Significance
It is thought that evolution of antiviral genes has been shaped over the long term by antagonistic interactions with viruses, but in most cases this is challenging to investigate. In this study we examine the evolution of A3 genes—antiviral genes that target retroviruses by inducing mutations in their genomes. We demonstrate that ancient, fossilized retrovirus sequences in mammalian genomes contain clear signatures of A3-mediated mutation and provide several additional lines of evidence that A3 evolution has been driven by long-running conflicts with ancient retroviruses.
Keywords: mammal, APOBEC3, gene amplification, endogenous retrovirus, evolutionary arms race
Abstract
APOBEC3 (A3) genes are members of the AID/APOBEC gene family that are found exclusively in mammals. A3 genes encode antiviral proteins that restrict the replication of retroviruses by inducing G-to-A mutations in their genomes and have undergone extensive amplification and diversification during mammalian evolution. Endogenous retroviruses (ERVs) are sequences derived from ancient retroviruses that are widespread mammalian genomes. In this study we characterize the A3 repertoire and use the ERV fossil record to explore the long-term history of coevolutionary interaction between A3s and retroviruses. We examine the genomes of 160 mammalian species and identify 1,420 AID/APOBEC-related genes, including representatives of previously uncharacterized lineages. We show that A3 genes have been amplified in mammals and that amplification is positively correlated with the extent of germline colonization by ERVs. Moreover, we demonstrate that the signatures of A3-mediated mutation can be detected in ERVs found throughout mammalian genomes and show that in mammalian species with expanded A3 repertoires, ERVs are significantly enriched for G-to-A mutations. Finally, we show that A3 amplification occurred concurrently with prominent ERV invasions in primates. Our findings establish that conflict with retroviruses is a major driving force for the rapid evolution of mammalian A3 genes.
Activation-induced cytidine deaminase/apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (AID/APOBEC) superfamily proteins are cellular cytosine deaminases that catalyze cytosine-to-uracil (C-to-U) mutations. AID/APOBEC family proteins contain a conserved zinc-dependent catalytic domain (Z domain) with the HxE/PCxxC motif and are closely associated with important phenomena found in vertebrates such as immunity, malignancy, metabolism, and infectious diseases (reviewed in refs. 1 and 2). For instance, AID induces somatic hypermutation in B cells and promotes antibody diversification (2), and APOBEC1 (A1) regulates lipid metabolism by enzymatically editing the mRNA of apolipoprotein B gene (3). The physiological roles of APOBEC2 (A2) and APOBEC4 (A4) remain unknown, but APOBEC3 (A3) genes are known to encode antiviral factors that restrict the replication of retroviruses (4) and other viruses (5–7).
While most AID/APOBEC family genes are conserved in vertebrates, A3 genes are specific to placental mammals (1). Furthermore, whereas AID, A1, A2, and A4 genes are singly encoded in each vertebrate including mammals, dramatic expansion of the A3 repertoire occurred in many mammalian lineages, including primates (8). A3 genes are grouped into 3 classes (A3Z1, A3Z2, and A3Z3) on the basis of their conserved Z domain sequences (4, 8, 9). For example, human A3 genes are composed of 7 paralogs (A3A, A3B, A3C, A3D, A3F, A3G, and A3H). Of these, A3A, A3C, and A3H (which in other mammals are referred to as A3Z1, A3Z2, and A3Z3, respectively) contain a single Z domain, while the other 4 genes harbor double Z domains: A3Z2-A3Z1 for A3B and A3G and A3Z2-A3Z2 for A3D and A3F (8, 9).
The conflict between human A3G protein and HIV type 1 (HIV-1) has been studied particularly intensively. Human A3G proteins are incorporated into HIV-1 particles and enzymatically induce C-to-U mutations in viral cDNA, causing guanine-to-adenine (G-to-A) mutations in the viral genome (10, 11). A3G-mediated mutations lead to the accumulation of lethal mutations and ultimately abolish viral replication. On the other hand, an HIV-1–encoding protein, viral infectivity factor (Vif), counteracts this antiviral action by degrading A3G in a ubiquitin-proteasome–dependent manner (4). Such conflicts between A3 proteins and modern viruses (particularly retroviruses) have been reported in a broad range of mammalian species and viruses infecting them (reviewed in ref. 9), and consistent with this, A3 genes contain strong signatures of diversifying selection (12–14).
Endogenous retroviruses (ERVs) are retrotransposon lineages that are thought to have originated from ancient exogenous retroviruses via infection of germline cells (15, 16). ERVs occupy a substantial fraction of mammalian genomes, demonstrating extensive germline invasion by retroviruses. To combat ERVs and other intragenomic parasites, mammals have developed defense systems such as Krüppel-associated box domain-containing (KRAB) zinc finger proteins (17) and PIWI-interacting RNAs (18). A3 proteins have been shown to suppress the replication of reconstructed ERVs in cell cultures (15, 19) and in a transgenic mouse model (20). Furthermore, previous studies identified the signature of A3-mediated G-to-A mutations in ERVs indicating that ancient retroviruses experience attacks by A3 proteins (15, 16, 19, 21). In this study, we examine the history of evolutionary interaction between ERVs and A3 genes via genomic analysis of 160 mammalian species.
Results
Identification and Classification of Mammalian AID/APOBEC Family Genes.
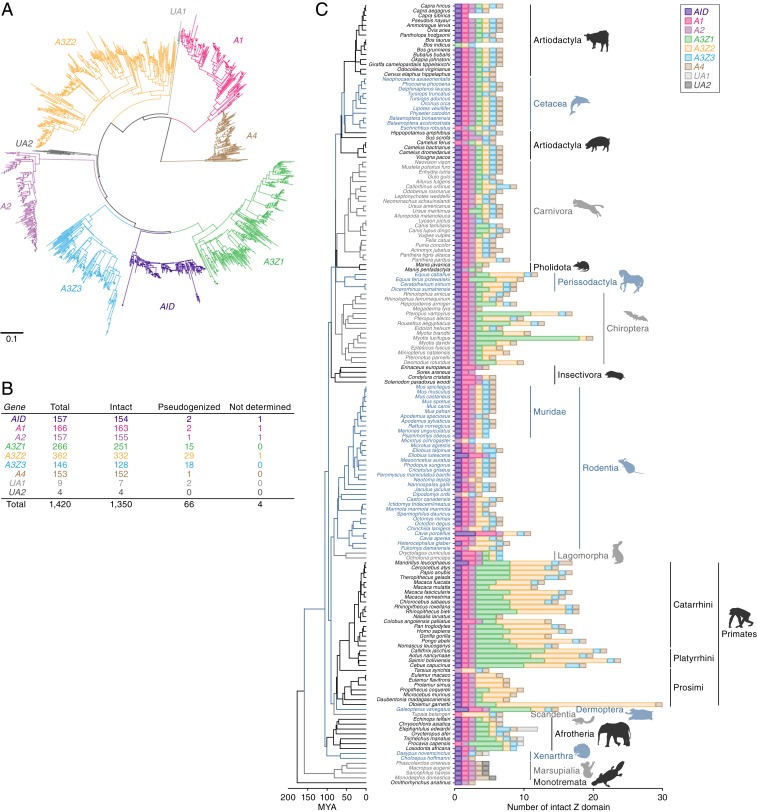
We screened whole genome sequence (WGS) data of 160 mammalian species in silico and extracted 1,420 sequences disclosing homology to the conserved Z domains of AID/APOBEC family genes (8) (SI Appendix, Fig. S1 and Datasets S1–S3). Phylogenetic reconstructions revealed that these Z domain loci group into 9 clades, 7 of which represent the canonical AID/APOBEC lineages (AID, A1, A2, A3Z1, A3Z2, A3Z3, and A4) (Fig. 1 A and B). We also identified additional, previously uncharacterized lineages, designated UA1 and UA2 (Fig. 1 A and B). UA1 genes were only found in basal eutherian mammal groups: afrotherians (elephants, tenrecs, and sea cows) and xenarthrans (armadillos). UA2 genes were only found in marsupials (infraclass Marsupialia) (Fig. 1C). These phylogenetic relationships were supported by multiple methods (Fig. 1A and SI Appendix, Fig. S2A). In addition, HxE and PCxxC motifs corresponding to the canonical catalytic domain of AID/APOBEC proteins were found in UA1 and UA2 gene sequences (SI Appendix, Fig. S2B). The UA1 and UA2 genes contain signatures of purifying selection (SI Appendix, Fig. S2C) indicating they are protein-coding members of the AID/APOBEC family. Indeed, the UA2 gene in opossum (Monodelphis domestica) was annotated as APOBEC5 in a previous study (22).
Fig. 1.
Distribution and diversity of AID/APOBEC Z domains in mammalian genomes. (A) A phylogenetic tree of AID/APOBEC Z domains identified via in silico screening of 160 mammalian genomes. The tree shown here was based on an alignment of nucleic acid sequences and was reconstructed using the NJ method (63). Scale bar indicates the genetic distance. (B) Number of AID/APOBEC Z domains. Those labeled “intact” contain no premature stop codons, while the remainder are labeled as “pseudogenized.” Z domain sequences that contained unresolved regions were labeled “not determined.” (C) Number of the intact AID/APOBEC Z domains identified in each mammal species. See SI Appendix, Fig. S3, for further details. The species tree shown here was derived from the TimeTree database (73).
As summarized in Fig. 1B, we detected 157 AID, 166 A1, 157 A2, 266 A3Z1, 362 A3Z2, 146 A3Z3, 153 A4, 9 UA1, and 4 UA2 genes in 160 species of mammalian genomes. Interestingly, A3Z1 and A3Z2 genes were highly amplified, while the other family genes were not (Fig. 1 B and C). We also found that some sequences, particularly those of A3 genes, were pseudogenized (Fig. 1B). The numbers of A3 Z domains were different among species. In particular, A3Z1 and A3Z2 genes in Perissodactyla, Chiroptera, Primates, and Afrotheria were highly amplified (Fig. 1C and SI Appendix, Fig. S3). Consistent with previous reports (12, 23, 24), canonical A3 genes were not detected in marsupials or monotremes (order Monotremata). Furthermore, A3Z1 was commonly absent in Rodentia, while A3Z3 was absent in Strepsirrhini and Microchiroptera. Amplification of A3Z3 genes was not detected in any mammalian groups except for Carnivora (carnivores), in which duplicated A3Z3 genes were almost entirely pseudogenized (SI Appendix, Fig. S4).
Evolution of Mammalian A3 Genes Under Strong Selection Pressures.
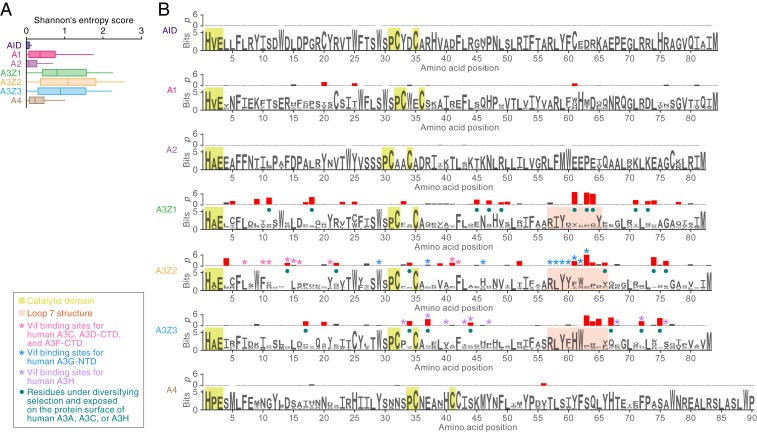
We used comparative genomic approaches to investigate the evolutionary history of mammalian A3 genes. As shown in Fig. 2A, the positional conservation (Shannon entropy) scores in A3Z1, A3Z2, and A3Z3 genes tended to be much higher than those found in other AID/APOBEC family genes, indicating strong diversifying selection. We detected codon sites evolving under diversifying selection by calculating dN/dS ratios using the branch-site model (25). Although the catalytic domains, which are composed of HxE and PCxxC motifs (1, 2, 4), were highly conserved among the 7 AID/APOBEC family proteins, we detected the signature of diversifying selection at numerous sites (Fig. 2B). Comparisons to human A3A (A3Z1 ortholog in primates) (26), A3C (A3Z2 ortholog in primates) (27), and A3H (A3Z3 ortholog in primates) (28) revealed that these sites are preferentially detected in a structural region called loop 7, which recognizes substrate nucleic acids (Fig. 2B). Furthermore, most of the sites under diversifying selection are located on the protein surface (Fig. 2B).
Fig. 2.
Evolutionary features of AID/APOBEC Z domains. The analyses are based on the MSAs of respective classes of AID/APOBEC Z domains. The MSAs of intact Z domains of AID (n = 163), A1 (n = 155), A2 (n = 251), A3Z1 (n = 332), A3Z2 (n = 132), A3Z3 (n = 152), and A4 (n = 154) (listed in Dataset S3) were used. (A) Difference in the sequence conservations among 7 classes of AID/APOBEC Z domains. Positional sequence conservation scores (Shannon’s entropy scores) were calculated in respective amino acid sites of the MSA (shown as logo plots in B). (B) Top rows show the P values (−log10) in dN/dS ratio test [with branch-site model (25)] at each codon site. The sites under diversifying selection with statistically significance (P < 0.05) are indicated by red bars. Bottom rows show logo plots of the conserved sequences of the AID/APOBEC Z domains. Yellow square indicates the amino acid residues comprising the catalytic domain of AID/APOBEC proteins. Pink square indicates the amino acid residues corresponding to the structure loop 7. The other characteristics on each amino acid residue [e.g., Vif binding sites for human A3C (27), human A3D-CTD (27), human A3F-CTD (27, 74, 75), human A3G-NTD (41, 42), and human A3H (28, 76)] are summarized in the box to the lower left of the panel. CTD, C-terminal domain; NTD, N-terminal domain.
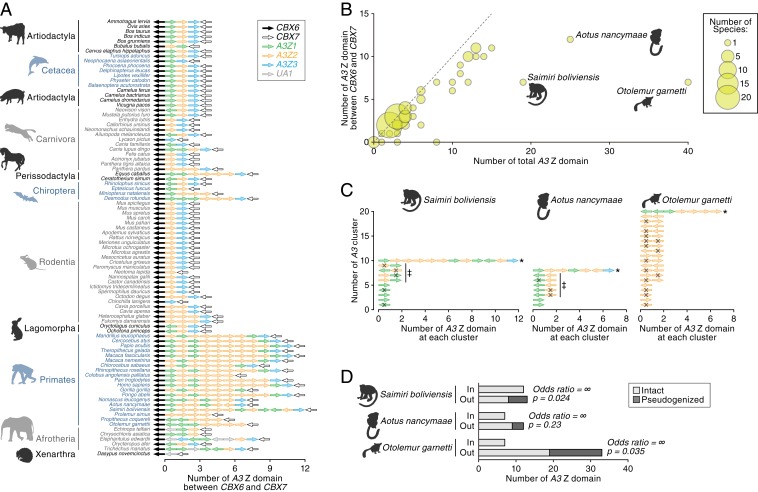
Investigation of amplified A3 loci revealed that the majority of A3 genes are encoded in the canonical A3 genomic locus (8, 9), flanked by the CBX6 and CBX7 genes (Fig. 3A and Dataset S4), indicating that amplification of A3 genes has mainly occurred via tandem gene duplication. However, there are exceptions to this rule: 3 primate species, Saimiri boliviensis, Aotus nancymaae, and Otolemur garnettii, were found to encode more A3 loci outside the canonical locus than within it (Fig. 3B). The A3 genes in these 3 primates were mostly encoded at entirely distinct loci (Fig. 3C) and exhibit double-domain (A3Z2–A3Z1) and intronless structures (SI Appendix, Fig. S5A and Dataset S5) indicating they likely originated via retrotransposition of spliced mRNA (29). These retrotransposed A3 genes in New World monkeys were more closely related to the human A3G gene than the other double-domain A3 genes in humans (SI Appendix, Fig. S5B). Although most were pseudogenized (Fig. 3D), some retain relatively long ORFs (SI Appendix, Fig. S5C). In particular, 1 of the retrotransposed A3 genes in A. nancymaae (referred to as “outside #3”) retains a full-length ORF (SI Appendix, Fig. S5C). Indeed, this gene is annotated in the Ensembl gene database (http://www.ensembl.org; Release 97; ENSANAG00000031271). Moreover, analysis of public RNA-sequencing (RNA-Seq) data revealed that mRNA of outside #3 is expressed in a broad range of tissues in A. nancymaae (SI Appendix, Fig. S5D). Taken together, these data show that A3G-like genes have been amplified via retrotransposition in New World monkeys, and some of these amplified genes are likely functional.
Fig. 3.
Genomic location of A3 genes. (A) Genomic order of the AID/APOBEC Z domains within the canonical A3 gene locus, which is sandwiched by CBX6 and CBX7 genes. Mammalian genomes in which CBX6 and CBX7 genes were detected in the same scaffold were only analyzed. The arrows indicate the direction of respective loci. (B) Bubble plot of the number of A3 Z domains in mammals. The number of the A3 Z domains in the whole genome (x axis) and that within the canonical A3 gene locus (y axis) in each mammal are plotted. Dot size is proportional to the number of species. (C) Genomic locations of A3 Z domains in S. boliviensis, A. nancymaae, and O. garnetti. A3 Z domains within 100 kb of each other were clustered. An asterisk denotes the A3 cluster corresponding to the canonical A3 gene locus. The arrows indicate the direction of respective loci. Pseudogenized sequences are indicated with an X. The sequences indicated by double daggers are intronless sequences and correspond to those described in SI Appendix, Fig. S5A. (D) The association between the genomic location of A3 genes and pseudogenization. The labels “in” and “out” denote the numbers of A3 Z domains located inside or outside the canonical A3 gene locus, respectively. Results for S. boliviensis, A. nancymaae, and O. garnetti are shown. Odds ratio and P value, calculated with Fisher’s exact test, are shown.
ERVs Evidence a Long-Running Conflict Between Retroviruses and A3 Genes.
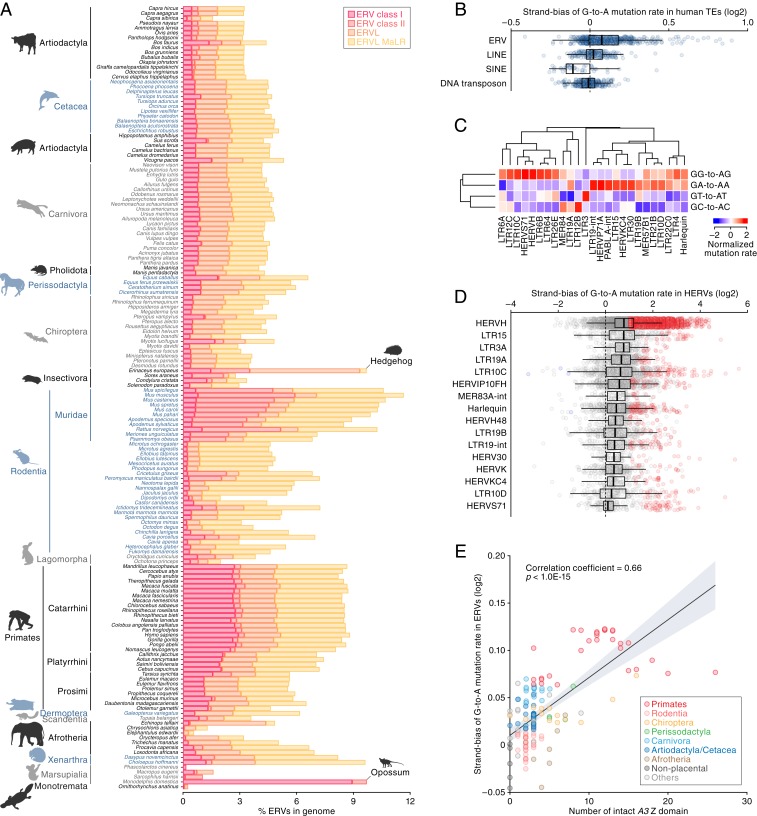
To explore the impact of A3 activity on ERVs and their ancient exogenous ancestors, we performed comparative analysis of transposable elements (TEs) in 160 mammalian genomes. As shown in Fig. 4A and SI Appendix, Fig. S6, the TE composition of mammalian species varies with respect to the proportions of DNA transposons, SINEs, LINEs, and ERVs. To investigate the accumulation level of G-to-A mutations in ERVs, we measured the strand bias of the G-to-A mutation rate in ERVs and other TEs. Since A3 proteins selectively induce G-to-A mutations on the positive strand of retroviruses, strand bias can be an indicator of A3 attack on retroviruses. Consistent with previous reports (30–32), preferential accumulation of G-to-A mutations was observed in human ERVs but not in other human TEs (Fig. 4B). We next classified mutation patterns based on the dinucleotide context. As shown in Fig. 4C, ERVs in the human genome preferentially exhibited GG-to-AG or GA-to-AA mutations, consistent with the reported preferences of human A3G (GG-to-AG) and A3D, A3F, and A3H (GA-to-AA mutations) (10, 33–39). Additionally, some ERVs exhibited G-to-A hypermutation (Fig. 4D).
Fig. 4.
Signatures of A3 activity in ERV sequences and its association with A3 amplification. (A) Proportions of ERV sequences in the genomes of mammalian species. For proportions of LINE, SINE, and DNA transposon sequences, see SI Appendix, Fig. S6. (B) Strand bias scores of G-to-A mutation rates in human TEs (log2-transformed). The strand bias score is calculated as the G-to-A mutation rate ratio between the positive and negative strands. Dots indicate the strand bias scores of respective TE groups. (C) Dinucleotide sequence composition of G-to-A mutation sites in human ERV subfamilies. Of the top 50 ERV subfamilies with respect to the strand bias score, the top 25 ERV subfamilies with respect to the variation (i.e., coefficient of variation) among the 4 G-to-A mutation sites (GA, GT, GG, and GC) are shown. (D) ERV copies presenting the G-to-A hypermutation signature. ERV copies with >1 log2-transformed strand bias score and <0.1 false discovery rate are indicated as red. (E) Association of the number of A3 Z domains with the accumulation level of G-to-A mutations in ERVs in mammals. The x axis indicates the number of intact A3 Z domains, and the y axis indicates the mean value of the log2-transformed strand bias scores among ERVs in the genome. Correlation coefficient and P value are calculated by Pearson’s correlation.
To explore the potential impact of A3 gene amplification on ERVs, we first assessed the accumulation level of G-to-A mutations across all mammalian ERVs (SI Appendix, Fig. S7), then examined the association between 1) accumulation of G-to-A mutations in ERVs and 2) the number of A3 Z domains. This revealed a strong positive correlation (Fig. 4E) (Pearson’s correlation coefficient = 0.69, P < 1.0E-15) wherein the possession of fewer A3 genes (e.g., nonplacental mammals and rodents) is associated with lower accumulation levels, and a higher number of A3 genes (e.g., simiiformes and some chiropterans) is associated with higher accumulation levels.
Correlation of A3 Gene Amplification and Diversification with ERV Activity.
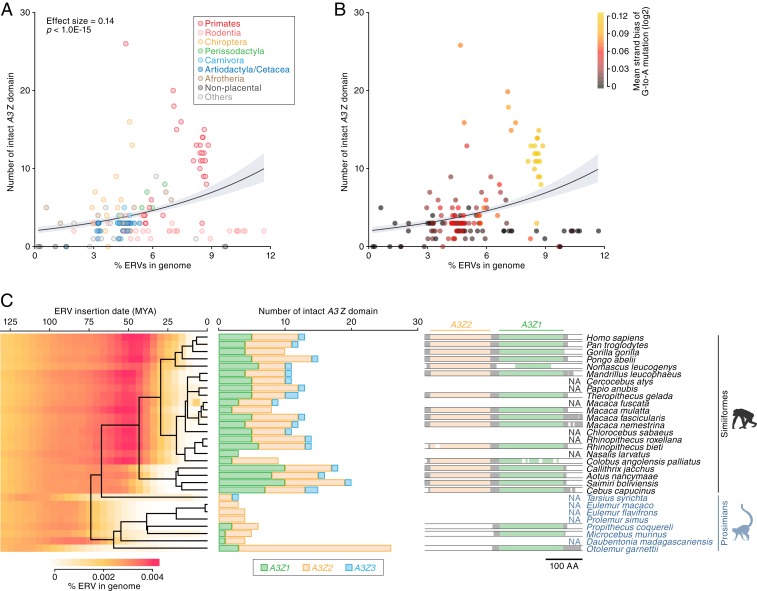
We examined the association between ERV invasions and A3 gene family expansion. As shown in Fig. 5 A and B, we found that the number of A3 Z domains was positively associated with the percentage of ERVs in mammalian genome (in Poisson regression, coefficient = 0.14, P < 1.0E-15). Thus, species in which a greater proportion of the genome is composed of ERVs tend to have a higher number of A3 genes. Exceptions occur in the rodent family Muridae, as well as in 2 other species, hedgehog (Erinaceus europaeus) and opossum (M. domestica). In all of these outlier species, a large proportion of the genome is composed of ERV sequences, but relatively few or no A3 genes appear to be present (SI Appendix, Fig. S8A). As might be expected, ERVs in these outlier species exhibited lower accumulation levels of G-to-A mutations overall (Fig. 5B). In addition, many of the ERVs identified in these species are relatively young (SI Appendix, Fig. S8 B–D) indicating that they derive from recent genome colonization events and have been incorporated into the germline without encountering A3-mediated mutation.
Fig. 5.
Association between A3 gene family expansion and ERV invasion. (A and B) Association of the number of A3 Z domains with the amount of ERV insertions in the genome. Dots are colored according to the species taxa (A) or the accumulation level of G-to-A mutations in ERVs (B). The association was evaluated under the Poisson regression with log link function. (C) Temporal association of ERV invasion with A3 gene amplification in primates. (Left) Amount of ERV insertions in each age category in distinct primate species. ERV insertion date was estimated based on the genetic distance of each ERV integrant from the consensus sequence under the molecular clock assumption [2.2 × 10−9 mutations per site per year (68)]. (Middle) Number of intact A3 Z domains. (Right) Schematic of the MSA of A3G (A3Z2-Z3Z1 type) gene. Sequences of A3G genes in primates recorded in the Ensembl gene database (http://www.ensembl.org) were used. NA, not applicable (no available data).
To investigate the association of A3 gene family expansion with ERV activity, we focused on primates because the evolutionary history of primate ERVs has been explored in depth and is relatively well characterized. We assessed the age of ERV invasions in each species using a genomic distance-based method and found that ERVs prominently invaded in the common ancestors of Simiiformes (including Hominoidea, Old World monkeys, and New World monkeys) around 50 million years ago (Fig. 5 C, Left). In contrast, ancestors of prosimians (including Lemurs, Lorisoids, and Tarsiers) did not experience prominent ERV invasion in this period. Furthermore, simians encoded higher numbers of A3 genes than prosimians (except for O. garnettii), suggesting that A3 gene amplification occurred early in the divergence of simian species (Fig. 5 C, Middle).
We investigated the timing of the formation of the double-domain A3G gene (i.e., A3G gene with A3Z2-A3Z1 structure) using the Ensembl gene database (www.ensembl.org/). We found that simian primates encoded the double-domain (A3Z2-A3Z1) A3G gene, whereas prosimians did not, suggesting that the emergence of double-domain A3G genes also occurred during this period (Fig. 5 C, Right). Absence of a double-domain A3G gene in prosimians is supported by the finding that no A3Z2-A3Z1 genetic structures were observed in prosimian genomes (Fig. 3A). Overall, the timing of A3 gene amplification and diversification in primates was highly concordant with the timing of the prominent ERV invasions.
Discussion
Mammalian A3 family genes possess potent antiviral activities and are thought to have diversified during their evolution to allow targeting of a broader range of viruses (8, 12–14). ERVs provide a rich fossil record for retroviruses, enabling unique insights into the long-term coevolutionary interactions between retroviruses and their hosts. In the present study, we used the ERV fossil record to explore the coevolutionary history of A3 genes and ERVs.
When examining the ERV fossil record, it is vital to keep in mind that it is necessarily an incomplete record of retrovirus evolution. The vast majority of ERV sequences are fixed in the gene pool of host species, but since 1) fixation of any novel allele is extremely unlikely in the absence of strong selection and 2) most ERV insertions are likely to be selectively neutral at best, it is reasonable to assume that the fixed ERVs we observe in the genomes of contemporary species represent a tiny subset of all of the ERVs that colonized their ancestors genomes. Furthermore, the ERV fossil record is presumably heavily biased toward retrovirus lineages that target germline cells, and there may have been many ancestral retrovirus lineages that never generated germline copies. Nonetheless, the fixed ERVs that are found in contemporary genomes are a unique source of retrospective information about the ancestral interactions between retroviruses and their hosts. Furthermore, because A3 genes restrict retrovirus replication via DNA editing, ERV sequences can contain genomic signatures that reveal information about their interactions with this particular group of restriction factors.
We show a strong positive correlation between A3 Z copy number and the extent to which G-to-A mutations have accumulated in ERV sequences (Fig. 4E). This finding reinforces the previously proposed concept (15, 16, 19, 21) that the accumulation of G-to-A mutations in ERVs reflects the antiviral activity of A3 proteins. We further show that mammalian species that have accumulated more ERVs (measured as a proportion of their genome) tend to have higher A3 Z copy numbers (Fig. 5 A and B). In addition, our analysis revealed that A3 amplification occurred concurrently with prominent ERV invasions in primates. Overall, our findings provide evidence that the evolution of mammalian A3 genes has been shaped by a long-running evolutionary conflict with retroviruses, including those retroviruses that have actively invaded mammalian genomes during their evolution, leading to the generation of fixed ERV loci.
The loop 7 region of A3 proteins is thought to determine the sequence specificity of viral nucleotide substrates (40). Our analysis indicates that this region has evolved under strong diversifying selection (Fig. 2B), consistent with the idea that rapid evolution in mammalian A3 genes has been driven by interaction with viruses. Since the genes examined are not orthologous, the variation we observed may reflect diversification that occurred following gene duplication. In addition, it is well established that HIV-1 Vif, an antagonist of A3G activity, specifically binds to loop 7, leading to its degradation (41, 42). This raises the possibility that Vif-like proteins encoded by ancestral retroviruses and/or ERVs may have exerted diversifying selective pressure on A3s. Indeed, remnants of vif gene-like ORFs have been identified in endogenous lentiviruses (43–45). In addition, it has recently been reported that herpesviruses encode ribonucleotide reductase large subunits that degrade human A3 proteins (5, 46, 47) and that the A3 antagonists of Epstein–Barr virus and Kaposi’s sarcoma-associated herpesvirus specifically recognize the loop 7 structure of A3B (5). Therefore, A3 antagonists encoded by viruses other than retroviruses may also have exerted selective pressure on the loop 7 structures of A3 genes.
Most A3 genes are encoded in the canonical A3 locus and have been amplified by tandem gene duplication (Fig. 3 A and B). However, we also detected duplicated A3 genes outside this region in 3 primate species (S. boliviensis, A. nancymaae, and O. garnetti) (Fig. 3 B and C and SI Appendix, Fig. S5). All of these intronless A3G-like genes were amplified by retrotransposition. Furthermore, some are transcribed and may be functional (SI Appendix, Fig. S5).
A3 genes have been amplified in multiple lineages of mammals, but in addition, many A3 genes have been lost or pseudogenized (Fig. 1C and SI Appendix, Fig. S3). For example, the A3Z1 gene was lost in Rodentia, and the A3Z3 gene was lost in Strepsirrhini and Microchiroptera. These findings might be attributed to genotoxic potential of these A3 genes: uncontrolled A3 expression can be harmful, and exogenous expression of human A3A (A3Z1 ortholog) in cell cultures triggers cytotoxic effects (48–50). Similarly, the aberrant expression of some human A3 proteins, particularly A3A (51, 52), A3B (A3Z2–A3Z1 ortholog) (51–54), and A3H (A3Z3 ortholog) (55), can contribute to cancer development by inducing somatic G-to-A mutations in the human genome.
Unlike the A3Z1 and A3Z2 genes, A3Z3 is highly conserved in most mammals and is not amplified in most mammalian lineages. Exceptions occur in carnivores and some other species; however, almost all duplicated A3Z3 genes identified in these species were pseudogenized (SI Appendix, Fig. S4). Moreover, phylogenetic relationships and the pattern of the premature stop codon positions (SI Appendix, Fig. S4) indicate that the duplication–pseudogenization events have happened twice independently during carnivore evolution. These observations support that while the A3Z3 gene is indispensable for the hosts, its duplication might be genotoxic.
A3 proteins can suppress retroviral replication in a G-to-A mutation-independent fashion (e.g., inhibition of reverse transcription) (56–59). We could not address this dimension of ERV–A3 interaction because of the technical difficulty of assessing the mutation-independent effect of A3 proteins on retroviruses using only genomic information. It should also be noted that the number of A3 genes counted in this study might underestimate the true value because of relatively low resolution of many whole genome sequences. Moreover, we particularly focused on the numbers and sequences of the Z domain of AID/APOBEC family genes, and we could not fully address whether 1) some 2 Z domains compose a double domain gene and 2) there are splicing variants. Nevertheless, this is to our knowledge the most comprehensive investigation of A3 gene evolution performed to date.
Materials and Methods
Sequence Data.
WGS assemblies and RNA-Seq data analyzed in this study are summarized in Datasets S1 and S6, respectively. Mammalian TE sequences were obtained using RepeatMasker (version open-4-0-9) (http://repeatmasker.org) with Repbase RepeatMasker libraries (version 20181026) (60). RMBlast was selected as the search engine, and RepeatMasker was run with the options “-q xsmall -a -species <species>” where <species> denotes the species name of the analyzed genome (Dataset S7).
Genome Screening.
Similarity search-based screens of sequence databanks were performed using the database-integrated genome-screening (DIGS) tool (61) which provides a relational database framework for performing systematic tBLASTn-based screening of WGS databanks (61). We used AID/APOBEC polypeptide sequences of 5 species (human, mouse, cow, megabat, and cat) as queries for DIGS (SI Appendix, Fig. S1 A–C and Dataset S2). The resultant list of hits (i.e., sequences disclosing homology to AID/APOBEC family genes) was filtered to remove short and low-similarity matches (tBLASTn bitscore < 50). In the DIGS hit sequences, a partial sequence region [referred to as conserved region (8)] of AID/APOBEC family genes was extracted and used in downstream analyses (SI Appendix, Fig. S1A). Because the conserved regions of AID/APOBEC family genes are located on a single exon (SI Appendix, Fig. S1C) the set of loci identified via DIGS could readily be interrogated using phylogenetic approaches. We selected sequences that covered >70% of the conserved region (SI Appendix, Fig. S1D) and constructed multiple sequence alignments (MSAs) using the L-INS-I algorithm as implemented in MAFFT (version 7.407) (62). A phylogenetic tree was reconstructed using the neighbor-joining (NJ) method (63) as implemented in MEGAX (64). Only alignment sites with the >85% site coverage were used for phylogenetic construction. Additional tree-based filtering of the underlying dataset was performed prior to construction of a final tree: a preliminary tree was constructed, and subsequently, phylogenetic outlier sequences, which have extremely long external branches (i.e., standardized external branch length > 5), were detected and discarded from downstream analyses. The final set of AID/APOBEC-related loci is summarized in Dataset S3.
To investigate the genomic context of AID/APOBEC-related loci, the polypeptide sequences of genes flanking the canonical A3 locus (i.e., CBX6 and CBX7) were used as queries for DIGS. Genomic synteny was illustrated using ggplot2 (https://ggplot2.tidyverse.org/) with the R library ggquiver (https://github.com/mitchelloharawild/ggquiver).
Sequence Analysis.
In-frame MSAs of nucleotide sequences were constructed using the codon-based alignment algorithm implemented in MUSCLE (65). Codon sites with >50% site coverages were used for downstream analyses. Logo plots of the amino acid sequences were generated using weblogo3 (66). Positional Shannon’s entropy score was calculated for amino acid MSAs using tools available via the Los Alamos HIV-1 sequence database website (www.hiv.lanl.gov/content/sequence/ENTROPY/entropy_one.html). A dN/dS ratio test using the branch-site model as implemented in Hyphy MEME (25) was used to detect codon sites under diversifying selection. The phylogenetic tree for this test was constructed using maximum likelihood method as implemented in MEGAX (64).
Mutation Strand Bias Analysis.
To assess the accumulation level of G-to-A mutations in ERVs and other TEs, the strand bias of the G-to-A mutation rate was calculated. First, we calculated the number of nucleotide changes relative to consensus for each TE integrant using the pairwise sequence alignment generated by RepeatMasker. TE integrants with low-confidence alignments (<1,000 Smith–Waterman score) were excluded from the analysis. Next, G-to-A mutation rates in the positive and negative strands of each TE were calculated. Finally, the strand bias score was defined as a ratio of the G-to-A mutation rate between the positive and negative strands (i.e., the mutation rate in the positive strand was divided by the one in the negative strand). The strand bias score was calculated for each TE integrant or each TE group. Statistical significance of the strand bias was evaluated by Fisher’s exact test. False discovery rate was calculated according to the Benjamini–Hochberg method (67).
Estimation of Insertion Dates of ERVs.
Insertion dates of ERV loci were estimated using both 1) ortholog distribution-based and 2) genetic distance-based methods. Ortholog distribution-based estimation was performed for ERVs in human and mouse genomes. Liftover chain files were downloaded from UCSC genome browser (https://genome.ucsc.edu/) (Dataset S8). The Liftover program (http://genome.ucsc.edu/cgi-bin/hgLiftOver) and chain file were used as the basis for attempting to convert the genomic coordinates of ERV integrants in one species genome to those found in another species using the option “minMatch=0.5.” If conversion succeeded, we inferred that the orthologous copy of the ERV integrant was likely present in the corresponding genome. In the case of mouse ERVs, we first converted genomic coordinates of ERVs in Mm9 to those in Mm10, which is the latest version of the mouse reference genome. Subsequently, the genomic coordinates in Mm10 converted to those in the genomes of increasingly distantly related species. Insertion dates of ERVs were estimated from the ortholog distributions according to the scheme summarized in SI Appendix, Fig. S9.
Genetic distance-based estimation of insertion dates was performed for ERVs by calculating the genetic distance of each ERV integrant from a consensus sequence representing the specific lineage the ERV derived from. The distribution of genetic distances was summarized using the Landscape function implemented in RepeatMasker. Genetic distances were converted to the age estimations under the assumption of a neutral molecular clock. For Primates, Insectivora, and Marsupialia a neutral rate of 2.2 × 10−9 mutations per year per site (68) was used. For Rodents, which experience relatively rapid rates of neutral change (69), a rate of 7.0 × 10−9 mutations per year per site was used. For each of these 2 groups, the estimated insertion dates using these rates were highly concordant between the genetic distance-based and ortholog distribution-based methods (SI Appendix, Fig. S9).
RNA-Seq Analysis of AID/APOBEC Family Genes.
RNA-Seq dataset used in the present study is summarized in Dataset S6. RNA-Seq reads were trimmed by Trimmomatic (version 0.36) (70) and subsequently mapped to the reference genomes using STAR (version 020201) (71). Reads mapped on the identified loci of AID/APOBEC family genes were counted using featureCounts (version 1.6.4) (72). Only reads mapped to unique genomic regions were counted. Read counts were normalized to the total number of uniquely mapped reads, and expression levels were measured as fragments per kilobase per million mapped fragments.
Data Availability.
The data, associated protocols, code, and materials in this study are available at https://giffordlabcvr.github.io/A3-Evolution/.
Supplementary Material
Acknowledgments
We thank Mai Suganami (Division of Systems Virology, Institute of Medical Science, The University of Tokyo, Japan) for technical support and Daniel Sauter (Institute of Molecular Virology, Ulm University Medical Center, Germany) for thoughtful comments and suggestions for the manuscript. The supercomputing resource, SHIROKANE, was provided by Human Genome Center, The Institute of Medical Science, the University of Tokyo, Japan. This study was supported in part by Japan Agency for Medical Research and Development (AMED) Japanese Initiative for Progress of Research on Infectious Disease for Global Epidemic (J-PRIDE) 19fm0208006h0003 (K.S.); AMED Research Program on HIV/AIDS 19fk0410014h0002 (to K.S.) and 19fk0410019h0002 (to K.S.); Japan Science and Technology Agency CREST (to K.S.); Grants-in-Aid for Scientific Research (KAKENHI) Scientific Research B 18H02662 (to K.S.), Scientific Research on Innovative Areas 16H06429 (to K.S.), 16K21723 (to K.S.), 17H05813 (to K.S.), and 19H04826 (to K.S.), and Fund for the Promotion of Joint International Research (Fostering Joint International Research) 18KK0447 (to K.S.); Japan Society for the Promotion of Science (JSPS) Research Fellow PD 19J01713 (to J.I.); Takeda Science Foundation (to K.S.); ONO Medical Research Foundation (to K.S.); Ichiro Kanehara Foundation (to K.S.); Lotte Foundation (to K.S.); Joint Usage/Research Center program of Institute for Frontier Life and Medical Sciences, Kyoto University (to K.S.); International Joint Research Project of the Institute of Medical Science, the University of Tokyo, 2019-K3003 (to R.J.G. and K.S.); and JSPS Core-to-Core program (A. Advanced Research Networks) (to R.J.G. and K.S.). R.J.G. was supported by a grant from the UK Medical Research Council (MC_UU_12014/10).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data, associated protocols, code, and materials in this study are available at https://giffordlabcvr.github.io/A3-Evolution/.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914183116/-/DCSupplemental.
References
- 1.Conticello S. G., The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conticello S. G., Langlois M. A., Yang Z., Neuberger M. S., DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 94, 37–73 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Teng B., Burant C. F., Davidson N. O., Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260, 1816–1819 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Harris R. S., Dudley J. P., APOBECs and virus restriction. Virology 479–480, 131–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng A. Z., et al. , Epstein-Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 4, 78–88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouzidi M. S., et al. , APOBEC3DE antagonizes hepatitis B virus restriction factors APOBEC3F and APOBEC3G. J. Mol. Biol. 428, 3514–3528 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Köck J., Blum H. E., Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J. Gen. Virol. 89, 1184–1191 (2008). [DOI] [PubMed] [Google Scholar]
- 8.LaRue R. S., et al. , Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83, 494–497 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakano Y., et al. , A conflict of interest: The evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology 14, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangeat B., et al. , Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H., Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Münk C., Willemsen A., Bravo I. G., An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol. 12, 71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duggal N. K., Malik H. S., Emerman M., The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J. Virol. 85, 11361–11371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawyer S. L., Emerman M., Malik H. S., Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, E275 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y. N., Malim M. H., Bieniasz P. D., Hypermutation of an ancient human retrovirus by APOBEC3G. J. Virol. 82, 8762–8770 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jern P., Stoye J. P., Coffin J. M., Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 3, 2014–2022 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ecco G., Imbeault M., Trono D., KRAB zinc finger proteins. Development 144, 2719–2729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishizu H., Siomi H., Siomi M. C., Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev. 26, 2361–2373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esnault C., Priet S., Ribet D., Heidmann O., Heidmann T., Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: Ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology 5, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treger R. S., et al. , Human APOBEC3G prevents emergence of infectious endogenous retrovirus in mice. J. Virol. 93, e00728-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knisbacher B. A., Levanon E. Y., DNA editing of LTR retrotransposons reveals the impact of APOBECs on vertebrate genomes. Mol. Biol. Evol. 33, 554–567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Severi F., Chicca A., Conticello S. G., Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol. Biol. Evol. 28, 1125–1129 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Ikeda T., et al. , Opossum APOBEC1 is a DNA mutator with retrovirus and retroelement restriction activity. Sci. Rep. 7, 46719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaRue R. S., et al. , The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 9, 104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murrell B., et al. , Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouno T., et al. , Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat. Commun. 8, 15024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura S., et al. , The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 19, 1005–1010 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Shaban N. M., et al. , The antiviral and cancer genomic DNA deaminase APOBEC3H is regulated by an RNA-mediated dimerization mechanism. Mol. Cell 69, 75–86.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison P. M., Zheng D., Zhang Z., Carriero N., Gerstein M., Transcribed processed pseudogenes in the human genome: An intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 33, 2374–2383 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anwar F., Davenport M. P., Ebrahimi D., Footprint of APOBEC3 on the genome of human retroelements. J. Virol. 87, 8195–8204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinomoto M., et al. , All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 35, 2955–2964 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hulme A. E., Bogerd H. P., Cullen B. R., Moran J. V., Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390, 199–205 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Refsland E. W., Hultquist J. F., Harris R. S., Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 8, e1002800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gourraud P. A., et al. , APOBEC3H haplotypes and HIV-1 pro-viral vif DNA sequence diversity in early untreated human immunodeficiency virus-1 infection. Hum. Immunol. 72, 207–212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop K. N., et al. , Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14, 1392–1396 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S., APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14, 1385–1391 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R., A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23, 2451–2458 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Y. H., et al. , Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78, 6073–6076 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Q., et al. , Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11, 435–442 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Rathore A., et al. , The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J. Mol. Biol. 425, 4442–4454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouno T., et al. , Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nat. Struct. Mol. Biol. 22, 485–491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavens D., et al. , Definition of the interacting interfaces of Apobec3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic Acids Res. 38, 1902–1912 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hron T., Farkašová H., Padhi A., Pačes J., Elleder D., Life history of the oldest lentivirus: Characterization of ELVgv integrations in the dermopteran genome. Mol. Biol. Evol. 33, 2659–2669 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Han G. Z., Worobey M., Endogenous lentiviral elements in the weasel family (Mustelidae). Mol. Biol. Evol. 29, 2905–2908 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gifford R. J., Viral evolution in deep time: Lentiviruses and mammals. Trends Genet. 28, 89–100 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Cheng A. Z., et al. , A conserved mechanism of APOBEC3 relocalization by herpesviral ribonucleotide reductase large subunits. J. Virol., 10.1128/JVI.01539-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart J. A., Holland T. C., Bhagwat A. S., Human herpes simplex virus-1 depletes APOBEC3A from nuclei. Virology 537, 104–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landry S., Narvaiza I., Linfesty D. C., Weitzman M. D., APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 12, 444–450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suspène R., et al. , Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc. Natl. Acad. Sci. U.S.A. 108, 4858–4863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Land A. M., et al. , Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J. Biol. Chem. 288, 17253–17260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nik-Zainal S., et al. , Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 46, 487–491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor B. J., et al. , DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2, e00534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burns M. B., Temiz N. A., Harris R. S., Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45, 977–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burns M. B., et al. , APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starrett G. J., et al. , The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 7, 12918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi T., et al. , Quantification of deaminase activity-dependent and -independent restriction of HIV-1 replication mediated by APOBEC3F and APOBEC3G through experimental-mathematical investigation. J. Virol. 88, 5881–5887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bishop K. N., Verma M., Kim E. Y., Wolinsky S. M., Malim M. H., APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4, e1000231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holmes R. K., Koning F. A., Bishop K. N., Malim M. H., APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282, 2587–2595 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Bishop K. N., Holmes R. K., Malim M. H., Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80, 8450–8458 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bao W., Kojima K. K., Kohany O., Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu H., Dennis T., Hughes J., Gifford R. J., Database-integrated genome screening (DIGS): Exploring genomes heuristically using sequence similarity search tools and a relational database. 10.1101/246835 (25 April 2018). [DOI]
- 62.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 64.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 68.Kumar S., Subramanian S., Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. U.S.A. 99, 803–808 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bulmer M., Wolfe K. H., Sharp P. M., Synonymous nucleotide substitution rates in mammalian genes: Implications for the molecular clock and the relationship of mammalian orders. Proc. Natl. Acad. Sci. U.S.A. 88, 5974–5978 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Albin J. S., et al. , A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J. Biol. Chem. 285, 40785–40792 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith J. L., Pathak V. K., Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J. Virol. 84, 12599–12608 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakashima M., et al. , Mapping region of human restriction factor APOBEC3H critical for interaction with HIV-1 Vif. J. Mol. Biol. 429, 1262–1276 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data, associated protocols, code, and materials in this study are available at https://giffordlabcvr.github.io/A3-Evolution/.