypoxia Is Time-of-Day–Dependent and Tissue-Specific.
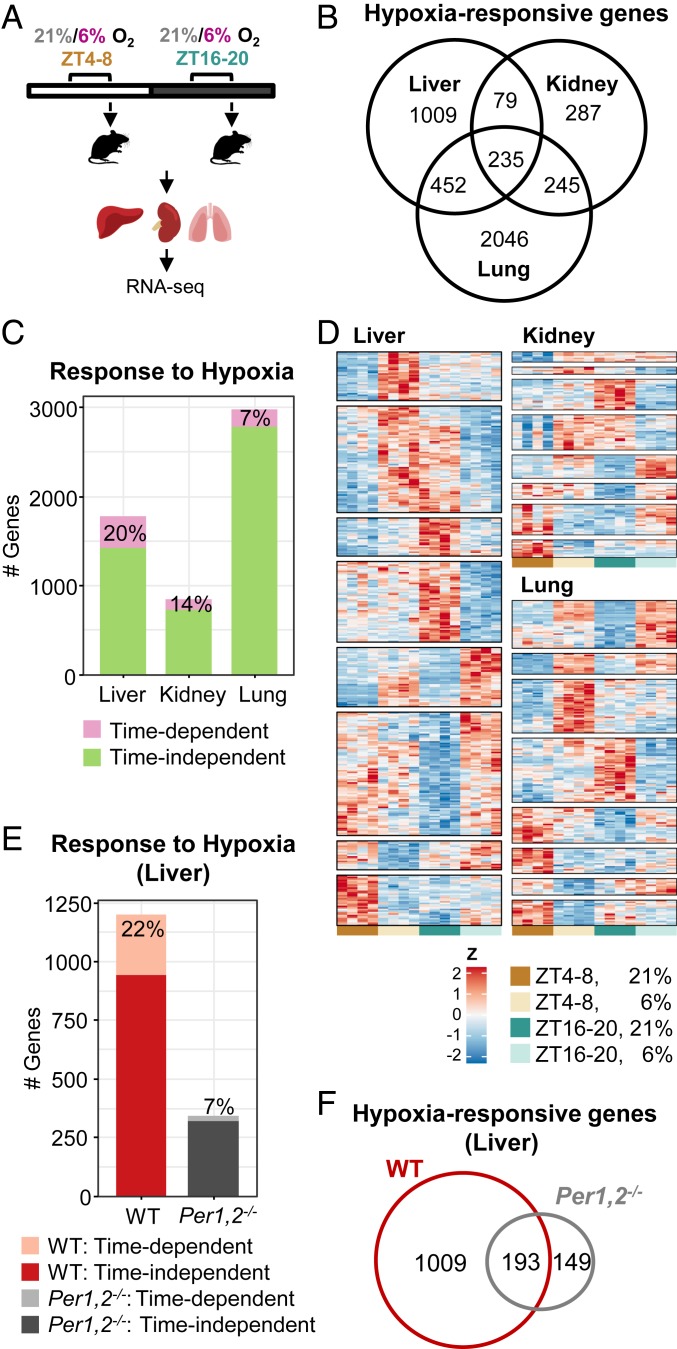
We first examined whether the transcriptional response to hypoxia differs throughout the day. To this end, mice were housed under a 12-h light–dark regimen and were exposed to hypoxia (6% O2) or normoxia (21% O2) for 4 h, during either the light (zeitgeber time [ZT] 4–8) or the dark phase (ZT 16–20). Subsequently, animals were killed, selected organs (liver, kidney, and lung) were harvested, and their transcriptome was analyzed by RNA-Seq (Fig. 1A) (16). Overall, the response to hypoxia (hypoxia effect) was more prominent compared to the basal changes in gene expression between the two time points (basal time effect) in each tissue (SI Appendix, Fig. S1A). The extent of the transcriptional response to hypoxia differed between the tissues, with the lung exhibiting the highest effect on gene expression followed by the liver and kidney (2,978, 1,775, and 846 genes, respectively) regardless of time of day. Furthermore, the transcriptional response to hypoxia was highly tissue-specific with a relatively small overlap (235 genes) between the different tissues (Fig. 1B) despite high overlap in detected genes (SI Appendix, Fig. S1B). Next, we screened for genes that exhibit a significant interaction between their hypoxic response and time of day, hence exhibiting a time-dependent response to hypoxia. We found that a substantial fraction of the transcriptional response to hypoxia in the different tissues was time-dependent, with the highest propensity in the liver (≈20%) followed by the kidney (≈14%) and the lung (≈7%) (Fig. 1C). This time-dependent response was tissue-specific as well (SI Appendix, Fig. S1C). Intriguingly, the lung exhibited the highest transcriptional response to hypoxia, with the lowest time-dependent fraction. We clustered the time-dependent responsive genes according to their expression patterns—that is, genes that responded exclusively at one time point but not at the other, or responded at both time points to a different degree or in different directions (e.g., up-regulated at one time point and down-regulated at the other) (Fig. 1D). The response of several canonical hypoxia target genes (e.g., Slc2a1 [also known as Glut1], Egln3) to dimethyloxallyl glycine (DMOG), an HIF1α activator, was previously reported to be more pronounced during the light phase (13). However, we found that the effect of hypoxia in vivo is far more complex and gene-specific (Fig. 1D and SI Appendix, Fig. S1D). Additionally, motif analysis of promoters of the hypoxia-regulated genes showed a significant enrichment of HIF1-binding sites in the liver (SI Appendix, Fig. S1E). Other transcription factors were overrepresented on the genes’ promoters (SI Appendix, Fig. S1E and Dataset S7), hinting that the response to hypoxia in vivo might not exclusively rely on HIF1α.
Fig. 1.
The transcriptional response to hypoxia is time- and tissue-dependent. (A) Schematic representation; mice were exposed to 4 h of either hypoxia (6% O2) or normoxia (21% O2) in the light (ZT 4–8) or dark (ZT 16–20) phase. Animals were killed, tissues were harvested, and RNA was prepared and sequenced. (B) Venn diagram representing the number of genes that significantly responded to hypoxia in each tissue, including response at either of the time points or a significant interaction (stage-wise analysis, Overall False Discovery Rate [OFDR] < 0.05, adjusted P < 0.05, n = 4 per condition) (see gene lists in Dataset S3). (C) Number of genes that responded in a time-dependent manner (significant interaction: adjusted P < 0.05) and a time-independent manner (significant response in either of the time points and nonsignificant interaction). (D) Heatmap representation of time-dependent responsive genes in each tissue, clustered by their expression pattern. (E) Number of genes that responded to hypoxia in a time-dependent and a time-independent manner in the livers of WT and Per1,2−/− mice housed in constant dark. (OFDR < 0.05, n = 3 per condition). (F) Venn diagram representing the number of genes that significantly responded to hypoxia (both time-dependent and time-independent) in the livers of WT or Per1,2−/− mice (see gene lists in Dataset S5).
To further characterize this time- and tissue-dependent response to hypoxia, we performed a pathway enrichment analysis and found that some pathways were enriched in all three tissues at both time points (e.g., HIF- and circadian-clock–related), while others were enriched exclusively at a specific time or in a specific tissue (SI Appendix, Fig. S2). Overall, we detected a functional signature that is time- and tissue-specific. This suggests that a different genetic program is activated in response to hypoxia at different times of the day, presumably allowing an adaptive response that addresses tissue-specific metabolic needs at that particular time.
The Time-Dependent Transcriptional Response to Hypoxia Is Circadian-Clock–Controlled.
The time-dependent response to hypoxia can be a mere reaction to environmental changes, primarily the light–dark cycle, or endogenously driven by the circadian clock. To discriminate between the two scenarios, we examined the transcriptional response to hypoxia at the same respective times of day but in constant dark (i.e., circadian time [CT] 4–8 and CT 16–20) (16). Overall, a similar fraction (≈22%) of hypoxia-responsive genes in the liver maintained time dependency in constant dark (Fig. 1E). We therefore concluded that the time-dependent transcriptional response to hypoxia is mostly endogenously driven. This prompted us to test whether the molecular circadian oscillator plays a role in the observed time-dependent response. To this end, we repeated the experiment in constant dark, this time with whole-body clock mutant Per1,2−/− mice. Per1,2−/− mice exhibit arrhythmic behavior, physiology, and gene expression in constant dark (9, 17, 18). Consistently, the basal time-of-day variance in gene expression observed for the livers of wild-type (WT) mice in constant dark was obliterated in Per1,2−/− mice (SI Appendix, Fig. S3A). Furthermore, in line with recent reports on interaction between PER2 and HIF1α (11, 19, 20), we found that the overall response to hypoxia in the liver was markedly attenuated and largely different in Per1,2−/− mice compared to WT mice (Fig. 1 E and F and SI Appendix, Fig. S3B). Notably, the time-dependent response to hypoxia was largely abolished in Per1,2−/− mice (Fig. 1E).
Given that the time-dependent response to hypoxia was largely preserved in constant dark and markedly eliminated in circadian-clock–deficient mice, we concluded that the time-dependent response to hypoxia, at least in the liver, is circadian-clock–controlled.
The Immediate Response of Core Clock Components to Hypoxia Is Regulated Both Transcriptionally and Posttranscriptionally.
Previous reports revealed that clock genes respond to hypoxia and HIF1α-activating drugs in a variety of cultured cell lines, and identified the presence of the HIF-responsive element (HRE) in their promoter regions (10, 13, 21–23). Along this line, our analysis evinced that, in all tissues examined, circadian-clock–related pathways were enriched among the hypoxia-responsive genes (SI Appendix, Fig. S2).
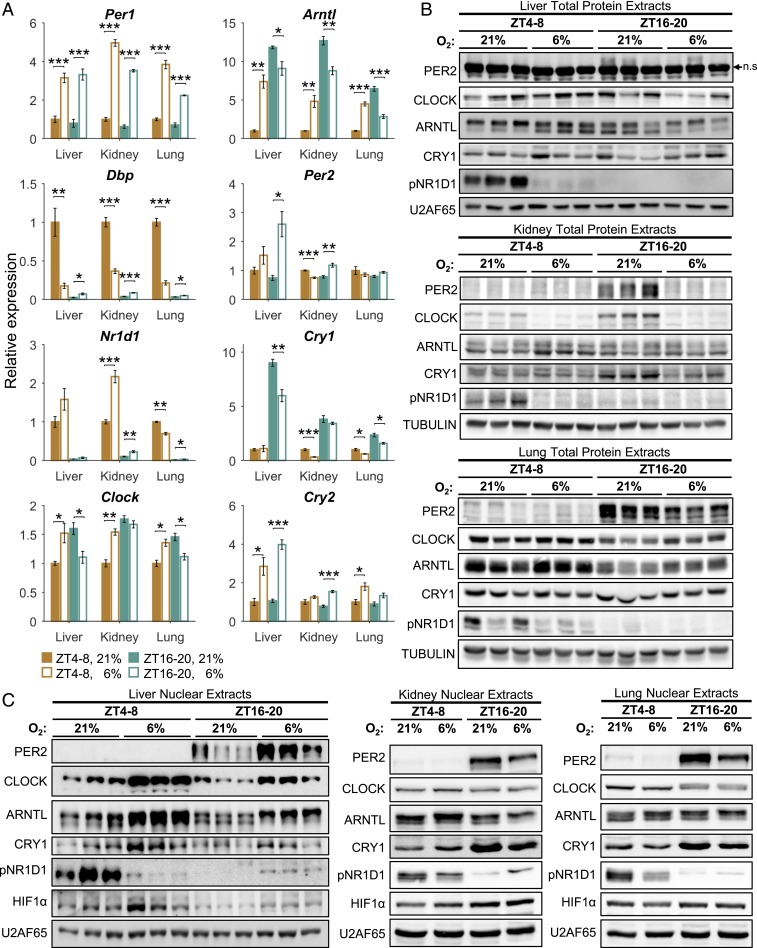
Although the majority of core clock genes responded to hypoxia, the transcriptional response was far more complex than foreseen. We categorized it into three main groups as follows: 1) tissue- and time-independent (e.g., Per1); 2) tissue-independent but time-dependent (e.g., Arntl, Dbp); and, the most common, 3) tissue- and time-dependent (e.g., Cry1, Nr1d1, Per2) (Fig. 2A).
Fig. 2.
Clock-associated genes respond to acute hypoxia in a time- and tissue-specific manner. (A) qPCR analysis of clock-associated transcript levels under normoxia (21% O2) or hypoxia (6% O2) in the light (ZT 4–8) or dark (ZT 16–20) phase in different tissues (mean ± SE, n = 4 per condition; *P < 0.05, **P < 0.01, ***P < 0.001, two-sample Student’s t test). (B) Immunoblot analysis of total protein extracts from normoxic and hypoxic mice as in A (n = 3; n.s., nonspecific band). (C) Immunoblot analysis of nuclear extracts from normoxic and hypoxic mice as in A (liver: n = 3; kidney and lung: pools of n = 4).
Core clock components are extensively controlled posttranscriptionally (e.g., protein stability, modifications, and localization) (24). Analysis of whole- and nuclear-protein extracts from the different tissues revealed that the time-dependent and tissue-specific response to hypoxia is regulated posttranscriptionally as well (i.e., protein accumulation and nuclear localization) (Fig. 2 B and C). Intriguingly, the response observed in the protein level did not directly correspond to the changes observed in their transcript levels. For example, liver Cry1 transcript levels were down-regulated upon hypoxia exclusively in the dark phase (Fig. 2A), whereas CRY1 total protein levels were mildly affected in both dark and light phases (Fig. 2B). However, CRY1’s nuclear protein levels were strongly elevated, particularly in the dark phase (Fig. 2C). Interestingly, the nuclear accumulation of HIF1α in response to hypoxia differed between tissues and time of day (Fig. 2C). Collectively, our results suggest that the tissue-specific and time-of-day–dependent response to hypoxia of core clock components is regulated at multiple levels, from gene transcription to protein accumulation and localization.
The Response of Clock Components to Hypoxia Is Circadian-Clock–Controlled.
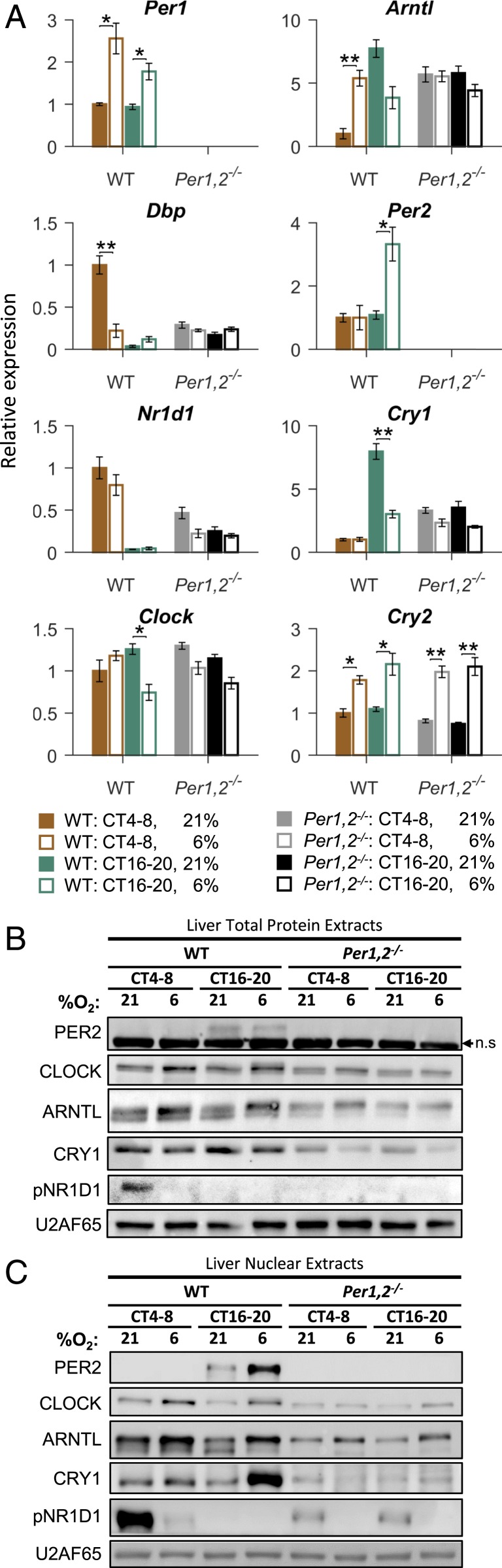
We next examined whether the response of clock components to hypoxia is circadian-clock–controlled. Experiments conducted with the clock mutant Per1,2−/− mice revealed that the time-dependent effect of hypoxia on clock gene expression requires a functional clock (Fig. 3A and SI Appendix, Fig. S4). For example, in the liver several clock genes lost their time-dependent response and did not respond to hypoxia at all (e.g., Dbp, Arntl), whereas other genes retained their time-independent hypoxic response (e.g., Cry2) (Fig. 3A). Consistently, we did not observe any significant time-dependent effects at the protein level in the livers of Per1,2−/− mice (Fig. 3 B and C). The clock components that retained their hypoxic response are potential participants in the input pathway to the core clock, while the components that lost their response are probably downstream effectors.
Fig. 3.
Time-dependent response of liver core clock components to hypoxia is circadian-clock–controlled. (A) qPCR analysis of clock-associated transcript levels in the livers of WT or Per1,2−/− mice housed in constant dark and exposed to hypoxia (6% O2) or normoxia (21% O2) in the respective light (CT 4–8) or dark (CT 16–20) phase (mean ± SE, n = 4 per condition; *P < 0.05, **P < 0.01, two-sample Student’s t test). (B) Immunoblot analysis of total liver protein extracts from WT or Per1,2−/− mice as in A (pools of n = 4; n.s., nonspecific band). (C) Immunoblot analysis of liver nuclear extracts from WT or Per1,2−/− mice as in A (pools of n = 4).
Next, we asked whether the time-dependent effect of hypoxia on clock gene expression in the liver requires a functional liver clock or can be systemically driven by other clocks (e.g., the SCN). To this end, we performed the experiment described previously, this time with mice deficient in hepatocyte clocks—namely, Bmal1 liver-specific knockout (BLKO; Alb-Cre+ Bmal1fl/fl) mice (25, 26)—alongside Alb-Cre control mice. As in the Per1,2−/− mice, in the BLKO mice some core clock genes ceased to respond to hypoxia (e.g., Dbp) while others retained their hypoxic response yet lost the time-dependent effect (e.g., Cry1) (SI Appendix, Fig. S5A). These effects were also apparent to some degree at the protein level (SI Appendix, Fig. S5B). Notably, we detected some systemically driven temporal changes both in basal PER2’s protein levels [consistent with previous reports (27)] and in its hypoxic response (SI Appendix, Fig. S5B). We concluded that the tissue-specific and time-dependent response of clock components to hypoxia is circadian-clock–controlled, mostly by tissue-endogenous clocks.
Hypoxia In Vivo Can Elicit Intertissue Circadian Clock Misalignment.
The immediate response of clock genes to hypoxia prompted us to examine whether these changes can phase-shift the clock. Moreover, since the response of clock genes varied between different tissues, we posited that the extent or direction of the phase shift might differ as well.
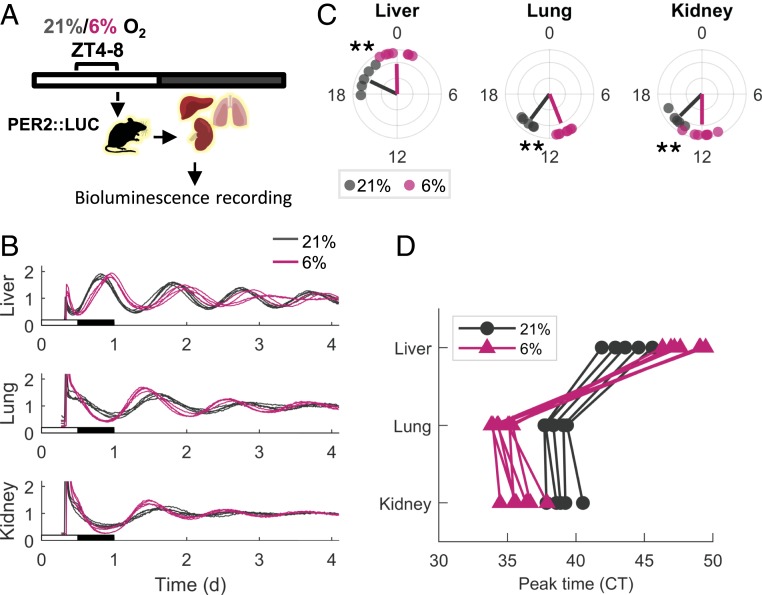
To test our hypothesis, we employed PER2::LUC mice, which express a PERIOD2::LUCIFERASE fusion protein that can be used as a real-time reporter of circadian dynamics in mice (28). Tissues were harvested from PER2::LUC mice following 4-h exposure to hypoxia (6% O2) or normoxia (21% O2) during the light phase (ZT 4–8) (Fig. 4A). Organotypic slices were prepared, and their bioluminescence was recorded for several consecutive days. The phase was determined based on the time of the first peak in the bioluminescence recordings ex vivo. Strikingly, the liver traces were phase-delayed in the hypoxia-treated animals compared to the controls while the kidney and the lung were phase-advanced (Fig. 4 B and C). Thus, the phase relationship between the tissues was derailed (Fig. 4D). Intriguingly, when the tissues were harvested 24 h following hypoxia treatment, rather than immediately after exposure (i.e., ZT 8 on the following day) (SI Appendix, Fig. S6A), both the kidney and the lung were still phase-advanced whereas the liver returned to its original phase (SI Appendix, Fig. S6B). This suggested the presence of mechanisms that specifically restore the phase of the liver but not the clock in the kidney or the lung.
Fig. 4.
Hypoxia phase-shifts the clock in a tissue-specific manner based on PER2::LUC bioluminescence recordings. (A) PER2::LUC mice were exposed to 4 h of either hypoxia (6% O2) or normoxia (21% O2) in the light phase (ZT 4–8). Animals were killed, and tissues were harvested and sliced for bioluminescence recordings. (B) Representative relative bioluminescence plots of the PER2::LUC tissue slices (three to five slices from each mouse are shown). The x-axes are aligned with the original light–dark schedule of the mice. (C) Polar plot of the phase distribution of PER2::LUC bioluminescence. Each point represents the CT of the first peak on the second day of recordings of a single mouse (mean of three to five technical replicates). Lines’ angle represents the circular mean of each condition, and lines’ radius anticorrelates with the circular variance (n = 5 for normoxia, n = 7 for hypoxia; **P < 0.01, Watson–Williams test). (D) Phase map representation of the phase relation between tissues. Each line connects the phases of tissues from the same animal.
Feeding is widely considered a dominant zeitgeber for clocks in peripheral tissues (29). Previous reports demonstrated that the liver clock is highly responsive to feeding time (30–32) while the response of other peripheral tissues to feeding is in general less characterized. Since the mice had access to food following exposure to hypoxia, we posited that food ingestion might predominantly affect the liver clock and to a lesser extent the kidney and lung clocks. In this conjecture, mice were exposed to a single episode of daytime-restricted feeding and the effect of feeding on the clock in the liver, lung, and kidney was assayed using PER2::LUC organotypic slices (SI Appendix, Fig. S6 C and D). While the liver clock was considerably shifted, both the lung and the kidney clocks were hardly affected by a single bout of daytime feeding.
Overall, these results indicated that in vivo hypoxia can phase-shift the clock in a tissue-specific manner and elicit intertissue circadian clock desynchrony. Notably, feeding predominantly affected the liver clock and likely restored its original phase. However, it had little effect on the lung and kidney clocks, which retained their hypoxia-induced phase. Thus, different timing cues (e.g., oxygen versus feeding) vary in their dominance over circadian clocks in various tissues.
Tissue-Intrinsic Components Differentially Control the Circadian Clock Response to Hypoxia.
The observed differences in circadian clock responses to hypoxia in the various tissues raised the question of whether these disparities stem from extrinsic (e.g., different oxygen tensions in each organ) or intrinsic tissue-specific properties.
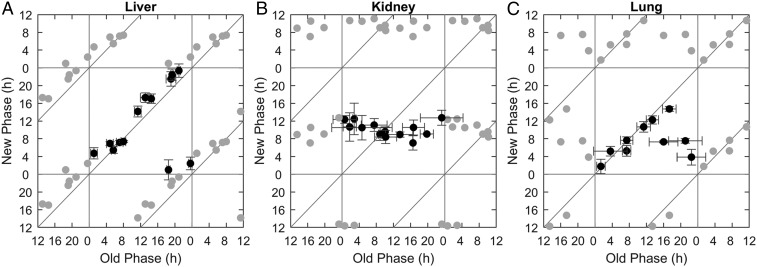
To explicitly test whether the clock response to hypoxia can be intrinsically driven in a tissue-specific manner, we exposed PER2::LUC organotypic slices to hypoxia ex vivo (4-h 2.5% O2) at different times of the day. This enabled us to generate phase transition curves (PTCs) (33), in which the new phase was plotted against the old phase at high temporal resolution for each tissue. The PTCs largely differed between the different tissues (Fig. 5). The liver PTC had a slope of 1 (Fig. 5A) and hence qualified as, in chronobiological parlance, type 1 resetting, which is considered weak since it elicits only modest phase shifts (34). By contrast, the PTC slope was zero in the kidney (Fig. 5B), type 0 resetting, which is regarded as strong (i.e., the new phase is similar irrespective of the old phase). The lung PTC was qualified as type 0 with a large “dead zone” (where no response was observed) (Fig. 5C). Based on these qualitative differences between the PTCs of each tissue upon hypoxia ex vivo, we concluded that tissue-intrinsic components are sufficient to differentially shift the clock in response to hypoxia and may account for hypoxia-induced intertissue clock misalignment in vivo.
Fig. 5.
Tissue-intrinsic components differentially control the circadian clock response to hypoxia. PTCs for ex vivo hypoxia in the liver (A) the kidney (B), and the lung (C). PER2::LUC organotypic slices were cultured and exposed to hypoxia (4-h 2.5% O2) ex vivo at different times throughout the day, and their phases posttreatment were compared to those of untreated control slices (21% O2). The x-axis represents the initial phase in hours relative to the hypoxia pulse, retrieved from the control slices, and the y-axis represents the posttreatment phase, retrieved from the treated slices. Each point represents the results of one initial phase from one mouse (some mice were used for two different treatment times). Gray points are duplications of the original data for the sake of presentation (n = 9 mice for liver, n = 8 for kidney, and n = 6 for lung; mean ± SD for each axis).
Sustained and Intermittent Hypoxia, as in OSA, Elicits Intertissue Circadian Clock Desynchrony.
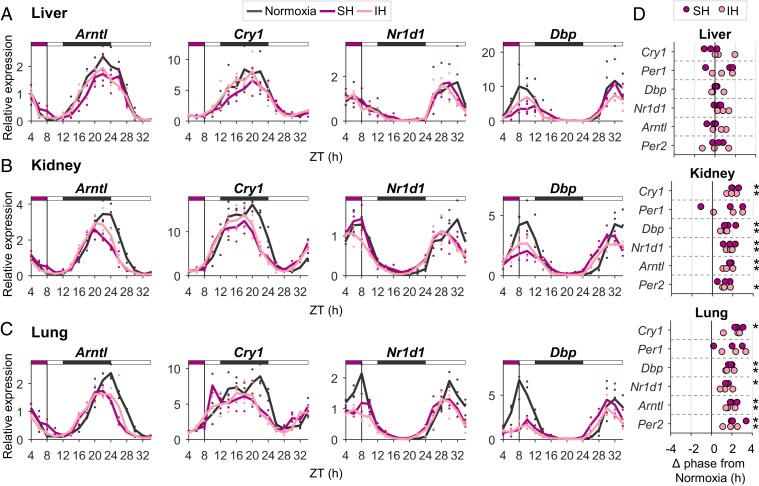
The above-described experiments using organotypic slices from PER2::LUC mice revealed variance in the capacity of the oscillator in different tissues to respond to hypoxia. To unequivocally determine the effect of hypoxia on the circadian clock of different tissues in vivo, mice were exposed to 4-h hypoxia (6% O2) or normoxia (21% O2) during the light phase (ZT 4–8) and tissues were harvested at 2-h intervals during and following the treatment for 24 h. RNA was prepared, and transcript levels of the core clock genes were determined by qPCR (Fig. 6 A–C; see SI Appendix, Fig. S7E). Upon hypoxia, the clock in the kidney and the clock in the lung were phase-advanced, whereas the liver clock was not affected (Fig. 6D), consistent with the above-described PER2::LUC organotypic slices data (SI Appendix, Fig. S6B). Thus, while the lung and kidney maintained their phase shift throughout the time course, the liver clock rapidly restored its original phase, likely in response to food ingestion at the beginning of the dark phase. Importantly, the phase relationship between the kidney, lung, and liver clocks was derailed following exposure to hypoxia. Thus, we concluded that hypoxia elicits intertissue circadian clock desynchrony in vivo.
Fig. 6.
Sustained and intermittent hypoxia, as in OSA, elicits intertissue circadian clock desynchrony in vivo. Mice were exposed to 4-h sustained hypoxia (SH; 6% O2), intermittent hypoxia (IH), or normoxia (21% O2) in the light phase (ZT 4–8). Animals were killed at 2-h intervals, from ZT 4 until ZT 10 on the day after. (A–C) qPCR analysis of transcript levels of representative clock genes in the liver (A), kidney (B), and lung (C). Solid lines are averages of three biological replicates; individual replicates are marked by dots; the upper bar represents the light–dark regimen; the purple box marks the time of hypoxia. (D) Analysis of phase differences (Δ phases) in clock gene expression between SH and IH relative to normoxia. Phases were obtained using a cosine fit (*P < 0.05, paired Student’s t test, n = 3).
OSA is a sleep disorder characterized by pauses in breathing or periods of shallow breathing during sleep. It affects about 3% to 7% of men and 2% to 5% of women in the Western population and is associated with obesity, diabetes, and metabolic syndrome (35). To examine the pathophysiological relevance of our findings, mice were exposed during the light phase (ZT 4–8) to intermittent hypoxia (21% to 6%, ≈30 cycles per h for 4 h), a widely used protocol for mimicking sleep apnea (36, 37). Very similarly to sustained hypoxia, the intermittent hypoxia protocol shifted the clocks of the liver and lung in opposing directions in organotypic slices (SI Appendix, Fig. S7 A–D) and elicited sustainable intertissue circadian clock desynchrony in vivo (Fig. 6 A–D and SI Appendix, Fig. S7E), whereby the kidney and lung clocks were phase-advanced and the liver clock maintained its original phase (Fig. 6D).
Discussion
In recent years, there has been growing interest in the relationship between circadian clocks and human health, from molecular mechanisms to therapeutic interventions. As aforementioned, hypoxia plays a critical role in a wide variety of common pathologies, yet whether and how the interaction between the circadian makeup and the hypoxic response plays a role in their pathophysiology is far from being resolved (38, 39). In the current study, we found that the transcriptional response to hypoxia is tissue-specific and time-of-day–dependent. Of note, our experimental design depicted only two opposing time points within the day, namely the middle of the light and dark phases. Therefore, it likely underestimates the extent of time dependency, as it favors genes with peaks and troughs of responsiveness within the sampling times. Likewise, our study was limited to three different tissues. To gain a comprehensive view of the phenomenon, one would need to increase the temporal resolution and test additional tissues. At any rate, the spatial and temporal variance in the response to hypoxia needs to be taken into consideration when studying both the deleterious and the beneficial effects of hypoxia (40, 41). Intriguingly, our motif analysis of hypoxia-responsive genes, and our finding that HIF1α nuclear accumulation upon hypoxia differs between tissues and is scant in some cases, might hint at the involvement of other transcription factors in the time- and tissue-specific response to hypoxia. Loss-of-function experiments are necessary to clearly determine the contribution of HIF1α in this regard.
The fact that the genetic response to hypoxia is time-dependent can be explained in several ways. One possibility is that a given tissue has different needs when coping with hypoxia at different times of the day and therefore different genetic programs are activated as a response. Another possibility is that the tissue is more customized for coping with hypoxic stress at one time point than the other. Both scenarios must stem from some basal differences between the time points which generate a different milieu (transcriptional, metabolic) for the hypoxic signal to be received. A similar reasoning is relevant to the observed tissue specificity. Importantly, we showed that, at least in the liver, determinants for the clock hypoxic response are largely controlled by tissue-endogenous factors rather than environmental cues or signals emerging from clocks in other tissues.
Hypoxia was studied as a zeitgeber in Drosophila from early on in the chronobiology field (42, 43). The main impetus for these early experiments was to show that circadian rhythms are endogenously driven and specifically that they are essentially metabolism-dependent. More recently, hypoxia was identified as a zeitgeber in vertebrates (10, 12, 13, 44). We previously showed that daily cycles of oxygen concentration, similar to those observed in tissues in vivo, are sufficient to synchronize clocks in a nonsynchronized cell population in cell culture (10). This finding suggests that oxygen levels can entrain peripheral clocks under physiological conditions. The current study expands on this view by demonstrating that hypoxia phase-shifts circadian clocks in vivo and that this effect is tissue-specific. This differential response has a tissue-intrinsic origin, as demonstrated by the ex vivo experiments. Different tissues differentially responded also to feeding signals in our experiments. It is noteworthy that experiments using organotypic slices should be interpreted cautiously (e.g., potential confounding effects due to tissue handling and ex vivo culture). Yet they are widely acceptable in the field (45) and, more importantly, the results are confirmed by an independent assay (qPCR data). Taken together, our results point toward differences in the dominance of different timing cues over each organ. While differences in the strength/rigidity of the oscillators themselves (46, 47) can explain why one tissue would be more sensitive to all cues compared to another tissue, it cannot explain differences that are signal-specific. Therefore, this type of tissue and signal specificity is likely to stem from differences in the mechanisms of input to the clock. Notably, the kidney is known to be particularly sensitive to oxygen levels and regulates blood circulation (48), while the liver is a metabolic hub for nutrient processing from food ingestion. Hence, in view of the dominance of feeding over the liver clock and that of oxygen over the kidney clock, it appears that tissue “specialization” corresponds to the degree of the tissue’s clock response to different systemic cues.
A consequence of the difference in zeitgeber dominance is the ability of a single cue to cause intertissue misalignment, as indeed demonstrated in this work for hypoxia. Hitherto, circadian misalignment was mostly investigated between body clocks and the environment (e.g., light–dark cycle, feeding) and was implicated in a wide variety of pathologies (49). We show that internal desynchrony between peripheral tissues can be induced and is associated with pathological conditions such as OSA.
Virtually every cell in the body has its own autonomous circadian clock. Clocks in various tissues regulate different processes. If we assume that these rhythmic processes need to act in concert, this arrangement imposes a major challenge on the system: the need to keep clocks in different tissues, which experience different environments and express different tissue-specific genes, in stable phase relations between one another. It seems that, under normal conditions in healthy animals, this is achieved (50) but, under certain circumstances, the intertissue alignment might be interrupted (51).
In animal models, feeding activity misalignment (e.g., daytime feeding) causes phase desynchrony between the central SCN clock and the peripheral clocks (30, 32) and is associated with metabolic dysregulation (52, 53). In humans, night eating syndrome, a condition in which people eat more than 25% of their calories during their sleep phase, correlates with metabolic diseases (54, 55).
Notably, although in a different context (56), this notion was raised long before we knew of cell- and tissue-specific clocks. Recently, the possibility that health consequences of circadian disruption are predominantly caused by internal misalignment was more formally described (51). Thus, loss of peripheral clock synchrony upon hypoxia is likely to play a role in the pathophysiology and sequelae of OSA and other hypoxia-related morbidities.
Materials and Methods
All animal experiments and procedures were conducted in conformity with the Weizmann Institute Animal Care and Use Committee (IACUC) guidelines. Three to 4-mo-old male wild-type C57BL/6 mice (Envigo), Per1,2−/− (17), back-crossed to C57BL/6 and PER2::LUC mice (28), were used. Alb-Cre+ Bmal1fl/fl were generated by crossing Alb-Cre+ mice (Jackson Laboratories) with Bmalfl/fl mice (Jackson Laboratories) (26). The sustained hypoxia treatment described in Figs. 1–4 was conducted using a homemade constant-flow system. The sustained and intermittent hypoxia treatments described in Fig. 6 were conducted using the VelO2x in vivo hypoxia system (Baker Ruskinn).
Unless indicated otherwise, animals were killed immediately after treatment by cervical dislocation. The organotypic slice bioluminescence assay was performed as previously described (28). For RNA and protein, tissues were snap-frozen and then extracted and assayed according to standard protocols. A detailed description of the methods is provided in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Saar Ezagouri and Benjamin Leudouix for their assistance with the animal work, Jonathan Sobel for his help in the motif analysis, Hadas Keren-Shaul for her guidance on the RNA sequencing, John Hogenesch for valuable scientific discussions, and all members of the G.A. laboratory for their comments on the manuscript. G.A. is supported by the European Research Council (ERC-2017 CIRCOMMUNICATION 770869), the Abisch Frenkel Foundation for the Promotion of Life Sciences, the Adelis Foundation, and Susan and Michael Stern, and is a recipient of the EMBO Young Investigator award. R.A. is a recipient of a fellowship from the Azrieli Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data disposition: RNA-Seq data, including fastq files and raw counts, have been uploaded to the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo, under accession number GSE130613; the analyzed data are provided in Datasets S1 and S2.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914112117/-/DCSupplemental.
References
- 1.Semenza G. L., Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365, 537–547 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Montaigne D., Staels B., Time to check the clock in cardiovascular research and medicine. Circ. Res. 123, 648–650 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Truong K. K., Lam M. T., Grandner M. A., Sassoon C. S., Malhotra A., Timing matters: Circadian rhythm in sepsis, obstructive lung disease, obstructive sleep apnea, and cancer. Ann. Am. Thorac. Soc. 13, 1144–1154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partch C. L., Green C. B., Takahashi J. S., Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24, 90–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dibner C., Schibler U., Albrecht U., The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Reinke H., Asher G., Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20, 227–241 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Bass J., Lazar M. A., Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Panda S., Circadian physiology of metabolism. Science 354, 1008–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamovich Y., et al. , Oxygen and carbon dioxide rhythms are circadian clock controlled and differentially directed by behavioral signals. Cell Metab. 29, 1092–1103.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Adamovich Y., Ladeuix B., Golik M., Koeners M. P., Asher G., Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab. 25, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi M., et al. , A circadian clock gene, PER2, activates HIF-1 as an effector molecule for recruitment of HIF-1α to promoter regions of its downstream genes. FEBS J. 284, 3804–3816 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Peek C. B., et al. , Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. 25, 86–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y., et al. , Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 25, 73–85 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Dimova E. Y., et al. , The circadian clock protein CRY1 is a negative regulator of HIF-1alpha. iScience 13, 284–304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogenesch J. B., Gu Y. Z., Jain S., Bradfield C. A., The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U.S.A. 95, 5474–5479 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manella G., Asher G., The effect of acute sustained hypoxia on gene expression in liver, kidney and lung on different times-of-day. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130613. Deposited 2 May 2019.
- 17.Zheng B., et al. , Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Adamovich Y., et al. , Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 19, 319–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang-Verslues W. W., et al. , Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl. Acad. Sci. U.S.A. 110, 12331–12336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckle T., et al. , Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia. Nat. Med. 18, 774–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okabe T., et al. , The impact of HIF1α on the Per2 circadian rhythm in renal cancer cell lines. PLoS One 9, e109693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki K., et al. , Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J. Biol. Chem. 277, 47014–47021 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Yu C., et al. , Hypoxia disrupts the expression levels of circadian rhythm genes in hepatocellular carcinoma. Mol. Med. Rep. 11, 4002–4008 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Hirano A., Fu Y. H., Ptáček L. J., The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 23, 1053–1060 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Lamia K. A., Storch K. F., Weitz C. J., Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storch K. F., et al. , Intrinsic circadian clock of the mammalian retina: Importance for retinal processing of visual information. Cell 130, 730–741 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornmann B., Schaad O., Bujard H., Takahashi J. S., Schibler U., System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo S. H., et al. , PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher G., Sassone-Corsi P., Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Stokkan K. A., Yamazaki S., Tei H., Sakaki Y., Menaker M., Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Asher G., et al. , Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142, 943–953 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Damiola F., et al. , Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winfree A. T., The Geometry of Biological Time (Springer, New York, ed. 2, 2001), pp. xxvi, 777 pp.
- 34.Daan S., Pittendrigh C. S., A functional analysis of circadian pacemakers in nocturnal rodents. J. Comp. Physiol. 106, 253–266 (1976). [Google Scholar]
- 35.Garvey J. F., Pengo M. F., Drakatos P., Kent B. D., Epidemiological aspects of obstructive sleep apnea. J. Thorac. Dis. 7, 920–929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chopra S., Polotsky V. Y., Jun J. C., Sleep apnea research in animals. Past, present, and future. Am. J. Respir. Cell Mol. Biol. 54, 299–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunyor I., Cook K. M., Models of intermittent hypoxia and obstructive sleep apnea: Molecular pathways and their contribution to cancer. Am. J. Physiol. Regul. Integr. Comp. Physiol. 315, R669–R687 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Andreas S., Eichele G., Sleep apnoea: Time to consider clock genes. Eur. Respir. J. 32, 1–2 (2008). [DOI] [PubMed] [Google Scholar]
- 39.von Allmen D. C., et al. , Circadian dysregulation: The next frontier in obstructive sleep apnea research. Otolaryngol. Head Neck Surg. 159, 948–955 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Navarrete-Opazo A., Mitchell G. S., Therapeutic potential of intermittent hypoxia: A matter of dose. Am. J. Physiol. Regul. Integr. Comp. Physiol. 307, R1181–R1197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain I. H., et al. , Hypoxia as a therapy for mitochondrial disease. Science 352, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalmus H., Periodizität und Autochronie (Ideochronie) als zeitregelnde Eigenschaffen der Organismen. Biol. Gen. 11, 93–114 (1935). [Google Scholar]
- 43.Pittendrigh C. S., On temperature independence in the clock system controlling emergence time in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 40, 1018–1029 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egg M., et al. , Linking oxygen to time: The bidirectional interaction between the hypoxic signaling pathway and the circadian clock. Chronobiol. Int. 30, 510–529 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Yamazaki S., Takahashi J. S., Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 393, 288–301 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abraham U., et al. , Coupling governs entrainment range of circadian clocks. Mol. Syst. Biol. 6, 438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pett J. P., Kondoff M., Bordyugov G., Kramer A., Herzel H., Co-existing feedback loops generate tissue-specific circadian rhythms. Life Sci Alliance 1, e201800078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haase V. H., Mechanisms of hypoxia responses in renal tissue. J. Am. Soc. Nephrol. 24, 537–541 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Baron K. G., Reid K. J., Circadian misalignment and health. Int. Rev. Psychiatry 26, 139–154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B., A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roenneberg T., Merrow M., The circadian clock and human health. Curr. Biol. 26, R432–R443 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Zarrinpar A., Chaix A., Panda S., Daily eating patterns and their impact on health and disease. Trends Endocrinol. Metab. 27, 69–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherji A., et al. , Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E6691–E6698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallant A., et al. , Night eating behavior and metabolic heath in mothers and fathers enrolled in the QUALITY cohort study. Eat. Behav. 15, 186–191 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Stunkard A. J., Grace W. J., Wolff H. G., The night-eating syndrome; A pattern of food intake among certain obese patients. Am. J. Med. 19, 78–86 (1955). [DOI] [PubMed] [Google Scholar]
- 56.Aschoff J., Circadian rhythms in man. Science 148, 1427–1432 (1965). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.