Significance
Tight regulation is required to control biochemical reactions in cells so that these reactions can be activated precisely when and where needed. Our work focuses on actin, a protein that polymerizes into filaments in at least 20 distinct processes in mammalian cells. We have previously shown that lysine acetylation, a relatively minor modification on actin, is crucial for regulating a specific population of actin filaments through a protein called INF2. Here we identify key acetylated lysines that control INF2 activity. INF2 mutations link to 2 human diseases, focal segmental glomerulosclerosis (a kidney disease) and Charcot–Marie–Tooth disease (a neuropathy), and our findings further the mechanistic understanding of these diseases.
Keywords: cyclase-associated protein, WH2 motif, nucleation, U2OS, mitochondria
Abstract
INF2 is a formin protein that accelerates actin polymerization. A common mechanism for formin regulation is autoinhibition, through interaction between the N-terminal diaphanous inhibitory domain (DID) and C-terminal diaphanous autoregulatory domain (DAD). We recently showed that INF2 uses a variant of this mechanism that we term “facilitated autoinhibition,” whereby a complex consisting of cyclase-associated protein (CAP) bound to lysine-acetylated actin (KAc-actin) is required for INF2 inhibition, in a manner requiring INF2-DID. Deacetylation of actin in the CAP/KAc-actin complex activates INF2. Here we use lysine-to-glutamine mutations as acetylmimetics to map the relevant lysines on actin for INF2 regulation, focusing on K50, K61, and K328. Biochemically, K50Q- and K61Q-actin, when bound to CAP2, inhibit full-length INF2 but not INF2 lacking DID. When not bound to CAP, these mutant actins polymerize similarly to WT-actin in the presence or absence of INF2, suggesting that the effect of the mutation is directly on INF2 regulation. In U2OS cells, K50Q- and K61Q-actin inhibit INF2-mediated actin polymerization when expressed at low levels. Direct-binding studies show that the CAP WH2 domain binds INF2-DID with submicromolar affinity but has weak affinity for actin monomers, while INF2-DAD binds CAP/K50Q-actin 5-fold better than CAP/WT-actin. Actin in complex with full-length CAP2 is predominately ATP-bound. These interactions suggest an inhibition model whereby CAP/KAc-actin serves as a bridge between INF2 DID and DAD. In U2OS cells, INF2 is 90-fold and 5-fold less abundant than CAP1 and CAP2, respectively, suggesting that there is sufficient CAP for full INF2 inhibition.
A common mechanism for protein regulation is autoinhibition, whereby intramolecular contacts inhibit the protein’s biological function. Formin proteins are actin assembly factors. Several of the 15 mammalian formins are autoinhibited through interaction between the N-terminal diaphanous inhibitory domain (DID) and the C-terminal diaphanous autoregulatory domain (DAD), which blocks the ability of the formin homology 2 (FH2) domain to accelerate actin polymerization (1, 2). One formin, INF2, uses a variation of this mechanism. Despite having both DID and DAD sequences, which are necessary for INF2 regulation in cells (Fig. 1A), purified INF2 is fully active in biochemical assays. Direct-binding studies show that INF2’s DID/DAD interaction is at least 10-fold weaker than that of mDia1 (3, 4), providing an explanation for the lack of autoinhibition. In addition, INF2’s DAD is similar to an actin-binding WH2 motif (5) and binds actin monomers with high affinity (6, 7). These findings led us to predict that an additional protein is required to inhibit INF2 in a DID/DAD-dependent manner.
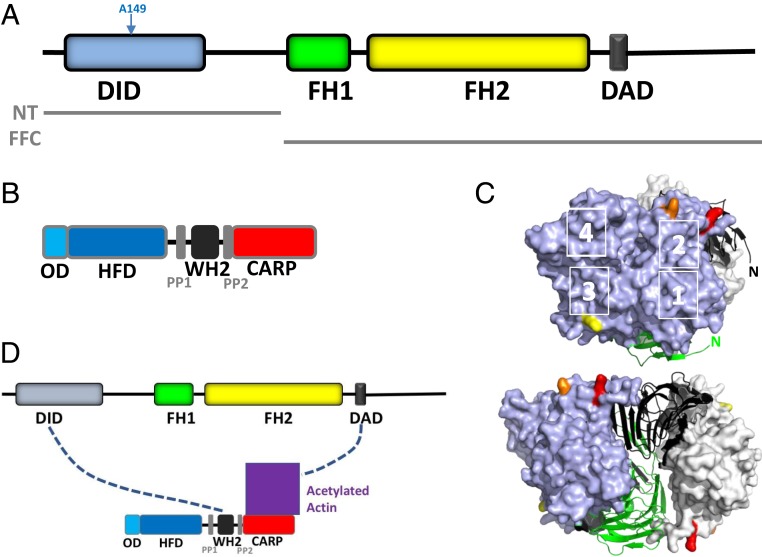
Fig. 1.
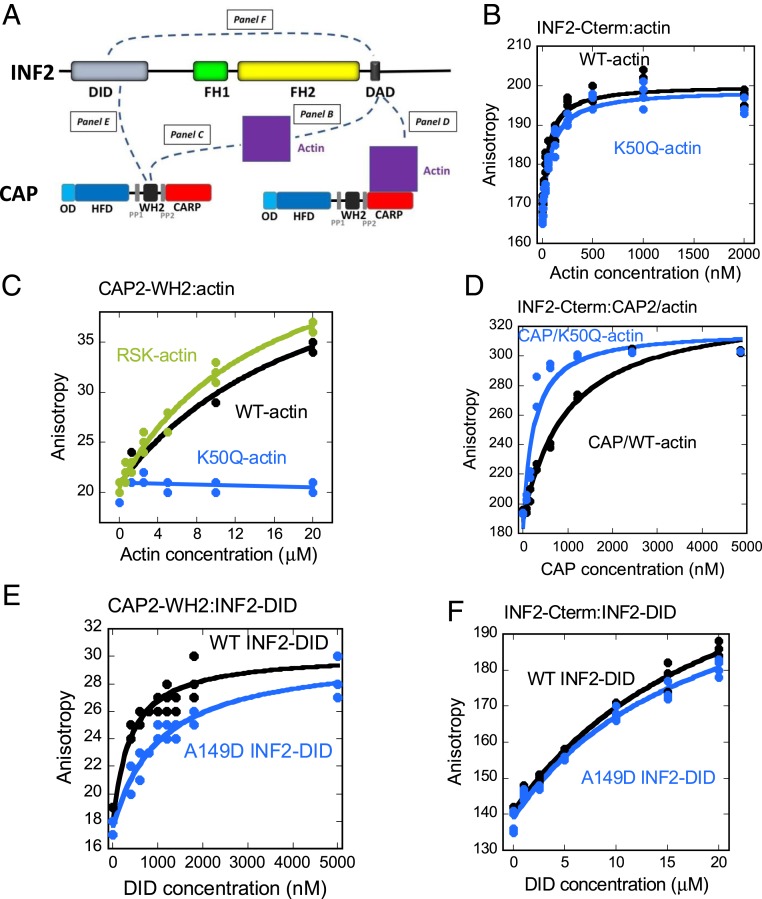
INF2, CAP, and the bridge model of INF2 inhibition. (A) Schematic of human INF2-nonCAAX (1,240 amino acids), including DID (amino acids 32 to 261), formin homology 1 domain (FH1; amino acids 421 to 520), formin homology 2 domain (FH2; amino acids 554 to 940), and DAD/WH2 (amino acids 971 to 1,000). Boundaries of the N-terminal construct (NT) and FH1-FH2-C (FFC) construct used in this study are also shown. (B) Schematic of human CAP2 (477 amino acids), including oligomerization domain (OD; amino acids 1 to 42), HFD (amino acids 43 to 217), proline-rich region 1 (PP1; amino acids 229 to 245), WH2 motif (amino acids 254 to 297), proline-rich region 2 (PP2; amino acids 308 to 323), and CARP domain (amino acids 324 to 477). (C) Actin monomers (blue, gray surfaces) bound to the dimeric CARP domain of CAP1 (black, green ribbons). K50, K61, and K328 on actin are highlighted in red, orange, and yellow, respectively. Actin subdomains are indicated by white numbers. N, amino-termini of CARP subunits. Adapted from Protein Data Bank ID code 6FM2, data from ref. 12. (Bottom) Structure rotated 90° to the left. (D) Bridge model of INF2 inhibition by CAP/actin, whereby INF2-DID interacts with CAP-WH2 while INF2-DAD interacts with acetylated actin, which is bound to the CARP domain of CAP.
We isolated a protein complex capable of inhibiting purified INF2 in a manner that requires INF2’s DID (8). The complex consists of cyclase-associated protein (CAP) bound to actin itself. Both mammalian CAP proteins, CAP1 and CAP2, can inhibit INF2. CAP consists of an N-terminal region containing an oligomerization domain (OD) and a helical folded domain (HFD), a middle region consisting of a WH2 motif flanked by proline-rich sequences, and a C-terminal CARP domain (Fig. 1B). The N-terminal OD from both budding yeast CAP and human CAP1 has been shown to hexamerize (9, 10), consistent with our results on full-length CAP2 (8).
Two regions of CAP can bind actin monomers: the WH2 motif and the CARP domain. The CARP domain binds ADP-actin with high affinity, while the WH2 domain binds ATP-actin with lower affinity (11–13). The structure of the CARP/actin complex shows that dimeric CARP binds 2 actin monomers in a manner that exposes actin’s WH2-binding site (12) (Fig. 1C). CAP has multiple potential functions in actin dynamics, including accelerating actin nucleotide exchange and enhancing cofilin-mediated depolymerization (14). This latter function requires the HFD, which can bind the cofilin-actin complex (15–18).
A key feature of INF2 inhibition by the CAP/actin complex is that lysine acetylation of the actin is required, as supported by the following findings (8). First, pretreatment of CAP/actin with histone deacetylase 6 (HDAC6) virtually eliminates INF2 inhibition. Second, isolation of the CAP/actin complex from mammalian cells treated with HDAC6 inhibitor increases both the amount of acetylated actin bound to CAP and the inhibitory potency of the CAP/actin complex. Third, transient activation of INF2 in cells causes a decrease in lysine-acetylated actin in CAP/actin complex on a similar time course as INF2 activation. Fourth, HDAC6 inhibition blocks INF2-mediated actin polymerization in cells.
Lysine acetylation of histones and other nuclear proteins has long been known to serve as an important regulatory mechanism for gene expression, but multiple cytoplasmic proteins are also known to be lysine-acetylated (19, 20), including tubulin, cortactin, and the formin mDia2 (21–23). Actin itself can be acetylated on multiple lysines (24, 25). The functional significance of actin acetylation has not been studied extensively, with one published study suggesting that acetylation of K326 or K328 in Drosophila flight muscle causes defects in morphology and performance (26).
In the present work, we addressed 2 questions. First, which lysine(s) on actin is/are relevant for acetylation-mediated INF2 regulation? Using lysine-to-glutamine mutations to mimic acetylation, we find that K50 and K61 are key residues, whereas K328 is not. Second, what interactions between CAP, lysine-acetylated actin (KAc-actin) and INF2 mediate the inhibitory interaction? Two proposed mechanisms (8) are “facilitated autoinhibition,” whereby CAP/KAc-actin binds the DID/DAD complex to secure the low-affinity DID/DAD interaction, and the “bridge” model, whereby CAP/KAc-actin binds between the DID and the DAD. Our findings provide support for the bridge model (Fig. 1D).
Results
K-to-Q Mutants of β-Actin Retain Similar Polymerization Properties as WT-Actin.
We reasoned that lysine-to-glutamine mutations might functionally mimic acetylation at specific sites on actin. Similar mutations have been used to mimic acetylation of actin (26) and other proteins (27–29). Here we examined 3 mutants to human β-actin biochemically and in cells: K50Q, K61Q, and K328Q. These residues are surface-exposed in the filament structure, with K50 and K61 in subdomain 2 and K328 in subdomain 3.
To produce β-actin amenable for biochemical tests, we used a system similar to those used previously (30, 31), in which actin is expressed as an N-terminal fusion with the monomer-binding protein thymosin β4 (Tβ4) and then cleaved with chymotrypsin after purification, resulting in actin with no additional amino acids. The initial affinity purification step results in actin-Tβ4 without detectable free actin (SI Appendix, Fig. S1A), suggesting that no endogenous actin copurifies. The use of this system results in actin without other major protein bands for WT, K50Q, K61Q, and K328Q (SI Appendix, Fig. S1B).
We next conducted tests to assess the functionality of the recombinant actins compared with rabbit muscle (RSK) actin. All recombinant actins are polymerization-competent. By high-speed pelleting assay (32), <0.17 μM actin was in the supernatant after overnight polymerization (Fig. 2A and SI Appendix, Fig. S1C), suggesting that the critical concentrations are similar to that of RSK-actin (33). By pyrene-actin polymerization, all actins polymerize with similar kinetics (Fig. 2B), with 2 variations. First, WT β-actin has a shorter polymerization lag than either RSK-actin or any of the mutants. Second, the plateau fluorescence for all mutant actins is ∼10% lower than that for WT β-actin or RSK-actin. This difference in plateau likely represents a difference in pyrene fluorescence change rather than a difference in polymerization ability, given the similar critical concentrations. We quantified the polymerization rates of the actins by determining the time to 1/2 maximum polymerization (Fig. 2D), and found that the rates of WT, K61Q, and K328Q β-actin are faster than RSK-actin, whereas K50Q β-actin is indistinguishable from RSK-actin.
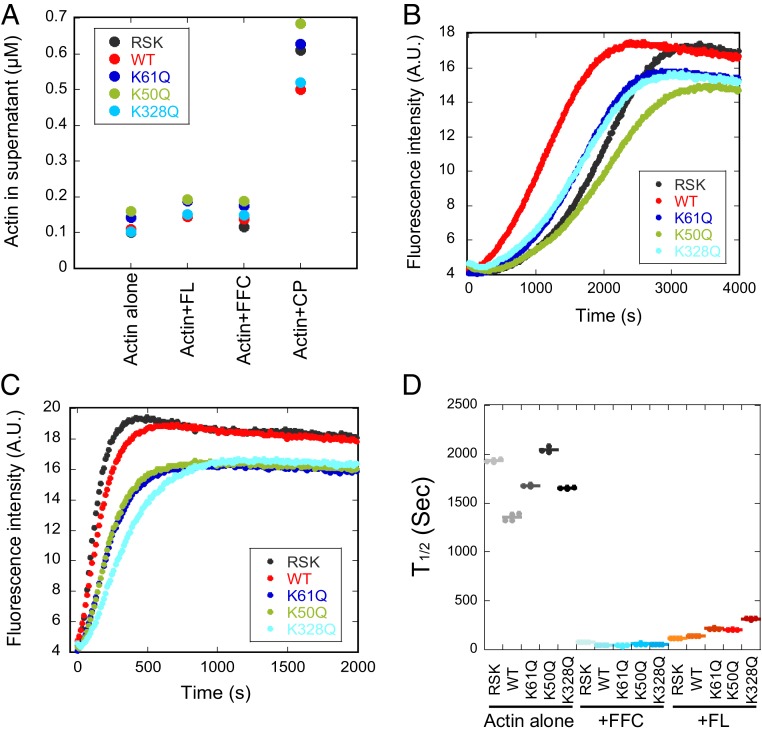
Fig. 2.
K-to-Q mutant actins are polymerization-competent. (A) Quantification of actin concentration (μM) in supernatant from high-speed pelleting assays. Actins were polymerized for 18 h at 23 °C (2 μM actin), then ultracentrifuged to separate polymerized actin (pellet) from monomeric actin (supernatant). Four conditions were tested: actin alone (−), +20 nM INF2-FL (FL), +20 nM INF2-FFC (FFC), and +10 nM capping protein (CP). Sample gels are shown in SI Appendix, Fig. S1C. (B and C) Pyrene-actin polymerization assays of 2 μM actin alone (B) or +20 nM INF2-FL (C). Actin composition: 1.9 μM of the indicated actin +0.1 μM pyrene-labeled RSK actin. (D) Time to half-maximum polymerization, measured from pyrene-actin curves similar to those in B and C and SI Appendix, Fig. S1D. Sec, seconds; A.U., arbitrary units.
We also assessed the effects of INF2 on the polymerization of recombinant actins, testing both full-length INF2 and INF2-FFC. The INF2 isoform used in this study is the nonCAAX variant, which is largely cytosolic and lacks the C-terminal prenylation site of the INF2-CAAX isoform (34, 35). By high-speed pelleting assay, both INF2 constructs cause a slight increase in the amount of actin recovered in the supernatant (Fig. 2A and SI Appendix, Fig. S1C), suggestive of a shift in critical concentration due to INF2’s effects on barbed end dynamics (36, 37). The degree of this shift is similar for all actins tested and is <0.2 μM. As a control, polymerization of all actins in the presence of barbed end capping protein results in a shift in supernatant actin to a value similar to the pointed end critical concentration (Fig. 2A and SI Appendix, Fig. S1C), suggesting that capping protein interaction is not fundamentally affected by the mutations.
By a pyrene-actin assay in the presence of INF2-FFC or INF2-FL, all 3 actin mutants display a lower polymerization plateau than WT β-actin or RSK-actin (Fig. 2C and SI Appendix, Fig. S1D). This effect is similar to that found for actin alone, again suggesting a change in pyrene-actin fluorescence rather than in polymerization properties. In terms of polymerization rate, the values for all mutants in the presence of INF2-FL are slightly slower than those for WT β-actin or for RSK actin (Fig. 2D).
In summary, these analyses suggest that the β-actins produced recombinantly have largely similar polymerization properties to RSK-actin, both in the absence and the presence of INF2. The minor differences in their polymerization rates in the presence of INF2-FL do not account for the effects of specific mutants on INF2-FL activity when in complex with CAP2, described next.
K50Q and K61Q Actin Are INF2 Inhibitors When in Complex with CAP2.
We assessed the ability of CAP-complexed β-actin mutants to inhibit actin polymerization by INF2-FL. We previously we found that purified CAP1 or CAP2 preparations from HEK293 cells, containing approximately equimolar actin, are poor INF2 inhibitors. However, CAP became a potent INF2 inhibitor when the bound actin (293A) was exchanged with certain types of actin (mouse brain or chicken skeletal muscle actin), but not with others (RSK-actin) (8). Here we used a similar approach to exchange WT-actin or K-to-Q β-actin mutants onto CAP2.
Exchange of CAP2/293A with either RSK-actin or any of the recombinantly expressed β-actins results in similar CAP/actin ratios (SI Appendix, Fig. S2A). We also examined the nucleotide state of the bound actin before and after exchange. Purified CAP2/actin contains both ATP and ADP, with ATP being ∼65% of the total (SI Appendix, Fig. S2B). Interestingly, the “mock” exchange reaction, in which CAP2/actin is incubated with buffer containing 0.2 mM ATP, causes the ATP:ADP ratio to increase, suggesting nucleotide exchange on the bound actin. Exchange with either WT- or K50Q-actin results in a similar increase in ATP:ADP ratio (SI Appendix, Fig. S2C).
We assessed the effects of exchanged CAP2/actin complexes on actin polymerization by full-length INF2-nonCAAX in pyrene-actin assays. At 1 μM CAP, the CAP/K50Q and CAP/K61Q complexes display strong inhibition of INF2 activity (Fig. 3A). In contrast, the CAP/K328Q, CAP/WT, CAP/RSK, and CAP/293A complexes display much less inhibition. Concentration curves show that CAP/K50Q has an IC50 of 212 nM, while CAP/K61Q has an approximate IC50 of 770 nM and does not reach an inhibition plateau at the highest concentration tested (Fig. 3B). The other constructs display minimal inhibition at all concentrations tested (Fig. 3B).
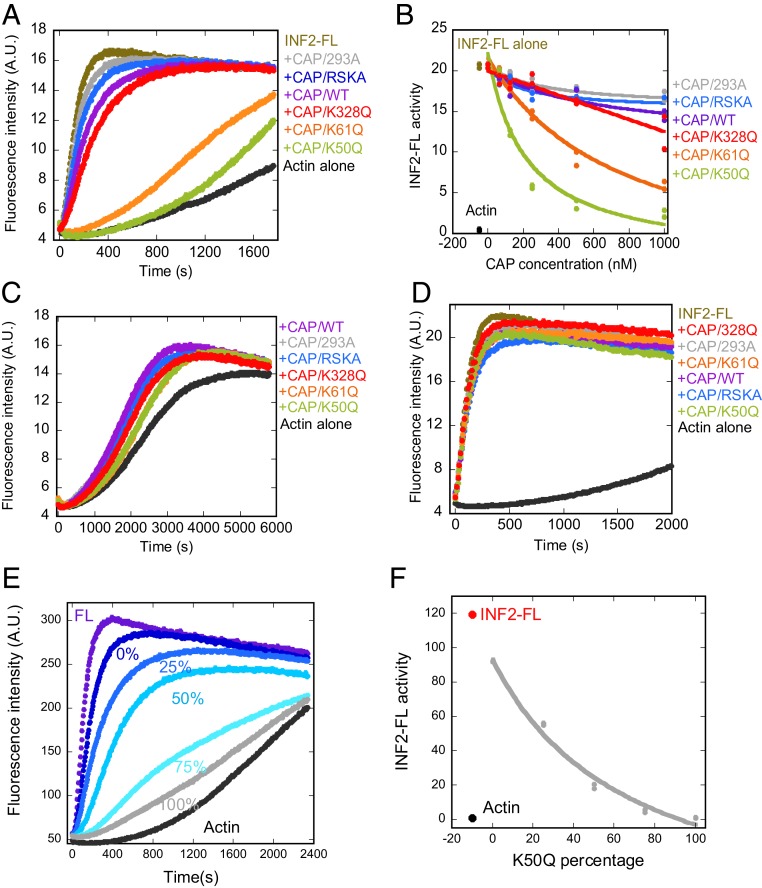
Fig. 3.
β-actin mutants K50Q and K61Q, when bound to CAP2, inhibit INF2-mediated actin polymerization in biochemical assays. (A) Pyrene-actin polymerization assays (2 μM RSK-actin, 5% pyrene-labeled) containing 20 nM INF2-FL and 1 μM of the indicated CAP/actin complex. (B) Concentration curves of CAP/actin inhibition of INF2-FL polymerization activity, from assays similar to panel A. Polymerization activity of actin alone (black) and with INF2-FL (brown) shown to left of the curves. (C) Pyrene-actin polymerization assays of actin alone (2 μM RSK-actin, 5% pyrene-labeled) or in the presence of 1 μM of the indicated CAP/actin complex. (D) Pyrene-actin polymerization assays (2 μM RSK-actin, 5% pyrene-labeled) with 20 nM INF2-FFC and 1 μM of the indicated CAP/actin complex. (E) Pyrene-actin polymerization assays similar to those in D, in which varying ratios of K50Q:WT-β-actin were exchanged onto CAP2, then assayed for inhibition of INF2-FL. The percentages listed represent the percentage of K50Q-actin in the exchange reaction. Concentrations in pyrene-actin assays: 2 μM RSK-actin (5% pyrene), 20 nM INF2-FL, and 1 μM CAP/actin. (F) Graph of INF2-FL activity in the presence of 1 μM CAP/actin as a function of percent K50Q-actin in exchange reactions, from data similar to those in E. A.U., arbitrary units.
We tested the specificity of this CAP/actin inhibition for full-length INF2 in 2 ways. First, we examined the effects of the CAP/actin complexes on polymerization of actin alone. At 1 μM CAP, all complexes slightly accelerate actin polymerization (Fig. 3C), similar to our past results on CAP/actin complexes (8). Next, we examined the effects of CAP/actin complexes on actin polymerization acceleration by INF2-FFC, which lacks the N-terminal DID (Fig. 1A). At 1 μM CAP, none of the complexes affect INF2-FFC activity (Fig. 3D).
Finally, we asked whether increasing the percentage of K50Q-actin to WT-actin in the CAP/actin complex caused a progressive change in INF2 inhibition, by altering the K50Q:WT ratio in exchange reactions. Increasing the K50Q:WT percentage causes a progressive increase in INF2 inhibition (Fig. 3 E and F). Since the amount of actin bound to CAP is not different between K50Q- and WT-actin exchanged CAP (SI Appendix, Fig. S2A), this suggests that an increased proportion of Ac-actin bound to CAP increases inhibitory potency.
These results suggest that the K50Q and K61Q actin mutants, when complexed with CAP2, act as potent inhibitors of INF2 activity, presumably by acting as acetylation mimics. This inhibition requires INF2’s DID, suggesting that the CAP/actin complexes aid INF2 autoinhibition. In contrast, the K328Q mutant is a poor actin inhibitor.
We also attempted to load a previously described C-terminal construct of CAP1 (15), containing the WH2 motif and the CARP domain, with ATP-actin. Interestingly, while full-length CAP2 can remain stably bound to ATP-actin, we obtain only ∼15% binding of ATP-actin to CAP1-Cterm for RSK-actin, WT-, or K50Q-β-actin (SI Appendix, Fig. S2D). This result agrees with previous studies showing low affinity of this construct for ATP-actin (15).
Acetyl-Mimetic Actin Mutants Inhibit INF2-Mediated Actin Polymerization in Cells.
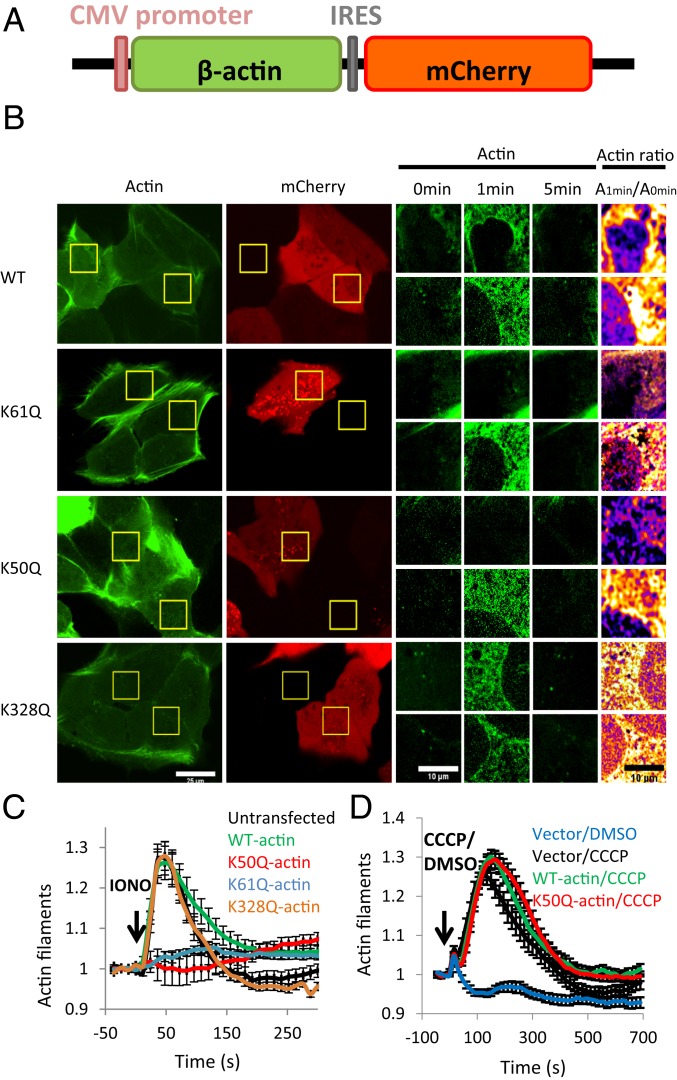
To test the effect of K-to-Q mutant actins in cells, we constructed a bis-cistronic vector coexpressing untagged β-actin along with mCherry (Fig. 4A). We transiently transfected this actin/mCherry vector into U2OS cells, along with a vector expressing GFP-F-tractin, an actin filament probe. We then assessed cytosolic actin filament levels induced by ionomycin stimulation, which we and others have previously shown to be INF2-dependent (38–41). We imaged the cells in a medial z-section, to avoid abundant basal actin structures. In control cells without the actin/mCherry vector or expressing mCherry alone, ionomycin causes a transient increase in cytoplasmic actin filaments, as detected by an increase in filamentous GFP-F-tractin (SI Appendix, Fig. S3 A and B). Cells expressing WT- or K328Q-actin display a similar ionomycin-induced actin burst (Fig. 4 B and C). In contrast, cells expressing the K50Q or K61Q mutants display greatly reduced responses to ionomycin (Fig. 4 B and C). These results suggest that acetylation at K50 or K61 is sufficient for potent INF2 inhibition in cells.
Fig. 4.
β-actin K50Q and K61Q mutants inhibit the calcium-induced actin burst in U2OS cells. (A) Schematic of mammalian expression construct coexpressing β-actin (untagged) and mCherry. (B) Micrographs of calcium-induced actin burst for cells expressing 4 β-actin constructs: WT, K61Q, K50Q, and K328Q. U2OS cells were cotransfected with the β-actin/mCherry expression plasmid along with a plasmid containing GFP-F-tractin (to label actin filaments). Live cells were imaged for GFP and mCherry before and during stimulation with ionomycin (4 μM in serum-containing medium). The micrographs at left are full-field views of GFP and mCherry, and those in the center are zoom-in views of GFP-F-tractin at 3 time points of ionomycin stimulation (0, 1, and 5 min) for 2 cells in the field: an mCherry-expressing cell (Top) and a cell not expressing detectable mCherry (Bottom). At the right is a differential heat map showing the ratio of the actin signal at 1 min to the signal at 0 min (red/yellow colors denote a higher 1 min:0 min ratio). (Scale bars: 25 μm at left and 10 μm in the zoom-in views). (C) Quantification of the ionomycin-induced actin burst for cells expressing WT β-actin, K50Q β-actin, K61Q β-actin, and K328Q β-actin, compared with cells not expressing an actin/mCherry construct (untransfected). Results are from 3 experiments, with a total of 17 untransfected, 38 WT, 42 K61Q, 29 K50Q, and 34 K328Q cells analyzed. (D) Quantification of the CCCP-induced actin burst for cells transfected with the mCherry vector with no actin (vector/CCCP, black, 61 cells), the mCherry/WT-actin vector (WT-actin/CCCP, green, 71 cells), or the mCherry/K50Q-actin vector (K50Q-actin/CCCP, red, 72 cells). Mock-stimulation of mCherry/no actin-transfected cells (Vector/DMSO, blue, 40 cells) shown for comparison. Results are from 3 independent experiments.
To test whether the actin mutants affect other types of stimulus-induced actin polymerization, we examined U2OS response to the mitochondrial uncoupler CCCP, which induces a rapid and transient actin polymerization burst around mitochondria that is INF2-independent (42). Expression of either WT-actin or K50Q-actin construct in U2OS cells does not affect the CCCP-induced actin burst (Fig. 4D and SI Appendix, Fig. S3C). This result suggests that the effect of the K50Q mutant on ionomycin-induced actin polymerization is INF2-specific, not an effect on actin polymerization in general.
We conducted additional experiments to test the validity of the actin expression system. To test the effect of exogenous actin expression on overall actin protein levels, we sorted transfected cells for low and high mCherry expression by flow cytometry and then conducted Western blot analysis for actin, mCherry, and calnexin (loading control). None of the actin/mCherry vectors causes an increase in the actin:calnexin ratio in either the low- or high-mCherry pools (SI Appendix, Fig. S4 A and B), suggesting that the level of exogenous actin is well below that of the total cellular actin pool.
We next assessed the possibility that expression of actin mutants changes overall actin filament distribution, by analyzing fixed cells stained with fluorescein isothiocyanate (FITC)-phalloidin. Visual examination of either the basal region or a medial region shows no major differences in FITC staining between transfected and untransfected cells for any of the constructs (SI Appendix, Fig. S4C). We also examined the levels of GFP-F-tractin and mCherry expressed in the cells used for live-cell analysis of ionomycin response. None of the constructs displayed aberrantly high expression of F-tractin or mCherry (SI Appendix, Fig. S4 D and E), suggesting that effects of the K50Q and K61Q constructs are not due to overexpression of these constructs. In fact, mCherry expression levels are consistently lower in the K50Q mutant-expressing cells compared with the other samples. Finally, we examined the effect of F-tractin expression level on the ionomycin-induced actin burst response and found a linear relationship between initial F-tractin intensity and peak ionomycin-induced F-tractin intensity (SI Appendix, Fig. S4F). Since F-tractin levels are similar among the constructs tested, this result shows that the system is not saturated for F-tractin.
Our overall conclusion is that 2 actin mutants, K50Q and K61Q, significantly compromise INF2 activation in U2OS cells. In contrast, K328Q has little effect on INF2 activation.
Direct-Binding Studies Suggest That the CAP-WH2 Domain Binds INF2-DID and CAP/Ac-Actin Binds INF2-DAD.
We used fluorescence anisotropy to examine binding between the WH2 motifs of CAP1 or CAP2 (SI Appendix, Fig. S5A) with both WT and K50Q β-actin monomers, as well as with INF2’s DID-containing N-terminal region. In parallel, we conducted similar binding studies for INF2’s DAD-containing C-terminal region. A schematic of the interactions tested here is provided in Fig. 5A.
Fig. 5.
CAP’s WH2 binds INF2 DID, while INF2’s DAD binds CAP/KAc-actin. (A) Schematic of binding interactions measured in B–F, showing bars for INF2 (top bar) and CAP (bottom bars) and a purple square for actin. (B–F) Fluorescence anisotropy measurements using TAMRA-INF2-Cterm (50 nM) or FITC-CAP2-WH2 (100 nM). (B) Interaction between INF2-Cterm and WT-β-actin (black) or K50Q-β-actin (blue). (C) Interaction between CAP2-WH2 and RSK-actin (green), WT-β-actin (black) or K50Q-β-actin (blue). CAP1-WH2 results are presented in SI Appendix, Fig. S5B. (D) Interaction between INF2-Cterm and CAP/WT-β-actin (black) or CAP/K50Q-β-actin (blue) complex. (E) Interaction between CAP2-WH2 and INF2-Nterm (black) or INF2-A149D-Nterm (blue). CAP1-WH2 results are presented in SI Appendix, Fig. S5C. (F) Interaction between INF2-Cterm (right) and INF2-Nterm (black) or INF2-A149D-Nterm (blue). All anisotropy values are in milli-anisotropy units.
Similar to previous results using muscle actin (6, 7), INF2-Cterm binds both WT-actin and K50Q-actin with Kdapp values of 78 nM and 63 nM, respectively (Fig. 5B). In contrast, neither CAP1-WH2 nor CAP2-WH2 binds RSK-actin or WT-actin with sufficient affinity to determine an accurate dissociation constant, with estimated Kdapp values of 10, 16, 25, and 22 μM, respectively (Fig. 5C and SI Appendix, Fig. S5B). Interestingly, the WH2 motifs from both CAP proteins display no detectable binding to K50Q-actin (Fig. 5C and SI Appendix, Fig. S5B).
We previously showed that INF2-Cterm displays moderate affinity for CAP/actin (8). To test whether actin acetylation affects this interaction, we conducted anisotropy experiments for INF2-Cterm with either CAP/WT-actin or CAP/K50Q-actin. Interestingly, the affinity increases by ∼5-fold with K50Q-actin (Fig. 5D; Kdapp of 1,066 nM for CAP/WT-actin and 211 nM for CAP/K50Q-actin).
Finally, we tested the possibility that CAP-WH2 might interact with INF2-DID. Interestingly, both CAP1-WH2 and CAP2-WH2 display significant affinity for the N-terminal region of INF2 (Fig. 5E and SI Appendix, Fig. S5C), with Kdapp of 480 nM and 390 nM, respectively. We have previously shown that the A149D mutation in INF2-DID, which is analogous to a mutant that disrupts DID/DAD binding in mDia1, causes constitutive activation of INF2 in cells (6, 8). Interestingly, both CAP1-WH2 and CAP2-WH2 display reduced affinity for A149D-mutant INF2-DID (Kdapp of 1,723 nM and 1,148 nM, respectively). In contrast, INF2-Cterm displays low affinity for both WT- and A419D-INF2-Nterm (estimated Kdapp of 22 and 18 μM respectively; Fig. 5F), similar to previous findings (3).
CAP1 and CAP2 Are Present in Excess Over INF2 in U2OS Cells.
We used quantitative Western blot analysis to determine levels of CAP1, CAP2, and INF2 in U2OS cells (SI Appendix, Fig. S6). We first raised polyclonal antibodies against bacterially expressed human CAP1 and CAP2 and then determined the specificity of the antibodies against their respective CAP-GFP purified from HEK293 cells. Anti-CAP2 displays no observable cross-reactivity to CAP1 at the protein amounts tested, whereas anti-CAP1 displays weak CAP2 cross-reactivity, ∼30-fold less than that of CAP1 from band densities. Interestingly, the band of untagged CAP in both the CAP1-GFP and CAP2-GFP preparations appears to be predominately CAP1, since it is detected by anti-CAP1 but not by anti-CAP2.
We determined a linear range of detection for purified CAP1-GFP, CAP2-GFP, and GFP-INF2-nonCAAX using anti-INF2 raised previously (35) in the presence of fixed concentrations of U2OS cell extract. We then used U2OS extracts of well-defined cell and protein concentrations to analyze CAP1, CAP2, and INF2 signal intensity in the linear detection range. Both CAP1-GFP and CAP2-GFP display doublet bands on Western blot analysis, and we used both bands for quantification. From extracts, CAP2 runs as a doublet, and we used the lower band for quantification.
U20S cells contain 7.95 × 106, 4.44 × 105, and 8.79 × 104 molecules/cell of CAP1, CAP2, and INF2, respectively (Table 1). From the same extract, the total actin concentration is 1.96 × 108 molecules/cell (Table 1), similar to that determined previously (43). To estimate cytoplasmic concentrations, we used cell and organelle volumes determined from an in-depth lattice light sheet study of Cos7 cells (44), with the resulting cytoplasmic volume (3.14 pL) approximating that determined for NIH 3T3 cells (2.26 pL) using different methods (45). These estimates are provided in Table 1. Overall, CAP1 and CAP2 are ∼90-fold and 5-fold more abundant, respectively, than INF2 in U2OS cells.
Table 1.
Concentrations of CAP1, CAP2, INF2, and actin in U2OS cells
| Protein | Molecules/cell, ×106, mean ± SD | Cytoplasmic concentration, μM | N |
| CAP1 | 7.95 ± 1.23 | 4.21 | 8 |
| CAP2 | 0.444 ± 0.089 | 0.24 | 6 |
| INF2 | 0.0879 ± 0.0105 | 0.05 | 6 |
| Actin | 196 ± 12 | 104.00 | 6 |
Concentrations reflect CAP and INF2 monomers.
Discussion
This study makes several advances in understanding the mechanism of INF2 regulation by CAP/KAc-actin. First, we show that acetyl-mimetic mutants of 2 residues, K50 and K61, confer inhibitory activity to β-actin when coupled to CAP both in biochemical assays and in cells. Second, we provide evidence that the DID of INF2 does not engage in an autoinhibitory interaction with its DAD but binds the WH2 of CAP, while INF2-DAD binds at another site on the CAP/KAc-actin complex in a manner influenced by actin acetylation. These findings support a bridge model for INF2 inhibition (Fig. 1D).
We purified recombinant β-actin as well as 3 mutants—K50Q, K61Q, and K328Q—for biochemical assays. The positions of these 3 residues suggest that the mutations do not affect the basic polymerization properties of actin. Indeed, the critical concentrations of all β-actins made here appear similar to those of RSK-actin, while there are minor differences in polymerization kinetics. Most importantly, all of the mutant actins are efficiently polymerized by INF2, suggesting that acetylated actin alone is not an INF2 inhibitor.
When in complex with CAP2, 2 acetyl-mimetic mutants, K50Q and K61Q, inhibit INF2 with significantly higher potency than WT-actin. The inhibition potency of CAP2/K50Q-actin (IC50 of 212 nM) is comparable to that of the inhibitory CAP/actin complexes that we identified previously, CAP2/brain actin and CAP2/chicken muscle actin, with IC50 values of 54 nM and 254 nM, respectively. CAP2/K61Q-actin is not as potent, with an estimated IC50 of 772 nM. In contrast, CAP2/K328Q-actin displays negligible INF2 inhibitory activity.
These results suggest that acetylation in subdomain 2 of the actin monomer is key to INF2 inhibition. K50 is in the “D-loop”, which is unstructured in many actin structures, whereas K61 is in a helical region. Both residues face away from CAP in the CARP/actin crystal structure (Fig. 1C). Intriguingly, this region of subdomain 2 gets pulled back toward CARP (12), further exposing these residues (Fig. 1C).
This study represents an initial mechanistic evaluation of actin acetylation effects and, as such, raises interesting questions. First, might dual acetylation of K50 and K61 increase INF2 inhibition, as suggested for K326 and K328 acetylation in Drosophila flight muscle (26)? Second, how does actin acetylation interface with other actin posttranslational modifications? Interestingly, K50 and K61 reside near M44 and M47, which are substrates for MICAL-mediated oxidation (46). Third, do other acetylated lysines play roles in INF2 regulation? We have previously identified several other acetylated lysines (8), and a number of additional acetylation sites have been identified in proteomic screens (24).
As an aside, both INF2 full-length and INF2-FFC do increase the apparent critical concentration slightly for all actins tested. This change might reflect INF2’s severing/depolymerization ability (7, 36, 37) or the relative effects of the FH2 on on-rate and off-rate at the barbed end, which vary between formins (47–50). We have previously found that INF2-FFC slows barbed end elongation by ∼60% (37).
Actin binding by full-length CAP reveals several interesting features. First, full-length CAP2 can bind ATP-actin with high affinity, since significant ATP-actin remains on CAP after extensive purification in nucleotide-free buffer. Given that previous results have shown the CARP domain to have much higher affinity for ADP-actin than for ATP-actin (11–13), this result is surprising and suggests that the full-length protein varies somewhat. The WH2 motif is not the major ATP-actin binder, since it has low affinity for ATP-actin (Fig. 5). However, the situation might be different in the context of the full-length CAP. Second, full-length CAP2 can allow nucleotide exchange on the bound actin without causing its release. Third, ATP-actin can replace bound actin on CAP2. In view of the fact that the “mock” exchange reactions do not result in loss of actin from CAP2, this result raises interesting questions as to how this exchange takes place.
The present work also provides insight into the relevant interactions regulating INF2 activity. Two pieces of evidence suggest that INF2-DAD binds primarily to actin in the CAP/actin complex in a manner enhanced by actin acetylation. First, INF2-Cterm binds actin monomers with much higher affinity than it binds INF2-DID. While INF2’s DID/DAD interaction would be significantly enhanced by their presence in the same polypeptide, the high affinity of INF2-DAD for actin disfavors DID/DAD interaction even in the full-length protein (6). Second, INF2-Cterm binds CAP/K50Q-actin with 5-fold higher affinity than CAP/WT-actin. We propose that INF2-DAD binds actin in an analogous manner to established WH2/actin interactions (5). We also hypothesize that the acetylated actin residues are not part of the binding interface, but that their acetylation results in conformational changes that enhance WH2 binding. Given their relative affinities for DAD, free actin monomers would be expected to outcompete CAP/K50Q-actin; however, the high concentration of profilin in mammalian cells reduces free actin levels substantially.
Our data also suggest that the relevant interaction of INF2-DID is with CAP-WH2, based on the following data. First, both CAP1-WH2 and CAP2-WH2 bind the DID-containing INF2-Nterm with submicromolar affinity, which is appreciably tighter than INF2’s own DID/DAD interaction. Second, neither WH2 motif binds actin monomers with appreciable affinity, and binding to K50Q-actin is undetectable. While it has been shown that actin monomer binding by the WH2 motif might be relevant for CAP’s other biochemical functions (11–13), the lack of binding to K50Q-actin makes it unlikely to interact with actin in INF2 regulation.
Based on these results, we propose that CAP/KAc-actin inhibits INF2 by serving as a bridge between INF2-DID and INF2-DAD/WH2 (Fig. 1D), rather than as a facilitator of INF2’s DID/DAD interaction. KAc-actin is likely bound to CAP’s CARP domain, since the HFD appears to bind only cofilin-bound actin monomers (15–18). One interesting structural feature is that CAP’s CARP forms a back-to-back dimer with actins on the outer surfaces (12), so that a 2:2 complex of CARP with KAc-actin might inhibit 1 INF2 dimer (8). Another interesting feature is that the N-terminal region of CAP has been shown to hexamerize (9, 10). Our previous studies (8) suggested that both purified human CAP1 and CAP2 form hexameric complexes, raising the possibility that 3 INF2 dimers could be inhibited by 1 CAP hexamer bound to 6 acetylated actins. However, our fractionation results from mouse brain suggest that smaller CAP-containing complexes might also exist (8), so the stoichiometry of the inhibitory complex in cells remains to be determined.
In our model, INF2 activation would occur through HDAC6-mediated deacetylation of lysines in subdomain 2, which would weaken the affinity of INF2-DAD for actin in the CAP/actin complex. Our proposal that the acetylated lysines do not form part of the DAD-binding interface would allow for HDAC6 access to the acetylated groups, resulting in the rapid stimulus-induced INF2 activation that we observe in cells (8, 41). The fate of the CAP/actin complex after deacetylation is interesting to consider. The complex could remain bound to INF2-DID and influence INF2’s activity on actin in a positive manner, possibly by directing ATP-bound actin monomers to FH1-bound profilin.
Our work here focuses on the INF2-nonCAAX splice variant, which is predominantly cytosolic. It is likely, however, that the ER-bound INF2-CAAX variant is subject to similar regulation because it is strongly inhibited in cells, similar to INF2-nonCAAX (6, 38, 40, 41). In addition, HDAC6 inhibition blocks the mitochondrial calcium increase that occurs downstream of both histamine and ionomycin stimulation in U2OS cells (8), an effect dependent on INF2-CAAX (41).
Finally, we show that the cellular concentrations of CAP1 and CAP2 are in excess to that of INF2. These results suggest that CAP is in sufficient excess to inhibit INF2, in addition to its other cellular roles (14). CAP1 is nearly 20-fold more abundant than CAP2 in U2OS cells, so is likely the primary regulator of INF2 in this cell type. One question is how CAP’s role in INF2 regulation is balanced with its other cellular roles. In addition, it will be important to determine the differential roles of CAP1 and CAP2 in INF2 regulation, as well as their own regulatory mechanisms. CAP1 has known phosphorylation sites just N-terminal to the CARP domain (51) that have been shown to influence its cellular effects, and CAP2 has sites that are possibly analogous. It is interesting that CAP2 consistently runs as a doublet both from cell lysates and as the purified protein, possibly due to posttranslational modification.
Materials and Methods
Plasmids.
Human INF2 full-length nonCAAX and CAP2 constructs have been described previously (8). For the actin purification construct, the entire human WT β-actin-thymosin β4-6xHis tag cDNA (including stop codon 3′ to 6xHis) was PCR-amplified from an expression plasmid designed for insect cells (a gift from Kathy Trybus) and subcloned into Xho1/EcoR1 sites of the eGFP-N1 vector (Clontech). For bis-cistronic β-actin expression plasmid, human WT β-actin alone was PCR- amplified from the Trybus β-actin expression plasmid and cloned into Xho1/EcoR1 sites of PICherryNeo (Addgene; 52119). For bacterial expression, CAP1 and CAP2 WH2 motifs (amino acids 247 to 292 and 254 to 297, respectively) were PCR-amplified from human CAP1 and CAP2 cDNA (NovoPro; 710829-5 [NM-006367] and 710470-11 [NM-006366]) and subcloned into BamH1/EcoR1 sites of pGEX-KT (52) with the HRV3C protease site introduced between the GST tag and WH2 motif. Human INF2-CAAX C-term (amino acids 941 to 1,249) and DID-containing construct (amino acids 1 to 420) described previously (6). GFP-F-tractin plasmid has been described previously (41).
Protein Expression, Purification, and Labeling.
Rabbit skeletal muscle actin was purified from acetone powder (53) and labeled with pyrenyliodoacetamide (54). Both labeled and unlabeled actin gel-filtered on Superdex 75 16/60 columns (GE Healthcare) and stored in G buffer (2 mM Tris⋅HCl pH 8, 0.5 mM DTT, 0.2 mM ATP, 0.1 mM CaCl2, and 0.01% wt/vol sodium azide) at 4 °C. Expression and purification of INF2-FL non-CAAX, CAP1-GFP, and CAP2-GFP in Expi293-F cells (Thermo Fisher Scientific; A14527) was described previously (8) and is described in detail in SI Appendix. For recombinant β-actin expression and purification in Expi293-F cells, previously described methods (30, 31) were adapted, as described in detail in SI Appendix. Protein expression and purification of INF2-Nterm, INF2 C-term, and the GST-fusion of human CAP1-Cterm from Escherichia coli have been described previously (6, 12). Labeling of INF2-Cterm with tetramethylrhodamine-succinimide (Molecular Probes; C1171) as described previously (6). Labeling of CAP1-WH2 and CAP2-WH2 with FITC-maleimide (Thermo Fisher Scientific; 62245) is described in SI Appendix.
Antibodies.
CAP1 and CAP2 rabbit polyclonal against GST-human CAP1 and GST-human CAP2 were produced by Cocalico Biologicals. Antibodies were affinity-purified using the GST-fusion proteins attached to SulfoLink (Thermo Fisher Scientific; 20404) in the presence of 10 mg/mL GST, eluted in 200 mM glycine-HCl pH 1.9, then dialyzed into PBS. Anti-actin (mouse monoclonal, clone C4; EMD Millipore; MAB1501), anti-calnexin (rabbit monoclonal; Cell Signaling; 2679), anti-mCherry (rat monoclonal; Invitrogen; M11217). Anti-INF2 rabbit polyclonal has been described previously (35). Secondary antibodies were IRDye 680RD goat anti-mouse (LiCor; 926-68070), and IRDye 800CW goat anti-rabbit (LiCor; 926-32211).
Actin Biochemical Assays.
Pyrene actin polymerization assay and high-speed sedimentation assay are described in detail in SI Appendix. Actin exchange onto CAP was described previously (8) and in SI Appendix. Fluorescence anisotropy measurements have been described previously (6) and in detail in SI Appendix. The units reported here are milli-anisotropy units. For CAP/actin nucleotide content analysis, CAP2/293A was loaded on Strep-Tactin beads and then incubated in G-buffer alone (containing 0.2 mM ATP) or in G-buffer containing the indicated actin overnight at 4 °C with end-over-end rotation. Beads were washed with 20 CV of G-buffer and then 20 CV of K50MEHD (10 mM Hepes pH 7.4, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT), followed by elution with K50MEHD containing 2.5 mM dethiobiotin. Samples were balanced for CAP2 protein mass by Coomassie-stained SDS/PAGE gel and then boiled for 7 min. Protein was removed by centrifugation at 13,000 × g for 5 min, after which supernatant was loaded onto 1 mL of SourceQ, followed by salt gradient (0–150 mM NaCl, 10 mM Tris pH 8.0) elution. Unexchanged samples were in K50MEHD and were never exposed to ATP during purification or mock exchange.
Cellular Assays.
Live-cell and fixed-cell imaging methods have been described in detail previously (41, 42) and are explained in SI Appendix. Flow cytometry and quantification of CAP1, CAP2, INF2, and actin levels from U2OS cell extracts are described in detail in SI Appendix. Cytosolic concentrations were estimated using a cytoplasmic volume calculated for Cos7 cells obtained from (44) except for the nuclear volume, which was provided by Sarah Cohen (University of North Carolina at Chapel Hill).
Statistical Analysis.
Errors (as SEM, except for Table 1, in which they are SD) were calculated using Microsoft Excel, version 2007 or 2010. Binding curves were fit and dissociation constants (Kd) were calculated using KaleidaGraph 4.5.3. Linear standard curves were fit using Microsoft Excel, version 2007 or 2010.
Data Availability.
All data are available in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank Sarah Cohen for advice on nuclear volume in Cos7 cells, Roberto Dominguez for discussions and advice on WH2 motifs, Pekka Lappelainen for advice and C-terminal CAP protein, Nate Glurio for keeping us balanced, Dave Sept for his idea that CAP-WH2 might interact with INF2-DID, Chris Shoemaker for help with flow sorting, and Kathy Trybus for help in establishing the actin expression/purification system. This work was supported by NIH Grants R01 GM069818 and R35 GM122545 to H.N.H., R01 DK088826 to M. Pollak (an H.N.H. subcontract), and P20 GM113132 to the Institute for Biomolecular Targeting Centers of Biomedical Research Excellence (COBRE).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914072117/-/DCSupplemental.
References
- 1.Higgs H. N., Formin proteins: A domain-based approach. Trends Biochem. Sci. 30, 342–353 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Goode B. L., Eck M. J., Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Sun H., Schlondorff J. S., Brown E. J., Higgs H. N., Pollak M. R., Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2). Proc. Natl. Acad. Sci. U.S.A. 108, 2933–2938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F., Higgs H. N., The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Dominguez R., The WH2 domain and actin nucleation: Necessary but insufficient. Trends Biochem. Sci. 41, 478–490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramabhadran V., Hatch A. L., Higgs H. N., Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction. J. Biol. Chem. 288, 26847–26855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chhabra E. S., Higgs H. N., INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 281, 26754–26767 (2006). [DOI] [PubMed] [Google Scholar]
- 8.A M., Fung T. S., Kettenbach A. N., Chakrabarti R., Higgs H. N., A complex containing lysine-acetylated actin inhibits the formin INF2. Nat. Cell Biol. 21, 592–602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen S., Collins A., Golden L., Sokolova O., Goode B. L., Structure and mechanism of mouse cyclase-associated protein (CAP1) in regulating actin dynamics. J. Biol. Chem. 289, 30732–30742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhry F., et al. , Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol. Biol. Cell 24, 31–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattila P. K., et al. , A high-affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein. Mol. Biol. Cell 15, 5158–5171 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotila T., et al. , Structural basis of actin monomer re-charging by cyclase-associated protein. Nat. Commun. 9, 1892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhry F., Little K., Talarico L., Quintero-Monzon O., Goode B. L., A central role for the WH2 domain of Srv2/CAP in recharging actin monomers to drive actin turnover in vitro and in vivo. Cytoskeleton (Hoboken) 67, 120–133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono S., The role of cyclase-associated protein in regulating actin filament dynamics—More than a monomer-sequestration factor. J. Cell Sci. 126, 3249–3258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotila T., et al. , Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat. Commun. 10, 5320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makkonen M., Bertling E., Chebotareva N. A., Baum J., Lappalainen P., Mammalian and malaria parasite cyclase-associated proteins catalyze nucleotide exchange on G-actin through a conserved mechanism. J. Biol. Chem. 288, 984–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriyama K., Yahara I., Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 115, 1591–1601 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Quintero-Monzon O., et al. , Reconstitution and dissection of the 600-kDa Srv2/CAP complex: Roles for oligomerization and cofilin-actin binding in driving actin turnover. J. Biol. Chem. 284, 10923–10934 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narita T., Weinert B. T., Choudhary C., Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Allfrey V. G., Faulkner R., Mirsky A. E., Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X., et al. , HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 27, 197–213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., et al. , Histone deacetylase 6 regulates cytokinesis and erythrocyte enucleation through deacetylation of formin protein mDia2. Haematologica 102, 984–994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palazzo A., Ackerman B., Gundersen G. G., Cell biology: Tubulin acetylation and cell motility. Nature 421, 230 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Hornbeck P. V., et al. , PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terman J. R., Kashina A., Post-translational modification and regulation of actin. Curr. Opin. Cell Biol. 25, 30–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viswanathan M. C., Blice-Baum A. C., Schmidt W., Foster D. B., Cammarato A., Pseudo-acetylation of K326 and K328 of actin disrupts Drosophila melanogaster indirect flight muscle structure and performance. Front. Physiol. 6, 116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D. H., et al. , A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. EMBO J. 34, 184–199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heidinger-Pauli J. M., Unal E., Koshland D., Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell 34, 311–321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorsky M. K., Burnouf S., Dols J., Mandelkow E., Partridge L., Acetylation mimic of lysine 280 exacerbates human Tau neurotoxicity in vivo. Sci. Rep. 6, 22685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noguchi T. Q., Kanzaki N., Ueno H., Hirose K., Uyeda T. Q., A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. J. Biol. Chem. 282, 27721–27727 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Lu H., Fagnant P. M., Bookwalter C. S., Joel P., Trybus K. M., Vascular disease-causing mutation R258C in ACTA2 disrupts actin dynamics and interaction with myosin. Proc. Natl. Acad. Sci. U.S.A. 112, E4168–E4177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris E. S., Higgs H. N., Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 406, 190–214 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Pollard T. D., Cooper J. A., Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55, 987–1035 (1986). [DOI] [PubMed] [Google Scholar]
- 34.Chhabra E. S., Ramabhadran V., Gerber S. A., Higgs H. N., INF2 is an endoplasmic reticulum-associated formin protein. J. Cell Sci. 122, 1430–1440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramabhadran V., Korobova F., Rahme G. J., Higgs H. N., Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell 22, 4822–4833 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurel P. S., et al. , INF2-mediated severing through actin filament encirclement and disruption. Curr. Biol. 24, 156–164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurel P. S., et al. , Assembly and turnover of short actin filaments by the formin INF2 and profilin. J. Biol. Chem. 290, 22494–22506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wales P., et al. , Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife 5, e19850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao X., Li Q., Mogilner A., Bershadsky A. D., Shivashankar G. V., Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc. Natl. Acad. Sci. U.S.A. 112, E2595–E2601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji W. K., Hatch A. L., Merrill R. A., Strack S., Higgs H. N., Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakrabarti R., et al. , INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol. 217, 251–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fung T. S., Ji W. K., Higgs H. N., Chakrabarti R., Two distinct actin filament populations have effects on mitochondria, with differences in stimuli and assembly factors. J. Cell Sci. 132, jcs234435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatch A. L., Ji W. K., Merrill R. A., Strack S., Higgs H. N., Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol. Biol. Cell 27, 3109–3121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valm A. M., et al. , Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicholson-Dykstra S. M., Higgs H. N., Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil. Cytoskeleton 65, 904–922 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alto L. T., Terman J. R., MICALs. Curr. Biol. 28, R538–R541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pring M., Evangelista M., Boone C., Yang C., Zigmond S. H., Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42, 486–496 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Kovar D. R., Kuhn J. R., Tichy A. L., Pollard T. D., The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161, 875–887 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D., Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Pernier J., Shekhar S., Jegou A., Guichard B., Carlier M. F., Profilin interaction with actin filament barbed end controls dynamic instability, capping, branching, and motility. Dev. Cell 36, 201–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou G. L., Zhang H., Wu H., Ghai P., Field J., Phosphorylation of the cytoskeletal protein CAP1 controls its association with cofilin and actin. J. Cell Sci. 127, 5052–5065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hakes D. J., Dixon J. E., New vectors for high level expression of recombinant proteins in bacteria. Anal. Biochem. 202, 293–298 (1992). [DOI] [PubMed] [Google Scholar]
- 53.Spudich J. A., Watt S., The regulation of rabbit skeletal muscle contraction, I: Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 (1971). [PubMed] [Google Scholar]
- 54.Cooper J. A., Walker S. B., Pollard T. D., Pyrene actin: Documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4, 253–262 (1983). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript and SI Appendix.