Significance
Enzyme complexes consist of several protein subunits that often catalyze sequential reactions in the cell. The activities of the individual subunits must be kept in phase, which requires a sophisticated communication mechanism that is mediated by certain residues. We wanted to elucidate the communication mechanisms between the α-subunits and β-subunits of the tryptophan synthase (TS) complex, which catalyzes the last 2 steps of the biosynthesis of the essential amino acid tryptophan. To this end, an approach involving the reconstruction of TSs from extinct species was followed. The results of our experimental and computational analyses identified 4 residues that contribute to the communication between the α-subunits and β-subunits and provide an explanation of how they act.
Keywords: allostery, ancestral sequence reconstruction, vertical approach, tryptophan synthase
Abstract
Tryptophan synthase (TS) is a heterotetrameric αββα complex. It is characterized by the channeling of the reaction intermediate indole and the mutual activation of the α-subunit TrpA and the β-subunit TrpB via a complex allosteric network. We have analyzed this allosteric network by means of ancestral sequence reconstruction (ASR), which is an in silico method to resurrect extinct ancestors of modern proteins. Previously, the sequences of TrpA and TrpB from the last bacterial common ancestor (LBCA) have been computed by means of ASR and characterized. LBCA-TS is similar to modern TS by forming a αββα complex with indole channeling taking place. However, LBCA-TrpA allosterically decreases the activity of LBCA-TrpB, whereas, for example, the modern ncTrpA from Neptuniibacter caesariensis allosterically increases the activity of ncTrpB. To identify amino acid residues that are responsible for this inversion of the allosteric effect, all 6 evolutionary TrpA and TrpB intermediates that stepwise link LBCA-TS with ncTS were characterized. Remarkably, the switching from TrpB inhibition to TrpB activation by TrpA occurred between 2 successive TS intermediates. Sequence comparison of these 2 intermediates and iterative rounds of site-directed mutagenesis allowed us to identify 4 of 413 residues from TrpB that are crucial for its allosteric activation by TrpA. The effect of our mutational studies was rationalized by a community analysis based on molecular dynamics simulations. Our findings demonstrate that ancestral sequence reconstruction can efficiently identify residues contributing to allosteric signal propagation in multienzyme complexes.
Allostery describes the phenomenon that protein function can be altered by the binding of an effector ligand at a site that is topographically distinct from the active site (1). Allosteric communication is central for the regulation of the activity of many enzyme complexes and has been intensely studied with various approaches over the past decades. Nevertheless, due to the complexity of the studied systems, there are still plenty of questions open with respect to the structural basis of allostery (2). Previous studies have indicated that allostery is based on dynamic effects of the protein backbone and of side chains that can be understood by a combination of computational and experimental approaches (3, 4). However, allosteric communication between distant sites is likely mediated via multiple paths, which makes it difficult to identify the role of individual structural elements for signal propagation (2). One possibility to identify such elements is the analysis of specific mutations that disturb communication between the allosteric site and the active site (1). It has been shown, for instance, that allosteric signals in protein kinase A (4), in imidazole glycerol phosphate synthase (5), and in aminodeoxychorismate synthase (6) can be disrupted with single point mutations.
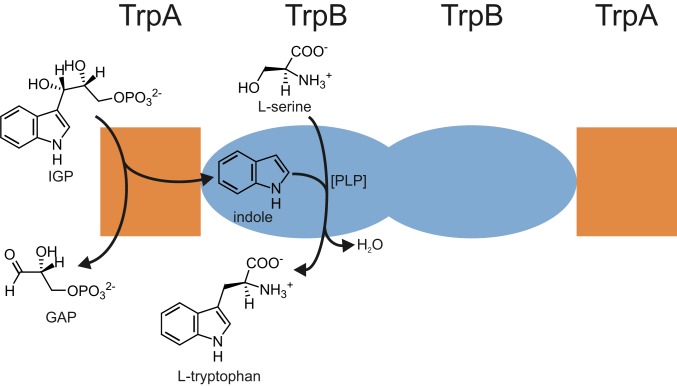
Tryptophan synthase (TS) is a heterotetrameric αββα complex where a central dimer of 2 β-subunits (TrpB) is surrounded by 2 α-subunits (TrpA) (7). These subunits catalyze the last 2 steps in the biosynthesis of the essential amino acid tryptophan: TrpA cleaves indole-3-glycerol phosphate (IGP) into glyceraldehyde-3-phosphate (GAP) and the reaction intermediate indole, which is subsequently channeled to the active site of TrpB where it reacts in a pyridoxal phosphate (PLP)-mediated reaction with l-serine to l-tryptophan (Fig. 1). In order to coordinate these reactions, the TrpA and TrpB subunits mutually activate each other in an allosteric manner (8). These properties and a long history of research make the TS an excellent model system for the analysis of allosteric communication within multienzyme complexes (9–11). However, many aspects of allostery in TS are still not understood and only a fraction of residues involved in signal propagation from the active site of TrpA to the active site of TrpB (and vice versa) has been identified.
Fig. 1.
Structure and function of TS. The αββα TS complex is constituted of a central dimer of β-subunits (TrpB; colored light blue) with two adjacent α-subunits (TrpA, colored orange). TrpA catalyzes the cleavage of IGP into GAP and indole. The latter is channeled to the active site of TrpB where it condenses in a PLP-dependent manner with l-serine, yielding l-tryptophan.
Ancestral sequence reconstruction (ASR) is an in silico method that allows one to calculate the most probable sequences of proteins from extinct species with the help of a multiple sequence alignment of extant species and a phylogenetic model (12, 13). We have previously used ASR to determine the amino acid sequences of TrpA and TrpB from the last bacterial common ancestor (LBCA-TrpA and LBCA-TrpB). A crucial element of ASR is a reliable phylogenetic tree, which was made robust by the concatenation of the extant TrpA and TrpB sequences and did not signal cases of lateral gene transfer (14). We have then shown that both LBCA-TrpA and LBCA-TrpB were catalytically active and assembled to a αββα complex within which indole was channeled with similar efficiency as in modern TS complexes. However, although IGP cleavage of LBCA-TrpA was elevated by 2 orders of magnitude in the presence of LBCA-TrpB similar as in modern TS, the condensation of l-serine and indole catalyzed by LBCA-TrpB was considerably slowed down by LBCA-TrpA. These differences offer the opportunity to use a “vertical” approach for the identification of structural elements and specific amino acids that are crucial for the communication between TrpA and TrpB. In contrast to the conventional “horizontal” approach in which extant homologous proteins are compared, in a vertical approach reconstructed proteins representing extinct species of the phylogenetic tree are compared with each other and with their modern homologs. Vertical approaches are much more straightforward for the analysis of sequence-function relationship of proteins than horizontal approaches, especially because functional differences can be traced back to much fewer differences in sequence (12).
Specifically, in the case of TS, starting from the root of the tree (identical to LBCA-TS), all internal TS nodes leading to a specific leaf (a modern TS) are available and sequence differences can be correlated with functional differences. Hence, we experimentally characterized 6 phylogenetically intermediate TS sequences (Anc1nc-TS–Anc6nc-TS) that link LBCA-TS with the modern TS from the γ-proteobacterium Neptuniibacter caesariensis (ncTS), in which ncTrpB is strongly stimulated by ncTrpA. We found that on the way from LBCA-TS to ncTS, the transition from allosteric inhibition to allosteric activation occurs between the adjacent Anc2nc-TS and Anc3nc-TS and is entirely based on sequences differences in TrpB. Subsequent site-directed mutagenesis showed that only 4 (of 413) TrpB residues in this lineage are crucial for the inversion of allostery from inhibitory to activating, and the effect of these residues could be rationalized by a computational community analysis (15) that is based on molecular dynamics (MD) simulations.
Results
A Vertical Approach Identifies 10 TrpB Positions That Determine the Direction of the Allosteric Effect.
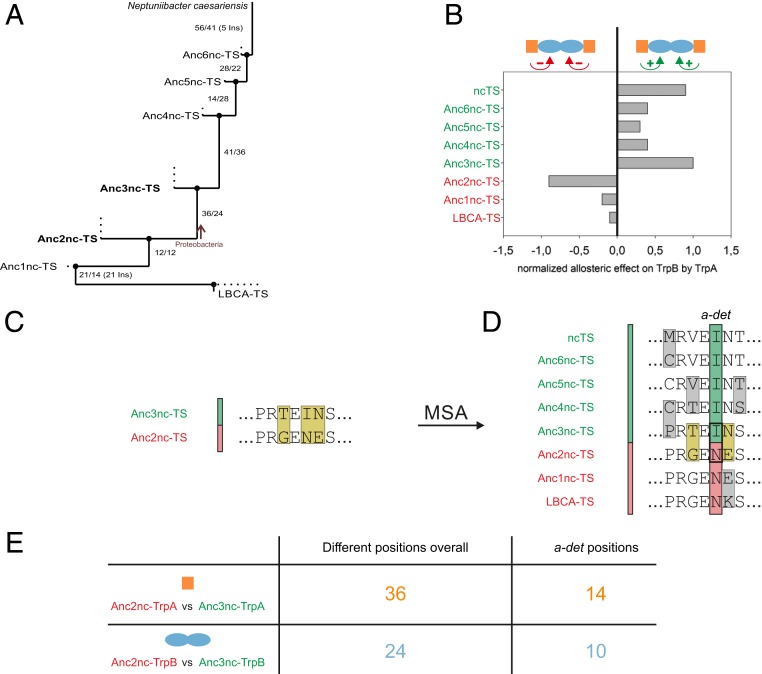
The reconstructed LBCA-TS displays similar features as the well-studied TS complexes from Salmonella typhimurium or Escherichia coli (stTS/ecTS) except for the “inversed” allosteric effect of TrpA on TrpB, which similarly has been observed in a “stand-alone” TrpB variant from Pyrococcus furiosus (PfTrpB0B2) that was produced by directed evolution (9). While in all characterized extant TS the TrpB subunit is activated by the respective TrpA subunit (11, 16), in LBCA-TS the LBCA-TrpB subunit is inhibited by LBCA-TrpA (14) (Table 1). In order to identify residues that are responsible for these different allosteric effects of LCBA-TS and extant TS complexes, a vertical approach was applied. In the first step, a path within the phylogenetic tree connecting LBCA-TS (root of the tree) with the extant ncTS (one leaf of the tree), which shows an activating allosteric effect (Table 1), was selected. LBCA-TrpA and ncTrpA share a sequence identity of 48%, while LBCA-TrpB and ncTrpB share a sequence identity of 72%. This means that LBCA-TrpA and ncTrpA differ in 137 residues, while LBCA-TrpB and ncTrpB differ in 111 residues; moreover, compared to LBCA-TrpB, the ncTrpB sequence contains 13 additional residues. These drastic sequence variations make it impossible to test experimentally which residues are responsible for the different allosteric effects. Therefore, in a second step, all internal nodes of the selected path linking LBCA-TS and ncTS (Anc1nc-TS–Anc6nc-TS) were analyzed (Fig. 2A). For this purpose, the genes coding for the studied TrpA and TrpB proteins were cloned (see SI Appendix, Table S1 for primers), and the recombinant proteins (see SI Appendix, Table S2 for sequences) were generated in E. coli and purified. Screening by a high-performance liquid chromatography (HPLC)-based assay revealed that not only LBCA-TS but also Anc1nc-TS and Anc2nc-TS display an inhibitory allosteric effect, whereas Anc3nc-TS–Anc6-ncTS and ncTS show an activating allosteric effect (Fig. 2B). These findings demonstrate that a discrete transition from an allosterically inhibited to an allosterically activated enzyme occurs between Anc2nc-TS and Anc3nc-TS.
Table 1.
Steady-state kinetic parameters for the reactions of various TrpB proteins
| l-serine | Indole | |||||
| kcat, s−1 | Kml-ser, mM | kcat/Kml-ser, M−1s−1 | kcat, s−1 | KmIndole, µM | kcat/KmIndole, M−1s−1 | |
| ncTrpB | 0.37 ± 0.07 | 991.2 ± 373.5 | 3.8 × 10−1 ± 2.2 × 10−1 | 0.27 ± 0.03 | 28.3 ± 7.3 | 9.6 × 100 ± 3.4 × 100 |
| ncTrpB + ncTrpA | 2.3 ± 0.07 | 3.1 ± 0.5 | 7.5 × 102 ± 1.5 × 102 | 1.8 ± 0.2 | 51.7 ± 11.2 | 3.4 × 101 ± 1.1 × 101 |
| LBCA-TrpB | 4.8 ± 1.4* | 1.3 ± 0.4* | 3.7 × 103 ± 1.1 × 103* | 5.7 ± 1.7 | 6.7 ± 2.0 | 8.5 × 102 ± 5.08 × 102 |
| LBCA-TrpB + LBCA-TrpA | 0.95 ± 0.29* | 1.2 ± 0.4* | 7.9 × 102 ± 2.4 × 102* | 0.76 ± 0.23 | 15.0 ± 4.5 | 5.1 × 101 ± 3.1 × 101 |
| Anc2nc-TrpB | 0.35 ± 0.005 | 0.73 ± 0.06 | 4.82 × 102 ± 4.40 × 101 | 0.38 ± 0.016 | 2.52 ± 0.76 | 1.53 × 102 ± 5.24 × 101 |
| Anc2nc-TrpB + Anc2nc-TrpA | 0.0055 ± 0.0003 | 1.30 ± 0.50 | 4.24 × 100± 1.87 × 100 | 0.0055 ± 0.0004 | 6.14 ± 2.20 | 8.91 × 10−1 ± 3.81 × 10−1 |
| Anc2nc-TrpB_AM4 | 0.24 ± 0.016 | 3.99 ± 1.03 | 6.18 × 101 ± 1.97 × 101 | 0.23 ± 0.020 | 16.6 ± 4.16 | 1.43 × 101 ± 4.74 × 100 |
| Anc2nc-TrpB_AM4 + Anc2nc-TrpA | 0.47 ± 0.021 | 3.79 ± 0.85 | 1.25 × 102 ± 3.38 × 101 | 0.50 ± 0.032 | 26.00 ± 5.20 | 1.93 × 101 ± 5.08 × 100 |
Experimental conditions: For l-serine dependent kinetics: 100 mM potassium phosphate buffer pH 7.5, 180 mM KCl, 40 µM PLP, 200 µM indole, and varying concentrations of l-serine in presence of 2 µM ncTrpB alone, or 0.5 µM ncTrpB in presence of 8 µM ncTrpA; 1 µM Anc2nc-TrpB alone, or 4 µM Anc2nc-TrpB and 10 µM Anc2nc-TrpA; or 0.25 µM Anc2nc-TrpB_AM4 alone or with 2.5 µM Anc2nc-TrpA. For indole-dependent kinetics: 100 mM potassium phosphate buffer pH 7.5, 180 mM KCl, 40 µM PLP, 50 mM l-serine, and varying concentrations of indole in presence of 2 µM ncTrpB alone, or 0.5 µM ncTrpB in presence of 8 µM ncTrpA; 0.05 µM LBCA-TrpB alone, or 0.03 µM LBCA-TrpB and 0.03 µM LBCA-TrpA; 0.5 µM Anc2nc-TrpB alone, or 4 µM Anc2nc-TrpB and 10 µM Anc2nc-TrpA; or 0.25 µM Anc2nc-TrpB_AM4 alone or with 2.5 µM Anc2nc-TrpA. All enzyme concentrations are monomer concentrations. The reactions for LBCA-TrpB were followed at 60 °C, while all other reactions were followed at 30 °C by monitoring the condensation of l-serine and indole to l-tryptophan at 290 nm. The mean and the SEs were calculated from at least 3 independent measurements.
Data taken from ref. 14.
Fig. 2.
Identification of a-det positions in TrpA and TrpB by a vertical approach. (A) Path within the phylogenetic tree of TS connecting LBCA-TS with ncTS via the intermediate nodes Anc1nc-TS–Anc6nc-TS. The numbers of differing amino acids between the connected sequences are indicated. (Number of differences in TrpA sequence/number of differences in TrpB sequence.) Ins, insertions. (B) Allosteric effect of TrpA subunits on their respective TrpB subunits. The TrpB reactions were performed at 30 °C in presence of 100 mM EPPS/KOH pH 7.5, 180 mM KCl, 40 µM PLP, 45 mM l-serine, 300 µM indole, 0.5 µM TrpB (monomer), and 1 µM TrpA (monomer). The resulting reaction mixture was analyzed by reversed-phase HPLC. The normalized allosteric effect is defined as the decadic logarithm of the TrpB activity in the presence of TrpA divided by the TrpB activity in absence of TrpA. Raw data are given in SI Appendix, Table S4. TS complexes with allosteric activation of TrpB by TrpA are colored green, TS complexes with allosteric inhibition are colored red. (C) Scheme of the pairwise sequence comparison between Anc2nc-TS and Anc3nc-TS. The shown sequences are randomly composed examples with differing amino acids framed. (D) Scheme of the multiple sequence alignment (MSA) of the concatenated TS amino acid sequences. The sequences were grouped according to the allosteric effect on the TrpB subunit by the TrpA subunit. Sequences, where the TrpB subunit is activated, are colored green. Sequences, where the TrpB subunit is inhibited, are colored red. Positions occupied with residues that differ between the 2 groups, but are conserved within each group, were designated as “allostery-determining” (a-det) positions. The shown sequences are random examples. (E) Table listing the number of “different overall positions” identified in C and of a-det positions identified in D.
Pairwise sequence comparisons identified 36 residue differences between Anc2nc-TrpA and Anc3nc-TrpA and 24 residue differences between Anc2nc-TrpB and Anc3nc-TrpB (Fig. 2 C and E). The detailed experimental assessment of these 60 residue differences would still not be possible with reasonable effort. To further narrow down the number of essential residues, a multiple sequence alignment (MSA) for the concatenated TrpA and TrpB sequences was created, which contained all sequences of the selected path from LBCA-TS up to ncTS. Within the MSA, the sequences were grouped according to the allosteric effect of TrpA on TrpB, whether activating or inhibitory. Positions occupied with residues that differ between the 2 groups but are conserved within each group were designated as allostery-determining (a-det) positions (Fig. 2D). Within the TrpA sequences 14 a-det positions and within the TrpB sequences 10 a-det positions were identified (Fig. 2E).
We next asked whether the identity of the TrpA subunit is at all significant for the activation or inhibition of the TrpB subunit. To this end, cross-wise activity titrations between Anc2nc-TrpB and Anc2nc-TrpA (SI Appendix, Fig. S1A) or Anc3nc-TrpA (SI Appendix, Fig. S1B) were performed, as well as between Anc3nc-TrpB and Anc2nc-TrpA (SI Appendix, Fig. S1C) or Anc3nc-TrpA (SI Appendix, Fig. S1D). The results revealed that Anc2nc-TrpB is inhibited by both Anc2nc-TrpA and Anc3nc-TrpA, while Anc3nc-TrpB is activated by both Anc2nc-TrpA and Anc3nc-TrpA. This led to the conclusion that the specific identity of the TrpA subunit is unimportant for the allosteric effect on the TrpB subunit. It follows that the 10 a-det positions identified for the TrpB subunit are sufficient for the inversion of the allosteric effect of TrpA on TrpB.
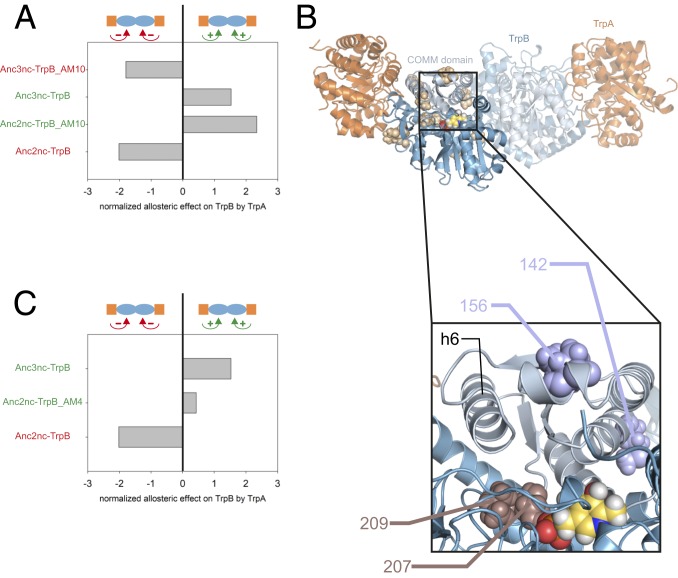
To confirm this conclusion, 2 variants were created. In variant Anc2nc-TrpB_AM10, the 10 a-det residues as present in Anc2nc-TrpB were mutated toward the Anc3nc-TrpB residues. Inversely, in the variant Anc3nc-TrpB_AM10, the 10 a-det residues as present in Anc3nc-TrpB were mutated toward the Anc2nc-TrpB residues. These variants were assayed for the allosteric effect of Anc2nc-TrpA (for Anc2nc-TrpB_AM10) or Anc3nc-TrpA (for Anc3nc-TrpB_AM10) on the TrpB subunit. The results obtained with both variants confirmed that the exchange of the 10 a-det residues are sufficient to inverse the allosteric effect of TrpA on TrpB in both ways, from an inhibited TrpB to an activated TrpB (Anc2nc-TrpB vs. Anc2nc-TrpB_AM10) and from an activated TrpB to an inhibited TrpB (Anc3nc-TrpB vs. Anc3nc-TrpB_AM10) (Fig. 3A).
Fig. 3.
Mutational exchange of a-det and a-ess residues between Anc2nc-TrpB and Anc3nc-TrpB. (A) The transfer of the 10 a-det positions from Anc2nc-TrpB into Anc3nc-TrpB leads to an inhibitory allosteric effect in Anc3nc-TrpB_AM10. Vice versa, the transfer of the 10 a-det positions from Anc3nc-TrpB into Anc2nc-TrpB leads to an activating allosteric effect in Anc2nc-TrpB_AM10. The normalized allosteric effect is defined as the decadic logarithm of the TrpB activity in presence of TrpA divided by the TrpB activity in absence of TrpA. Raw data are given in SI Appendix, Table S5. (B) Homology model of Anc2nc-TS. The 10 a-det positions are shown as beige spheres. The active site is indicated by the spherical representation of the PLP cofactor colored yellow. (Lower) Detail of the homology model of Anc2nc-TrpB. The 4 a-ess positions responsible for the inversion of the allosteric effect are numbered and shown as spheres. The 2 positions from cluster B (cf. SI Appendix, Fig. S2) located in the COMM domain are colored in light blue, and the 2 positions from cluster C close to the active site (indicated by the PLP cofactor) are colored in brown. α-Helix 6 (h6) in TrpB is also marked. (C) The transfer of the 4 a-ess residues from Anc3nc-TrpB into Anc2nc-TrpB leads to an activating allosteric effect in Anc2nc-TrpB_AM4. The a-ess residues were identified by sequential back-mutation experiments (see the text for details). The normalized allosteric effect is defined as the decadic logarithm of the TrpB activity in presence of TrpA divided by the TrpB activity in absence of TrpA. Raw data are given in SI Appendix, Table S6.
Site-Directed Mutagenesis Identifies 4 TrpB Positions That Determine the Direction of the Allosteric Effect.
We next wanted to identify among the 10 a-det positions in Anc2nc-TrpB_AM10 those that are necessary for the conversion of the allosteric effect from inhibitory to activating. The 10 a-det positions are distributed all over the 3-dimensional (3D) structural model of Anc2nc-TrpB (Fig. 3B), but belong to 4 spatial clusters (A–D; SI Appendix, Fig. S2A). Cluster A comprises 2 residues close to the TrpA-TrpB interface. Cluster B contains 4 positions in the COMM domain, which has been shown to be significant for the allosteric communication between TrpA and TrpB (17). The 2 positions of cluster C are located close to the active site, and the 2 positions of cluster D are located next to the TrpB homodimer interface. Starting from Anc2nc-TrpB_AM10, first residues of clusters A, C, and D were individually mutated back to their identities in Anc2nc-TrpB (SI Appendix, Table S3). The protein variants with back-mutated cluster A (Anc2nc-TrpB_AM8-A) and cluster D (Anc2nc-TrpB_AM8-D) residues retained the activating allosteric effect, whereas the variant with back mutated cluster C (Anc2nc-TrpB_AM8-C) residues showed an inhibitory allosteric effect (SI Appendix, Fig. S2B). These results indicated that cluster A and D residues are not required for the conversion of the allosteric effect from inhibitory to activating. A variant with the combined back mutations of cluster A and D (Anc2nc-TrpB_AM6) residues confirmed this conclusion. In contrast, the residues from cluster C seem to contribute to the inversion of the allosteric effect. In a next step, the 4 residues of cluster B were individually back-mutated. The resulting variants Anc2nc-TrpB_AM5_M187I and Anc2nc-TrpB_AM5_D199E retained the activating allosteric effect as observed in Anc2-ncTrpB_AM10, whereas Anc2nc-TrpB_AM5_R142M and Anc2nc-TrpB_AM5_V156I showed an inhibitory allosteric effect.
The 4 residues of Anc2nc-TrpB_AM10 that were shown to be essential for the inversion of the allosteric effect from inhibitory to activating (a-ess) were then combined in the variant Anc2nc-TrpB_AM4. The activating residues R142 and V156 from cluster B are located within the COMM domain, while the activating residues T207 and A209 from cluster C are close to the active site (Fig. 3 B, Lower). As expected, Anc2nc-TrpB_AM4 showed an activating allosteric effect (Fig. 3C and SI Appendix, Fig. S3). Further studies with Anc2nc-TrpB_AM3 variants where 1 of the 4 positions was back-mutated suggested that all mutations in Anc2nc-TrpB_AM4 contribute to the observed activating allosteric effect (SI Appendix, Table S3).
The Effect of the 4 Identified Residues Is Independent of a Specific TrpB Background.
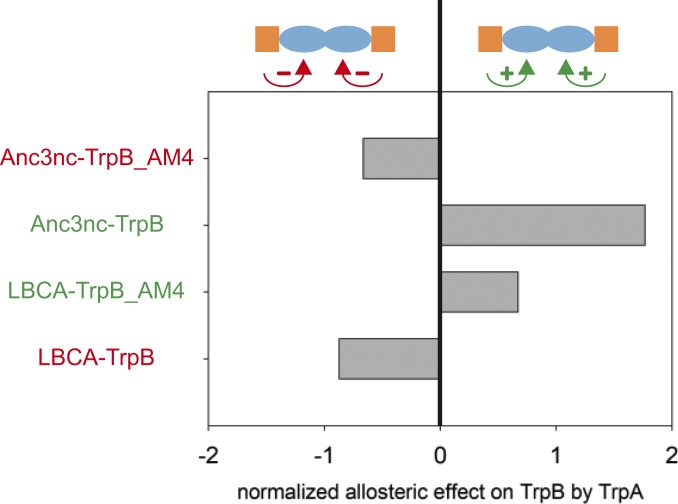
To test whether the 4 identified residues are also effective in TrpB backgrounds other than Anc2nc-TrpB, variants of LBCA-TrpB, and of Anc3nc-TrpB were created. In LBCA-TrpB_AM4, the 4 a-ess residues were mutated to their respective Anc3nc-TrpB identities. In Anc3nc-TrpB_AM4, the 4 a-ess residues were mutated to their respective Anc2nc-TrpB identities. For both variants, we could show the inversion of the allosteric effect by the introduced mutations. While LBCA-TrpB is inhibited by LBCA-TrpA, the variant LBCA-TrpB_AM4 is activated upon binding of LBCA-TrpA. Along the same lines, Anc3nc-TrpB is activated by Anc3nc-TrpA, whereas the variant Anc3nc-TrpB_AM4 is inhibited upon binding of Anc3nc-TrpA (Fig. 4). These results show that the inversion of the allosteric effect of TrpA on TrpB is also possible in other TrpB subunits than Anc2nc-TrpB, meaning that the role of the 4 positions does not depend on a specific TrpB background.
Fig. 4.
Mutational exchange of a-ess residues between LBCA-TrpB and Anc3nc-TrpB. The transfer of the 4 a-ess residues from Anc3nc-TrpB into LBCA-TrpB leads to an activating allosteric effect in LBCA-TrpB_AM4. Vice versa, the transfer of the 4 a-ess residues from LBCA-TrpB into Anc3nc-TrpB leads to an inhibitory allosteric effect in Anc3nc-TrpB_AM4. The normalized allosteric effect is defined as the decadic logarithm of the TrpB activity in the presence of TrpA divided by the TrpB activity in the absence of TrpA. Raw data are given in SI Appendix, Table S7.
Biochemical Characterization of the a-ess TrpB Positions.
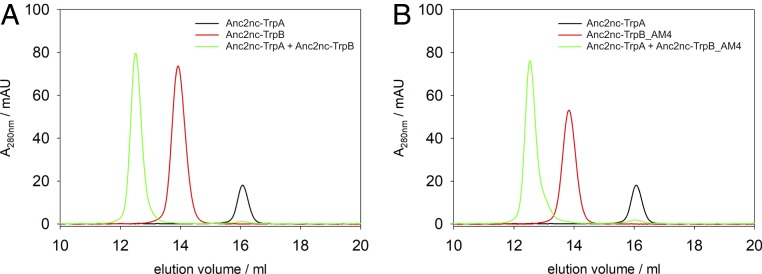
To unravel the mechanism by which the residues in Anc2nc-TrpB_AM4 inverse the allosteric effect compared to Anc2nc-TrpB, comparative experimental and computational studies were performed. Analytical size-exclusion chromatography showed that the 4 residue differences between Anc2nc-TrpB and Anc2nc-TrpB_AM4 do not influence their ability to form a stable complex with Anc2nc-TrpA (Fig. 5). For a more detailed study of the allosteric effect, steady-state kinetic data of Anc2nc-TrpB and Anc2nc-TrpB_AM4 in absence and presence of Anc2nc-TrpA were recorded (Table 1). Anc2nc-TrpA reduces the turnover number (kcat value) of Anc2nc-TrpB about 35- to 65-fold but does not change significantly the Michaelis constants Kml-ser and KmIndole. In contrast, Anc2nc-TrpA increases the kcat value of Anc2nc-TrpB_AM4 about 2-fold, whereas Kml-ser and KmIndole also do not change much. Moreover, absorbance spectra of the PLP cofactor, which are a sensitive measure for the operating mode of the TS complex (18), were monitored. Steady-state spectra of Anc2nc-TrpB (inhibitory allosteric effect) and Anc2nc-TrpB_AM4 (activating allosteric effect) were recorded in their holo-forms, after addition of l-serine, and after addition of Anc2nc-TrpA. The spectral shifts observed for Anc2nc-TrpB_AM4 as a consequence of the addition of the various ligands were identical to those of well-characterized TrpB proteins from extant species. Hence, the spectra indicated that the addition of l-serine resulted in the conversion of the internal to the external aldimine, which after the addition of Anc2nc-TrpA further reacts to the aminoacrylate (SI Appendix, Fig. S4A). In contrast, the addition of l-serine to Anc2nc-TrpB did not result in the generation of the external aldimine but did directly lead to the formation of the aminoacrylate, which was not further populated by the addition of Anc2nc-TrpA (SI Appendix, Fig. S4B). The population of the aminoacrylate is indicative of the degree of closure of the COMM domain and, hence, TrpB activation (10). Therefore, this data suggests that the energetics of the conformational transitions of the COMM domain is altered in Anc2nc-TrpB compared to Anc2nc-TrpB_AM4.
Fig. 5.
Structural complex formation of Anc2nc-TrpA with Anc2nc-TrpB and Anc2nc-TrpB_AM4 as observed by analytical gel filtration chromatography. (A) Elution profiles of Anc2nc-TrpA, Anc2nc-TrpB, and a mixture of the 2 proteins. (B) Elution profiles of Anc2nc-TrpA, Anc2nc-TrpB_AM4, and a mixture of the 2 proteins. The proteins (30 µM monomer concentration for each sample) were loaded on a Superdex S200 column that was equilibrated with 50 mM potassium phosphate (pH 7.5) and 300 mM KCl and eluted at a flow rate of 0.3 mL/min. Elution was followed by monitoring the absorbance at 280 nm.
In Silico Analysis of the a-ess Mutations.
Enzymes that catalyze multistep reactions require the population of several, discrete conformational states associated with the different reaction steps. To obtain insights into differences between the COMM domain configurations in Anc2nc-TrpB and Anc2nc-TrpB_AM4, we employed standard MD simulations. Due to the lack of crystal structures, we derived a homology model for the full complex Anc2nc-TS by means of Yasara (19) and introduced the corresponding mutations by means of PyMOL (20) to create a model of the Anc2nc-TS_AM4 complex. For both models, which adopt the open configuration of the COMM domain as observed in LBCA-TS, 3 μs of accumulated MD simulations were performed. We compared all snapshots with crystal structures representing open (O), partially closed (PC), and closed (C) conformations of the COMM domain. The results indicate that the relative fraction of the O and PC conformations were virtually identical for both proteins and that the closed conformation was not sampled (Anc2nc-TrpB: 62% O/38% PC/0% C; Anc2nc-TrpB_AM4: 66% O/34% PC/0% C). This is in accordance with the relatively slow O-PC-C transition that usually takes place on time scales larger than the currently accessible simulation times (21).
An advanced in silico approach to assess conformational heterogeneities is based on the dynamic network model (15), which assumes that the propagation of allosteric signals in a protein complex depends on the communication between “communities.” These structures of subdomain size can be identified by a correlation analysis of the motions observed during a MD simulation. We were interested to find out how the a-ess mutations affect the organization of communities in Anc2nc-TrpB and Anc2nc-TrpB_AM4. We utilized a standardized protocol (22) to identify for the 2 TrpB variants all communities consisting of more than 2 residues.
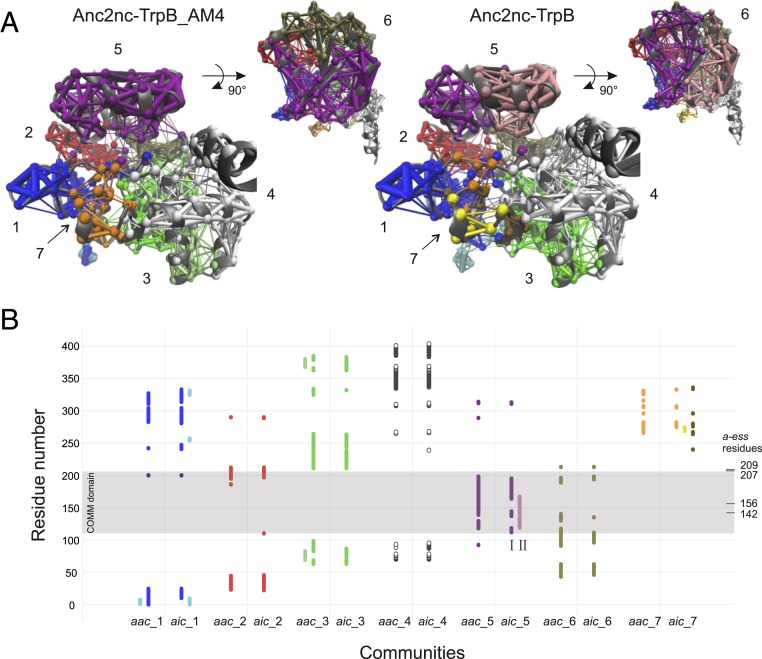
Generally, networks that are well suited for topology analysis contain 2–10 communities (23). This requirement is met by our MD simulations, because the dynamics of the allosterically activated Anc2nc-TrpB_AM4 gave rise to 7 dominant communities named aac_1 to aac_7 (Fig. 6 A, Left). Interestingly, aac_5 contains nearly all residues of the COMM domain (residues 110–206 in Anc2nc-TrpB_AM4). Thus, the residue composition of aac_5 confirmed that this analysis of our MD simulations is biochemically plausible. The comparison of the communities aic_1 to aic_7 of the allosterically inhibited Anc2nc-TrpB (Fig. 6 A, Right) with aac_1 to aac_7 indicated a high degree of overlap in their residue composition. However, we also noticed the existence of some smaller communities whose residue compositions were specific for the activated or the inhibited TrpB (Fig. 6B).
Fig. 6.
Identification of TrpB communities induced by correlated motions in MD simulations. (A) Communities identified by analyzing MD simulations of the allosterically activated Anc2nc-TrpB_AM4 and the allosterically inhibited Anc2nc-TrpB. Communities, whose residue composition overlap to a great extent are shown in the same color. The diameter of edges interconnecting residue positions i and j corresponds to the value -log|cij|, where cij is the correlation between residue i and j. (B) Residue-specific representation of community compositions. Communities observed in the allosterically activated (aac) and the allosterically inhibited TrpBs (aic) are color coded as in A. Note that the disintegration of communities gave rise to smaller communities that are positioned next to the major ones. The localization of the COMM domain and the a-ess residues is indicated. The COMM domain residues constitute in the allosterically activated Anc2nc-TrpB_AM4 the community aac_5 and disintegrate in the allosterically inhibited Anc2nc-TrpB into the 2 smaller communities aic_5-I and aic_5-II.
One of the most pronounced differences is the separation of aac_5 of Anc2nc-TrpB_AM4 into 2 smaller communities aic_5 (named I and II in Fig. 6B) in Anc2nc-TrpB. This finding suggests that the 4 a-ess residues affect the rigidity and conformational space of the COMM domain. Hence, TrpA binding splits the COMM domain of Anc2nc-TrpB into 2 less-coordinated structural elements, suggesting the perturbance of the productive conformational ensemble required for multistep catalysis. Similarly, enhanced sampling MD techniques have indicated that the inhibitory effect of PfTrpA on the stand-alone PfTrpB0B2, which also shows an inversed allosteric effect, can be explained by the truncation of the conformational ensemble and the population of unproductive closed states of the COMM domain (21). In contrast, the COMM domain of the allosterically activated Anc2nc-TrpB_AM4 seems rigidified upon TrpA binding and to recover the conformational heterogeneity required for tryptophan production.
Discussion
Vertical versus Horizontal Approaches for the Analysis of Sequence-Function Relationships.
One of the central objectives in biochemistry is to uncover the relationship between amino acid sequence and protein function. A conventional approach to achieve this goal is to align the sequences of homologous proteins and to use site-directed mutagenesis to test the effect of individual residue differences for functional divergence. However, such an approach based on the horizontal comparison of extant proteins is facing 2 major problems: First, due to neutral drift, even closely related proteins often share sequence identities of less than 50%, which corresponds to 100 different residues in a small protein of 200 amino acids. Second, the effect of certain residues depends on the context within the protein, a property that is called epistasis (24). Both neutral drift and epistasis significantly impair the identification of essential residues by site-directed mutagenesis. This is especially true for complex functional properties such as allostery as observed in metabolic enzyme complexes, which is most probably realized by the interplay of networks of interconnected residues. An elegant alternative to such cumbersome classical horizontal approaches is a vertical comparison of sequences that represent the nodes of a phylogenetic tree of a given protein. These sequences are determined with the help of ASR (12, 13), followed by the expression of the corresponding synthetic genes and the characterization of the recombinant proteins. Since the sequences of adjacent nodes in a phylogenetic tree are generally more similar to each other than the sequences of extant proteins, the identification of residues crucial for functional differences is more straightforward (25, 26).
In our case of TS, the vertical approach proved to be of great value for the identification of residues that are essential for the allosteric regulation of TrpB by TrpA. We started with 2 variants in which the allosteric effect was either inhibitory (LBCA-TS) or activating (ncTS) and faced the practically insolvable problem to assign this functional disparity to 1 or more of 261 amino acid differences (137 differences in the TrpA sequences/111 differences and 13 indels in the TrpB sequences). Moreover, since known positions within TrpB that affect allostery are conserved (for example, R141 in stTrpB/R153 in ncTrpB/R138 in LBCA-TrpB or D305 in stTrpB/D317 in ncTrpB/D302 in LBCA-TrpB) (11), there was no straightforward way to identify crucial positions responsible for the inversion of the allosteric effect of TrpA on TrpB. However, our vertical approach allowed us to narrow down the number of amino acids responsible for this functional difference to only 4 residues, all of which were located in TrpB.
Function of the 4 a-ess Residues that Are Sufficient for the Inversion of the Allosteric Effect.
The 4 positions within the TrpB subunit that exert a drastic effect on the allosteric manipulation by TrpA have not been analyzed so far. In order to elucidate the molecular function of these a-ess positions, analytical gel filtration, steady-state enzyme kinetics, absorbance spectroscopy of the PLP cofactor, and in silico analysis were performed. In terms of structural complex formation the 4 a-ess positions seem to be neutral as Anc2nc-TrpB (inhibitory allosteric effect) and Anc2nc-TrpB_AM4 (activating allosteric effect) both assemble with Anc2nc-TrpA to the regular αββα TS. Steady-state enzyme kinetics confirmed that the kcat value of Anc2nc-TrpB is decreased 35- to 65-fold by Anc2nc-TrpA, while the kcat value of Anc2nc-TrpB_AM4 is enhanced 2-fold in the presence of Anc2nc-TrpA. In contrast, the Kml-ser and KmIndole values are not influenced significantly by Anc2nc-TrpA, neither for Anc2nc-TrpB nor for Anc2nc-TrpB_AM4. These results show that the residues at the 4 a-ess positions do not change the way by which substrate binding by TrpB is modulated by TrpA. However, these residues drastically change the way by which catalysis by TrpB is influenced by TrpA.
One might speculate that the allosteric inversion observed in our oldest TrpB predecessors indicates that TrpB evolved to work independently before functioning in a complex with TrpA. However, we cannot definitely exclude that a slight uncertainty of ASR leads to the inhibitory allosteric effect observed in LBCA-TS, Anc1nc-TS, and Anc2nc-TS. In addition, the LBCA-TS sequence might not be the most likely predecessor of all TS sequences in the tree, because midpoint rooting is only a rough estimate of the unknown root position. However, these uncertainties do not compromise the utility of these proteins for the analysis of the structure-function relationship of TS described in this study. Interestingly, the stand-alone variant PfTrpB0B2, which has been generated by directed evolution, has virtually identical biochemical properties as Anc2nc-TrpB, including inhibition by PfTrpA (9). In line with these findings, our community analysis of Anc2nc-TrpB and extended MD simulations of PfTrpB0B2 (21) indicate that allosterically inhibited (or stand-alone) TrpB proteins display an extended conformational heterogeneity of the COMM domain that is normally observed in TS complexes.
Taken together, the vertical approach used in this study provided unexpected insights into the allosteric communication within TS. Specifically, 4 residues in TrpB could be identified that had escaped attention despite intense research on this fascinating enzyme complex during the past decades. We trust that vertical approaches will prove to be highly valuable for the analyses of sequence-function relationship of other proteins in the future.
Materials and Methods
Cloning.
The genes LBCA-trpA und LBCA-trpB were previously cloned into pET21a(+) using NdeI/XhoI restriction sites for expression with a C-terminal His6-tag (14). The genes anc1nc-trpA, anc1nc-trpB, anc2nc-trpA, anc2nc-trpB, anc2nc-trpB_AM10, anc3nc-trpA, anc3nc-trpB, anc3nc-trpB_AM10, anc4nc-trpA, anc4nc-trpB, anc5nc-trpA, anc5nc-trpB, anc6nc-trpA, anc6nc-trpB, nctrpA (GenBank accession no. EAR61762.1), and nctrpB (GenBank accession no. EAR61761.1) were codon-optimized for expression in E. coli and synthesized by Thermo Fisher Scientific (GeneArt Strings DNA Fragments). Anc1nc-trpA, anc2nc-trpA, anc3nc-trpA, anc4nc-trpA, and nctrpB were cloned into pUR28_BsaI using BsaI restriction sites (27) for expression with a N-terminal His6-tag. Anc1nc-trpB, anc2nc-trpB, anc2nc-trpB_AM10, anc3nc-trpB, anc3nc-trpB_AM10, anc4nc-trpB, anc5nc-trpB, anc6nc-trpB, anc3nc-trpB_AM4, and LBCA-trpB_AM4 were cloned into pET21a_BsaI using BsaI restriction sites for expression with a C-terminal His6-tag. Anc5nc-trpA, anc6nc-trpA, and nctrpA were cloned into pMAL-c5T_BsaI using BsaI restriction sites for expression with a N-terminal His6-maltose binding protein/MBP tag.
Site-Directed Mutagenesis.
The variants studied in this work were generated starting from anc2nc-trpB_AM10 using QuikChange site-directed mutagenesis (28) or the method according to the NEB Q5 site-directed mutagenesis kit (PNK). The templates, primer pairs, and the methods for the generation of all variants are listed in SI Appendix, Table S1. For all constructs, Sanger sequencing was applied to confirm the desired sequence.
Gene Expression and Protein Purification.
E. coli BL21 (DE3) Gold cells were transformed with respective plasmids. The cells were grown in lysogeny broth medium supplemented with 150 mg/mL ampicillin in case of pET21 and pMAL-c5T plasmids or 75 mg/mL kanamycin in case of pUR28 plasmids; 20 mM potassium phosphate buffer (pH 7.5) was added in case of trpA constructs; and 20 µM PLP was added in case of trpB constructs. At a cell density of OD600 = 0.6, protein expression was induced by addition of 0.5 mM isopropyl-β-thiogalactopyranoside. After growth overnight at 20 °C, the cells were harvested by centrifugation (Beckman Coulter Avanti J-26S XP, JLA-8.1, 15 min, 4,000 rpm, 4 °C). Cell pellets were suspended in 100 mM potassium phosphate buffer (pH 7.5), 300 mM KCl, and 10 mM imidazole for purification of TrpA proteins or 50 mM potassium phosphate buffer (pH 7.5), 300 mM KCl, 10 mM imidazole, and 40 µM PLP for purification of TrpB proteins. The cells were disrupted by sonification (Branson Sonifier W-250D, amplitude 50%, 2 × 2 min, 2 s pulse/2 s pause). The insoluble cell fraction was removed by centrifugation (Beckman Coulter Avanti J-26S XP, JA-25.50, 45 min, 14,000 rpm, 4 °C). The soluble fractions of Anc1nc-TrpA, Anc1nc-TrpB, Anc4nc-TrpA, and Anc4nc-TrpB underwent heat treatment (75 °C for 20 min), denatured proteins were removed by centrifugation (Beckman Coulter Avanti J-26S XP, JA-25.50, 45 min, 14,000 rpm, 4 °C). The supernatants were subjected to purification by immobilized metal ion affinity chromatography (GE Healthcare, HisTrap FF Crude). The proteins of interest were eluted in 100 mM potassium phosphate buffer (pH 7.5) in case of TrpA proteins or 50 mM potassium phosphate buffer (pH 7.5) in case of TrpB proteins, with 300 mM KCl and a linear gradient of 10–1,000 mM imidazole. The fractions with enriched protein of interest were pooled and subjected to preparative size-exclusion chromatography (GE Healthcare, HiLoad Superdex 75-pg column for TrpA proteins or GE Healthcare, HiLoad Superdex 200-pg column for TrpB proteins). The proteins were eluted in 100 mM potassium phosphate (pH 7.5) in case of TrpA proteins or in 50 mM potassium phosphate (pH 7.5) in case of TrpB proteins, with 300 mM KCl. The soluble fractions of MBP-Anc5nc-TrpA, MBP-Anc6nc-TrpA, and MBP-ncTrpA were subjected to purification by immobilized metal ion affinity chromatography (GE Healthcare, HisTrap FF Crude). The fusion proteins consisting of MBP and the proteins of interest were eluted in 100 mM potassium phosphate buffer (pH 7.5) with 300 mM KCl and a linear gradient of 10–500 mM imidazole. The fractions with enriched fusion protein were pooled, and 5 U/mL thrombin was added. For protease cleavage, the solution was subjected to dialysis against 100 mM potassium phosphate buffer (pH 7.5) with 300 mM KCl overnight at 4 °C. After dialysis the protein solution was subjected to immobilized metal ion affinity chromatography (GE Healthcare, HisTrap FF Crude). The flow-through fraction contained the proteins of interest, was collected, and subjected to preparative size-exclusion chromatography (GE Healthcare, HiLoad Superdex 75-pg column). The proteins were eluted in 100 mM potassium phosphate (pH 7.5). The fractions containing purified protein were pooled and used for further experiments. Protein concentrations in case of TrpA proteins were determined by absorbance spectroscopy at 280 nm (Thermo Scientific, NanoDrop One) using molar extinction coefficients that were calculated from the amino acid sequence (29), and by using a commercial Bradford reagent (Bio-Rad, Bradford protein assay) for TrpB proteins.
HPLC Assay for Recording of TrpB Activity.
The conversion of l-serine and indole to l-tryptophan by TrpB in the presence or absence of TrpA was observed by a discontinuous assay. The reactions contained 0.5 µM TrpB and 0 or 1 µM TrpA, 45 mM l-serine, 100 mM EPPS/KOH (pH 7.5), 180 mM KCl, and 40 µM PLP. The reactions were initiated by addition of 300 µM indole and incubated for 2 min at 30 °C. Reactions were stopped by the addition of 0.5 volume of potassium hydroxide solution and 1 volume of methanol. The reaction products were subsequently analyzed by reversed-phase HPLC using an Agilent system (1200 series) with a C18 column [Phenomenex, Luna 5 µm C18(2) 100 Å, 150 × 3 mm]. The separation was performed according to ref. 14. The elution time of l-tryptophan was 16.86 min. l-tryptophan was quantified according to a calibration curve with different concentrations of l-tryptophan detected by fluorescence (λex = 278 nm, λem = 350 nm).
Steady-State Enzyme Kinetics of TrpB Enzymes and Activity Titrations by Detection of TrpB Activity.
The conversion of l-serine and indole to l-tryptophan by TrpB was directly observed at 30 °C by absorbance spectroscopy (V-650 spectrophotometer, JASCO) using Δε290(tryptophan-indole) = 1.89 mM−1cm−1 (30). Initial velocities were recorded in 100 mM potassium phosphate (pH 7.5), 180 mM KCl, and 40 µM PLP with saturating concentrations of l-serine or indole. For steady-state kinetics, the initial velocities were plotted as a function of the l-serine or indole concentration and fitting a hyperbolic function to the obtained data points (SigmaPlot version 12.5). For activity titration experiments, the initial velocities were plotted as a function of the added TrpA concentration. The resulting data points were fitted with a hyperbolic function with an offset for the TrpB activity in absence of TrpA.
Analytical Size-Exclusion Chromatography.
Analytical size-exclusion chromatography was performed at room temperature with a chromatography system (GE Healthcare, ÄKTAmicro) coupled with an autosampler (Spark Holland, Alias GE Bio Cool), providing a cooled sample compartment at 4 °C. Proteins were eluted from an analytical size exclusion column (GE Healthcare, Superdex S200, 10/300 GL) in 50 mM potassium phosphate (pH 7.5) and 300 mM KCl at a flow rate of 0.3 mL/min. Elution was followed by an absorbance detector at 280 nm.
Absorbance Spectroscopy.
The formation of TrpB-reaction intermediates was recorded by absorbance spectroscopy with 10 µM (monomer concentration) of the respective TrpB and TrpA subunits in 50 mM potassium phosphate buffer (pH 7.5) in absence or presence of 20 mM l-serine (V-650 spectrophotometer, JASCO).
Ancestral Sequence Reconstruction.
The ancestral sequences were deduced from a previously compiled dataset (14) consisting of concatenated TrpA and TrpB sequences and the corresponding phylogenetic tree (SI Appendix, Fig. S5). TS underwent a complicated evolutionary history, indicated by the fact that the crenarcheal TS differs from the bacterial and euryarcheal TS, both with respect to sequence (31) and complex structure (32). Most likely, the TS from bacteria and euryarchaeota share a common evolutionary history (31). This is why we concentrated on bacterial and euryarcheal sequences, and our midpoint rooting reflects the most plausible history of bacterial and euryarcheal TS sequences.
This tree and the recent full-length sequences were the basis for a marginal sequence reconstruction by means of FastML (33). The chosen substitution model was JTT, and the method for indel reconstruction was “maximum likelihood.” The options “Optimize Branch Lengths” and “Use Gamma Distribution” were selected as well as the option “Probability cutoff to prefer indel over character” with the default value of 0.5. For each internal node of the tree used for ASR, FastML reconstructs 2 sequences without or with indels. The latter ones were taken for the experimental validation after the manual curation of their N and C termini. The chosen FastML parameters tend to generate shortened sequences; to correct their length, the ancestral sequences resulting from the marginal reconstruction with indels were aligned to the recent sequences, and the lacking N- and C-terminal residues were taken from the corresponding FastML sequences reconstructed without indels. The resulting sequences and the posteriori probabilities of all residues are listed in the Dataset S1.
Homology Modeling.
To compute a 3D structure of Anc2ncTS, the homology modeling module of YASARA (version 17.4.17) (19) was used. In brief, the corresponding protocol consisted of the following steps: To begin with, 2 templates were added manually, namely LBCA-TS (Protein Data Bank [PDB] ID 5EY5) and stTS (PDB ID 2J9X). An additional 3 templates were identified by PSI-BLASTing (34) the target sequence against UniRef90. The resulting position-specific scoring matrices were then used to search the PDB, which resulted in the 3 templates with PDB ID 5EY5, 5KIN, and 5TCH. All 5 templates were ranked based on the alignment score and their structural quality assessed by means of WHAT_CHECK (35). Analogously, a profile was deduced. Target and template proteins were aligned based on the corresponding profiles and additional structural information contained in the templates. Then, initial side-chain rotamer configurations were determined by using dead-end elimination in the context of a repulsive function (36). YASARA optimized loops by testing a large number of different conformations and by fine-tuning the orientation of side chains based on electrostatic and knowledge-based interactions. Subsequently, for each of the models, the hydrogen bonding network was optimized and a high-resolution energy minimization was run to confirm the stability of each structure. Finally, YASARA determined model quality based on scores determined for each residue; the structural model had an acceptable Z-score of −0.253.
The homology model for Anc2nc-TS_AM4 was created by introducing the 4 Anc2nc-TS_AM4-specific mutations into Anc2nc-TS via PyMOL (20). For each substitution, the rotamer was chosen that induced the fewest steric clashes.
MD Simulations and Their Analysis.
MD simulations with the homology models of Anc2nc-TS and Anc2nc-TS_AM4 were performed with GROMACS (37) (version 2016.4) and the AMBER03 force field. The complex was placed in a rectangular water box; the simulation cell was at least 5 Å larger than the solute along each axis and the system was neutralized by adding NaCl ions. To prepare a production run, the solvated system was at first energy minimized by a maximum of 5,000 steps of steepest descent MD minimization followed by a 2-part equilibration phase (each lasted 50 ps) that began with an NVT simulation followed by an NPT simulation. The symbols N (number of particles), V (volume), T (temperature), and P (pressure) indicate the parameters that were kept constant during the simulation. Finally, the production run (NVT conditions, time step 1 fs) was started at 298 K and ran for 100 ns, generating a snapshot every 0.1 ns. During the simulation, the temperature was kept constant at 298 K by using a Berendsen thermostat; for details see GROMACS documentation. Thirty replicates were generated for each complex resulting in a total of 3 μs of simulation time each.
All snapshots were compared to crystal structures representing an open (O, PDB ID 5DVZ chain A), a partially closed (PC, PDB ID 6AMH chain A), and a closed (C, PDB ID 5VM5 chain D) conformation of the COMM domain (10). In all cases, the coordinates of residues 81–186 were superimposed and by means of the GROMACS rms function, the rmsd of the corresponding backbone atoms was determined. Each snapshot was assigned to 1 of the 3 conformations that gave the smallest rmsd value; the resulting distribution was normalized to 100%.
Community Analysis.
All replicates from the MD simulation were pooled, and a cross correlation matrix for Anc2nc-TrpB and Anc2nc-TrpB_AM4 based on the Cα trajectories was generated with the help of the VMD (38) plugin NetworkView as described in ref. 15. The content of this matrix specifies a network of nodes and edges and contains substructures (communities of nodes) that are more densely interconnected to each other than to the other nodes of the network. Per definition, residues belonging to the same community can communicate with each other through multiple routes. The structure of the communities was determined by means of the Girwan–Newman (39) algorithm implemented in NetworkView. Communities with less than 3 residues were discarded.
Data Availability.
Raw data of biochemical characterization and the sequences of all TrpB constructs can be found in SI Appendix and Dataset S1, respectively. Requests for materials should be directed to the corresponding author.
Supplementary Material
Acknowledgments
We thank Christiane Endres and Sonja Fuchs for excellent technical assistance and Silvia Osuna for commenting on our MD simulations. This work is part of the cumulative doctoral thesis of Michael Schupfner (https://epub.uni-regensburg.de/40708/1/Thesis.pdf).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912132117/-/DCSupplemental.
References
- 1.Lu S., Shen Q., Zhang J., Allosteric methods and their applications: Facilitating the discovery of allosteric drugs and the investigation of allosteric mechanisms. Acc. Chem. Res. 52, 492–500 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Wodak S. J., et al. , Allostery in its many disguises: From theory to applications. Structure 27, 566–578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang Z., Verkhivker G. M., Hu G., Integration of network models and evolutionary analysis into high-throughput modeling of protein dynamics and allosteric regulation: Theory, tools and applications. Brief. Bioinform. bbz029 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Lisi G. P., Loria J. P., Allostery in enzyme catalysis. Curr. Opin. Struct. Biol. 47, 123–130 (2017). [DOI] [PubMed] [Google Scholar]
- 5.List F., et al. , Catalysis uncoupling in a glutamine amidotransferase bienzyme by unblocking the glutaminase active site. Chem. Biol. 19, 1589–1599 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Semmelmann F., et al. , Mapping the allosteric communication network of aminodeoxychorismate synthase. J. Mol. Biol. 431, 2718–2728 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R., Three-dimensional structure of the tryptophan synthase α2β2 multienzyme complex from Salmonella typhimurium. J. Biol. Chem. 263, 17857–17871 (1988). [PubMed] [Google Scholar]
- 8.Miles E. W., Tryptophan synthase: A multienzyme complex with an intramolecular tunnel. Chem. Rec. 1, 140–151 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Buller A. R., et al. , Directed evolution of the tryptophan synthase β-subunit for stand-alone function recapitulates allosteric activation. Proc. Natl. Acad. Sci. U.S.A. 112, 14599–14604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buller A. R., et al. , Directed evolution mimics allosteric activation by stepwise tuning of the conformational ensemble. J. Am. Chem. Soc. 140, 7256–7266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn M. F., Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Arch. Biochem. Biophys. 519, 154–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochberg G. K. A., Thornton J. W., Reconstructing ancient proteins to understand the causes of structure and function. Annu. Rev. Biophys. 46, 247–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merkl R., Sterner R., Ancestral protein reconstruction: Techniques and applications. Biol. Chem. 397, 1–21 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Busch F., et al. , Ancestral tryptophan synthase reveals functional sophistication of primordial enzyme complexes. Cell Chem. Biol. 23, 709–715 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Sethi A., Eargle J., Black A. A., Luthey-Schulten Z., Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. U.S.A. 106, 6620–6625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miles E. W., The tryptophan synthase α2β2 complex: A model for substrate channeling, allosteric communication, and pyridoxal phosphate catalysis. J. Biol. Chem. 288, 10084–10091 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider T. R., et al. , Loop closure and intersubunit communication in tryptophan synthase. Biochemistry 37, 5394–5406 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Schiaretti F., Bettati S., Viappiani C., Mozzarelli A., pH dependence of tryptophan synthase catalytic mechanism: I. The first stage, the beta-elimination reaction. J. Biol. Chem. 279, 29572–29582 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Krieger E., et al. , Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77 (suppl. 9), 114–122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrödinger, LLC , The PyMOL molecular graphics system (Version 1.7, 2015). https://www.schrodinger.com/pymol. Accessed 19 July 2018.
- 21.Maria-Solano M. A., Iglesias-Fernández J., Osuna S., Deciphering the allosterically driven conformational ensemble in tryptophan synthase evolution. J. Am. Chem. Soc. 141, 13049–13056 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Vanwart A. T., Eargle J., Luthey-Schulten Z., Amaro R. E., Exploring residue component contributions to dynamical network models of allostery. J. Chem. Theory Comput. 8, 2949–2961 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao X. Q., Momin M., Hamelberg D., Establishing a framework of using residue-residue interactions in protein difference network analysis. J. Chem. Inf. Model. 59, 3222–3228 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Starr T. N., Thornton J. W., Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holinski A., Heyn K., Merkl R., Sterner R., Combining ancestral sequence reconstruction with protein design to identify an interface hotspot in a key metabolic enzyme complex. Proteins 85, 312–321 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Siddiq M. A., Hochberg G. K., Thornton J. W., Evolution of protein specificity: Insights from ancestral protein reconstruction. Curr. Opin. Struct. Biol. 47, 113–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohweder B., Semmelmann F., Endres C., Sterner R., Standardized cloning vectors for protein production and generation of large gene libraries in Escherichia coli. Biotechniques 64, 24–26 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Wang W., Malcolm B. A., Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques 26, 680–682 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Gasteiger E., et al. , “Protein identification and analysis tools on the ExPASy Server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Humana Press, Totowa, NJ, 2005), pp. 571–607. [Google Scholar]
- 30.Faeder E. J., Hammes G. G., Kinetic studies of tryptophan synthetase. Interaction of substrates with the B subunit. Biochemistry 9, 4043–4049 (1970). [DOI] [PubMed] [Google Scholar]
- 31.Merkl R., Modelling the evolution of the archeal tryptophan synthase. BMC Evol. Biol. 7, 59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming J. R., et al. , Evolutionary morphing of tryptophan synthase: Functional mechanisms for the enzymatic channeling of indole. J. Mol. Biol. 430, 5066–5079 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Ashkenazy H., et al. , FastML: A web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hooft R. W. W., Vriend G., Sander C., Abola E. E., Errors in protein structures. Nature 381, 272 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Canutescu A. A., Shelenkov A. A., Dunbrack R. L. Jr, A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12, 2001–2014 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berendsen H. J. C., Vanderspoel D., Vandrunen R., Gromacs–A message-passing parallel molecular-dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995). [Google Scholar]
- 38.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graphics 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Girvan M., Newman M. E., Community structure in social and biological networks. Proc. Natl. Acad. Sci. U.S.A. 99, 7821–7826 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of biochemical characterization and the sequences of all TrpB constructs can be found in SI Appendix and Dataset S1, respectively. Requests for materials should be directed to the corresponding author.