Significance
Gut bacteria-associated metabolites pivotally maintain host immune and developmental homeostasis. Here we demonstrate that the cbiMCbl riboswitch regulates expression of the cobA operon controlling bacterial VB12 biosynthesis. Molecular modification of this riboswitch significantly enhances VB12 production by the commensal Propionibacterium strain UF1. These findings outline the potential to further develop this bacterium, which has already demonstrated healthful benefits by repelling intestinal diseases, as an improved next-generation probiotic.
Keywords: riboswitch, vitamin B12, probiotic, Propionibacterium
Abstract
Vitamin B12 (VB12) is a critical micronutrient that controls DNA metabolic pathways to maintain the host genomic stability and tissue homeostasis. We recently reported that the newly discovered commensal Propionibacterium, P. UF1, regulates the intestinal immunity to resist pathogen infection, which may be attributed in part to VB12 produced by this bacterium. Here we demonstrate that VB12 synthesized by P. UF1 is highly dependent on cobA gene-encoding uroporphyrinogen III methyltransferase, and that this vitamin distinctively regulates the cobA operon through its 5′ untranslated region (5′ UTR). Furthermore, conserved secondary structure and mutagenesis analyses revealed a VB12-riboswitch, cbiMCbl (140 bp), within the 5′ UTR that controls the expression of downstream genes. Intriguingly, ablation of the cbiMCbl significantly dysregulates the biosynthesis of VB12, illuminating the significance of this riboswitch for bacterial VB12 biosynthesis. Collectively, our finding is an in-depth report underscoring the regulation of VB12 within the beneficial P. UF1 bacterium, through which the commensal metabolic network may improve gut bacterial cross-feeding and human health.
Vitamin B12 (VB12), also known as cobalamin, is uniquely synthesized by few bacteria and archaebacteria and is a crucial cofactor for critical enzymes catalyzing numerous transmethylation and biochemical reactions (1, 2). Structurally, VB12 consists of a corrin ring in which the cobalt is positioned centrally and coordinated with upper and lower ligands composed of 5, 6-dimethylbenzimidazole (DMB) (1, 3, 4). In both bacterial and mammalian cells, methionine synthase for biosynthesis of S-adenosylmethionine (SAM) and methylmalonyl-CoA mutase converting methylmalonyl-CoA to succinyl-CoA are dependent on VB12 for their metabolic activities (5), making VB12 essentially required for the biosynthesis of nucleic acids, amino acids, and fatty acids. Thus, VB12 deficiency is critically associated with micronuclei formation and chromosomal abnormalities (6–8) and may contribute to adverse pregnancies along with neurologic morbidity and death in neonates (9–14).
In humans, VB12 can be absorbed from food of animal origin or may be provided by some gut microbes through fermentation of complex carbohydrates (14). However, only a few bacterial and archaeal species have the genetic complexity for VB12 biosynthesis via aerobic and/or anaerobic pathways depending on the nature of microorganisms (3, 15). For example, Pseudomonas species produce VB12 only under aerobic condition (5, 16), whereas Propionibacteria use both aerobic and anaerobic routes for its biosynthesis. It has been shown that VB12 biosynthetic pathways involve nearly 30 different enzymes, including hemBCD, cbi, and cob genes (17). CobA serves as a rate-limiting enzyme that converts uroporphyrinogen II to precorrin-2, which is eventually incorporated with DMB to form cobalamin (17). Interestingly, the cobA gene was initially annotated as cysG in some bacteria, including Propionibacterium spp. (18, 19); however, the specificity of this gene in VB12 biosynthesis remains elusive.
Propionibacteria are Gram-positive, facultative anaerobic, and nonmotile microorganisms with a high GC content that can be taxonomically classified into cutaneous (e.g., Propionibacterium acnes) and dairy (e.g., Propionibacterium freudenreichii) species (20, 21). Although P. freudenreichii, along with Pseudomonas species, is a major producer of VB12 and is widely used for industrial fermentation (2, 17), the regulatory elements that restrict VB12 biosynthesis in these bacteria, are not fully understood. It has been reported that VB12 biosynthesis is tightly regulated by noncoding RNAs (ncRNA), known as riboswitches (22–24), which are embedded within the 5′ UTR of VB12-synthesizing operons. However, the elucidation of the identity and mechanisms by which these riboswitches regulate gene expression and VB12 biosynthesis within P. UF1, which was recently isolated from gut microbiota of preterm infants fed human breast milk (25), still require further rigorous investigation.
P. UF1 shares 90% sequence identity with P. freudenreichii (25). We have recently reported that P. UF1 not only regulates the innate and T cell response to intestinal infection (25–27), but also controls the maturation of neonatal protective T cell immunity to resist pathogen infection (28). Here, to further elucidate the regulatory mechanisms exerted by this bacterium, we demonstrate that P. UF1 abundantly produces VB12, which in turn regulates expression of the cobA operon through a riboswitch, cbiMCbl, within this beneficial bacterium that may affect the host milieu and contribute to gut homeostasis.
Results
CobA as the Key Enzyme for Bacterial VB12 Biosynthesis.
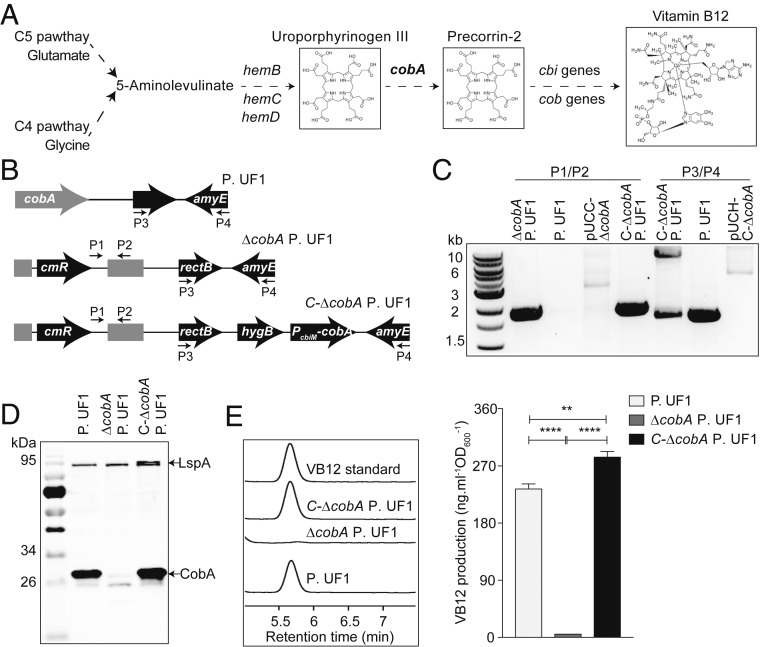
Biochemical studies demonstrate that CobA is essential for the conversion of uroporphyrinogen III to precorrin-2 (Fig. 1A), which is the branch point between the biosynthesis of VB12 and siroheme (18). However, whether CobA plays a critical role in the biosynthesis of VB12 within P. UF1 is currently unknown. Thus, the cobA gene was deleted from the bacterial chromosome by homologous recombination with a single crossover event, resulting in ΔcobA P. UF1 (Fig. 1B and SI Appendix, Table S1). For complementation of the cobA-deficient bacterial strain, the cobA gene, along with its native promoter, was integrated into the chromosome of ΔcobA P. UF1 (Fig. 1B and SI Appendix, Table S1). PCR and Western blot analysis demonstrated cobA expression in P. UF1 and C-ΔcobA P. UF1 but not in ΔcobA P. UF1 (Fig. 1 C and D). HPLC analysis demonstrated that cobA deficiency led to complete abrogation of intracellular VB12 within ΔcobA P. UF1, referring to the VB12 standard, and the complementation of cobA mutation restored VB12 biosynthesis in C-ΔcobA P. UF1 (Fig. 1E). Furthermore, deletion of cobA significantly decreased VB12 production over time when cultured in either MRS medium or Poznan medium, a minimal medium containing no VB12 (SI Appendix, Fig. S1). These data highlight the pivotal role of CobA in critically directing VB12 biosynthesis within P. UF1.
Fig. 1.
cobA is essential for VB12 biosynthesis within P.UF1 (A) Proposed biosynthetic pathway for VB12 produced by P. UF1 in which cobA is responsible for converting uroporphyrinogen III to precorrin-2. (B) Genetic organization of P. UF1, ΔcobA P. UF1, and C-ΔcobA P. UF1 strains. cmR, chloramphenicol-resistant gene. hygB, hygromycin B. (C) PCR identification of P. UF1, ΔcobA P. UF1, and C-ΔcobA P. UF1 strains using primers P1/P2 and P3/P4 as shown in B. (D) Western blot (WB) analysis of cobA expression in P. UF1, ΔcobA P. UF1, and C-ΔcobA P. UF1 strains using mouse serum antibodies against CobA. The large surface layer protein (LspA) served as a reference control. (E) HPLC chromatograms of VB12 extracted from P. UF1, ΔcobA P. UF1, and C-ΔcobA P. UF1 strains. The bar graph shows the intracellular levels of VB12 in the indicated strains. Data are representative of 3 independent experiments. Error bars indicate SEM. **P < 0.01; ****P < 0.00001, 2-tailed unpaired t test.
Controlling cobA Operon Expression by VB12.
In bacteria such as Escherichia coli and Salmonella Typhimurium, VB12 interacts with the 5′ UTR of the VB12 biosynthesis operon to repress translation of the corresponding genes, including the cob and btuB operons (29, 30). Thus, a demonstration of the central role of cobA in controlling VB12 biosynthesis within P. UF1 prompted us to assess the feedback regulation of cobA operon by VB12. We cloned the 5′ UTR of the cobA operon (PcbiM) into the first gene of the operon, cbiM, with His-tag or the flavin mononucleotide-binding fluorescent protein (FbFP) gene (31) to construct the CbiM–wild-type (WT) and FbFP-WT reporter bacterial strains, respectively (S1 Appendix, Table S1). Here, when adding cobalt and DMB (2 substrates required for VB12 biosynthesis) or VB12 alone into FbFP-WT and CbiM-WT bacterial cultures, the expression levels of FbFP and cbiM were significantly decreased (Fig. 2 A and B).
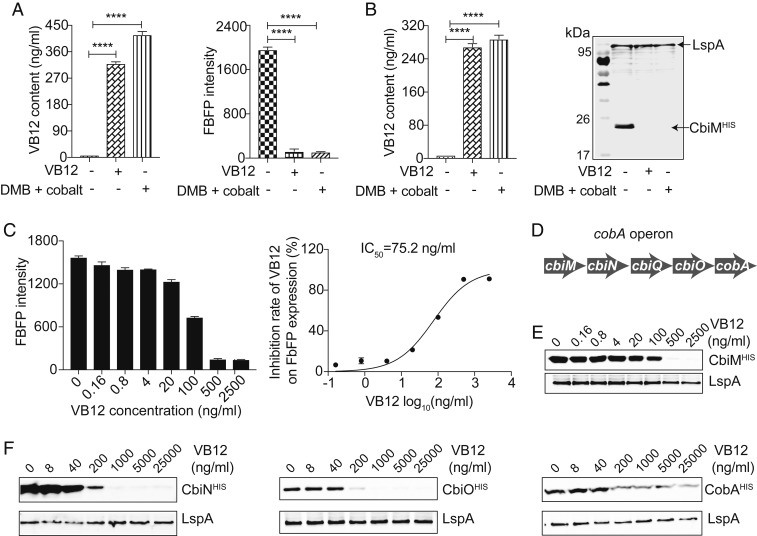
Fig. 2.
The cobA operon is feedback-regulated by VB12. (A) The intracellular levels of VB12 and FbFP fluorescence intensity in the FbFP-WT P. UF1 strain cultured with VB12 or DMB plus cobalt at day 10. (B) The intracellular levels of VB12 and cbiM expression in the CbiM-WT P. UF1 strain cultured with VB12 or DMB plus cobalt at day 10. (C, Left) FbFP fluorescence intensity of the FbFP-ΔcobA P. UF1 strain incubated with increasing VB12 (0 to 2,500 ng/mL). (C, Right) Dose-response curve used to generate the IC50 of VB12 on FbFP expression by the FbFP-ΔcobA P. UF1 strain. (D) Genetic organization of the cobA operon. (E) WB analysis of cbiM expression of the CbiM-ΔcobA P. UF1 strain incubated with increasing VB12 (0 to 2,500 ng/mL) using anti-His antibody. (F) WB analysis of cbiN, cbiO, and cobA expression of the CbiM-ΔcobA P. UF1 strain responding to different concentrations of VB12 (0 to 25,000 ng/mL) using anti-His antibody (Bottom). In A–C, data are representative of 3 independent experiments. Error bars indicate SEM. ****P < 0.0001, 2-tailed unpaired t test.
To further explore for a correlation between VB12 concentration and the expression of these reporter genes, we first examined the effect of VB12 on FbFP expression. As shown in Fig. 2C, VB12 down-regulated the FbFP expression in a VB12 dose-dependent manner. The half-maximal inhibitory concentration (IC50) of VB12 was 75.2 ng/mL, and the expression of FbFP was completely abated at 500 ng/mL (Fig. 2C). In addition, Western blot analyses consistently demonstrated the dose-dependent inhibition of cbiM expression by VB12 using His-tag antibody (Fig. 2E). Furthermore, the same VB12-mediated gene suppression was also observed in P. UF1 using mouse serum antibodies generated against CobA (SI Appendix, Fig. S2A). These data demonstrate that both endogenous and exogenous VB12 modulated the expression of cobA operon through its 5′ UTR.
The cobA operon harbors cbiMNQOA genes encoding proteins critical for cobalt transport and precorrin-2 biosynthesis (Fig. 2D). To investigate whether VB12 regulates the entire cobA operon, we labeled the C termini of respective cbiN, cbiO, and cobA genes with His-tag to assess their expression by Western blot analyses. Our data demonstrate that the expression of cbiN and cbiO was dampened by adding VB12 to CbiN-ΔcobA P. UF1 and CbiO-ΔcobA P. UF1 cultures (Fig. 2F and SI Appendix, Table S1), as observed for cbiM expression (Fig. 2E). Note that cobA expression displayed the dose-dependent inhibition but to a lesser extent in the CobA-ΔcobA P. UF1 strain (Fig. 2F and SI Appendix, Table S1). These data suggest that expression of the entire cobA operon is tightly controlled by VB12.
VB12 possesses analogs such as cyanocobalamin (manufactured form), methylcobalamin (active form), hydroxocobalamin (storage form), and adenosylcobalamin (active form) (14). To elucidate a possible differential regulation by the VB12 analogs, CbiM-ΔcobA P. UF1 and FbFP-ΔcobA P. UF1 cultures (SI Appendix, Table S1) were treated with various concentrations of the VB12 analogs to analyze the cbiM and FbFP expression in these bacterial strains. The expression of cbiM and FbFP was similarly down-regulated by all the analogs (SI Appendix, Fig. S2 B and C). These data demonstrate that VB12 and its analogs control expression of the cobA operon through its 5′ UTR within P. UF1.
Identifying a VB12 Riboswitch within 5′ UTR of the cobA Operon.
Riboswitches are noncoding RNA (ncRNA) regulatory elements that specifically bind small-molecular ligands such as VB12 to modulate gene expression (24). To identify potential regulatory elements, the 5′ UTR (309 bp) of the cobA operon was used to perform a comparative analysis. Conserved secondary structure and sequence homology analyses demonstrated the presence of a potential VB12 riboswitch (Rfam accession, RF00174; 140 bp), designated as cbiMCbl riboswitch, which contains 3 major stem loops (SLs): SL1, SL2, and SL3 (Fig. 3A).
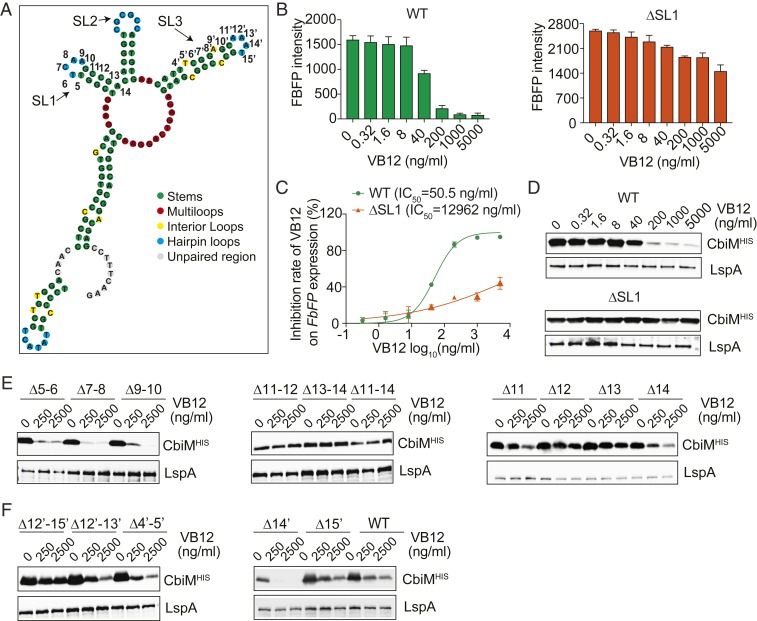
Fig. 3.
Stem loops of cbiMCbl riboswitch are key for VB12-mediated gene regulation. (A) Secondary structure of cbiMCbl riboswitch predicted using forna software. (B) FbFP fluorescence intensity in the FbFP-ΔcobA P. UF1 strain harboring WT or SL1-deleted riboswitch cultured with VB12 (0 to 5,000 ng/mL). (C) Dose-response curve used to generate the IC50 of VB12 on FbFP expression. (D) Anti-His WB analysis of cbiM expression in the CbiM-ΔcobA P. UF1 strain with WT or SL1-deleted riboswitch in the presence of VB12 (0 to 5,000 ng/mL). (E) cbiM expression driven by SL1-mutated riboswitches in response to VB12 (0, 250, and 2,500 ng/mL). (F) cbiM expression directed by SL3-mutated riboswitches with various concentrations of VB12. Data in B and C are representative of 3 independent experiments. Error bars indicate SEM.
Mechanistically, SL domains interact with ligands and the targeting RNA to regulate gene expression within various microorganisms (32). Thus, to determine the regulatory function of these SLs, we tested the effect of each SL domain on the expression of cbiM and FbFP by P. UF1. Here the FbFP reporter assays demonstrated that the SL1-deleted riboswitch (ΔSL1) lost the VB12 dose-dependent regulation, and the IC50 of VB12 was 12,962 ng/mL, which was 256-fold higher than that of the WT riboswitch (Fig. 3 B and C). Consistently, the repression of cbiM expression was observed for ΔSL1 compared with the WT riboswitch in P. UF1 (Fig. 3D). Furthermore, to precisely elucidate the sites or regions required for VB12-mediated gene regulation within P. UF1, site-directed mutations were introduced into SL1 of the cbiMCbl riboswitch. While the WT riboswitch exhibited ∼10-fold repression of FbFP expression at 250 ng/mL VB12, site mutation of 12 or 13 in SL1 only retained marginal reduction of protein expression even at 2,500 ng/mL VB12 (Fig. 3E and SI Appendix, Fig. S3C). In contrast, site mutations of 5 to 6, 7 to 8, or 9 to 10 did not impact the regulatory activity of this riboswitch (Fig. 3E and SI Appendix, Fig. S3C). Furthermore, deletion of SL3 resulted in a total loss of gene expression (SI Appendix, Fig. S2 A and B), whereas deletion of the SL3 loop region, sites 12′ to 15′, abolished the dose-dependent down-regulation of the downstream gene expression in P. UF1 (Fig. 3F and SI Appendix, Fig. S3D). In contrast, site mutation of the loop region did not down-regulate the downstream gene expression by VB12 (Fig. 3F and SI Appendix, Fig. S3D), indicating that these sites may cooperatively maintain the regulatory activity of the SL3 domain in this bacterium. These findings emphasize that VB12-mediated regulation is highly dependent on the structure of the cbiMCbl riboswitch within P. UF1.
Regulation of cbiM Expression by Ribosome-Binding Site–Mediated Base Pairing.
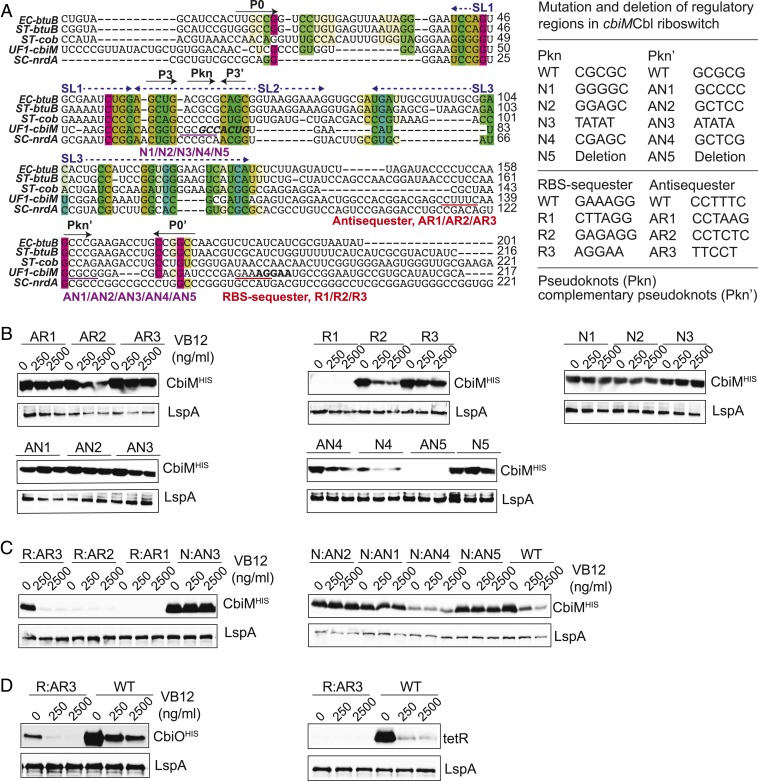
VB12-element exhibits a conserved RNA regulatory sequence in various VB12 riboswitches of numerous microorganisms (15, 33). To investigate whether the VB12 element exists within cbiMCbl riboswitch, the known VB12 riboswitches and cbiMCbl riboswitch were compared by sequence alignment analysis using LocARNA, whereby a conserved VB12 element and various secondary structures (e.g., Pkn) were identified within the cbiMCbl riboswitch (Fig. 4A). cbiMCbl riboswitch contained a conserved core region, 5′-GCCACUG-3′, which partially overlapped with the SL2 domain (Fig. 4A). Importantly, 2 groups of Watson–Crick base pairs, between Pkn and Pkn′ and between antisequester and ribosome-binding site (RBS) sequester, were found within the cbiMCbl riboswitch (Fig. 4A).
Fig. 4.
cbiMCbl riboswitch regulates cbiM expression by RBS-mediated base pairing. (A) Multiple alignment of VB12 riboswitches. The colored background denotes P0 and P3 stems and their complementary sequences (P0′ and P3′). The conserved core region is shown in italic, bold, and black letters. RBSs are shown in bold and black letters. Red underscores denote the proposed antisequester and RBS sequester. Purple underscores denote the candidate pseudoknots (Pkn) and complementary pseudoknots (Pkn′). EC, E. coli; ST, S. Typhimurium; SC, Streptomyces coelicolor. The summary table shows the sequences of respective mutations. (B) Western blot analysis of cbiM expression by a series of riboswitches mutated in Pkn, Pkn′, sequester, or antisequester. (C) Western blot analysis of cbiM expression by riboswitches with paired double mutations in Pkn and Pkn′ or sequester and antisequester. (D) Western blot analysis of cbiO and tetR expression by WT and the double-mutant riboswitch R:AR3.
To elucidate whether these base pairs are important for VB12-mediated regulatory activity, site mutation and region deletion were introduced into these sequences, and their impacts on cbiM and FbFP expression were analyzed (Fig. 4A). Here single-site (e.g., AR2) or multisite mutations (e.g., AR3) in the antisequester and RBS sequester distinctly weakened the regulatory activity of the cbiMCbl riboswitch, along with the increased levels of cbiM expression compared to the WT riboswitch (Fig. 4B). Furthermore, the regulatory activity of cbiMCbl riboswitch was abolished by mutated AR1 or R3 (Fig. 4B). In addition, the multisite mutation R1 led to complete loss of cbiM expression (Fig. 4B). Similarly, disruption of Pkn and Pkn′ base pairing differentially impacted the regulatory functions of cbiMCbl riboswitch. For example, the site mutations N1, N2, N3, N4, N5 and AN2 rendered the modified riboswitches unable to down-regulate cbiM expression (Fig. 4B). While mutations AN1 and AN3 decreased the regulatory activity of the riboswitches, deletion of Pkn′ (AN5) resulted in complete loss of regulation and cbiM expression (Fig. 4B). Furthermore, site mutation of Pkn (AN4) demonstrated comparable regulatory activity to the WT riboswitch (Fig. 4B). In addition, cbiM expression assays were largely consistent with FbFP reporter assays in these riboswitch-modified strains (Fig. 4B and SI Appendix, Fig. S4A). However, R3- and N4-modified strains demonstrated the VB12 dose-dependent down-regulation of cbiM expression but not of FbFP expression (Fig. 4B and SI Appendix, Fig. S4A). These data suggest that Pkn, Pkn′, antisequester, and RBS sequester are the key regulatory domains of the cbiMCbl riboswitch in P. UF1.
Thus far, it is unclear whether the regulatory activity of the cbiMCbl riboswitch depends on Watson–Crick base pairing within the regulatory (Pkn and antisequester) and their complementary (Pkn′ and RBS sequester) domains. Thus, to elaborate on this notion, we introduced a second mutation into these single mutant bacterial strains to rescue their base pairing (Fig. 4A). Here, most of the double-mutant strains were still unable to regulate cbiM and FbFP expression (Fig. 4C and SI Appendix, Fig. S4B). However, the double-mutant strain CbiM-R:AR3-ΔcobA P. UF1 regained the VB12 dose-dependent regulation of cbiM expression but not FbFP expression (Fig. 4C and SI Appendix, Fig. S4B and Table S1). These data led to the hypothesis that the base pair-mediated regulation may be influenced by the gene-specific signals. Thus, the expression of cbiO and tetR genes was further analyzed using the R:AR3 double-mutant strain (Fig. 4D). Obtained data thus consistently demonstrated regulatory activity for cbiO rather than for exogenous tetR (Fig. 4D), indicating that the cbiMCbl riboswitch specifically regulates the expression of cobA operon by RBS-mediated base pairing. Collectively, these data suggest that RBS-mediated Watson–Crick base pairing, as a pivotal factor, may facilitate the regulation of cobA operon expression through the cbiMCbl riboswitch in P. UF1.
Regulation of VB12 Biosynthesis by cbiMCbl Riboswitch.
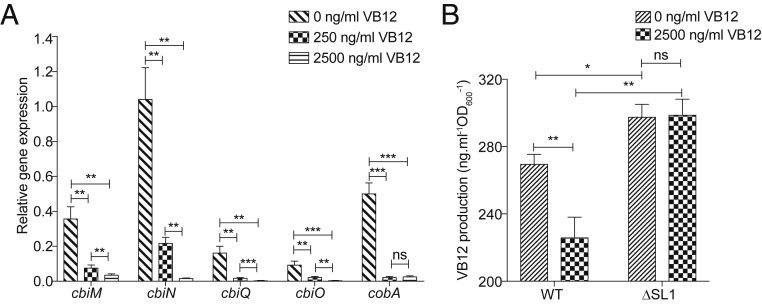
Knowing that VB12 riboswitch controls gene expression through transcriptional and/or translational modifications (24, 34), the impact of the cbiMCbl riboswitch on the transcription of cobA operon was further assessed by qRT-PCR. The data thus obtained demonstrated that the mRNA levels of most of the genes, including cbiM, cbiN, cbiO and cbiQ, were significantly repressed in a VB12-dependent manner within P. UF1 (Fig. 5A).
Fig. 5.
cbiMCbl riboswitch controls VB12 production. (A) qRT-PCR analysis of expression of cbiMNQO and cobA genes in P. UF1 incubated with various concentrations of VB12. (B) HPLC analysis of VB12 production by WT and SL1-deleted riboswitches within OW-operon-ΔcobA and OΔSL1-operon-ΔcobA strains. The bacteria-absorbed VB12 was excluded for this analysis. Data are representative of 3 independent experiments. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001, 2-tailed unpaired t test. ns, not significant: ns = P > 0.05.
To further investigate whether cbiMCbl riboswitch regulates VB12 biosynthesis within P. UF1, the cobA operon was overexpressed under WT and ΔSL1 riboswitches within OW-operon-ΔcobA P. UF1 and OΔSL1-operon-ΔcobA P. UF1 strains (SI Appendix, Table S1). The data demonstrated that disruption of SL1 significantly elevated VB12 production, and this difference was continuously extended in the presence of exogenous VB12 (Fig. 5B), while this effect was abrogated due to SL1 deficiency in this bacterium (Fig. 5B). Taken together, these data indicate that the cbiMCbl riboswitch negatively controls the bacterial VB12 biosynthesis through suppression of cobA operon expression.
Discussion
Mechanisms involved in bacterial molecular machinery critically direct the metabolic circuits that regulate bacterial homeostasis and their stable abundancy, which may contribute to human health (3, 35–37). One of these metabolites is VB12, which crucially impacts the cross-talk between gut microbes and the host (3, 38). Although previously reported data demonstrate how pathogens, including Salmonella (29, 39), regulate the biosynthesis of this vitamin, the control of VB12 biosynthesis in bacteria with bifidogenic properties, particularly probiotic bacteria such as P. UF1, is currently obscure. Thus, to characterize the mechanistic complexes potentially regulating VB12 in the newly discovered P. UF1 bacterium, we first focused on the genomic region of bacterial CobA. CobA catalyzes the SAM-dependent bismethylation of uroporphyrinogen III synthase (40) to form precorrin-2, the primary precursor of VB12 (18). Thus, deleting cobA from the bacterial chromosome abolished VB12 production in the ΔcobA P. UF1 strain. VB12 was fully restored by complementing ΔcobA P. UF1 with the WT cobA gene, illuminating the critical role of cobA in bacterial VB12 biosynthesis.
We also demonstrated that endogenous and exogenous VB12 tightly controlled the expression of cobA operon via its 5′ UTR within P. UF1 bacterium; a feedback regulatory mechanism via VB12 that was also observed in E. coli and S. Typhimurium (29, 30). Importantly, we documented that VB12 completely inhibited the expression of downstream genes by the cbiMCbl riboswitch at 750 μM, which was significantly higher than env8HyCbl in E. coli (34). This differential regulatory activity may be ascribed to differential scales of VB12 biosynthesis in different bacteria such as P. UF1.
VB12 analogs displayed similar regulatory activities in the riboswitch of P. UF1 but not in E. coli (32), possibly as a result of diverse sequences and secondary structures of VB12 riboswitches. Our structure-based analyses revealed that SL1 and SL3 were highly required for the regulatory function of the cbiMCbl riboswitch, which belong to receptor domains conserved within the VB12 riboswitches from E. coli and S. Typhimurium (32, 33, 41). Furthermore, we also demonstrated that Pkn in SL2 and the complementary Pkn’ were crucial regulatory domains within the cbiMCbl riboswitch, whose regulatory function was notably not dependent on Watson–Crick base pairing within the 2 domains. In contrast, the regulation of env8HyCbl depends on the Watson–Crick base pairing of the “kissing loop” between SL2 and the RBS region (32, 34), indicating that the cbiMCbl riboswitch within P. UF1 may use a different mechanism to control gene expression. Interestingly, we also identified a Watson–Crick base pairing between the RBS sequester and antisequester, whose base pairing was essentially required for regulatory activity of the riboswitch. In E. coli, it is well documented that the env8HyCbl riboswitch regulates gene expression at both transcriptional and translational levels (32, 34). In Listeria monocytogenes and S. Typhimurium, VB12 riboswitches are incapable of controlling the transcription of the downstream genes (24, 41), indicating that various VB12 riboswitches may exhibit a distinct regulatory model for gene expression in bacteria with different natures and functions, mainly pathogenic or beneficial bacteria.
The probiotics are defined as live microbial feed supplements with beneficial properties, which potentially benefit the host when administered in adequate amounts (42). Propionibacterium species are currently of great interest for their beneficial effects as probiotics and are being applied to human dietary consumption, including Swiss cheese (43). We previously demonstrated that P. UF1 increases the frequency of protective T cells involved in fortifying the mucosal barrier function and regulating the intestinal inflammation against pathogenic infection (25–27). Here we demonstrate that the cbiMCbl riboswitch controls VB12 biosynthesis, and that the genetic modification of this riboswitch can significantly enhance VB12 within P. UF1. It is conceivable that this bacterium synthesizing VB12 may critically contribute to the observed immune homeostasis in the host. Thus, mechanistic strategies are currently in place to further illuminate this critical notion that may provide critical insights into VB12 and its impact on the microbiome and the host immune homeostasis against induced detrimental signals inducing tissue damage.
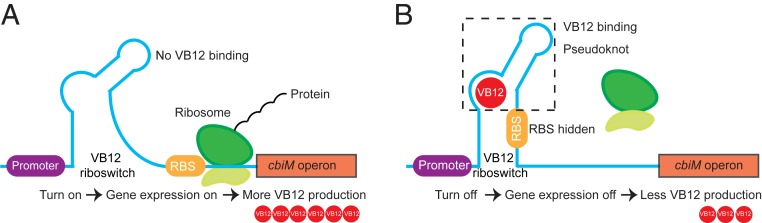
In summary, there is currently great interest in sustaining functional microbiota to promote human health (3). Accordingly, VB12 can be used by >80% of gut microbiota (3), suggesting that this vitamin can be used to support the healthy ecology of the gut microbiota involved in the intestinal homeostasis of the host (3, 44). Here we demonstrate how precisely VB12 biosynthesis is regulated within the cobA operon through the cbiMCbl riboswitch within P. UF1 (Fig. 6). This riboswitch may serve as an attractive target to increase the levels of VB12 by ingesting a probiotic bacterium with bifidogenic properties to perhaps enhance the cross-feeding of neighboring gut bacteria that might mitigate the symptoms of intestinal disorders (45) by reprogramming transcriptomic and metabolomic pathways of ileal epithelial cells to resist detrimental signals that may result in intestinal tissue damage and proinflammatory disease progression.
Fig. 6.
Proposed model for VB12 regulation via the cbiMCbl riboswitch. (A) Gene expression ON: in the absence of VB12, the riboswitch cannot form the pseudoknot, thereby allowing the RBS to initiate the biosynthesis of VB12. (B) Gene expression OFF: in the presence of VB12, the riboswitch binds to VB12 and forms the pseudoknot that prevents ribosome from binding with RBS, leading to blockage of VB12 biosynthesis.
Experimental Procedures
Bacterial Strains and Growth.
E. coli NEB 5-alpha (New England BioLabs) used for plasmid construction and E. coli Rosetta (DE3; Sigma-Aldrich) for protein expression were grown in Luria–Bertani medium at 37 °C. The P. UF1 and its genetically modified strains were grown at 30 °C in MRS medium (Difco) with 1% (wt/vol) sodium lactate (Thermo Fisher Scientific) or Poznan medium (46) in an anaerobic chamber (model AS-580; Anaerobe Systems). Antibiotics were added to the medium at the following final concentrations: 5 μg/mL chloramphenicol and/or 1 mg/mL hygromycin B for P. UF1 isogenic strains and 100 μg/mL ampicillin for E. coli isogenic strains.
For deleting the cobA gene from P. UF1, a 644-bp internal fragment of cobA was PCR amplified from the P. UF1 chromosome using primers cobAbam-F and cobAxba-R. The purified fragment was cloned into the pUCC plasmid (25), generating suicide plasmid pUCC-cobA. Following electroporation into P. UF1 (25), the chloramphenicol-resistant colonies were selected and the ΔcobA P. UF1 mutant was identified by PCR analysis using primers P1 and P2. For complementation of the cobA knockout bacterial strain, the cobA gene with the native promoter was integrated into the ΔcobA P. UF1 chromosome at the amyE locus, and the resultant C-ΔcobA P. UF1 strain was identified by PCR analysis using primers P3 and P4.
To generate the cbiMCbl riboswitch-FbFP fusion constructs, the 5′ UTR containing the promoter of the cobA operon, WT cbiMCbl riboswitch, and RBS was PCR-amplified from the P. UF1 genome (NCBI Genome database, CP018002) with primers S-cobA-1 and S-cobA22. The codon-optimized FbFP gene (31) was synthesized (GenScript) and PCR-amplified using primers S-cobA33 and S-cobA4. Overlapping PCR was used to ligate the 5′ UTR and FbFP gene using primers cobA-1 and S-cobA-4, which was subsequently cloned into plasmid pYMZ between the BamHI and XbaI sites. Mutations of the cbiMCbl riboswitch were generated by site-directed mutagenesis or overlapping PCR (SI Appendix, Table S1). cbiM, cbiN, cbiO, cobA, and tetR reporters were constructed using similar strategies. All plasmids were verified via Sanger sequencing (Genewiz) and introduced into P. UF1 or ΔcobA P. UF1 by electroporation.
To overexpress the cobA operon in ΔcobA P. UF1, the cobA operon with the 5′ UTR was PCR-amplified from P. UF1 genome with primers cbimop-sbf-F and cbimop-hid-R. After digestion with SbfI and HindIII, the purified PCR products were cloned into the pYMZ vector, and the resulting construct was electroporated into ΔcobA P. UF1 to obtain the OW-operon-ΔcobA P. UF1 strain. To investigate the effect of the SL1 deletion on VB12 biosynthesis, the SL1 region was deleted from the 5′ UTR, and the resulting fragment, along with the cobA operon, was ligated by overlapping PCR using primers cbimop-sbf-F/ΔSL1-R and ΔSL1-Fcbimop-hid-R. The plasmid thus constructed was electroporated into ΔcobA P. UF1 to obtain the OΔSL1-operon-ΔcobA P. UF1 strain. All obtained strains are listed in SI Appendix, Table S1.
VB12 Extraction and Analysis.
Bacterial cultures were centrifuged at 15,000 × g for 10 min at 4 °C and washed twice in PBS. The cells were disrupted by boiling for 15 min in 0.1 M phosphate buffer containing 0.01% potassium cyanide at pH 6.0. After centrifugation at 15,000 × g for 2 min, the supernatants were collected and passed through 0.22-μm filters (EMD Millipore). VB12 in the filtrates was quantified using the Agilent 1220 infinity II LC system composed of an automated sampler (G4282B), a gradient pump (G4281B), and a variable wavelength detector (G4284B).
Separations were performed using the following mobile phase (46) containing 0.25 M NaH2PO4 (pH 3.5) and methanol (75:25, vol/vol) in an Agilent C18 column (Eclipse Plus C18, 3.5 μm, 4.6 × 150 mm column) at 20 °C, with a flow rate of 1.0 mL/min. The total HPLC run time for each sample was 15 min, and the injection volume was 20 µL. The detector wavelength was set at 362 nm. Quantitation was based on peak area and the standard curve of VB12. Data acquisition and analysis were done using an Agilent ChemStation. To exclude the bacteria-absorbed VB12 for the analysis of VB12 biosynthesized by P. UF1, the absorbed VB12 was tested culturing P. UF1 without the substrates (DMB and cobalt) and with exogenous VB12. The biosynthesized VB12 equals the total extracellular VB12 content minus the bacteria-absorbed VB12.
FbFP Reporter Assays.
To measure FbFP fluorescence intensity, bacteria were cultivated for 3 d in triplicate, washed, and resuspended with PBS. The excitation and emission wavelengths of FbFP were set at 452 nm and 495 nm, respectively, on a microplate reader (BioTek) (47). FbFP intensity at 495 nm was normalized by corresponding OD600 values for each sample. A P. UF1 strain with an empty vector served as the baseline for the calculation of fluorescence intensity.
Preparation of Anti-CobA Serum Antibodies.
To generate CobA polyclonal antibodies, the pET21b-cobA expression plasmid was constructed by PCR amplification of the cobA gene using primers 21-cobA-bamF and 21-cobA-xhoR (SI Appendix, Table S2). Following transformation into E. coli Rosetta (DE3), CobA expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (Sigma-Aldrich). Cell lysates were separated by 12% SDS/PAGE, and the CobA proteins were excised from the gel and used to immunize C57BL/6 mice, resulting in anti-CobA serum antibodies. All animal studies were approved by the Institutional Animal Care and Use Committee of the University of Florida (protocol 201708484). Mice were maintained in accordance with the Animal Welfare Act and the Public Health Policy on Humane Care.
Western Blot Analysis.
Bacteria were cultivated for 3 d and then washed 3 times, followed by lysozyme digestion (10 mg/mL) for 2 h at 37 °C. Cell lysates were separated by 12% SDS/PAGE, transferred to PVDF membranes (Sigma-Aldrich), and blocked with Odyssey blocking buffer (Li-Cor Biosciences). Subsequently, membranes were incubated with anti–His-tag antibody (Thermo Fisher Scientific), anti-TetR antibody (Takara Bio), anti-CobA serum antibodies, or anti-LspA serum antibodies (available in our laboratory) for 2 h at room temperature in the blocking buffer with 0.1% Tween 20. After washing with TBST (20 mM Tris, 150 mM NaCl and 0.1% Tween 20, pH 7.4), membranes were incubated with IRDye 680RD goat anti-mouse secondary antibody (Li-Cor Biosciences) for 1 h at room temperature in the blocking buffer with 0.1% Tween 20. After washing with TBS, the proteins were detected using the Odyssey infrared imaging system (Li-Cor Biosciences). LspA served as an internal control.
qRT-PCR.
Total RNA was extracted from various bacterial cultures using TRIzol reagent (Thermo Fisher Scientific), and the RNA mixtures were further purified using the Aurum Total RNA Mini Kit (Bio-Rad). The cDNA was synthesized from total RNA using the iScript Advanced cDNA Synthesis Kit (Bio-Rad). qRT-PCR was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) on a CFX96 real-time PCR system (Bio-Rad) using the primers listed in SI Appendix, Table S2. Results were normalized to those obtained from the groL2 gene.
Data Availability.
All relevant data supporting the findings of this study are available in the paper and supplemental materials. The data that support the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 DK109560 (to M.M.).
Footnotes
Competing interest statement: R.C. is a founder, part owner, advisor, and member of the Board of Directors of Curtiss Healthcare, Inc., a startup biotech company in Gainesville, FL developing vaccines to control infectious diseases of farm animals. This company has no interest in products for human use. On behalf of M.M., the University of Florida has filed a provisional US patent application on the probiotic Propionibacterium UF1 strain, which would be used for humans and has yet to be licensed for commercial development.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916576116/-/DCSupplemental.
References
- 1.Crofts T. S., Seth E. C., Hazra A. B., Taga M. E., Cobamide structure depends on both lower ligand availability and CobT substrate specificity. Chem. Biol. 20, 1265–1274 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Fang H., Kang J., Zhang D., Microbial production of vitamin B12: A review and future perspectives. Microb. Cell Fact. 16, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degnan P. H., Taga M. E., Goodman A. L., Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obeid R., Fedosov S. N., Nexo E., Cobalamin coenzyme forms are not likely to be superior to cyano- and hydroxyl-cobalamin in prevention or treatment of cobalamin deficiency. Mol. Nutr. Food Res. 59, 1364–1372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth J. R., Lawrence J. G., Bobik T. A., Cobalamin (coenzyme B12): Synthesis and biological significance. Annu. Rev. Microbiol. 50, 137–181 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Fenech M., Ferguson L. R., Vitamins/minerals and genomic stability in humans. Mutat. Res. 475, 1–6 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Fenech M., The role of folic acid and vitamin B12 in genomic stability of human cells. Mutat. Res. 475, 57–67 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Fenech M., Aitken C., Rinaldi J., Folate, vitamin B12, homocysteine status and DNA damage in young Australian adults. Carcinogenesis 19, 1163–1171 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Dror D. K., Allen L. H., Effect of vitamin B12 deficiency on neurodevelopment in infants: Current knowledge and possible mechanisms. Nutr. Rev. 66, 250–255 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Clarke R., et al. , Folate and vitamin B12 status in relation to cognitive impairment and anaemia in the setting of voluntary fortification in the UK. Br. J. Nutr. 100, 1054–1059 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Mills J. L., et al. , Folate-related gene polymorphisms as risk factors for cleft lip and cleft palate. Birth Defects Res. A Clin. Mol. Teratol. 82, 636–643 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molloy A. M., Kirke P. N., Brody L. C., Scott J. M., Mills J. L., Effects of folate and vitamin B12 deficiencies during pregnancy on fetal, infant, and child development. Food Nutr. Bull. 29 (suppl. 2), S101–S111, discussion S112–S115 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Kvestad I., et al. , Vitamin B-12 status in infancy is positively associated with development and cognitive functioning 5 y later in Nepalese children. Am. J. Clin. Nutr. 105, 1122–1131 (2017). [DOI] [PubMed] [Google Scholar]
- 14.O’Leary F., Samman S., Vitamin B12 in health and disease. Nutrients 2, 299–316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S., Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278, 41148–41159 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Warren M. J., Raux E., Schubert H. L., Escalante-Semerena J. C., The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19, 390–412 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Piwowarek K., Lipińska E., Hać-Szymańczuk E., Kieliszek M., Ścibisz I., Propionibacterium spp—source of propionic acid, vitamin B12, and other metabolites important for the industry. Appl. Microbiol. Biotechnol. 102, 515–538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sattler I., et al. , Cloning, sequencing, and expression of the uroporphyrinogen III methyltransferase cobA gene of Propionibacterium freudenreichii (shermanii). J. Bacteriol. 177, 1564–1569 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escalante-Semerena J. C., Suh S. J., Roth J. R., cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J. Bacteriol. 172, 273–280 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falentin H., et al. , The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications. PLoS One 5, e11748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cousin F. J., et al. , The first dairy product exclusively fermented by Propionibacterium freudenreichii: A new vector to study probiotic potentialities in vivo. Food Microbiol. 32, 135–146 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S., Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9, 1084–1097 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrick J. E., Breaker R. R., The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8, R239 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellin J. R., et al. , Riboswitches: Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345, 940–943 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Colliou N., et al. , Commensal Propionibacterium strain UF1 mitigates intestinal inflammation via Th17 cell regulation. J. Clin. Invest. 127, 3970–3986 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colliou N., et al. , Regulation of Th17 cells by P. UF1 against systemic Listeria monocytogenes infection. Gut Microbes 9, 279–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge Y., et al. , Neonatal intestinal immune regulation by the commensal bacterium, P. UF1. Mucosal Immunol. 12, 434–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black M. M., Effects of vitamin B12 and folate deficiency on brain development in children. Food Nutr. Bull. 29 (suppl. 2), S126–S131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter-Dahlfors A. A., Ravnum S., Andersson D. I., Vitamin B12 repression of the cob operon in Salmonella typhimurium: Translational control of the cbiA gene. Mol. Microbiol. 13, 541–553 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Lundrigan M. D., Köster W., Kadner R. J., Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc. Natl. Acad. Sci. U.S.A. 88, 1479–1483 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobo L. A., Smith C. J., Rocha E. R., Flavin mononucleotide (FMN)-based fluorescent protein (FbFP) as reporter for gene expression in the anaerobe Bacteroides fragilis. FEMS Microbiol. Lett. 317, 67–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson J. E., Reyes F. E., Polaski J. T., Batey R. T., B-12 cofactors directly stabilize an mRNA regulatory switch. Nature 492, 133–137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nahvi A., Barrick J. E., Breaker R. R., Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32, 143–150 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polaski J. T., Holmstrom E. D., Nesbitt D. J., Batey R. T., Mechanistic insights into cofactor-dependent coupling of RNA folding and mRNA transcription/translation by a cobalamin riboswitch. Cell Rep. 15, 1100–1110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dantas G., Sommer M. O., Degnan P. H., Goodman A. L., Experimental approaches for defining functional roles of microbes in the human gut. Annu. Rev. Microbiol. 67, 459–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz Heijtz R., et al. , Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 3047–3052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman A. L., Gordon J. I., Our unindicted coconspirators: Human metabolism from a microbial perspective. Cell Metab. 12, 111–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupski J. R., Roth J. R., Weinstock G. M., Chromosomal duplications in bacteria, fruit flies, and humans. Am. J. Hum. Genet. 58, 21–27 (1996). [PMC free article] [PubMed] [Google Scholar]
- 39.Wu W. F., Urbanowski M. L., Stauffer G. V., Role of the MetR regulatory system in vitamin B12-mediated repression of the Salmonella typhimurium metE gene. J. Bacteriol. 174, 4833–4837 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathews M. A., et al. , Crystal structure of human uroporphyrinogen III synthase. EMBO J. 20, 5832–5839 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravnum S., Andersson D. I., Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol. Microbiol. 23, 35–42 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Hill C., et al. , Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Angelopoulou A., et al. , Production of probiotic feta cheese using Propionibacterium freudenreichii subsp shermanii as adjunct. Int. Dairy J. 66, 135–139 (2017). [Google Scholar]
- 44.Cordonnier C., et al. , Vitamin B12 uptake by the gut commensal bacteria Bacteroides thetaiotaomicron limits the production of shiga toxin by enterohemorrhagic Escherichia coli. Toxins (Basel) 8, E14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNulty N. P., et al. , The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 3, 106ra106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kośmider A., Białas W., Kubiak P., Drożdżyńska A., Czaczyk K., Vitamin B12 production from crude glycerol by Propionibacterium freudenreichii ssp. shermanii: Optimization of medium composition through statistical experimental designs. Bioresour. Technol. 105, 128–133 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Teng L., Wang K., Xu J., Xu C., Flavin mononucleotide (FMN)-based fluorescent protein (FbFP) as reporter for promoter screening in Clostridium cellulolyticum. J. Microbiol. Methods 119, 37–43 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data supporting the findings of this study are available in the paper and supplemental materials. The data that support the findings of this study are available from the corresponding author on reasonable request.