Significance
The physiological relevance of STAT2 (a member of STAT family) in melanoma formation is clearly shown using a human skin tissue array. Moreover, FBXW7-mediated STAT2 protein stability regulation via ubiquitination is shown to play an essential role in melanoma cell proliferation in monolayer and anchorage-independent 3D culture systems. The molecular mechanisms that regulate STAT2 protein stability by FBXW7 include the interaction between CCD and DBD domains of STAT2 and the WD40 domain of FBXW7. STAT2 phosphorylation at the putative degron motifs that contain Ser381, Thr385, and Ser393 might be mediated by GSK3β. These serve as critical amino acids that form hydrogen bonds with the WD40 domain of FBXW7. Thus, the FBXW7–STAT2 signaling axis is an important target for melanoma treatment.
Keywords: FBXW7, STAT2, ubiquitination, protein stability, melanoma
Abstract
In this study, we provide critical evidence that STAT2 stability regulation plays an essential role in melanoma cell proliferation and colony growth. We found that the interaction of FBXW7 and STAT2 induced STAT2 destabilization via a ubiquitination-mediated proteasomal degradation pathway. Notably, GSK3β-mediated STAT2 phosphorylation facilitated STAT2–FBXW7 interactions via the DNA binding domain of STAT2 and domains 1, 2, 6, and 7 of FBXW7 WD40. Importantly, the inverse correlation between protein levels of STAT2 and FBXW7 were observed not only in human melanoma cells but also in a human skin cancer tissue array. The relationship between protein levels of STAT2 and FBXW7, cell proliferation, and colony growth were similarly observed in the melanoma cell lines SK-MEL-2, -5, and -28. Moreover, STAT2 knockdown in melanoma cells suppressed melanoma cell proliferation and colony formation. These data demonstrated that FBXW7-mediated STAT2 stability regulation plays an essential role in melanoma cell proliferation and cancer growth.
The biological processes of the ubiquitin proteasome system (UPS) are mediated by 3 enzymatic reactions composed of the ubiquitin activating E1 enzyme, ubiquitin-conjugating E2 enzyme, and the ubiquitin-protein E3 enzyme (1). Among the E1–E3 enzymes, E3 ligases play a crucial role in determining the target proteins for ubiquitination and degradation (1). The cullin-RING E3 ligase (CRL) complex family—comprised of 8 members of CRL: CRL1, CRL2, CCRL3, CRL4A, CRL4B, CRL5, CRL7, and CRL9 (2, 3), and CRL1 (also named as S-phase kinase associated protein 1 [SKP1]-cullin 1-F-box protein [SCF] E3 ligase complex)—is the best characterized. The SCF E3 ligase complex is composed of SKP1, which is an adaptor protein for F-box protein and cullin 1; cullin 1, which is a scaffold protein for SKP1 and E3 ligase RING-BOX 1 (RBX1); and RBX1 (4, 5). The F-box protein that confers substrate selectivity for ubiquitination consists of 2 major functional domains: Various carboxyl-terminal domains that are involved in the binding to specific substrates, and the F-box motif that acts as a protein–protein interaction domain by directly binding with the adaptor protein SKP1 and recruiting the F-box protein into the SCF complex (5). To facilitate substrate protein selection and specificity, the F-box proteins target specific short and defined degron motifs within the substrates (6). Moreover, substrate proteins are required for proper posttranslational modifications that include phosphorylation (7) or glycosylation (8) and interaction with a respective F-box protein (9).
Some F-box proteins are currently categorized according to their potential as tumor suppressors (FBXW7, FBXO11, FBXW8, FBXL3, FBXO1, FBXO4, and FBXO18), oncogenes (SKP2, FBXO5, and FBXO9), and context-dependent different functions (βTrcP1 and βTrcP2) (10). Although some members have been suggested to have various functions in cancer development, there is little evidence available regarding the definitive role of F-box proteins in cancer development. The FBXW7 protein is a member of the FBXW subclasses, including FBXW7α, FBXW7β, and FBXW7γ, which has different subcellular localizations, such as the nucleoplasm, cytoplasm, and nucleoli (11); it is a well-characterized tumor suppressor. The tumor-suppressive function of FBXW7 was based on screening for FBXW7 mutations in human cancers, which showed that ∼6% of all primary human cancers contained FBXW7 mutations (12). In melanomas, the role of FBSW7 was recently addressed, indicating that nonsynonymous mutation rates of FBXW7 were about 8.1% in 77 tumor sample analysis (13). This group also reported that the nuclear FBXW7 content was decreased in melanomas (14.7%) vs. nevi (66.7%) by tissue microarray analysis consisting of benign melanocytic nevi (n = 21) and primary (n = 56) and metastatic (n = 12) melanoma samples (13). They further showed that 4 melanoma cell lines harbor FBXW7 mutations in a panel of 20 melanoma cell lines (13). Importantly, ∼43% of all mutations occur at Arg465 and Arg479, and these are critical for substrate recognition of FBXW7 (12). FBXW7 recognizes and binds to the phosphorylated degron sequence (Leu)-pThe/pSer-Pro-Pro-X-pSer/pThr/Glu/Asp to bind with a substrate, such as cyclin E, MYC, JUN, NOTCH, myeloid cell leukemia 1 (MCL1), SREBP, mammalian target of rapamycin, Krüpple-like factors, CCAAT/enhancer-binding proteins (C/EBPs), and mediator complex components MED13 and MED13L (7, 14). Cancer development from initiation to progression and malignancy are chronological and complicated cellular processes, and there may be more interactive partners with FBXW7.
STAT2 is a member of STAT family that shares a general structure, including N-terminal domain (ND), coiled-coil domain (CCD), DNA binding domain (DBD), linker domain (LD), SH2 domain (SH2D), and transactivation domain (TAD) (15). STAT2 is an essential component of the IFN-α/β signaling pathway (16), and IFN-α binding to IFNαR1-IFNαR2 leads to formation of the ternary IFN-stimulated gene factor 3 (ISGF3) complex that is composed of STAT1, STAT2, and IFN regulatory factor 9 (IEF9). The ISGF3 then localizes to the nucleus and initiates the transcription of target ISGs by binding to IFN-stimulated response element at the promoter region (17, 18). In addition to tyrosine phosphorylation, other posttranslational modifications of STATs, such as serine/threonine phosphorylation or lysine acetylation, play an essential role in cellular processes, including cell proliferation, transformation, apoptosis, and cancer development. For example, the acetylation of K390 of STAT2 induces the expression of antiviral genes by enhancing the interaction between STAT1 and STAT2 (19), and mutations of STAT2 S287 increase ISGF3’s DNA-binding ability (20). More recently, STAT2 T387 phosphorylation was identified by IFN-I stimulation, resulting in an inhibition of the signaling in response to IFN-I. Notably, this mutation to alanine enhances the antiviral and antiproliferative responses of cells treated with IFN-β. Therefore, a major fraction of STAT2 is constitutively phosphorylated on T387 in most untreated cell types (21). Thus, STAT2 was believed to likely harbor tumor-suppressive functions. A decade later, studies using STAT2-deficient cells and mice showed surprising results that STAT2 played an important role in promoting colorectal and skin carcinogenesis (22). Thus, STAT2 function is not concretely characterized in terms of carcinogenesis process, such as cell proliferation, cell cycle transition, transformation, or chemoresistance in malignant cancer cells.
Here, we found that STAT2 interacted with FBXW7. This interaction was based on GSK3β-mediated STAT2 phosphorylation at Ser381, Thr385, and Ser393, which are amino acids composing a degron motif with Glu389 for FBXW7. Notably, the interaction between FBXW7 and STAT2 induced by UVB treatment resulted in degradation of STAT2 via the proteasomal degradation pathway. Importantly, overexpression of STAT2 mutants to alanine at the degron motif suppressed cell proliferation and colony growth of melanoma cells and vice versa with knockdown of FBXW7. These results clearly demonstrated that FBXW7-mediated STAT2 destabilization suppresses melanoma cell proliferation and colony growth.
Methods
In brief, interaction of STAT2 and FBXW7 was identified by mammalian 2-hybrid assay screening in 293T cells. The participation of STAT2 and FBXW7 in the SCF complex was proved by immunoprecipitation (IP). The STAT2 ubiquitination and destabilization was determined by the IP/Western blotting using HA-Ubi– and K48-Ubi–specific antibodies, respectively. GSK3α/β-mediated STAT2 phosphorylation was conducted by in vitro kinase assay using [γ-32p]ATP and purified GST-STAT2. The roles of STAT2 on the cell proliferation and colony growth in melanoma cells was determined by the overexpression and/or knockdown systems. The physiological relevance of the STAT2 in melanoma formation was observed by immunohistofluorescence-based human skin cancer tissue array using STAT2- and FBXW7-specific antibodies. The structural prediction and docking between STAT2 DBD and FBXW7 WD40 domain was conducted using Discovery Studio v2018. The methodology is described in detail in SI Appendix.
Data Availability.
All of our data supporting the paper, including the information for the human skin tissue-array samples and the results obtained from Ampli-Seq RNA expression analysis is available in SI Appendix.
Results
STAT2 Is a Strong Binding Partner of FBXW7.
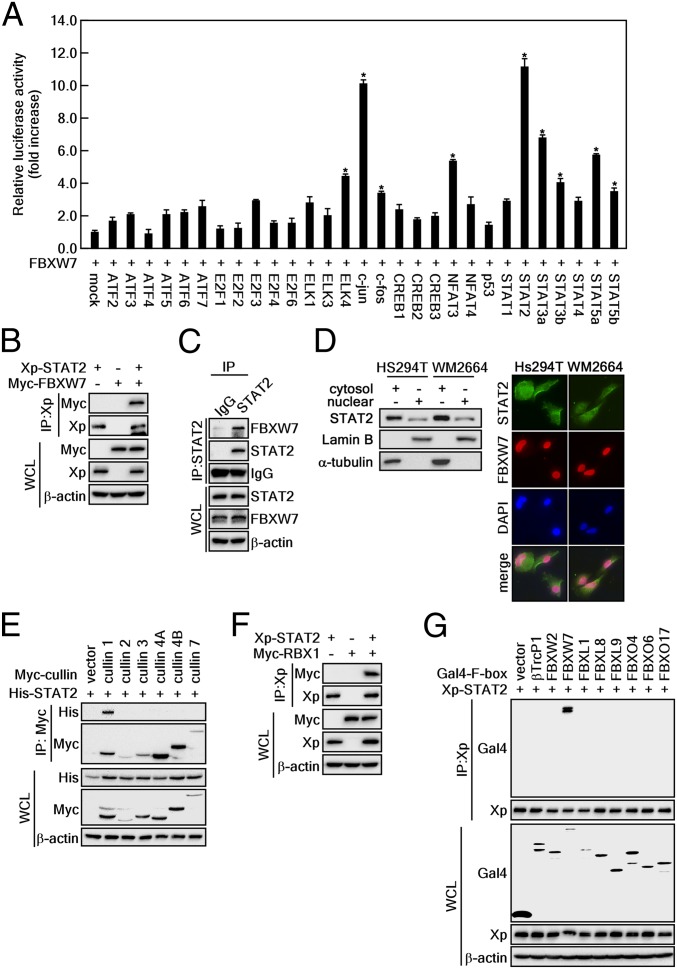
To identify FBXW7 binding partners, we conducted a mammalian 2-hybrid screening assay using 29 transcription factors as targets and FBXW7 as the bait (23). The relative luciferase activity indicated that FBXW7 had relatively strong binding with members of the STAT family, including STAT2, STAT3α, STAT3β, and STAT5α, as well as c-Jun, a well-known substrate of FBXW7 (24) (Fig. 1A). Mammalian 2-hybrid results showed that both c-Jun and STAT2 were transcription factors that harbored a strong affinity for FBXW7, but we focused on STAT2 because its role in carcinogenesis is largely unexplored. The interaction of FBXW7 and STAT2 was reconfirmed via IP using cell lysates that were obtained from cells overexpressing either STAT2 alone or STAT2 and FBXW7 together (Fig. 1B). The IP of endogenous STAT2 and FBXW7 indicated that the STAT2 and FBXW7 interaction can be observed in normal physiological conditions (Fig. 1C). We further found that STAT2 was localized in both the cytoplasm and the nucleus, although STAT2 was strongly detected in the cytoplasm rather than the nucleus (Fig. 1 D, Left). Interestingly, FBXW7 was mainly stained in the nucleus (Fig. 1 D, Right), indicating that FBXW7 and STAT2 interaction may occur at the nucleus. Moreover, STAT2 was immunoprecipitated with cullin 1 but not with cullin 2, 3, 4A, 4B, or 7 (Fig. 1E). Notably, STAT2 displayed binding affinity with RBX1 (Fig. 1F). Importantly, the specificity analysis of STAT2 binding to F-box protein family members, including βTrcP1, FBXW2 (βTrcP2), FBXW7, FBXL1, FBXL8, FBXL9, FBXO4, FBXO6, and FBXO17 showed that STAT2 only interacted with FBXW7 and not with other F-box family members (Fig. 1G). These results demonstrate that the interaction between STAT2 and FBXW7 is involved in the SCF complex containing cullin 1 and RBX1.
Fig. 1.
STAT2 is a binding partner of FBXW7. (A) Screening of FBXW7 binding partner via a mammalian 2-hybrid assay in 293T cells transfected with the indicated plasmids. The binding affinity was analyzed via a luciferase activity. Relative luciferase activity (fold-change) was normalized against control (pBIND-Gal4-FBXW7+pACT-VP16-mock). Data: Triplicate experiment; values: ±SEM; significance: *P < 0.01 versus control by Student’s t test. (B) Western blot (WB) analysis of whole-cell lysates (WCLs) and anti-Xp IPs derived from 293T cells transfected with Xp-STAT2 and Myc-FBXW7 plasmids. After 24 h of incubation, the cells were treated with 10 μM MG132 for 4 h before harvesting. (C) WB analysis of WCLs and co-IPs of endogenous STAT2 and FBXW7 was conducted in HeLa cells using anti-STAT2 antibody. (D) Subcellular localization of STAT2 and FBXW7 proteins. (Left) Western blotting using cytosol and nuclear fraction. (Right) Immunocytofluorescence assay using specific antibodies as indicated (400×). (E) WB analysis of WCLs and anti-Myc IPs derived from 293T cells transfected with His-STAT2 and Myc-cullins (cullin 1, 2, 3, 4A, 4B, and 7) plasmids. After 24-h incubation, the cells were treated with 10 μM MG132 for 4 h before harvesting. (F) WB analysis of WCLs and anti-Xp IPs derived from 293T cells transfected with Xp-STAT2 and Myc-RBX1 plasmids. After 24 h of incubation, the cells were treated with 10 μM MG132 for 4 h before harvesting. (G) WB analysis of WCLs and anti-Xp IPs derived from 293T cells transfected with Xp-STAT2 and Gal4-tagged F-box proteins. After 24 h of incubation, the cells were treated with 10 μM MG132 for 4 h before harvesting.
FBXW7 Plays a Critical Role in Ubiquitination-Mediated STAT2 Destabilization.
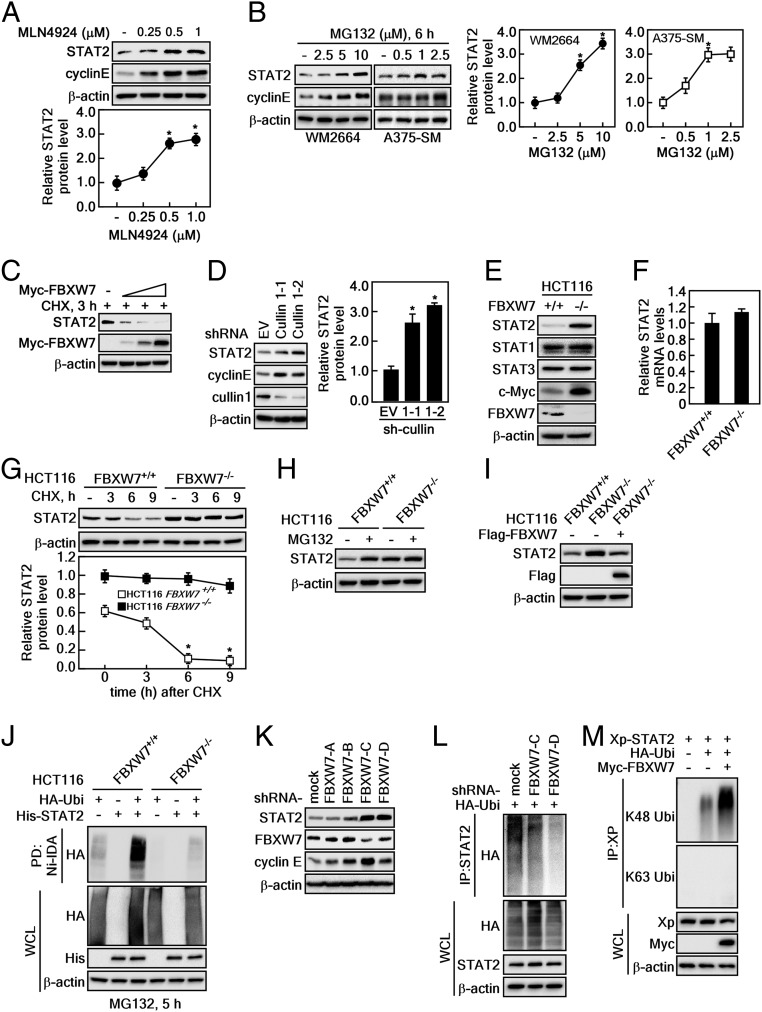
FBXW7 is known to regulate protein stability, and thus we examined the effects of FBXW7 on STAT2 protein stability. Based on our finding that STAT2 and FBXW7 are binding partners, we further found that recovery of STAT2 and cyclin E was observed during treatment with MLN4924, a small molecule inhibitor of NEDD8-activating enzymes that enhance CRL activity (Fig. 2A). Moreover, STAT2 protein levels were increased by proteasomal inhibitor MG132 treatment in melanoma cancer cell lines, such as WM2664 and A375-SM in a dose-dependent manner (Fig. 2B). Importantly, ectopic expression of FBXW7 in U2OS human osteosarcoma cells enhanced the endogenous STAT2 protein destabilization in a dose-dependent manner (Fig. 2C). The restoration of STAT2 was also observed by knockdown of cullin 1 (Fig. 2D). Importantly, the FBXW7 knockout HCT116 (HCT116FBXW7−/−) showed increased STAT2 and c-Myc protein levels than FBXW7 WT HCT116 cells (HCT116FBXW7+/+); however, STAT1 and STAT3 protein levels did not show a similar relationship (Fig. 2E). In contrast, STAT2 mRNA levels were not changed in HCT116FBXW7+/+ and HCT116FBXW7−/− cells (Fig. 2F), indicating that STAT2 protein levels are regulated by FBXW7. The STAT2 stability regulation by FBXW7 was confirmed by observing that the STAT2 protein content was only decreased via cycloheximide (CHX) treatment in HCT116FBXW7+/+ cells and not in HCT116FBXW7−/− cells over time (Fig. 2G). Notably, the FBXW7-mediated proteasomal degradation of STAT2 confirmed the observation that STAT2 protein levels were restored only in HCT116FBXW7+/+ cells; HCT116FBXW7−/− cells showed high and sustained levels of STAT2 compared to HCT116FBXW7+/+ cells (Fig. 2H). Importantly, reintroduction of the FBXW7 expression vectors into HCT116FBXW7−/− cells (HCT116FBXW7−/−/FBXW7) reduced the high STAT2 protein levels detected in HCT116FBXW7−/− cells (Fig. 2I). Strikingly, increased levels of ubiquitinated STAT2 in HCT116FBXW7+/+ cells were dramatically suppressed in HCT116FBXW7−/− cells (Fig. 2J). Additionally, increased STAT2 protein levels by FBXW7 knockdown (Fig. 2K) was inversely correlated with ubiquitinated STAT2 protein levels (Fig. 2L). We further confirmed that the ubiquitination pattern of STAT2 by FBXW7 was K48 ubiquitination but not K63 ubiquitination (Fig. 2M), suggesting that FBXW7-mediated ubiquitination regulates stability but not activity. These results demonstrate that FBXW7 regulates STAT2 protein stability through ubiquitination.
Fig. 2.
FBXW7 regulates STAT2 protein stability. (A and B) WB analysis of WCLs in HeLa (A), WM2664, and A375-SM (B) cells treated with the indicated dose of MLN4924 or MG132 for 6 h. Graphs: Band intensities of STAT2 by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus nontreated control by Student’s t test. (C) WB analysis of WCLs derived from U2OS cells transfected with Myc-FBXW7. The cells were treated with CHX for 3 h before harvesting. (D) WB analysis of WCLs derived from HeLa cells infected with sh-RNA lentiviral vectors against for cullin 1. After infection, the cells were treated with 2 μg/mL puromycin for 3 d to eliminate noninfected cells. Graph: Band intensities of STAT2 by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus empty vector (EV) by Student’s t test. (E) WB analysis of WCLs derived from FBXW7 WT (HCT116FBXW7+/+) and FBXW7 knockout (HCT116FBXW7−/−) HCT116 cells. (F) Real-time PCR analysis of STAT2 mRNA expression in HCT116FBXW7+/+ and HCT116FBXW7−/− cells. (G) WB analysis of WCLs derived from HCT116FBXW7+/+ and HCT116FBXW7−/− cells. The cells were treated with 10 μg/mL CHX and harvested at the indicated time points. Graph: Normalized band intensities of STAT2 by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus nontreated control by Student’s t test. (H) WB analysis of WCLs derived from HCT116FBXW7+/+ and HCT116FBXW7−/− cells. The cells were treated with 10 μM MG132 for 8 h before harvesting. (I) WB analysis of WCLs derived from HCT116FBXW7+/+ and HCT116FBXW7−/− cells. The HCT116FBXW7−/− cells were transfected with Flag-FBXW7 plasmids. (J) WB analysis of WCLs and IPs derived from HCT116FBXW7+/+ and HCT116FBXW7−/− cells. The cells were transfected with His-STAT2 and HA-Ubi. After incubation for 24 h, the cells were treated with 10 μM MG132 for 5 h before harvesting. (K) WB analysis of WCLs derived from 293T cells infected with sh-RNA lentiviral vectors against for FBXW7. After infection, the cells were treated with 2 μg/mL puromycin for 3 d to eliminate noninfected cells. (L) WB analysis of WCLs and IPs derived from the FBXW7 knocked down 293T cells transfected with HA-Ubi. After incubation for 24 h, the cells were treated with 10 μM MG132 before harvesting. (M) WB analysis of WCLs and IPs derived from 293T cells transfected with Xp-STAT2, HA-Ubi, and Myc-FBXW7. After incubation for 24 h, the cells were treated with 10 μM MG132 before harvesting. The specific K48-linked or K63-linked ubiquitination was also detected by WB using the specific antibodies.
GSK3α and -β Are Critical Regulators for FBXW7-Mediated STAT2 Protein Stability.
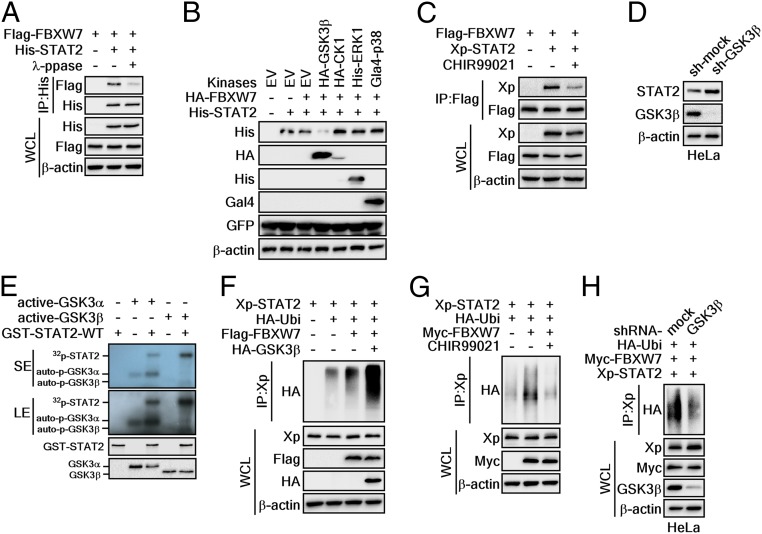
Phosphorylation of serine or threonine residues in a degron motif is required for FBXW7 to recognize specific substrate proteins (25). Therefore, we designed an experiment exploring whether λ-protein phosphatase (λ-PPase) treatment in protein extracts from cells transiently expressing FBXW7 and STAT2 influenced binding of FBXW7 and STAT2. The IP results clearly showed that strong binding of FBXW7 and STAT2 was observed for STAT2 using a FBXW7 and STAT2 coexpressing cell lysate, where the cell lysate was not treated with λ-PPase. In contrast, the binding of FBXW7 and STAT2 was dramatically reduced when the cell lysate was treated with λ-PPase (Fig. 3A). These results indicated that phosphorylation is required for the interaction of STAT2 and FBXW7. To identify the upstream kinase that phosphorylates the STAT2 degron motif, we selected several kinases that harbored an ability to phosphorylate FBXW7 substrates in the degron motif (26). These were coexpressed with FBXW7 and STAT2. We found that GSK3β dramatically reduced STAT2 protein levels (Fig. 3B). These were not observed from other kinases, including CK1, ERK1, and p38 kinase (Fig. 3B). The GSK3 inhibitor consistently decreased the interaction of FBXW7 and STAT2 (Fig. 3C). Moreover, GSK3β knockdown in HeLa cells led to STAT2 protein recovery (Fig. 3D). These results suggested that GSK3α and -β might be upstream kinases of STAT2 that regulate the FBXW7-mediated STAT2 protein stability.
Fig. 3.
GSK3α/β are critical kinases to regulate STAT2 protein stability mediated by FBXW7. (A) WB analysis of WCLs and IPs derived from 293T cells transfected with Flag-FBXW7 and His-STAT2. The IPs were treated (or not treated) with λ-phosphatase at 30 °C for 1 h. (B) WB analysis of WCLs derived from 293T cells transfected with the tagged-kinase, HA-FBXW7, and His-STAT2. (C) WB analysis of WCLs and IPs derived from 293T cells transfected with Flag-FBXW7 and Xp-STAT2 expression vectors. The cells were treated with the GSK3 inhibitor CHIR99021 before harvesting. (D) WB analysis of WCLs derived from HeLa cells infected with shRNA lentiviral vectors for GSK3β. After infection, the cells were treated with 2 μg/mL puromycin for 3 d to eliminate noninfected cells. (E) In vitro kinase assay using [γ-32P] ATP. Bacterially purified GST-STAT2-WT proteins were treated with active GSK3α and -β. The 32P-labeled STAT2-WT proteins were visualized via autoradiography. SE: short exposure, LE: long exposure. (F) WB analysis of WCLs and IPs derived from 293T cells transfected with Xp-STAT2, HA-Ubi, Flag-FBXW7, and HA-GSK3β. The cells were treated with 10 μM MG132 before harvesting. (G) WB analysis of WCLs and IPs derived from 293T cells transfected with Xp-STAT2, HA-Ubi, and Myc-FBXW7. The cells were treated with 10 μM MG132 and CHIR99021 before harvesting. (H) WB analysis of WCLs and IPs derived from HeLa cells infected with shRNA GSK3β. The cells were treated with 10 μM MG132 before harvesting.
To prove this hypothesis, we conducted an in vitro kinase assay using [γ-32P]-ATP, partially purified GST-STAT2-WT, and active GSK3α or -β. The results clearly demonstrated that STAT2 was phosphorylated by GSK3α or -β. Moreover, the band intensity from autoradiography indicated that GSK3β phosphorylated STAT2 more efficiently than GSK3α (Fig. 3E). In keeping with the observation that the ubiquitination of FBXW7 substrates is dependent upon degron motif phosphorylation, overexpression of GSK3β induced FBXW7-mediated STAT2 ubiquitination (Fig. 3F). Conversely, chemical and genetic inhibition of GSK3β activity by CHIR99021 treatment and GSK3β knockdown abolished FBXW7-mediated STAT2 ubiquitination (Fig. 3 G and H). These results collectively demonstrated that GSK3β is an upstream kinase of STAT2 and critical regulator for FBXW7-mediated STAT2 protein stability.
Determination of Binding Domains for FBXW7 and STAT2 Interaction.
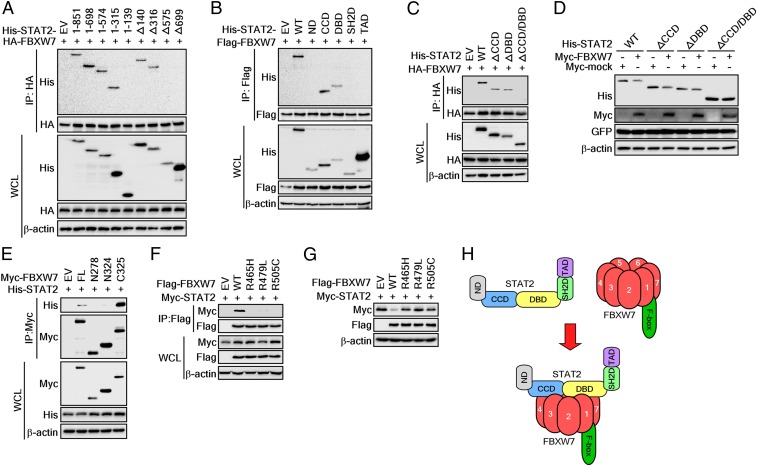
Our previous results demonstrated that FBXW7 plays a key role in STAT2 stability regulation by phosphorylation-dependent ubiquitination (Fig. 3). However, no molecular action mechanisms for STAT2 stability regulation by FBXW7 have been elucidated. We constructed STAT2 full-length and deletion constructs to decipher the interaction domains of STAT2 with FBXW7 (SI Appendix, Fig. S1A). Coexpression and IP experiments using serial deletion expression vectors from the C terminus of STAT2 showed that the binding of STAT2 and FBXW7 disappeared in the STAT2-1-138 deletion mutant that includes additional omissions of CCD amino acids (amino acids spanning 149 to 315) of STAT2-1-315 (Fig. 4A). These results suggested that CCD involved STAT2 and FBXW7 binding. We further confirmed that deletion mutants, Δ1–139 and Δ1–315, were bound with FBXW7 (Fig. 4A). Importantly, this binding disappeared with deletion of amino acid 575 (STAT2-Δ575) and amino acid 699 (STAT2-Δ699) from the N terminus of STAT2 (Fig. 4A). These results suggested that CCD and DBD might play a critical role in STAT2 and FBXW7 binding. This suggestion was confirmed by the IP of FBXW7 and the truncation of each domain of the STAT2 protein (SI Appendix, Fig. S1B): The CCD and DBD of STAT2 were coimmunoprecipitated with FBXW7 but not ND, SH2D, and TAD (Fig. 4B).
Fig. 4.
Identification of binding domains for STAT2 and FBXW7 interaction. (A–C) WB analysis of WCLs and IPs derived from 293T cells transfected with HA- or Flag-FBXW7 and His-STAT2 deletion constructs as indicated. The cells were treated with 10 μM MG132 for 4 h before harvesting. (D) WB analysis of WCLs derived from 293T cells transfected with Myc-FBXW7 and His-STAT2 domain constructs. (E) WB analysis of WCLs and IPs derived from 293T cells transfected with His-STAT2 and Myc-FBXW7 constructs. The cells were treated with 10 μM MG132 for 4 h before harvesting. (F) IP and WB analyses were used to determine the interaction between each FBXW7 mutant and STAT2. The cells were treated with 10 μM MG132 for 4 h before harvesting. (G) WB analysis of WCLs derived from 293T cells transfected with STAT2 and each FBXW7 mutant. (H) A proposed binding model between FBXW7 and STAT2.
To confirm that CCD and DBD of STAT2 were equally involved in the binding to FBXW7, we further constructed expression vectors that deleted only CCD (STAT2-ΔCCD) or only DBD (STAT2-ΔDBD) or both CCD and DBD (STAT2-ΔCCD/DBD) (SI Appendix, Fig. S1C). The essential involvement of CCD and DBD of STAT2 to interact with FBXW7 was confirmed by IP with FBXW7 and each truncated protein of STAT2 (Fig. 4C). Importantly, coexpression of FBXW7 with a STAT2-ΔCCD or STAT2-ΔDBD reduced the total protein levels of STAT2-ΔCCD and -ΔDBD compared to a mock control group, respectively (Fig. 4D). However, coexpression of a deletion construct for STAT2-ΔCCD/DBD with FBXW7 abrogated FBXW7-mediated reduction of the STAT2 protein compared to a mock control group (Fig. 4D). These results demonstrated that the CCD and DBD of STAT2 interact with FBXW7. We further examined which domain of FBXW7 can interact with the STAT2 protein by preparing deletion expression vectors of FBXW7 (SI Appendix, Fig. S1D). The IP of truncated deletion mutants of FBXW7 showed that the FBXW7-N278 (containing amino acids 1 to 278) and FBXW7-N324 (containing amino acids 1 to 324, including the F-box domain) of FBXW7 were not precipitated with STAT2 (Fig. 4E). In contrast, FBXW7-FL (harbors amino acids 1 to 707) and FBXW7-C325 (harbors amino acids 325 to 707) showed STAT2 bands by IP (Fig. 4E). Moreover, although ectopic expression of WT FBXW7 suppressed STAT2 protein levels, each FBXW7 mutant at Arg465His, Arg479Leu, and Arg505Cys—which were known to play an essential role in substrate binding (Fig. 4F)—did not affect the protein levels of STAT2 (Fig. 4G). These results suggested that the WD40 domain of FBXW7 can recognize CCD and DBD of STAT2 as a substrate (Fig. 4H).
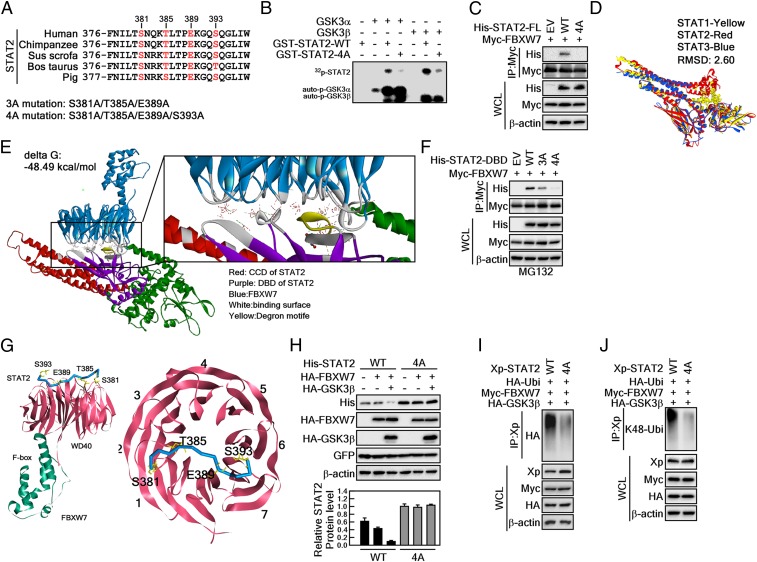
GSK3β-Mediated STAT2 Phosphorylation at Degron Motifs Plays a Key Role in FBXW7-Mediated K48 Ubiquitination of STAT2.
Based on our previous results (Fig. 4), we analyzed amino acid sequences of STAT2 spanning CCD and DBD to identify possible degron motifs for FBXW7. An amino acid alignment with conserved degron motifs for FBXW7 showed that the amino acids STAT2 at Ser381, Thr385, Glu389, and Ser393 were matched with the typical degron motif (pS/pT-X-X-X-pS/pT) for FBXW7 (Fig. 5A). Therefore, we compared the putative degron motifs of STAT2 in several different species. Interestingly, the putative degron motifs of FBXW7 were well conserved among other species (Fig. 5A). Thus, we constructed expression vectors harboring 4 point mutations at Ser381A, Thr385A, Glu389A, and Ser393A (designated as STAT2-4A) at the putative degron motif (Fig. 5A). An in vitro kinase assay using STAT2-WT and -4A mutant proteins demonstrated that the mutations of STAT2 almost abolished the GSK3α/β-mediated STAT2 phosphorylation (Fig. 5B). These results indicate that GSK3α/β-mediated STAT2 phosphorylation might enhance the interaction between FBXW7 and STAT2. IP of FBXW7 with STAT2-FL-WT or STAT2-FL-4A indicated that while STAT2-WT was coprecipitated with FBXW7, STAT2-4A was not (Fig. 5C).
Fig. 5.
GSK3β-mediated STAT2 phosphorylation at degron motifs plays a key role in FBXW7-mediated STAT2 protein stability. (A) Amino acid alignment of the putative degron motifs in the DNA binding domain of STAT2. (B) In vitro kinase assay using GSK3α and -β with STAT2-WT and STAT2-4A. The 4-point mutant (STAT2-4A) of STAT2 at S381A/T385A/E389A/S393A inhibited GSK3α/β-mediated STAT2 phosphorylation. The 32P-labeling of STAT2 was visualized by autoradiography. (C) The 4-point mutant of full-length STAT2 at S381A/T385A/E389A/S393A (STAT2-FL-4A) abolished interaction with FBXW7. (D) Superimposed structures of STAT1, STAT2, and STAT3. Yellow, STAT1; red, STAT2; blue, STAT3. (E) Protein–protein docking of STAT2 and FBXW7. The z-rank score implies the binding energy (kcal/mol). Red, CCD of STAT2; purple, DBD of STAT2; blue, FBXW7; gray, binding surface between STAT2 and FBXW7; yellow, degron motifs. FBXW7 X-ray crystal structure (PDB ID code 2OVP). The interacting amino acids between STAT2 and FBXW7 are provided in the SI Appendix, Fig. S2. (F) WB analysis of WCLs derived from 293T cells transfected with Myc-FBXW7 and His-STAT2-DBD-WT, His-STAT2-DBD-3A, or His-STAT2-DBD-4A. (G, Left) Interaction surface of STAT2 degron motifs containing pS381/pT385/E389/pS393 and WD40 domains of FBXW7. (Right) STAT2 bound across the narrow face of the FBXW7 β-propeller structure. (H) Comparison of the binding between 3- and 4-point mutants of STAT2-DBD on the FBXW7. STAT2-DBD-4A indicates the mutation at S381A/T385A/E389A/S393A. (I and J) WB analysis of WCLs and IPs derived from 293T cells transfected with Xp-STAT2-WT, Xp-STAT2-4A, Myc-FBXW7, or HA-GSK3β. The cells were treated with 10 μM MG132 for 4 h before harvesting.
Next, to determine the molecular mechanisms of the structures by which STAT2 and FBXW7 interact, we built a STAT2 structure via the SWISS modeling program (https://swissmodel.expasy.org/). We then confirmed that the built STAT2 structure showed overall similar structure with STAT1 and STAT3 (Fig. 5D). Using this STAT2 model, we performed protein docking experiments to predict protein–protein interactions between STAT2 and FBXW7 (PDB ID code 2OVP). We found that the models perfectly matched our previous interaction data, indicating that the CCD and DBD of STAT2 interacted with the WD40 domain of FBXW7 (Figs. 3 and 4). Refined structural interaction data suggested that the STAT2–FBXW7 interaction was characterized by 12 hydrogen bonds, 5 electrostatic interactions, and 1 hydrophobic interaction with a ∆G score (−48 kcal/mol) (Fig. 5E and SI Appendix, Fig. S2). We replaced the amino acids Ser381, Thr385, and Ser393 of STAT2 with the phosphorylated form because FBXW7 requires phosphorylation of amino acids in the degron motif; we then redocked with FBXW7. Surprisingly, we found that the ∆G score decreased to −90.16 kcal/mol by forming new interactions (SI Appendix, Fig. S3A). Analysis of the interaction interface between these 2 proteins indicated that the Ser393 of STAT2 was not involved in the interaction with FBXW7 (i.e., via protrusion of its side chain in the opposite direction) (SI Appendix, Fig. S3 B and C).
To confirm whether Ser393 of STAT2 was involved in the interaction between STAT2 and FBXW7, we constructed a 3-point mutant at Ser381A, Thr385A, and Glu389A (designated as STAT2-3A, as shown in Fig. 5A) and conducted IP. STAT2-3A showed reduced interactions with FBXW7 versus STAT2-WT, and STAT2-4A did not interact with FBXW7 (Fig. 5F). Four amino acids of STAT2 are required to interact with FBXW7, and we refined the phospho-STAT2 docking results with FBXW7. We found an interaction model indicating that Ser381, Thr385, Glu389, and Ser393 of STAT2 participated in the interaction with FBXW7 (SI Appendix, Fig. S4). The docking results indicated that Ser393 formed electrostatic interactions with Arg689 of FBXW7 (Fig. 5G, and SI Appendix, Fig. S4). The interaction angle between STAT2 with WD40 domains of FBXW7 was mediated via domains 1, 2, 6, and 7 (Fig. 5G). Notably, STAT2-4A constantly maintained the normal STAT2 protein level when STAT2 proteins were coexpressed with FBXW7 and/or GSK3β (Fig. 5H). Notably, there was increased STAT2 ubiquitination by coexpression of FBXW7 and GSK3β. This was abrogated in the presence of a STAT2-4A mutant protein (Fig. 5I). The ubiquitination pattern indicated that FBXW7-mediated STAT2 ubiquitination at K48 was abolished in the STAT2-4A mutation (Fig. 5J). These results indicated that GSK3β-mediated STAT2 phosphorylation at the putative degron motifs induces FBXW7-mediated K48 ubiquitination of STAT2, resulting in its enhanced destabilization.
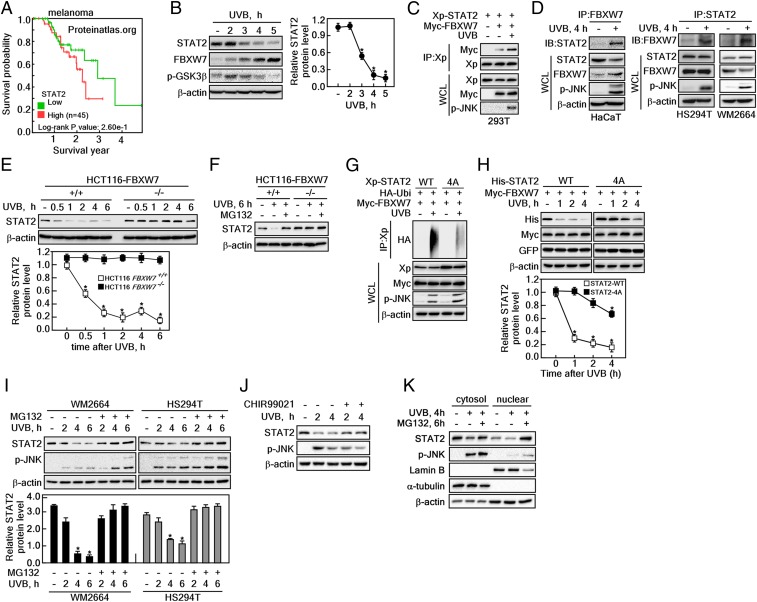
Involvement of STAT2 in Melanoma Susceptibility.
The Human Protein Atlas (https://www.proteinatlas.org) showed that melanoma cancer can be characterized in tissue expressing high levels of STAT2 mRNA and moderate levels of STAT2 protein (SI Appendix, Fig. S5). Moreover, 5-y survival probability data indicate that high levels of STAT2 protein were correlated with short survival rates (Fig. 6A). Furthermore, previous studies demonstrated that exposure of skin to UV produces genetic mutations and skin cancer, including melanoma (26). Therefore, we analyzed protein profiles of STAT2 and FBXW7 after UVB stimulation in HaCaT, a human skin keratinocyte cell line. We found that STAT2 protein levels initially increased and gradually decreased by UVB stimulation, whereas FBXW7 protein levels gradually increased in a time-dependent manner after UVB treatment (Fig. 6B). Simultaneously, the phosphorylation status of GSK3β phosphorylation at Ser9 was shown to have a similar pattern as STAT2 protein (Fig. 6B). These observations indicated that FBXW7 and STAT2 protein levels are inversely correlated to each other after UVB stimulation. The inverse correlation was supported by the interaction between FBXW7 and STAT2, as illustrated by the band intensity of co-IP STAT2 and FBXW7. These increased after UVB stimulation via overexpression of STAT2 and FBXW7 in HEK293T cells (Fig. 6C), as well as endogenous STAT2 and FBXW7 in HaCaT cells (Fig. 6 D, Left) as well as HS294T and WM2664 melanoma cells (Fig. 6 D, Right). Notably, HCT116FBXW7−/− cells did not alter the STAT2 protein levels after UVB stimulation, but HCT116FBXW7+/+ cells showed that UVB stimulation reduced STAT2 protein levels in a time-dependent manner (Fig. 6E). Moreover, the decreased STAT2 protein levels in UVB-treated HCT116FBXW7+/+ cells was restored to baseline by MG132 treatment (Fig. 6F). Decreased STAT2 protein levels after UVB stimulation was the cause of STAT2 ubiquitination, whereas STAT2-4A showed a dramatic reduction in STAT2 ubiquitination after UVB exposure (Fig. 6G).
Fig. 6.
UVB promotes destabilization of STAT2 by inducing the STAT2 and FBXW7 interaction. (A) Five-year survival probability of melanoma patients depending on STAT2 protein expression. (B) WB analysis of WCLs derived from HaCaT cells treated with UVB (100 J/m2). The cells were harvested at the indicated time point. (C) WB analysis of WCLs and IPs derived from 293T cells transfected with Xp-STAT2, Myc-FBXW7. The cells were treated with UVB (100 J/m2) and cultured for 6 h before harvesting. (D, Left) WB analysis of WCLs and endogenous STAT2 and FBXW7 IPs derived from HaCaT cells treated with UVB (100 J/m2) and cultured for 4 h before harvesting. (Right) WB analysis of WCLs and endogenous IPs to determine the interaction of STAT2 and FBXW7 by UVB stimulation in HS294T and WM2664 melanoma cells. (E) WB analysis of WCLs derived from HCT116FBXW7+/+ and HCT116FBXW7−/− cells treated with UVB (100 J/m2). The cells were harvested at the indicated time point after UVB treatment. Graph: Normalized band intensities of STAT2 by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus nontreated control by Student’s t test. (F) WB analysis of WCLs derived from HCT116FBXW7+/+ and HCT116FBXW7−/− cells treated with UVB (100 J/m2) and cultured for 6 h in the presence of 10 μM MG132 before harvesting. (G) WB analysis of WCLs and IPs derived from 293T cells transfected with HA-Ubi, and Myc-FBXW7 together with Xp-STAT2-WT, or Xp-STAT2-4A. The cells were treated with UVB (100 J/m2) and cultured for 6 h before harvesting. (H) WB analysis of WCLs derived from 293T cells transfected with His-STAT2-WT, His-STAT2-4A, and Myc-FBXW7. The cells were treated with UVB (100 J/m2) and harvested at the indicated time point. Graphs: Normalized band intensities of STAT2 by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus nontreated control by Student’s t test. (I) WB analysis of WCLs derived from 293T cells treated with UVB (100 J/m2) and/or 10 μM MG132. The cells were harvested at the indicated time point. Graph: Normalized band intensities of STAT2 by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus nontreated control by Student’s t test. (J) Effect of GSK3 inhibitor on the restoration of STAT2 protein levels decreased by UVB stimulation by Western blotting. The cells were harvested at the indicated time point after UVB stimulation either presence of CHIR99021 or absence. (K) WB analysis of cytosol and nuclear fraction to determine the subcellular organelle that mainly host the restoration of STAT2 by MG132 in HS294T melanoma cells.
We confirmed that UVB stimulation dramatically decreased STAT2-WT protein levels when cells were coexpressed with FBXW7 and STAT2-WT, but this was not seen with STAT2-4A (Fig. 6H). Similar results have also been seen in other melanoma cell lines, such as WM2664 and HS294T, indicating that the decreased STAT2 protein levels by UVB stimulation was restored to baseline by MG132 treatment (Fig. 6I). Notably, treatment with GSK3β inhibitor restored the reduced STAT2 protein levels by UVB irradiation in melanoma cells (Fig. 6J). Importantly, STAT2 protein levels were decreased by UVB stimulation in the cytoplasm and the nucleus, and MG132 treatment restored STAT2 protein levels in the cytoplasm and nucleus (Fig. 6K). The restoration ratio of STAT2 by MG132 was greater in the nucleus than in the cytoplasm, and thus we concluded that ubiquitination of STAT2 was mainly processed in the nucleus. These results suggested that STAT2 plays an important role in melanoma cancer susceptibility.
FBXW7-Mediated STAT2 Stability Regulation Modulates Melanoma Cell Proliferation and Colony Growth.
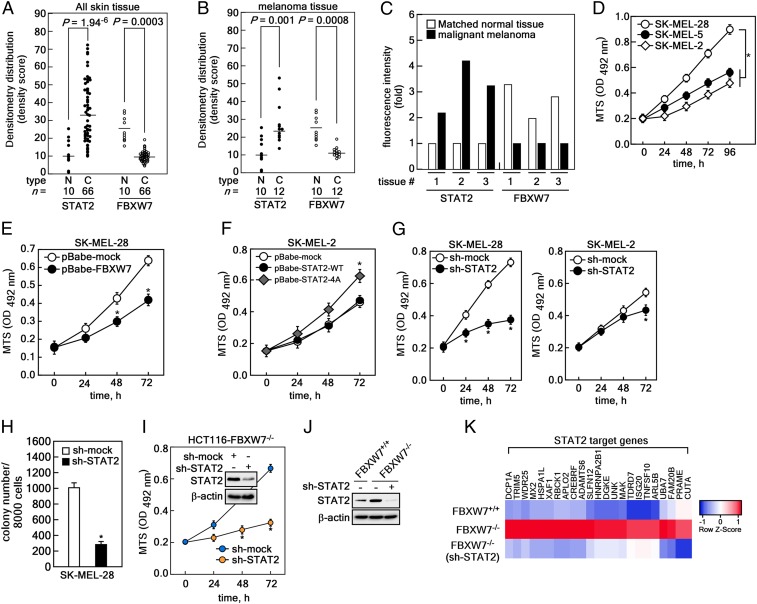
To examine the physiological relevance of STAT2 and FBXW7 in human skin cancer development, we used a human skin tissue array (https://www.biomax.us/) containing 70 skin cancer tissues. Samples were mainly squamous cell carcinoma, basal cell carcinoma, malignant melanoma, and 10 normal tissues (SI Appendix, Table S7) using STAT2 (green in SI Appendix, Fig. S6) and FBXW7 (red in SI Appendix, Fig. S7) antibodies. The statistical analysis of the fluorescence intensity showed that the average protein level of total STAT2 was significantly higher in skin cancer tissues than normal skin tissues (Fig. 7A). In contrast, the average FBXW7 protein levels were higher in normal skin tissues than skin cancer (Fig. 7A). Moreover, the fluorescence intensity using only malignant melanoma showed a similar inverse correlation of protein levels between STAT2 and FBXW7; this is also shown in normal skin tissues and skin cancer tissues (Fig. 7B). Importantly, the tissue array contained matched samples that biopsied melanoma and normal skin tissues from the same patients. Thus, we could compare STAT2 and FBXW7 protein levels. As expected, we found that STAT2 protein levels were higher in malignant melanoma, and FBXW7 protein levels were higher in normal skin tissues (Fig. 7C and SI Appendix, Fig. S8). These results indicate that the differences in STAT2 and FBXW7 protein levels are not due to differences between subjects and are physiologically relevant in human melanoma.
Fig. 7.
Involvement of FBXW7-mediated STAT2 stability in melanoma progression. (A) Human skin cancer tissue array against total STAT2 and FBXW7. (B) Distribution of STAT2 and FBXW7 in normal and melanoma tissue samples in human. (A and B) C, cancer tissues; N, normal tissues. The “n =” indicates numbers of tissue samples. Bars: Average fluorescence intensity; dots: Intensities for each sample; significance: Denoted as P = by Student’s t test. (C) Protein content of STAT2 and FBXW7 in matched normal and melanoma tissues biopsied from same patient. (D) Comparison of cell proliferation in SK-MEL-28, SK-MEL-5, and SK-MEL-2 by MTS. Data: OD 492 nm by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus SK-MEL-28 cells by Student’s t test. (E) Comparison of cell proliferation in SK-MEL-28 stably expressing mock or FBXW7 by MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. (F) Comparison of cell proliferation in SK-MEL-2 stably expressing mock, STAT2-WT, or STAT2-4A mutant by MTS. (G) STAT2 knockdown efficiently inhibited cell proliferation in SK-MEL-28 rather than SK-MEL-2. (H) Knockdown of STAT2 suppressed colony growth of SK-MEL-28 in soft agar. Data: Colony number in soft agar by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus sh-mock control by Student’s t test. (I) STAT2 knockdown suppressed cancer cell proliferation in HCT116-FBXW7−/− cells. (E–G and I) Data: OD 492 nm by 3 independent experiments; values: ±SEM; significance: *P < 0.01 versus nontreated control by Student’s t test. (J) WB analysis of WCLs derived from HCT116FBXW7+/+, HCT116FBXW7−/−, and HCT116FBXW7−/− knockdown of STAT2 cells. (K) Analysis of RNA-seq using mRNA from HCT116FBXW7+/+, HCT116FBXW7−/−, and HCT116FBXW7−/− knockdown of STAT2 cells.
Based on physiological relevance, we next selected 3 melanoma cancer cells to examine the role of STAT2 in melanoma proliferation and colony growth: SK-MEL-2, -5, and -28. Interestingly, we found that endogenous STAT2 and FBXW7 protein levels were inversely correlated in these 3 cells (SI Appendix, Fig. S9A). Moreover, the cell proliferation rate among these 3 melanoma cells was the highest in SK-MEL-28 (with the highest STAT2 expression and the lowest FBXW7 expression). The proliferation was the lowest in SK-MEL-2 containing the least STAT2 and the most FBXW7 (Fig. 7D). Notably, colony numbers formed in soft agar were enhanced in SK-MEL-28 cells compared to SK-MEL-2 cells (SI Appendix, Fig. S9B). We found that FBXW7 overexpression suppressed cell proliferation of SK-MEL-28 cells (Fig. 7E and SI Appendix, Fig. S9C), as well as formation of foci (SI Appendix, Fig. S9D). Notably, we found that ectopic expression of the STAT2-4A mutant induced proliferation and foci formation of SK-MEL-2 cells (Fig. 7F and SI Appendix, Fig. S9 E and F). Inhibition of cell proliferation by STAT2 knockdown was greater in SK-MEL-28 cells vs. SK-MEL-2 cells (Fig. 7G). Moreover, STAT2 knockdown suppressed colony growth of SK-MEL-28 in soft agar (Fig. 7H and SI Appendix, Fig. S9H). Importantly, knockdown of STAT2 in HCT116FBXW7−/− significantly suppressed cell proliferation compared to sh-mock control HCT116FBXW7−/− cells (Fig. 7I).
FBXW7-mediated regulation of STAT2 protein stability plays an essential role in melanoma cell proliferation in monolayer and anchorage-independent 3D culture systems. Thus, we further analyzed STAT2 downstream target genes through RNA-sequencing (RNA-seq) analysis using HCT116FBXW7+/+, HCT116FBXW7−/−, and HCT116FBXW7−/−/sh-STAT2 cells (Fig. 7J). Of the 20,813 genes, we selected 221 possible genes based on the average of normalized raw count (log2) values (SI Appendix, Table S8). We finally narrowed our selection down to 23 genes by comparing the genes with putative STAT2 signaling in melanoma proliferation (Fig. 7K). Unfortunately, the role of STAT2 target genes has been largely unexplored in melanoma cell proliferation. Our results demonstrated that FBXW7-mediated STAT2 stability regulation is physiologically relevant and plays a key role in melanoma cell proliferation and colony growth.
Discussion
Ubiquitination is a reversible process similar to phosphorylation and dephosphorylation. The phosphorylation of Tyr in the TAD domain in STATs plays an essential role in activation and nuclear localization by homo- and hetero-dimerization via the SH2 domain (27). Besides the recruitment of coactivators by serine phosphorylation in the TAD domain, STAT TAD also appears to regulate protein stability. For example, STAT4, STAT5, and STAT6 can be targeted for ubiquitin-dependent destruction, whereas STAT1, STAT2, and STAT3 are more stable (28). The phosphorylation-dependent STAT1 ubiquitination (29) emphasizes that the amount of protein regulation in STAT1 is an important activity regulation mechanism (via stability regulation). The V proteins of paramyxoviruses are largely attributed to host defense evasion and form a multisubunit E3 enzyme complex consisting of the cellular V-interaction proteins, DDB1 (a UV-damaged DNA binding protein), and members of the cullin family, especially cullin 4A (Cul4A) (30).
The ubiquitin-specificity of STATs was advanced by finding that skeletal muscle LIM domain containing a PDZ domain and a LIM domain interact with Tyr-phosphorylated STAT molecules in the nucleus to specifically inhibit gene expression mediated by either STAT1 or STAT4 (31). TNF receptor-associated factor 6 (TRAF6) intermediates for TNF receptor superfamily member and regulates NF-κB, MAPK, and Akt signaling pathways. This induces ubiquitination of STAT3 resulting in inactivation of STAT3 induced by IFN-α (32). Similarly, the Smad ubiquitination regulation factor 1 (Smurf1) was shown to bind a PY domain in STAT1 via a WW domain in Smurf1 to facilitate STAT1 proteasomal degradation (33). More recently, ubiquitome analysis reveals that there are 96 negative and 42 positive regulators induced by IFN-β stimulation. These provide evidence for the function of DC-STAMP domain containing 1 (DCST1) as an important negative regulator of IFN-I signaling. As an action mechanism, DCST1 could interact with both the N- and C-terminal halves of STAT2, and RING domain-deficient DCST1 could not degrade endogenous STAT2. These findings further demonstrated that DCST1 promoted conjugation of only the K48 lysine containing ubiquitin to STAT2. This conjugation indicates that it promotes K48 ubiquitination of STAT2 (34). Although recent studies have evaluated STAT stability regulation, the molecular mechanisms and E3 ubiquitin ligase for STATs are largely unknown.
The typical CRL family member is composed of SKP1, RBX1 (the E3 ligase and also known ROC1), cullin 1, and variable F-box proteins that confer substrate selectivity by targeting a distinct subset of substrates for ubiquitination (4, 5). In this study, we demonstrated that STAT2 is a substrate of FBXW7 and that the FBXW7–STAT2 signaling axis plays an essential role in melanoma formation and cell proliferation. Melanoma formation is mainly triggered by the stimulation of growth factors, such as EGF and UV irradiation (35). The involvement of FBXW7 in melanoma formation has been recently supported by the fact that the protein stability of c-Myc—a critical and important transcription factor to regulate carcinogenesis-related gene expression—is regulated by FBXW7 (36, 37). Although FBXW7 has been classified as a tumor suppressor based on the mutations and deletions in many human cancers (12, 38, 39), the involvement of FBXW7 in melanoma has recently emerged along with findings that FBXW7 mutations are present in about 8.1% melanoma patients and 40% of melanoma cell lines (13). The known substrates of FBXW7 in skin cancer include c-Jun, c-Myc, and Notch; these are all important regulators for cancer cell proliferation and carcinogenesis (24, 40). When cells are initially stimulated with UV, the total c-Jun levels were increased by protein stabilization (41). Our results showed that UVB stimulation suppressed FBXW7 protein levels in a time-dependent manner (Fig. 6B). We concurrently found that STAT2 protein levels were inversely correlated with FBXW7 (Fig. 6B). Moreover, the inverse correlation between STAT2 and FBXW7 protein levels were further supported by the human skin cancer tissue array, indicating that STAT2 protein levels increased in human skin cancer tissues when compared to normal skin tissues (Fig. 7 A and B). Together with a 5-y Kaplan–Meier analysis for survival probability (Fig. 6A), these results are critically important clues that the FBXW7–STAT2 signaling axis plays an essential role in skin cancer development in humans. The heterogeneity of the disease adds another layer of complexity. It is plausible that undefined genetic events representing novel potential targets are sequestered within the complex landscape of genetic events in melanoma. Thus, beyond recurrent mutated genes with high frequencies, these may be potential targets that are relevant to a subset of patients.
Melanomas generally have high metastatic potential with a high mutational load and complicated signaling networks (42, 43). Molecular epidemiology and oncology studies over the last 4 decades clearly demonstrated that these 2 genes are bona fide oncogenes frequently mutated in melanoma because human melanoma harbors about 30 to 45% constitutive active mutation in Ras and Raf genes (44). In addition to these 2 genes, recent advanced high-throughput sequencing provides an opportunity to examine substantial insight into the spectrum of genetic alterations of FBXW7 in melanoma (42–45). Recent data have also demonstrated that exome sequencing of metastatic melanoma (77 tumor samples and 26 cell lines) showed ∼8.1% nonsynonymous mutations (8 samples) in FBXW7, including 5 nonsense, 2 missense, and 1 frame-shift mutation (13). Importantly, the mutations of FBXW7 in metastatic melanoma were observed in WD40 domain with W486*, G423R, and R658* (13). Strikingly, the other half have recurrent heterozygous missense mutations in 3 “hotspots” that alter the core arginine residues (Arg465, Arg479, and Arg505) required for FBXW7 to interact with its substrates (46, 47). These 3 amino acids of FBXW7 were predicted to be involved in the interaction with the Thr385 of STAT2. This was predicted to be an amino acid that is phosphorylated by GSK3β (Fig. 5B) by forming hydrogen bonds (SI Appendix, Fig. S3). Unfortunately, although the FBXW7 gene has been found to have mutations in several exome-sequencing studies (42, 43), functional characterization of FBXW7 and its signaling axis is required for a complete understanding of its pathogenesis, experimental validation, functional characterization, and therapeutic implications.
During gene expression, 3 isoforms of FBXW7α, FBXW7β, and FBXW7γ are produced from a gene by sharing 10 C-terminal exons that encode the F-box and substrate-recognizing WD40 motifs (48). One of the key mechanisms for FBXW7’s specificity to select for particular substrates is the subcellular localization of FBXW7 and its substrates as well as cellular signaling. The FBXW7α isoform is mainly localized in the nucleus and has the highest protein levels in proliferating cells when compared to other isoforms. The FBXW7β and FBXW7γ isoforms are located in the cytoplasm and the nucleolus, respectively (11). The complexity of FBXW7 activity is due to the subcellular distribution of FBXW7 isotypes and the dimerization of FBXW7 isoforms, resulting in the alteration of subcellular localizations depending on heterodimerization (49). Isotype gene expression is also altered via specific stimulation and cellular contexts. For example, inhibition of DNA polymerization via aphidicolin (a reversible inhibitor of DNA polymerase) reduces FBXW7β mRNA levels without significant changes to FBXW7α and FBXW7γ mRNA levels (50). In WT nonmalignant cells such as foreskin fibroblasts, UV-radiation reduces the FBXW7α transcript and induces FBXW7β and c-Jun oncoproteins that are required for the development of skin cancer (51). However, this phenomenon was different from cancer cells, indicating that induction of FBXW7 mRNA and reduction of c-Jun protein levels were observed in HeLa cells due to UV stimulation (51). Moreover, UV radiation in SW480 colorectal cancer cells induced FBXW7α and FBXW7β transcripts in MCF breast cancer cells and the FBXW7α transcript but not the FBXW7β transcript (52). These results indicate that the role of FBXW7 in UV radiation has not yet been clearly elucidated. Our results indicate that FBXW7 was increased after UVB treatment over time in HaCaT human keratinocytes. Conversely, FBXW7 protein levels increased over time (Fig. 6B). Notably, induced GSK3β phosphorylation at Ser9 declined gradually over time (Fig. 6B). Importantly, interactions between STAT2 and FBXW7 were enhanced by UVB treatment (Fig. 6 C and D), indicating that FBXW7 has a tumor-suppressive role. Our results further indicate that ectopic expression of FBXW7 suppressed cell proliferation and foci formation and STAT2-4A mutant induced melanoma cell proliferation and foci formation (Fig. 7 E and F and SI Appendix, Fig. S9 C–F). These results demonstrate that FBXW7 plays a key role in melanoma progression and promotion.
Our results also showed that STAT2 was mainly stained in the cytoplasm with weak nucleoplasm staining (Fig. 1D). Similar results have been published elsewhere, and it is well known that the general distribution of STAT2 is concentrated in the cytoplasm. Importantly, nucleocytoplasmic shuttling kinetics demonstrated that STAT2 is permanently and rapidly shuttled between the cytoplasm and nucleus (53). The results indicate that the reason for the cytoplasmic staining of STAT2 is due to the strong and efficient export of STAT2 from the nucleus. Moreover, IFN type 1 stimulation completely abolished nuclear export of STAT2 (53). Importantly, UVB irradiation stimulates an inflammatory cell infiltrate and induction of type I IFN and inflammatory cytokines in skin in vivo (54). These data suggest that STAT2 and FBXW7 might mainly interact in the nucleus. Interestingly, we found that although the STAT2 protein was observed in the cytoplasm and the nucleus, STAT2 is mainly observed in cytoplasm (Fig. 1D). Importantly, when melanoma cells were treated with MG132 after UVB stimulation, STAT2 recovery was observed mainly in the nuclear fraction via MG132 treatment even though cytoplasmic STAT2 protein levels were also recovered (Fig. 6K). These results suggested that STAT2 and FBXW7 interactions mainly occur at the nucleus. Unfortunately, the molecular mechanisms selecting for specific isotype gene expression and cellular distribution of FBXW7 are unclear.
The phosphorylation status of STAT2 under physiological conditions in noncancer and cancer cells is critically important to understanding the roles of STAT2 in cell proliferation and colony growth. It is difficult to perfectly explain this because phospho-specific antibodies are not available. However, because FBXW7 is a well-known receptor protein recognizing only phosphorylated substrates, our results indirectly explain that the phosphorylation of STAT2 at Ser381, Thr385, and Ser393 is highly possible under physiological conditions. Our results showed that STAT2-FL-WT interacts with FBXW7 via IP. However, 4 site mutations of STAT2-FL at Ser381, Thr385, Glu389, and Ser393 to alanine (STAT2-FL-4A) totally abolished the interaction with FBXW7 (Fig. 5C). Interestingly, the 3 site mutations of STAT2-DBD at Ser381, Thr385, and Ser393 showed weak interactions, indicating that Glu389 might enhance the interaction of STAT2 and FBXW7 (Fig. 5F). Moreover, the GSK3 inhibitor CHIR99021 suppressed STAT2 and FBXW7 interactions (Fig. 3C), resulting in inhibition of STAT2 ubiquitination (Fig. 3G). Importantly, in vitro kinase assays demonstrated that phosphorylation of STAT2-WT by GSK3α or GSK3β was diminished by utilization of the STAT2 mutant protein at Ser381, Thr385, Glu389, and Ser393 to alanine (Fig. 5B). These results collectively, yet indirectly, demonstrated that STAT2 is phosphorylated by GSK3α and -β under physiological conditions.
The human skin cancer tissue array further showed that STAT2 protein levels were higher in cancer tissues than noncancer tissues; the opposite trend was seen for FBXW7 (Fig. 7 A and B and SI Appendix, Figs. S6 and S7). These results indicate that STAT2 proteins might be stabilized in cancer tissues. To enhance the STAT2 stability, the interaction between STAT2 and FBXW7 might be abolished. Since the STAT2 and FBXW7 interaction was phospho-dependent in STAT2 at Ser381, Thr385, and Ser393, the phosphorylation status of STAT2 at Ser381, Thr385, and Ser393 in cancer cells must be reduced. Although these results strongly suggested that the phospho-amino acids of STAT2 mediated by GSK3α/β acted as degron motifs for FBXW7, the subcellular organelle location where the interaction between FBXW7 and STAT2 takes place is not clear. The arguments for the interaction of FBXW7 and STAT2 taking place at the nucleus were based on our evidence, including the fact that endogenous IP shown in Fig. 6D was conducted with the same antibody, used for immunocytofluorescence shown in Fig. 1D. Moreover, because 1) STAT2 was detected in nucleus (Fig. 1D), 2) UVB irradiation induced the binding between STAT2 and FBXW7 (Fig. 6D), 3) UVB and MG132 preferentially recovered STAT2 protein level in the nucleus (Fig. 6K), and 4) FBXW7α is in the nucleoplasm and spared nucleoli (11), we believe that nuclear FBXW7 interacts with STAT2, despite having used whole cell lysate (Fig. 6D). Therefore, since there are reasons to suspect that transcriptionally active STAT2 would show higher intensity in tumor tissues than nontumor tissues, the phosphorylation status of STAT2 at Ser381, Thr385, and Ser393 might be higher in noncancer cells and lower in cancer cells. Thus, the increased STAT2 eventually leads to promotion of cell proliferation and carcinogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Bert Vogelstein of the Sidney Kimmel Comprehensive Cancer Center at the Johns Hopkins University for generously sharing HCT116-FBXW7+/+ and -FBXW7−/− and DLD-1-FBXW7+/+ and -FBXW7−/− cancer cell lines; and Dr. Michele Pagano, New York University School of Medicine, for generously sharing βTrcp1, FBXW2, and FBXW7. This study was supported by the Ministry of Science, Information and Communication Technology and Future Planning (NRF-2017R1A2B2002012, NRF-2017M3A9F5028608 and NRF-2017R1A4A1015036) and the Ministry of Education (BK21PLUS Grant NRF-22A20130012250).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909879116/-/DCSupplemental.
References
- 1.Pickart C. M., Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Deshaies R. J., Joazeiro C. A., RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Frescas D., Pagano M., Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai C., et al. , SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Mészáros B., Kumar M., Gibson T. J., Uyar B., Dosztányi Z., Degrons in cancer. Sci. Signal. 10, eaak9982 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Orlicky S., Tang X., Willems A., Tyers M., Sicheri F., Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Yoshida Y., et al. , E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438–442 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Skaar J. R., Pagan J. K., Pagano M., Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z., Liu P., Inuzuka H., Wei W., Roles of F-box proteins in cancer. Nat. Rev. Cancer 14, 233–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welcker M., Orian A., Grim J. E., Eisenman R. N., Clurman B. E., A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 14, 1852–1857 (2004). Curr Biol.15, 2285 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Akhoondi S., et al. , FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67, 9006–9012 (2007). Cancer Res.68, 1245 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Aydin I. T., et al. , FBXW7 mutations in melanoma and a new therapeutic paradigm. J. Natl. Cancer Inst. 106, dju107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nash P., et al. , Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Mitchell T. J., John S., Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas. Immunology 114, 301–312 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steen H. C., Gamero A. M., The role of signal transducer and activator of transcription-2 in the interferon response. J. Interferon Cytokine Res. 32, 103–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D., How cells respond to interferons. Annu. Rev. Biochem. 67, 227–264 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Schoggins J. W., Rice C. M., Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang X., et al. , Acetylation-dependent signal transduction for type I interferon receptor. Cell 131, 93–105 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Steen H. C., et al. , Identification of STAT2 serine 287 as a novel regulatory phosphorylation site in type I interferon-induced cellular responses. J. Biol. Chem. 288, 747–758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., et al. , Negative regulation of type I IFN signaling by phosphorylation of STAT2 on T387. EMBO J. 36, 202–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamero A. M., et al. , STAT2 contributes to promotion of colorectal and skin carcinogenesis. Cancer Prev. Res. (Phila.) 3, 495–504 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho Y. Y., et al. , The p53 protein is a novel substrate of ribosomal S6 kinase 2 and a critical intermediary for ribosomal S6 kinase 2 and histone H3 interaction. Cancer Res. 65, 3596–3603 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G. Jr, The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Koepp D. M., et al. , Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P., Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Velazquez L., Fellous M., Stark G. R., Pellegrini S., A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 70, 313–322 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Wang D., et al. , A small amphipathic alpha-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 19, 392–399 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T. K., Maniatis T., Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science 273, 1717–1719 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Horvath C. M., Silencing STATs: Lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 15, 117–127 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Tanaka T., Soriano M. A., Grusby M. J., SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity 22, 729–736 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Wei J., et al. , The ubiquitin ligase TRAF6 negatively regulates the JAK-STAT signaling pathway by binding to STAT3 and mediating its ubiquitination. PLoS One 7, e49567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan C., Qi J., Zhao X., Gao C., Smurf1 protein negatively regulates interferon-γ signaling through promoting STAT1 protein ubiquitination and degradation. J. Biol. Chem. 287, 17006–17015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair S., Bist P., Dikshit N., Krishnan M. N., Global functional profiling of human ubiquitome identifies E3 ubiquitin ligase DCST1 as a novel negative regulator of Type-I interferon signaling. Sci. Rep. 6, 36179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Abaseri T. B., Hansen L. A., EGFR activation and ultraviolet light-induced skin carcinogenesis. J. Biomed. Biotechnol. 2007, 97939 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welcker M., et al. , The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 (2004). Proc Natl Acad Sci U.S.A. 103, 504 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yada M., et al. , Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23, 2116–2125 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., et al. , Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 586, 1409–1418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welcker M., Clurman B. E., FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008). [DOI] [PubMed] [Google Scholar]
- 40.O’Neil J., et al. , FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204, 1813–1824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaulian E., et al. , The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell 103, 897–907 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Hodis E., et al. , A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krauthammer M., et al. , Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 44, 1006–1014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray-Schopfer V., Wellbrock C., Marais R., Melanoma biology and new targeted therapy. Nature 445, 851–857 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Berger M. F., et al. , Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502–506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis H., Tomlinson I., CDC4/FBXW7 and the ‘just enough’ model of tumourigenesis. J. Pathol. 227, 131–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kemp Z., et al. , CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 65, 11361–11366 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Spruck C. H., et al. , hCDC4 gene mutations in endometrial cancer. Cancer Res. 62, 4535–4539 (2002). [PubMed] [Google Scholar]
- 49.Welcker M., Clurman B. E., Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2, 7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sionov R. V., Netzer E., Shaulian E., Differential regulation of FBXW7 isoforms by various stress stimuli. Cell Cycle 12, 3547–3554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anzi S., Finkin S., Shaulian E., Transcriptional repression of c-Jun’s E3 ubiquitin ligases contributes to c-Jun induction by UV. Cell. Signal. 20, 862–871 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Kimura T., Gotoh M., Nakamura Y., Arakawa H., hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 94, 431–436 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frahm T., Hauser H., Köster M., IFN-type-I-mediated signaling is regulated by modulation of STAT2 nuclear export. J. Cell Sci. 119, 1092–1104 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Sontheimer C., Liggitt D., Elkon K. B., Ultraviolet B irradiation causes stimulator of interferon genes-dependent production of protective type I interferon in mouse skin by recruited inflammatory monocytes. Arthritis Rheumatol. 69, 826–836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of our data supporting the paper, including the information for the human skin tissue-array samples and the results obtained from Ampli-Seq RNA expression analysis is available in SI Appendix.