Significance
Life depends on the ability of organisms such as plants to convert the gas carbon dioxide into sugar molecules to produce energy and build biomass. Most of this activity is catalyzed by the CO2-fixing enzyme Rubisco. In spite of its crucial role, the enzyme can become blocked by its own substrate and other molecules. Specialized molecular chaperones (Rubisco activases) recognize inhibited Rubiscos and cause conformational rearrangements that lead to inhibitor release. Bacteria that derive energy from inorganic matter contain a complex, convergently evolved Rubisco activase. This molecular helper uses a cup-shaped motor to manipulate an adaptor protein. Changes are then passed from the adaptor to the inhibited Rubisco, resulting in inhibitor release. This understanding will enable precise synthetic biology applications.
Keywords: Rubisco activase, carbon fixation, AAA+ proteins
Abstract
The vast majority of biological carbon dioxide fixation relies on the function of ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). In most cases the enzyme exhibits a tendency to become inhibited by its substrate RuBP and other sugar phosphates. The inhibition is counteracted by diverse molecular chaperones known as Rubisco activases (Rcas). In some chemoautotrophic bacteria, the CbbQO-type Rca Q2O2 repairs inhibited active sites of hexameric form II Rubisco. The 2.2-Å crystal structure of the MoxR AAA+ protein CbbQ2 from Acidithiobacillus ferrooxidans reveals the helix 2 insert (H2I) that is critical for Rca function and forms the axial pore of the CbbQ hexamer. Negative-stain electron microscopy shows that the essential CbbO adaptor protein binds to the conserved, concave side of the CbbQ2 hexamer. Site-directed mutagenesis supports a model in which adenosine 5′-triphosphate (ATP)-powered movements of the H2I are transmitted to CbbO via the concave residue L85. The basal ATPase activity of Q2O2 Rca is repressed but strongly stimulated by inhibited Rubisco. The characterization of multiple variants where this repression is released indicates that binding of inhibited Rubisco to the C-terminal CbbO VWA domain initiates a signal toward the CbbQ active site that is propagated via elements that include the CbbQ α4-β4 loop, pore loop 1, and the presensor 1-β hairpin (PS1-βH). Detailed mechanistic insights into the enzyme repair chaperones of the highly diverse CO2 fixation machinery of Proteobacteria will facilitate their successful implementation in synthetic biology ventures.
Global biomass production is largely reliant on the function of the ubiquitous CO2-fixing enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco), which serves as the entry point of carbon dioxide into the Calvin–Benson cycle (1–4). The enzyme tends to present with modest catalytic properties, being slow and prone to catalyze a metabolite-damaging oxygenation reaction (5). In addition, Rubisco needs to be activated by a nonsubstrate CO2 and a Mg2+ cofactor (6). In their absence, it is prone to form inhibited complexes with its substrate ribulose 1,5-bisphosphate (RuBP), as well as other sugar phosphates (7). Even the activated holoenzyme will form inhibited complexes with a suite of molecules (8). These properties have prompted the evolution of at least 3 clades of Rubisco activases (Rcas) in different autotrophic lineages. They are all molecular chaperones, belonging to the AAA+ (ATPases associated with various cellular activities) superfamily (9, 10), that recognize inhibited Rubisco active sites and use the energy of adenosine 5′-triphosphate (ATP) hydrolysis to effect the release of the inhibitors. Three Rca classes have been discovered, and their mechanisms are understood to varying degrees (11–13). Both red- and green-type Rcas form hexameric ring-shaped particles and utilize a loop lining the central pore (pore-loop 1) to remodel the Rubisco substrate (12, 14, 15). Red-type Rca transiently threads the Rubisco large subunit C terminus whereas the moiety targeted by green-type Rca is not known. Recently, we have described the CbbQO-type Rubisco activase, which is found widely distributed in genomes of chemoautotrophic proteobacteria (16, 17). Different CbbQO Rca isoforms can activate either the hexadecameric form I or the hexameric form II Rubisco (18).
CbbQ belongs to the relatively poorly studied MoxR group of AAA+ proteins (19–21). The MoxR family is closely related to the AAA domains of midasin and dynein, with all belonging to the helix 2 insert (H2I) clade (22). CbbQ forms ring-shaped hexamers but cannot activate Rubisco unless copurified with the adaptor protein CbbO, with 1 monomer binding to a CbbQ hexamer (18, 23). CbbO is an ∼750-residue protein possessing a C-terminal von Willebrand factor A (VWA) domain that is essential for Rubisco activation. MoxR AAA+ proteins are frequently encoded in an operon with VWA domain containing proteins (20), and an adaptor function of the VWA protein has also been described for the RavA-ViaA chaperone, which modulates the activity of Escherichia coli fumarate reductase (24).
AfQ2O2 is the CbbQO-type Rca encoded by Acidithiobacillus ferrooxidans, which acts on hexameric form II Rubisco AfM as its substrate (18). Toward testing mechanistic hypotheses, we have solved the crystal structure of AfCbbQ2. Negative-stain electron microscopy (EM) data of AfQ2O2 show that the CbbO adaptor binds to the highly conserved concave surface of the AAA+ hexamer. Biochemical data are consistent with movements of the pore-forming H2I being propagated to AfM-Rubisco via CbbO. Similar to other AAA+ proteins (25), CbbQO ATPase activity is suppressed in the absence of its substrate, AfM-Rubisco. Mutagenesis revealed multiple residues on both CbbQ and CbbO involved in this suppression, outlining possible paths of communication from the inhibited AfM-Rubisco via CbbO to the CbbQ active site.
Results
The Structure of AfCbbQ2.
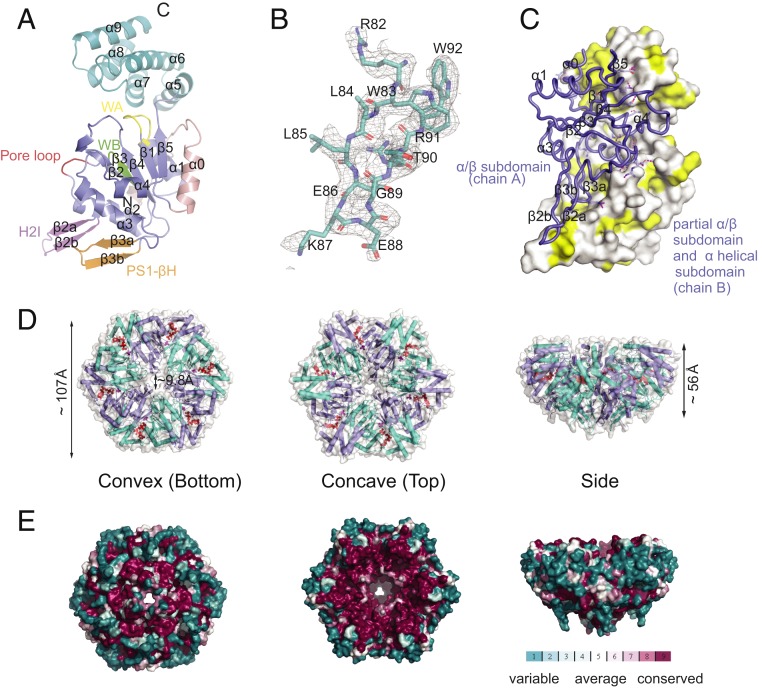
The crystal structure of AfCbbQ2 was determined at 2.2 Å. The asymmetric unit consists of 3 protomers (chains A, B, and C), which form a homotrimer (SI Appendix, Fig. S1). The structure is highly similar to the search model (PDB ID code 5C3C) from Halothiobacillus neapolitanus (HnCbbQ) (23), with a root-mean-square (RMSD) value of 0.63 for the aligned 213 Cα atoms (chain A) (SI Appendix, Fig. S2). Importantly 2 segments (residues 79 to 89 and 153 to 159) that are disordered in the HnCbbQ structure are well resolved in the AfCbbQ2 structure. The final model covers all residues, excluding the first 5, which are probably flexible. Chain A and B share the largest contact interface and form a hexamer, resembling the oligomeric state in solution (18), with 2 neighboring dimers from their crystallographic symmetry molecules.
The AfCbbQ2 protomer displays a typical AAA+ family protein architecture (10) and is composed of an N-terminal α/β subdomain (residues 39 to 177) and a C-terminal α-helical subdomain (residues 182 to 272), linked by a short loop (residues 178 to 181) (Fig. 1A). The α/β (large) subdomain consists of a central β-sheet with 5 parallel β-strands, ordered as 5-1-4-3-2, sandwiched between 6 α-helices. The α-helical (small) subdomain consists of 5 antiparallel α-helices (Fig. 1A). The large subdomain contains both a presensor 1-β hairpin (PS1-βH) (residues 132 to 143), which cradles a helix 2 insert (H2I) (residues 81 to 95), elements that, in AAA+ proteins, are frequently involved in substrate interactions (26, 27). The H2I was not resolved in the earlier HnCbbQ structure (23) and forms a β-hairpin (Fig. 1 A and B). The N-terminal large subdomain contains the canonical Walker A and B motifs (residues 44 to 50 and 107 to 111, respectively) and a pore loop with the sequence Asn71, Glu72, and Asp73. The sensor I and sensor 2 motifs are residues Ser153 and Arg226, respectively. The Arg172 is the arginine finger (Fig. 1A and SI Appendix, Fig. S1B) (28). As observed for HnCbbQ (23), each protomer was found to be bound to 1 adenosine 5′-diphosphate (ADP) and 1 PO4 molecule in the active site, presumably the posthydrolysis state derived from copurification of environmental ATP.
Fig. 1.
The crystal structure of AfCbbQ2 (A) Schematic representation of an AfCbbQ2 monomer. H2I, helix 2 insert; PS1-βH, presensor 1-β hairpin; WA, Walker A motif; WB, Walker B motif. (B) The Fo-Fc map of the AfCbbQ2 H2I. Residues are shown as sticks, with O, N, and C atom colored red, blue, and deep teal, respectively. (C) View of the dimer interface between the α/β subdomain of one subunit (chain A) and the α-helical subdomain and α/β subdomain of the adjacent subunit (chain B). The α/β and the α-helical subdomains of chain B are shown in surface, and the α/β subdomain of chain A is shown as ribbon. Hydrophobic side chains on the α/β subdomain of chain B are shown in yellow. (D) Surface representation of the AfCbbQ2 hexamer formed by chain A (colored slate) and chain B (colored cyan) dimer and its adjacent 2 dimers by expanding crystallographic symmetry of chain A and chain B. ADP and Pi of each subunit are shown as red spheres. (E) A sequence alignment of 100 CbbQ2 sequences was used to map surface conservation onto the AfCbbQ2 hexamer.
The C-terminal helices α6 and α7 in chain A are involved in an extensive contact with α3 and β5 in its neighboring chain B (Fig. 1C and SI Appendix, Fig. S1A). In addition, the H2I contacts its counterpart on chain B. Overall, an average interface area of 1,999.4 Å2 was buried between chain A and chain B, accounting for 14.6% of the solvent-exposed surface area (13,705.9 Å2) of each protomer.
The diameter of the hexameric ring is ∼107 Å while the height and the central pore size of the hexamer is ∼56 and 9.8 Å, respectively (Fig. 1D). The much smaller pore size compared to that reported by Sutter et al. (23) is caused by the H2I, which occludes the pore.
The CbbQ Hexamer Interacts with CbbO via Its Concave Surface.
One hundred CbbQ2 sequences (defined by being encoded directly downstream of a form II Rubisco gene) were mined from the JGI Integrated Microbial Genomes and Microbiomes system (29). The sequences (Dataset S1) were aligned with MultAlin (30), and a surface conservation score was mapped onto the AfCbbQ2 hexamer (Fig. 1E) (31). The convex surface is poorly conserved among CbbQ2 homologs, unlike red-type Rcas that engage Rubisco using the axial pore on that side (14). In contrast, the concave side of the AfCbbQ2 hexamer is highly conserved (23), which is indicative of a protein–protein interaction surface (Fig. 1E). We then visualized the AfQ2O2 complex using negative stain electron microscopy (Fig. 2 and SI Appendix, Fig. S3).
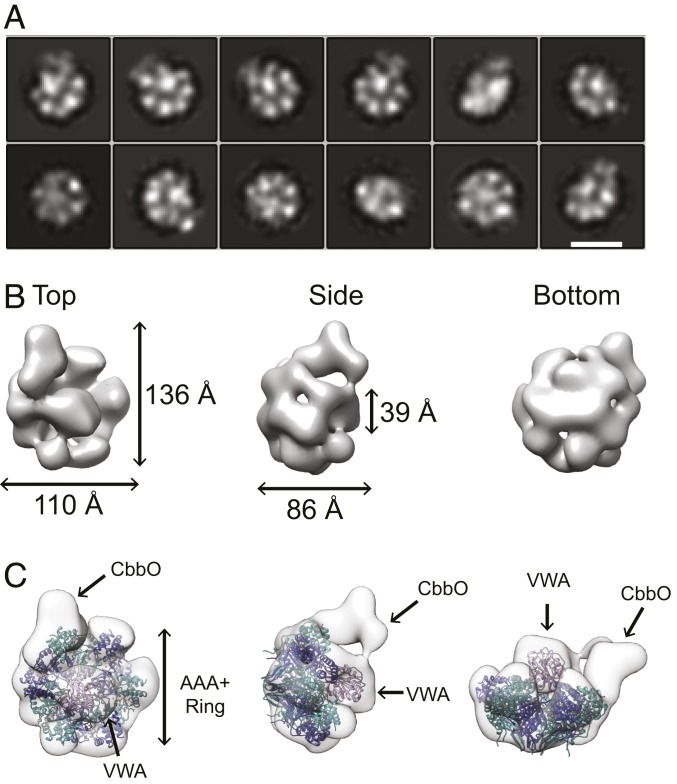
Fig. 2.
Negative-stain electron microscopy of AfQ2O2. (A) Selected reference free 2D class averages of AfQ2O2 derived from negative stain images in RELION-3. (Scale bar, 10 nm.) (B) A 3D reconstruction of AfQ2O2 and (C) 3D reconstruction of AfQ2O2 fit with the crystal structure of AfCbbQ2 (alternating light and dark blue ribbon) and the VWA domain of Integrin αL (53) (pink ribbon; PDB ID code 1T0P).
Although the micrographs suggested conformational heterogeneity, reference-free two-dimensional (2D) classification (32,879 particles) yielded clear density for some subunits of the AfCbbQ2 hexamer (Fig. 2A). Particles of selected 2D classes (15,386 particles) (Fig. 2A) were subjected to three-dimensional (3D) reconstruction and refined to 16 Å (Fig. 2B). When fitting the electron density of the AfCbbQ2 hexamer (18), additional density was apparent on the concave face, corresponding to CbbO2 (SI Appendix, Fig. S3D). The subunit extends from the rim to the hexamer to the central cavity, occluding the pore. Three subunits of the AfCbbQ2 hexamer contact the adaptor CbbO and are thus not clearly defined in the model. The other 3 subunits are adjacent to each other and clearly visible.
Recently, it has been found that the VWA domain of the ribosome maturation factor Rea1/Mdn1 docks in the center of the AAA+ ring (32, 33) at a similar position. The density occupying the center of our hexamer was not observed earlier when AfCbbQ2 complexed with a CbbO protein lacking the C-terminal VWA domain was imaged (18). Consistently the central density in our model accommodates a VWA domain well, and we thus interpret this density as the AfM-Rubisco interacting VWA domain of CbbO (Fig. 2C).
The Concave H2I Residue L85 Mediates Activase Activity.
To take advantage of the structural insights, we purified and biochemically characterized an assortment of AfQ2O2 complexes with amino acid substitutions in both CbbQ and CbbO (SI Appendix, Fig. S4). Complexes were assayed for the relative ability to remove the tight-binding transition-state analog carboxy-arabinitol bisphosphate (CABP) (34) from the activated form II Rubisco holoenzyme (ECM) (18). In addition, we assayed ATP hydrolysis rates of the complexes in the absence (basal) and presence (stimulated) of 3 µM inhibited AfM–Rubisco complexes. These were the enzyme–RuBP complex (ER) or the holoenzyme–CABP complex (ECMC) (18).
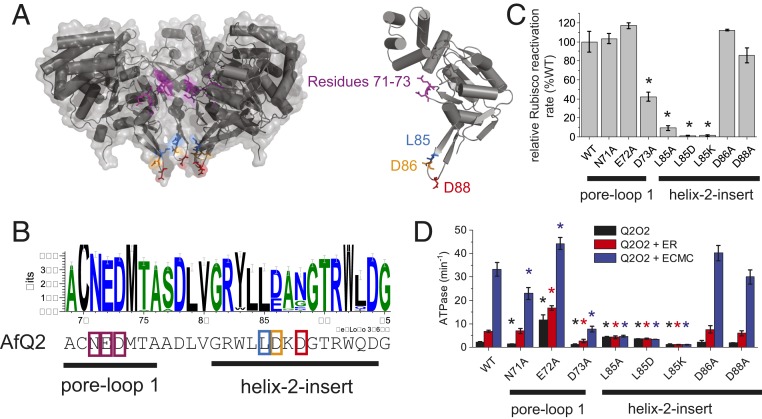
The current model for green- and red-type Rca function involves substrate engagement of the disk-shaped hexamer using its convex surface, followed by transient threading of a portion of Rubisco through the central pore of the hexamer via a pore-loop 1 (β2-α2 loop) tyrosine. Structural and biochemical data support this mechanism for the red-type Rubisco activases from proteobacteria and red algae, which thread the Rubisco large subunit C terminus (12, 14, 35). Pore loop 1 is also important for green (plant)-type Rubisco activase function, but the target moiety on Rubisco is unresolved (15). Unlike green- and red-type Rca, the axial pore of the AfCbbQ2 hexamer is formed by the H2I, and pore-loop 1 is not accessible from the convex surface (Fig. 3A). To constrain mechanistic models of CbbQO function, we therefore mutated residues in the vicinity of the pore. Convex face H2-insert residues D86 and D88 are poorly conserved (Fig. 3B) and were substituted by alanine. The activity of the mutant activases was not significantly different to wild-type (Fig. 3 C and D). Mutation of the adjacent L85 residue to alanine resulted in an uncoupled phenotype where basal ATPase was present at about 200% of the wild-type (Fig. 3D), but stimulation by inhibited AfM-Rubisco was abolished and Rubisco activase activity reduced to ∼10% (Fig. 3 C and D). L85 is part of H2I, but it faces the concave surface (Fig. 3A). Substitution of L85 with the charged residues lysine and aspartate resulted in complete abolishment of both activase function and stimulation of ATPase (Fig. 3 C and D).
Fig. 3.
Characterization of the helix 2 insert and pore-loop 1. (A) Transparent surface representation of a side view of the CbbQ hexamer (Left) and a single subunit (Right). Two subunits were removed to visualize the position of pore-loop1 residues 71 to 73 and the H2I on the concave surface with L85 in blue. Helices are shown as cylinders. (B) Logo motif of the pore-loop 1 and H2I generated using a sequence alignment of 100 CbbQ2 sequences. (C) Relative Rubisco activase function of AfQ2O2 variants. Rca assays contained 0.27 µM Q2O2 (oligomer) and 0.1 µM (active sites) CABP-inhibited AfM. (D) ATPase activity (turnover per CbbQ protomer) of 0.27 µM Q2O2 (oligomer) assayed in the absence or presence of 3 µM (active sites) inhibited Rubisco. ECMC, activated enzyme bound to CABP; ER, Enzyme-RuBP. Error bars indicate the mean and SD of at least 3 technical replicates. *P < Bonferroni-corrected α (0.05/8 = 0.00625), Welch’s t test compared to wild-type (WT) values.
This result implicates H2I in AfM-Rubisco remodeling via the concave L85 residue, instead of a typical (convex) pore-loop type function. We hypothesize that its side chain will form a hydrophobic interaction with CbbO and feature as a key contact mediating both transmission of mechanical force from CbbQ to the CbbO adaptor and transmit information regarding the presence of inhibited AfM-Rubisco from the adaptor to the motor. We then determined a temperature activity profile for Q2(L85A)O2. Whereas the wild-type Q2O2 ATPase activity was still increasing at 50 °C, Q2(L85A)O2 had its temperature optimum at 40 °C (SI Appendix, Fig. S5). Thus, the interaction between CbbQ and CbbO is compromised in this mutant.
Multiple Substitutions Can Relieve the Basal ATPase Suppression of CbbQO.
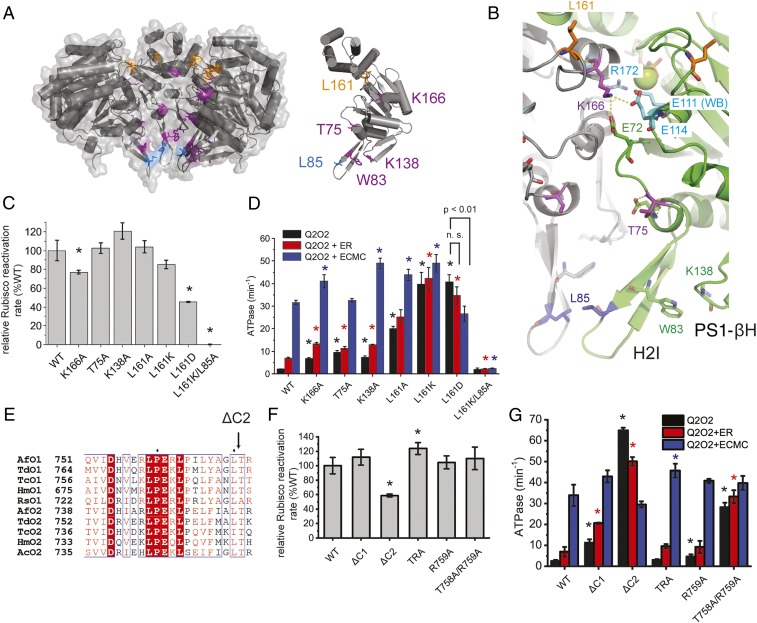
To assess whether pore-loop 1 in its concave location was also involved in AfM-Rubisco activation, multiple residues were substituted with alanine. Activase function for N71A and E72A variants was not significantly different to wild-type. D73A exhibited both significantly reduced ATPase and activase function (Fig. 3 C and D). Both the basal and ER-stimulated ATPase rates of E72A were increased (5- and 2.5-fold, respectively) compared to that of wild-type (Fig. 3D). This result indicates that E72 is involved in suppressing the ATP hydrolysis function of CbbQO to the low basal level, similar to the function of the glutamate switch residues in other AAA+ ATPases (25). In the active site formed by Chain A, the side chain of E72 forms a salt bridge with the amino group of K166 in the β4-α4 loop of the adjacent subunit, which also donates the trans-arginine finger (R172) to the active site (Fig. 4 A and B). K166 also interacts with E114, which is adjacent to the Walker B E111. It is possible that this interaction network constrains movement of the arginine finger and/or the Walker B glutamate and thus limits basal ATPase (Fig. 4B). The K166A mutant exhibited robust (∼80%) activase activity (Fig. 4C). As predicted, we found that basal ATPase activity was indeed 3-fold faster than the wild-type (Fig. 4D). Our structure did not reveal the presence of a “traditional” glutamate switch where the Walker B glutamate is hydrogen-bonded to a gate-keeper residue (25).
Fig. 4.
Residues involved in sensing or signaling the presence of inhibited Rubisco. (A) Visualization of CbbQ residues involved in substrate signaling in the hexamer (Left) or single subunit (Right) as in Fig. 3A. (B) Ribbon representation of adjacent subunits (green and gray) showing key CbbQ signaling residues in stick representation. L161 is located on the β4-α4 loop near the rim of the hexamer. Another residue on the same loop, K166, can form a salt bridge with E72 and E114. Mg2+ is shown as green sphere. (C) Relative Rubisco activase activities of CbbQ signaling mutants as described in Fig. 3C. (D) ATPase activity was measured in the absence and presence of inhibited Rubisco as described in Fig. 3D. n.s., not significant. (E) The length of CbbO C termini is highly conserved. The CbbO sequence alignment shown is that reported in ref. 15. Rca (F) and ATPase (G) activity assays of Q2O2 variants harboring mutations at the CbbO C terminus. Error bars indicate the mean and SD of at least 3 technical replicates. *P < Bonferroni-corrected α (C and D: 0.05/7; F and G: 0.05/5), Welch’s t test compared to WT values.
Multiple additional variants resulting in some loss of the basal ATPase suppression were encountered. These include F and L substitutions of the H2I insert residue W83 reported earlier (18), as well as the T75A mutant (Fig. 4D). K138, which is a conserved residue at the apex of the PS1-βH, also maintains the basal suppression of the ATP hydrolysis rate. These residues are all in the vicinity of the critical L85 moiety, and mutation likely favors conformations that mimic those elicited by AfM-Rubisco binding. Additional structural information, such as the conformation of CbbQ subunits while bound to the adaptor protein CbbO, will enable more accurate interpretations of these results.
The Concave Residue L161 Signals Rubisco Presence.
In order to test whether we could disrupt the interaction between the CbbQ hexamer and the CbbO adaptor, we mutated the conserved hydrophobic surface residue L161, located in the β4-α4 loop (part of the second region of homology [SRH]) (10) near the rim of the concave side (Fig. 4A). Q2O2 complexes with substitutions of L161 to alanine, lysine, and aspartate were readily purified, indicating that the interaction with CbbO was not significantly affected. In addition, the temperature activity profile of Q2(L161K)O2 indicated that the complex was stable up to at least 50 °C (SI Appendix, Fig. S5). Whereas Q2(L161A)O2 was a fully functional activase, its basal ATPase rate was 8-fold increased compared to the wild-type. Addition of stably inhibited ECMC AfM-Rubisco resulted in full stimulation of ATPase activity, as observed for the wild-type, indicating that AfM-Rubisco sensing was intact. In contrast, when L161 was substituted with lysine, the ATPase rate of the resultant activase was fully stimulated, irrespective of whether inhibited AfM-Rubisco was present (Fig. 4D). Rca function was mostly intact (85%) (Fig. 4C). Substitution of L161 with aspartate resulted in an intriguing inverse signaling phenotype. The basal ATPase of this variant was faster than that of the fully stimulated wild-type activase, but addition of inhibited ECMC AfM-Rubisco resulted in a 34% reduction of the ATP hydrolysis rate (Fig. 4D). Concomitantly, the activase activity was also reduced to half (Fig. 4C).
We were interested whether the high basal ATPase activity of Q2(L161K)O2 required the critical L85 residue and consequently produced and characterized a L85A/L161K double mutant. The double mutant was unable to activate AfM-Rubisco (Fig. 4C) and had a low ATP hydrolysis rate similar to Q2(L85A)O2 (Fig. 3D), indicating that the side chain of L85 is critically involved in the L161-mediated signaling pathway.
These data in summary suggest that L161 is involved in the signaling pathway from the site of AfM-Rubisco sensing to the CbbQ active site, with L85 an essential component of this pathway. Mutagenesis of L161 can uncouple and even reverse this signal.
The C Terminus of CbbO Communicates to the CbbQ Motor.
We have previously shown that both stimulation of ATPase and the activase function of the CbbQO complex depend on an intact metal ion dependent adhesion site (MIDAS) motif at the C-terminal VWA domain of CbbO. Our proposed model includes an interaction of this motif with the surface-exposed E75 residue of the AfM-Rubisco large subunit (18). When examining a large sequence alignment of CbbO sequences, we noticed that its C terminus was of conserved length, most frequently featuring an LTX motif (Fig. 4E). To assess the functional significance of this observation, we prepared a series of Q2O2 complexes mutated at the C terminus of CbbO. All variants generated were still functional at activating AfM-Rubisco although deleting the 2 C-terminal residues (ΔC2) resulted in a 40% reduction (Fig. 4F). In contrast, the ATPase function of CbbO variants was highly variable. Shortening CbbO by 1 residue (ΔC1) resulted in a 5-fold increase in basal and 3-fold increase in ER-stimulated ATPase activities (Fig. 4G). The additional removal of another amino acid in Q2O2(ΔC2) led to severe derepression of ATPase activity, with the basal ATPase rate increasing ∼30-fold to 65 min−1. As observed earlier for Q2(L161D)O2, the ΔC2 variant exhibited an inverse signaling phenotype, with inhibited Rubisco leading to a reduction of ATPase activity (Fig. 4G). Substituting the final 2 amino acids with alanine led to an activase (T758A/R759A) that was largely deregulated. Basal ATPase was similar to that of fully stimulated wild-type activase, and addition of inhibited AfM-Rubisco only led to slight increases in activity. These results demonstrate that the C terminus of CbbO, which forms part of the functionally critical VWA domain, is involved in sensing the presence of AfM-Rubisco, and this information is then transmitted to the CbbQ motor.
Discussion
In this work, we have used a crystal structure of AfCbbQ2 in conjunction with negative stain EM and site-directed mutagenesis to further develop our mechanistic model for this MoxR AAA+ ATPase (Fig. 5). A key difference to other Rubisco activases concerns the convex side of the axial pore, which is not involved in AfM-Rubisco activation and thus does not participate in a pore-loop threading mechanism. However, mutants of H2I residue L85, which is located on the conserved concave surface of the hexamer, exhibit uncoupled ATP hydrolysis and Rubisco activase activities. In addition, L85 mutants no longer respond to the presence of inhibited AfM-Rubisco. Since EM shows that the concave face of CbbQ is occupied by the adaptor protein CbbO, we reason that 1 or more L85 residues will bind the single adaptor and be critical to transmit force from the CbbQ hexamer to CbbO, which will then remodel the inhibited AfM-Rubisco substrate. We note that, in dynein, which also belongs to the H2I clade of AAA+ proteins (22), the H2I of AAA2 interacts with the dynein linker via a hydrophobic interaction, and H2I is involved in the linker-swing movement (36). In another member of the same clade, Mdn1/Rea1 (midasin), the H2I of AAA2 directly contacts the MIDAS domain, which, despite being 2,000 residues C-terminal to the AAA ring, occupies the central hole of the AAA ring (32, 33).
Fig. 5.
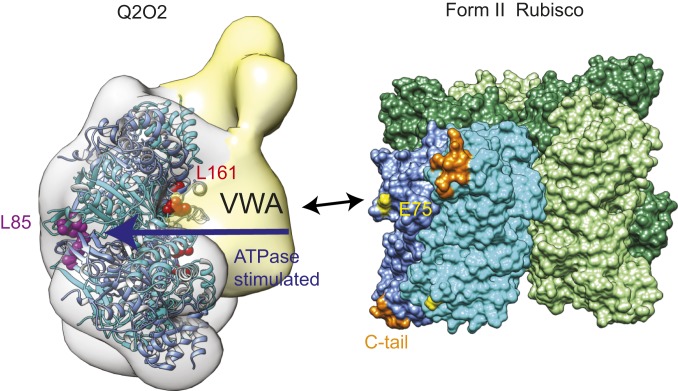
Model of CbbQO function. The VWA domain of the Rca’s CbbO adaptor protein engages the acidic surface residue E75 of the inhibited Rubisco. Rubisco binding results in a signal which originates at the VWA domain and is transferred toward the CbbQ active sites to release the suppressed ATPase activity. Both L161 (β4-α4 loop) and L85 are involved in signal transmission. L85 is likely involved in force transmission from the hexamer to the CbbO adaptor, which will remodel the inhibited Rubisco to trigger active site opening and inhibitor release. The Q2O2 model is an overlay of the EM reconstruction and the AfQ2 crystal structure shown in ribbon representation. L161 (red) and L85 (magenta) are shown as spheres. Rubisco is a surface representation of the hexameric Rhodopseudomonas palustris form II Rubisco (PDB ID code 4lf1).
The ATPase activity of AAA+ protein is commonly observed to be stimulated upon engaging their macromolecular substrates (14, 37, 38). In multiple clades, this has been linked to a glutamate switch mechanism where the Walker B glutamate is hydrogen-bonded to a gatekeeper residue (25, 39). Interaction with the substrate abolishes this interaction, leading to increased ATPase activity. We hypothesize that the interaction between the β4-α4 loop residue K166 and E72 and E114 may function as an analogous switch for CbbQO (Fig. 4B). Mutations of K166 and E72 to Ala increase ATPase activity. The loss of this interaction thus phenocopies AfM-Rubisco binding, and this may then position elements, such as the Walker B glutamate (E111) and the arginine finger (R172), into conformations more conducible to ATP hydrolysis.
Mutation of the PS1-βH K138 also released basal ATPase suppression. PS1-βH also buttresses the H2I of other AAA+ proteins and there participates in an intersubunit signaling (ISS) cascade (40) mediated by a conserved aspartate that interacts with the arginine finger (27, 41). The ISS aspartate is conserved in CbbQ (D128) and may also function in this role.
Biochemical data indicate that the MIDAS motif of the CbbO VWA domain interacts with the conserved acidic residue E75 on the AfM-Rubisco large subunit, which results in derepression of CbbQ’s basal ATPase rate (18) (Fig. 5). Here, we have shown that truncations of the VWA domain’s C-tail are sufficient to stimulate the Q2O2 ATPase rate. We hypothesize that the VWA domain of these mutants is in a conformation that mimics the Rubisco-bound state. Such conformational changes, accompanied by signaling, have been thoroughly described for the VWA domain of the integrin α I domain (42). The signal leading to derepression of the basal ATPase is then transmitted to the active site via the β4-α4 loop residue L161 on the concave rim of the CbbQ hexamer, but only if the key H2I residue L85 is intact.
In summary, here, we significantly expand the structural and biochemical information available on this MoxR AAA+ chaperone. We envisage that the underlying principles will hold true for this large, but mostly uncharacterized, group of prokaryotic molecular chaperones (20, 21), such as the NorQ/NorD maturation machinery for nitric oxide reductase (43). In addition, more detailed information on the regulation and maintenance of microbial carbon dioxide fixation machinery will be essential to inform synthetic biology ventures aiming to incorporate such modules (44, 45).
Materials and Methods
Molecular Biology and Proteins.
AfQ2O2 and all variants were produced using E. coli BL21 (DE3) cells harboring pHueAfCbbQ2 and pBAD33UbAfCbbO2. The relevant mutations in the plasmids were generated using the QuikChange protocol. The purification protocol was exactly as described (18). The form II Rubisco AfM was produced in BL21 (DE3) cells transformed with pHueAfcbbM and purified as described (18). For crystallization, the AfcbbQ2 gene from pHueAfCbbQ2 was subcloned into a modified pOP vector (pOPTHis6-lipoyl) (http://hyvonen.bioc.cam.ac.uk/pop-vectors/). The His6-lipoyl-CbbQ2 was purified using Ni-NTA affinity and gel filtration chromatography. The protein was concentrated to 15 mg/mL and stored in 20 mM Hepes, pH7.4, 150 mM NaCl, 5 mM β-mercaptoethanol, and 10 mM MgCl2. Primers used in this study are given in SI Appendix, Table S2.
Crystallography.
His6-lipoyl-CbbQ2 crystals were obtained using the sitting-drop vapor-diffusion method in 25% (vol/vol) 1,2-propanediol, phosphate-citrate, pH 4.2, 5% (vol/vol) PEG3000, and 10% (vol/vol) glycerol. The crystals were fished from the screening drops and flash frozen by plunging into liquid nitrogen. Diffraction data were collected at beamline NSRRC 13B1, and the data were processed using XDS (46). The AfCbbQ2 structure was determined by molecular replacement using Phaser (47) with homolog HnCbbQ (PDB ID code 5C3C) as a search model. Automatic model building was employed using ARP/wARP (48). The model was further improved by manual model building with Coot (49) and refined with Refmac5 (50) and Phenix (51).
Electron Microscopy.
Purified proteins diluted to a final concentration of 50 μg/mL in 20 mM Tris⋅HCl, pH8.0, 50 mM NaCl were applied to a carbon-coated TEM grid and stained with 2% (wt/vol) uranyl acetate. A total of 49 micrographs were recorded on an FEI T12 transmission electron microscope equipped with a 4K CCD camera (FEI) at a magnification of 49,000× and electron dose of 20 to 25 e−/Å2, resulting in a pixel size of 2.11 Å per pixel under low-dose conditions. A defocus range of −1.0 to 1.5 μm was used. Single particles were selected automatically and processed with Relion-3 (52). A total of 32,879 particles were used for reference-free unbiased 2D classifications, after which 15,386 particles from selected 2D classes were subjected to 3D reconstructions using a 3D model generated in Relion. The 3D refinement resulted in a 3D EM map of Q2O2 resolved to 16 Å resolution using gold standard criteria (0.143 criterion).
Enzymatic Assays.
AfM-Rubisco (20 µM active sites in 20 mM Tris⋅HCl, pH 8.0, 50 mM NaCl) was activated to form the active ECM by incubation with 40 mM NaHCO3 and 10 mM MgCl2. CABP was added to 80 μM to form inactive ECMC complexes. RuBP-inhibited AfM-Rubisco (ER) was produced by incubating 20 µM AfM-Rubisco with 0.8 mM RuBP in the presence of 4 mM EDTA.
Rubisco activase assays and ATPase assays were performed at 25 °C using the coupled spectrophotometric assays described (18). ATPase assays contained 0.27 µM activase (oligomer), 3 µM inhibited AfM-Rubisco (active sites) if applicable, and 1 mM ATP. Activities are reported as turnovers per active site. Activase assays contained 0.27 µM activase (oligomer) and 0.1 µM (active sites) CABP-inhibited AfM-Rubisco (ECMC). Substrate concentrations were 1 mM RuBP and 5 mM NaHCO3. To quantify the activase activity of AfQ2O2 variants, the relative activity of ECMC 1 min after Rca addition was measured.
Statistical Analysis.
Statistical comparisons were performed between the biochemical activity of wild-type and mutant proteins using Welch’s t tests and Bonferroni corrected α in OriginPro 2018b (OriginLab).
Supplementary Material
Acknowledgments
This work was funded by a Nanyang Technological University startup grant (to O.M.-C.) and by Ministry of Education (MOE) of Singapore Tier 2 grants to O.M.-C. (MOE2016-T2-2-088), Y.-G.G. (MOE2015-T2-1-078), and S.B. (MOE2017-T2-2-089). We thank Dr. Wilson Wen Bin Goh for advice on statistical analysis.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID code 6L1Q). The electron microscopy (EM) density map of AfQ2O2 has been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession no. EMD-0789). The biochemical data generated and analyzed in this study are available at https://researchdata.ntu.edu.sg/dataverse/cajar.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911123117/-/DCSupplemental.
References
- 1.Bracher A., Whitney S. M., Hartl F. U., Hayer-Hartl M., Biogenesis and metabolic maintenance of Rubisco. Annu. Rev. Plant Biol. 68, 29–60 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Tabita F. R., Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: A different perspective. Photosynth. Res. 60, 1–28 (1999). [Google Scholar]
- 3.Andrews T. J., Lorimer G. H.,“Rubisco: Structure, mechanisms, and prospects for improvement” in The Biochemistry of Plants: A Comprehensive Treatise, Hatch M. D., Boardman N. K., Eds. (Photosynthesis, Academic Press, New York, 1987), pp. 131–218, vol. 10. [Google Scholar]
- 4.Erb T. J., Zarzycki J., A short history of RubisCO: The rise and fall (?) of nature’s predominant CO2 fixing enzyme. Curr. Opin. Biotechnol. 49, 100–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tcherkez G., The mechanism of Rubisco-catalysed oxygenation. Plant Cell Environ. 39, 983–997 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Lorimer G. H., Badger M. R., Andrews T. J., The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions. Equilibria, kinetics, a suggested mechanism, and physiological implications. Biochemistry 15, 529–536 (1976). [DOI] [PubMed] [Google Scholar]
- 7.Jordan D. B., Chollet R., Inhibition of ribulose bisphosphate carboxylase by substrate ribulose 1,5-bisphosphate. J. Biol. Chem. 258, 13752–13758 (1983). [PubMed] [Google Scholar]
- 8.Parry M. A. J., Keys A. J., Madgwick P. J., Carmo-Silva A. E., Andralojc P. J., Rubisco regulation: A role for inhibitors. J. Exp. Bot. 59, 1569–1580 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V., AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43 (1999). [PubMed] [Google Scholar]
- 10.Hanson P. I., Whiteheart S. W., AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Portis A. R., Jr, Rubisco activase–Rubisco’s catalytic chaperone. Photosynth. Res. 75, 11–27 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Bhat J. Y., et al. , Mechanism of enzyme repair by the AAA+ chaperone Rubisco activase. Mol. Cell 67, 744–756.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Mueller-Cajar O., The diverse AAA+ machines that repair inhibited Rubisco active sites. Front. Mol. Biosci. 4, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller-Cajar O., et al. , Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Stotz M., et al. , Structure of green-type Rubisco activase from tobacco. Nat. Struct. Mol. Biol. 18, 1366–1370 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Badger M. R., Bek E. J., Multiple rubisco forms in proteobacteria: Their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Hayashi N. R., Arai H., Kodama T., Igarashi Y., The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem. Biophys. Res. Commun. 241, 565–569 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Tsai Y. C., Lapina M. C., Bhushan S., Mueller-Cajar O., Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat. Commun. 6, 8883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Bakkouri M., et al. , Structure of RavA MoxR AAA+ protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity. Proc. Natl. Acad. Sci. U.S.A. 107, 22499–22504 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snider J., Houry W. A., MoxR AAA+ ATPases: A novel family of molecular chaperones? J. Struct. Biol. 156, 200–209 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Wong K. S., Houry W. A., Novel structural and functional insights into the MoxR family of AAA+ ATPases. J. Struct. Biol. 179, 211–221 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Iyer L. M., Leipe D. D., Koonin E. V., Aravind L., Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Sutter M., et al. , Structural characterization of a newly identified component of α-Carboxysomes: The AAA+ domain protein CsoCbbQ. Sci. Rep. 5, 16243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong K. S., Bhandari V., Janga S. C., Houry W. A., The RavA-ViaA chaperone-like system interacts with and modulates the activity of the fumarate reductase respiratory complex. J. Mol. Biol. 429, 324–344 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Wigley D. B., The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat. Struct. Mol. Biol. 15, 1223–1227 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erzberger J. P., Berger J. M., Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Chang C. W., Lee S., Tsai F. T. F., Structural elements regulating AAA+ protein quality control machines. Front. Mol. Biosci. 4, 27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendler P., Ciniawsky S., Kock M., Kube S., Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta 1823, 2–14 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Chen I. A., et al. , IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corpet F., Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sosnowski P., et al. , The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1. eLife 7, e39163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z., et al. , Structural insights into Mdn1, an essential AAA protein required for ribosome biogenesis. Cell 175, 822–834.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierce J., Tolbert N. E., Barker R., Interaction of ribulosebisphosphate carboxylase/oxygenase with transition-state analogues. Biochemistry 19, 934–942 (1980). [DOI] [PubMed] [Google Scholar]
- 35.Loganathan N., Tsai Y.-C. C., Mueller-Cajar O., Characterization of the heterooligomeric red-type rubisco activase from red algae. Proc. Natl. Acad. Sci. U.S.A. 113, 14019–14024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kon T., et al. , The 2.8 Å crystal structure of the dynein motor domain. Nature 484, 345–350 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Weibezahn J., Schlieker C., Bukau B., Mogk A., Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 278, 32608–32617 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Burton R. E., Baker T. A., Sauer R. T., Energy-dependent degradation: Linkage between ClpX-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Sci. 12, 893–902 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mogni M. E., Costa A., Ioannou C., Bell S. D., The glutamate switch is present in all seven clades of AAA+ protein. Biochemistry 48, 8774–8775 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Augustin S., et al. , An intersubunit signaling network coordinates ATP hydrolysis by m-AAA proteases. Mol. Cell 35, 574–585 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biter A. B., Lee S., Sung N., Tsai F. T., Structural basis for intersubunit signaling in a protein disaggregating machine. Proc. Natl. Acad. Sci. U.S.A. 109, 12515–12520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo B. H., Carman C. V., Springer T. A., Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kahle M., Ter Beek J., Hosler J. P., Adelroth P., The insertion of the non-heme FeB Cofactor into nitric oxide reductase from P. Denitrificans depends on NorQ and NorD accessory proteins. Biochim. Biophys. Acta Bioenerg., 10.1016/j.bbabio.2018.05.020 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Claassens N. J., Sousa D. Z., Dos Santos V. A., de Vos W. M., van der Oost J., Harnessing the power of microbial autotrophy. Nat. Rev. Microbiol. 14, 692–706 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Liu D., Ramya R. C. S., Mueller-Cajar O., Surveying the expanding prokaryotic Rubisco multiverse. FEMS Microbiol. Lett. 364, fnx156 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langer G., Cohen S. X., Lamzin V. S., Perrakis A., Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song G., et al. , An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin alphaLbeta2. Proc. Natl. Acad. Sci. U.S.A. 102, 3366–3371 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.