Significance
Small cell carcinoma of the bladder (SCCB) is a lethal variant of bladder cancer with no effective treatment. A lack of available preclinical models and clinical cohorts impedes our understanding of its molecular pathogenesis. In this study, we provided a tumor model as functional evidence showing that SCCB and other bladder cancer phenotypes can be derived from normal human urothelial cells. We further demonstrated that SCCB has a distinct transcriptome and identified SCCB-associated cell surface proteins (CSPs) that can be further evaluated as potential therapeutic targets. We show that our model shares CSP profile with clinical SCCB samples. Our findings create a foundation to understand the molecular underpinnings of SCCB and provide tools for developing therapeutic strategies.
Keywords: cancer phenotypes, cell surface protein, urothelial cell, preclinical model
Abstract
Small cell carcinoma of the bladder (SCCB) is a rare and lethal phenotype of bladder cancer. The pathogenesis and molecular features are unknown. Here, we established a genetically engineered SCCB model and a cohort of patient SCCB and urothelial carcinoma samples to characterize molecular similarities and differences between bladder cancer phenotypes. We demonstrate that SCCB shares a urothelial origin with other bladder cancer phenotypes by showing that urothelial cells driven by a set of defined oncogenic factors give rise to a mixture of tumor phenotypes, including small cell carcinoma, urothelial carcinoma, and squamous cell carcinoma. Tumor-derived single-cell clones also give rise to both SCCB and urothelial carcinoma in xenografts. Despite this shared urothelial origin, clinical SCCB samples have a distinct transcriptional profile and a unique transcriptional regulatory network. Using the transcriptional profile from our cohort, we identified cell surface proteins (CSPs) associated with the SCCB phenotype. We found that the majority of SCCB samples have PD-L1 expression in both tumor cells and tumor-infiltrating lymphocytes, suggesting that immune checkpoint inhibitors could be a treatment option for SCCB. We further demonstrate that our genetically engineered tumor model is a representative tool for investigating CSPs in SCCB by showing that it shares a similar a CSP profile with clinical samples and expresses SCCB–up-regulated CSPs at both the mRNA and protein levels. Our findings reveal distinct molecular features of SCCB and provide a transcriptional dataset and a preclinical model for further investigating SCCB biology.
Carcinoma of the urinary bladder affects more than 80,000 people in the United States annually. While urothelial carcinoma is the predominant phenotype of bladder cancer, histological variants, such as squamous, glandular, plasmacytoid, sacromatoid, micropapillary, and small cell carcinoma are also described in clinical bladder cancer samples (1). Small cell carcinoma of the bladder (SCCB) is rare and accounts for only 0.5% of bladder cancer cases (2). As one of the most aggressive types of bladder cancer, SCCB is usually diagnosed at a late stage with a dismal prognosis of 1.7 y overall median survival (3). Histologically, SCCB shows a small round tumor cells with pyknotic round to oval nuclei and high mitotic rate, frequent tumor necrosis, and the expression of neuroendocrine differentiation (NED) markers, including synaptophysin (SYP), chromogranin (CHGA), and enolase 2 (ENO2) (2). Current management of SCCB is derived from the clinical experience with small cell lung cancer and includes surgery, chemotherapy, and radiotherapy (2). However, the outcomes of these treatments in SCCB patients have not improved for more than 10 y (4). Immunotherapy in the form of Bacillus Calmette–Guerin has been used to treat nonmuscle invasive bladder cancer for decades (5). Recently, immune checkpoint inhibitors targeting PD-1 or PD-L1 have been approved for the treatment of metastatic bladder cancer (6). The benefit of these innovative treatments in SCCB patients is still unknown. Efforts have been made to identify potential immune-therapeutic targets, such as DLL3 in SCCB (7). A better understanding of the distinguishing biology of SCCB is needed to guide the optimal clinical management and identify potential therapeutic targets for this aggressive disease.
Bladder cancer histological phenotypes have diverse clinical manifestations. The 5-y survival rate for in situ urothelial carcinoma is 95.7% and is 35.2% when tumors spread to regional lymph node (8), whereas for SCCB it is only 21.8% (9). In clinical samples, SCCB is found frequently in combination with other bladder cancer phenotypes (10). A recent genetic study comparing genetic alterations in small-cell lung cancer and SCCB suggests that SCCB originates from urothelial cells (11). However, the mechanisms underlying its development are largely unknown. Bladder cancer subtypes defined by gene-expression profiles are associated with different histological features, treatment responses, and distinct patient outcomes (12–14). Understanding the pathogenesis and molecular differences between SCCB and other bladder cancer histological phenotypes may serve an entry point for studying their diverse clinical consequences.
A lack of tumor models and patient samples limits our ability to study the pathogenesis and molecular features of SCCB. SCCB tumors can be generated using patient-derived xenograft models (7). However, the establishment of a patient-derived xenograft model relies on clinical SCCB samples and thus cannot provide enough biological replicates partly due to the rarity of SCCB cases (15). Genetically engineering noncancerous cells into subtype-specific tumors is an alternative strategy to establish tumor models (16). A recent study successfully initiated small cell carcinoma in prostate and lung epithelial cells using a set of defined genetic factors and established small cell carcinoma cell lines from different tissues of origin (17). Applying this strategy could provide novel SCCB models. There is also an unmet need for establishing larger clinical cohorts with SCCB samples that can be used for genomic and transcriptomic analyses. Given the rarity of fresh SCCB samples, identifying SCCB samples in previously archived formalin-fixed paraffin-embedded (FFPE) tissues could be a valuable resource.
In the present study, we establish a genetically defined SCCB model and a new cohort of clinical muscle-invasive bladder cancer (MIBC) samples with SCCB or non-SCCB histologies to characterize SCCB. Using these tools, we show that SCCB shares a urothelial origin with non-SCCB phenotypes but has a distinctive transcriptome and a unique cell surface protein (CSP) profile. We further demonstrate our tumor model as a representative tool for investigating CSPs in SCCB.
Results
SCCB and Other Bladder Cancer Phenotypes Can Be Initiated from Urothelial Cells by Defined Oncogenic Factors.
SCCB is histologically indistinguishable from other small cell carcinomas (11). This suggests shared pathogenesis among small cell carcinomas from different tissues. Therefore, we used an epithelial transformation system that successfully induced small cell carcinoma from prostate and lung epithelial cells to recapitulate the development of SCCB (17). In this system, a set of defined genetic factors initiated tumors in epithelial cells. These factors are composed of a dominant-negative form of TP53 (TP53-DN), myristoylated AKT1 (myr-AKT1), RB1 short-hairpin RNA, C-MYC, and BCL2 (termed PARCB). Genetic alterations mimicked by PARCB factors are relevant to bladder cancer. Mutations in TP53 and loss of RB1 are frequently found in SCCB samples (11, 18). Chromosome deletion at 10q and 13q that carrying PTEN (10q23) and RB1 (13q14) are common in SCCB (19). High-level amplifications are found at 8q24 in SCCB samples. This locus harbors MYC (20). A recent mutation study showed that mutations on the PTEN, RB1, TERT, promoter, and TP53 can present concurrently in clinical SCCB samples (11). BCL2 overexpression is associated with bladder cancer progression (21, 22). Thus, we hypothesized that urothelial cells (the epithelial cells in urinary tract) can be driven to SCCB by PARCB.
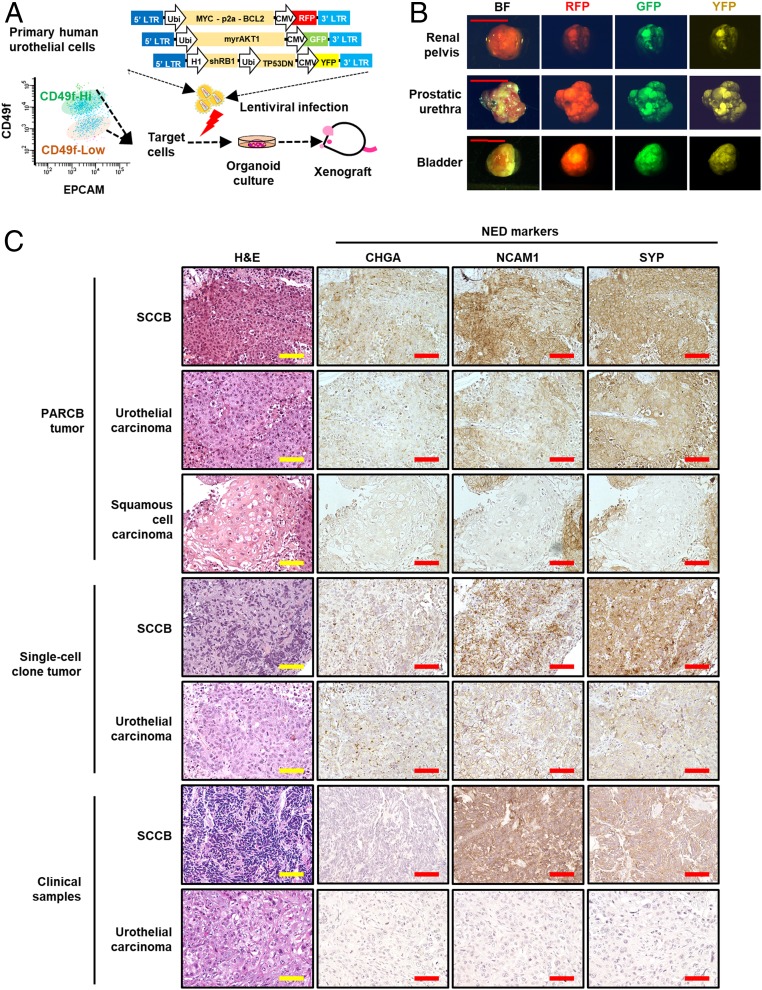
To address our hypothesis, we isolated benign epithelial cells from the renal pelvis, prostatic urethra, and bladder using an epithelial marker, EPCAM, and a basal urothelial marker, CD49f (23, 24). We found that primary human urothelial cells can be divided into 2 populations (CD49f high and CD49f low) (Fig. 1A). PARCB-transduced EPCAM+/CD49f high urothelial cells formed spherical structures in the organoid culture step of our transformation assay while the EPCAM+/CD49f low population did not (SI Appendix, Fig. S1). PARCB-transduced EPCAM+/CD49f high urothelial cells from renal pelvis and prostatic urethra, but not from bladder, form tumors in immune-deficient mice (NOD.CgPrkdcscid Il2rgtm1Wjl/SzJ [NSG] mice) over 3 to 4 mo after subcutaneous in vivo grafting (Fig. 1 B, Top and Middle). PARCB-transduced EPCAM+/CD49f low urothelial cells or control vector-infected cells did not initiate tumors. We also found that a combination of myr-AKT, c-Myc, and Bcl-2 did not initiated tumors in urothelial cells in multiple attempts, suggesting the essentiality of losing TP53 and RB1 functions in transforming urothelial cells. To further confirm whether bladder-derived epithelial cells can be transformed by the PARCB factors, we used commercially available primary bladder epithelial cells that are EPCAM+ and CD49f+ (SI Appendix, Fig. S2) for the PARCB transformation assay. These epithelial cells also generated tumors in NSG mice after the transduction of the PARCB factors (Fig. 1 B, Bottom). We noticed that the urothelial cells from prostatic urethra may be contaminated by prostate basal cells that also express CD49f and EPCAM. However, the tumors from renal pelvis and bladder urothelial cells are free of prostate contamination and have urothelial origin. Since these tumors expressed the PARCB factors (SI Appendix, Fig. S3), they are referred to as PARCB tumors.
Fig. 1.
Tumors derived from PARCB-transduced EPCAM+/CD49f high urothelial cells recapitulate divergent bladder cancer phenotypes. PARCB induce divergent bladder cancer phenotypes in normal human urothelial cells. (A) Schematic of human urothelial cells transformation assay with isolated EpCAM+/CD49f high cells and 3 lentiviruses delivering 5 genetic factors (PARCB). The urothelial cells were isolated from human urinary tract using enzymatic digestion and were sorted using flow cytometry. The isolated cells were infected with lentiviruses contain the 5 genetic factors and were cultured in organoid format followed by subcutaneous injection in NSG mice. CMV, cytomegalovirus promoter; GFP: green fluorescent protein; LTR, long-terminal repeats; RFP, red fluorescent protein; Ubi, ubiquitin promoter; YFP, yellow fluorescent protein. (B) Representative images of tumors formed by PARCB-transduced urothelial cells from different sites of human urinary tract. PARCB tumors were harvested from subcutaneous xenograft in NSG mice. Bright-field (BF) and fluorescent pictures were taken to show the tumor size and the expression of fluorescent markers. (Scale bars, 1 cm.) (C) Representative of H&E and IHC images using antibodies against NED markers CHGA, NCAM1, and SYP, in different areas of a PARCB tumor, a single-cell clone-derived tumor, and non-SCCB and SCCB clinical samples. IHC were performed using antibodies targeting NED markers on these samples. (Scale bars, 100 μm.)
PARCB tumors recapitulated clinical SCCB features, such as NED and high cell proliferation and apoptotic rates (2, 25), as shown by the expression of NED (CHGA, neural cell adhesion molecule 1 [NCAM1], and SYP) (Fig. 1C), cell proliferation (KI67), and apoptosis markers (cleaved CASPASE3) (SI Appendix, Fig. S4). Notably, we found that PARCB tumors exhibit diverse histological phenotypes, including small cell carcinoma, urothelial carcinoma, and squamous cell carcinoma (Fig. 1C and SI Appendix, Fig. S5). This is consistent with clinical observations that SCCB is frequently found in combination with other bladder cancer phenotypes (10). To further confirm these distinct histologies, we examined NED marker expression in different locations of PARCB tumors. Tumor lesions with SCCB histology expressed NED markers (Fig. 1C), whereas tumor lesions with non-SCCB histologies were weakly positive or negative for NED markers (Fig. 1C). This showed that PARCB tumors can display histological and molecular features of different bladder cancer phenotypes.
The mixed phenotypes of PARCB tumors may be derived from divergent expansion of a single-cell clone or from multiple cell clones that harbor different genetic alterations. To address this question, we established cell lines from PARCB tumors to perform single-cell clonal analyses. From trials of multiple PARCB tumors, we were able to generate 2 tumor cell lines (named bladder-PARCB1 and bladder-PARCB2), which continued to grow in 2D culture conditions (SI Appendix, Fig. S6A). These cell lines maintained clinical SCCB features including the expression of NED markers and were tumorigenic in NSG mice and (SI Appendix, Fig. S6 B and C). They also showed metastatic potential to lung and bone (SI Appendix, Fig. S7). We established single-cell clones from the bladder-PARCB1 cell line and show that these single-cell clones can form tumors in NSG mice (Fig. 1C). Both small cell carcinoma and urothelial carcinoma phenotypes can be found in an individual tumor derived from a single-cell clone (Fig. 1C). Therefore, both SCCB and urothelial carcinoma can be derived from the same single-cell clone.
A New Cohort of MIBC Samples with SCCB or Non-SCCB Phenotypes Was Established Using FFPE Samples.
Since our PARCB model demonstrated that SCCB shared similar genetic alterations and cells of origin with other bladder cancer phenotypes, we proposed that a distinct transcriptional profile of SCCB leads to its unique histology. The rarity of SCCB samples limits the availability of clinical cohorts that can be used to compare SCCB with other bladder cancer phenotypes. Therefore, we established a bladder cancer clinical cohort using archival FFPE samples from patients previously diagnosed with bladder cancer stage T2 or higher (indicating MIBC). The samples were reviewed by experienced pathologists and categorized into SCCB and non-SCCB groups based on their histological features. SCCB samples displayed typical small cell carcinoma histology features, such as small round cell shape, large nuclear to cytoplasm ratio, and pyknotic round to oval nuclei (SI Appendix, Fig. S8A), as well as the expression of NED markers (CHGA, ENO2, and SYP) (SI Appendix, Fig. S8B). Unlike SCCB, non-SCCB samples exhibited urothelial carcinoma phenotype, including infiltrative tumor cells in the muscle layer (SI Appendix, Fig. S8A) (26). In total, our cohort, which we refer to as University of California, Los Angeles–Bladder Cancer (UCLA-BLCA), contained 9 SCCB samples and 10 non-SCCB samples (SI Appendix, Fig. S8C).
SCCB Has a Distinct Transcriptional Profile and Transcriptional Regulatory Network from Non-SCCB Samples.
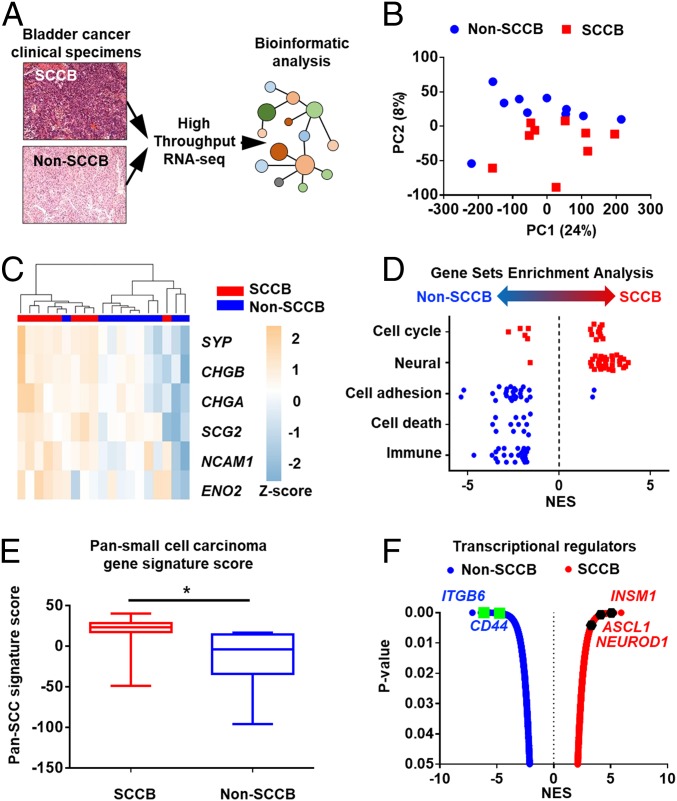
To characterize transcriptional similarities and differences between SCCB and non-SCCB samples, we performed high-throughput mRNA-sequencing and comprehensive bioinformatics analyses on the UCLA-BLCA cohort (Fig. 2A) (27). We found that SCCB and non-SCCB samples clustered into different groups that suggested fundamental differences in their transcriptomes (Fig. 2B). We sought to characterize these differences using various bioinformatics analyses. Since SCCB and non-SCCB samples have different histologies, we investigated whether SCCB can be discriminated from non-SCCB samples by gene sets that associated with SCCB histological features. Indeed, we found that a previously established bladder cancer NED gene signature (12) separates SCCB from non-SCCB samples in our cohort (Fig. 2C). We then performed gene set enrichment analysis (GSEA) to further identify functional gene sets that are associated with SCCB or non-SCCB phenotypes. SCCB samples were more enriched in cell-cycle and neural-associated gene sets, while non-SCCB samples were more enriched in cell death, cell adhesion, and immune-associated gene sets (Fig. 2D and Datasets S1 and S2). These differentially enriched gene sets in SCCB are consistent with its histological features including highly proliferative (19) (cell-cycle gene set) and NED (19) (neural gene sets). SCCB shares histological features with other small cell carcinomas, we thus tested whether transcriptional differences that distinguish small cell carcinoma in other tissues can also discriminate SCCB from non-SCCB samples. We applied a pan-small cell carcinoma gene signature that is derived from the comparison of small cell carcinomas from various tissues with their corresponding nonsmall cell carcinoma phenotypes (28). The SCCB samples had higher signature scores than non-SCCB samples (Fig. 2E and Dataset S3).
Fig. 2.
SCCB has a distinct transcriptional profile from non-SCCB phenotypes. Bioinformatics analyses revealed transcriptional differences between SCCB and non-SCCB samples in the UCLA-BLCA cohort. (A) Schematic of transcriptional analyses in the UCLA-BLCA cohort. FFPE samples were scratched and collected for RNA processing followed by high-throughput RNA-seq. (B) PCA performed using the TPM of mRNA expression of each sample from the UCLA-BLCA cohort. Figure shows that SCCB and non-SCCB samples are separated in this analysis. Each dot represents a sample with the corresponding phenotype (SCCB, red; non-SCCB, blue). (C) Unsupervised hierarchical clustering analysis using the TCGA neuroendocrine gene set separates SCCB from non-SCCB samples in the UCLA-BLCA cohort. Figure is shown by the z-score scaled by the TPM of each gene across samples. Each column represents a sample, each row represents a gene. Scale indicates the z-score. CHGB, chromogranin B; SCG2, Secretogranin II. (D) GSEA using differentially expressed gene profiles (normalized reads counts) identified gene sets that enriched in SCCB or non-SCCB samples. Each point represents a gene set associated with the category. Data are shown by normalized enrichment score (NES). Only gene sets that pass the P value filter <0.05 are shown. (E) Pan-small cell carcinoma gene-signature score of samples in the UCLA-BLCA cohort. SCCB samples has higher signature score than non-SCCB samples. Data are shown by the median and range of signature scores of all samples with corresponding phenotype (*P < 0.05, Student t test). (F) A plot shows the significance and enrichment score of transcriptional-regulators activated in corresponding phenotypes in the VIPER analysis. Each dot represents a master regulator gene that has a P < 0.05 in VIPER. NED regulators and cell-adhesion regulators are highlighted.
The distinctive transcriptional profile in SCCB suggests a unique gene-expression regulatory network. However, the gene-expression level of a transcriptional regulator is not always correlated with its activity (29). Thus, we performed VIPER (virtual inference of protein-activity by enriched regulon analysis) (30) on the UCLA-BLCA to identify phenotype-specific active transcriptional regulators. VIPER is an algorithm that infers the activities of proteins based on mRNA level of their activated or repressed genes. Therefore, VIPER provides information that cannot be uncovered by directly measuring gene-expression levels. VIPER analysis identified 968 transcriptional regulators with increased activity in SCCB samples and 602 regulators with increased activity in non-SCCB samples (Fig. 2F and Dataset S4). As we expected, NED regulators—such as ASCL1, NEUROD1, and INSM1—were more active in SCCB (Fig. 2F). These regulators are crucial for controlling transcriptional programs in small cell/neuroendocrine carcinomas from different tissues (31, 32). On the other hand, we identified a list of cell adhesion proteins, such as CD44 and ITGB6, with increased activity in non-SCCB samples (Fig. 2F) (33, 34). Altogether, these results showed that a substantial transcriptional program difference between SCCB and non-SCCB phenotypes reflects their histological features.
Profiling CSPs in UCLA-BLCA Cohort Predicted Brain-Expressed CSP Genes Associated with the SCCB Phenotype.
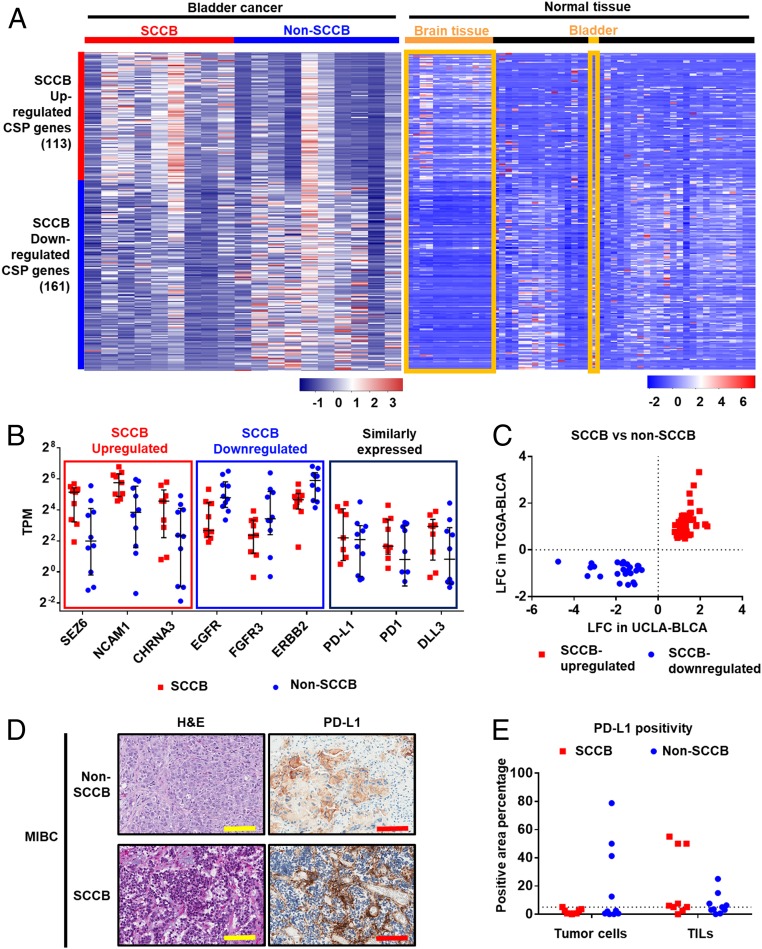
CSPs that are associated with specific cancer phenotypes can be used as diagnostic markers and therapeutic targets for immunotherapies (35, 36). Since the surfacesome in SCCB has never been profiled, we utilized our transcriptional dataset to predict CSPs that are associated with the SCCB phenotype. We identified 274 CSP genes that are differentially expressed in SCCB and non-SCCB samples (Fig. 3A and Datasets S5 and S6). Genes associated with neural functions—such as NCAM1, SEZ6, and CHRNA3 (37–39)—were up-regulated in SCCB samples. Conversely, CSP genes frequently altered in bladder cancers—such as ERBB2, FGFR3, and EGFR (40–42)—were down-regulated in SCCB samples (Fig. 3B). To examine the tissue specificity of these genes, we compared their mRNA expression in normal tissues using the Genotype-Tissue Expression (GTEx) database (43). The CSPs genes up-regulated in the SCCB phenotype were also highly expressed at the mRNA level in brain tissue compared to other tissues. The CSP genes down-regulated in the SCCB phenotype were highly expressed in the normal bladder tissue (Fig. 3A). This further supports that SCCB up-regulated CSPs are associated with neural functions. To test whether these differentially expressed CSPs have similar expression in other SCCB datasets, we compared our cohort with the The Cancer Genome Atlas (TCGA)-BLCA dataset (44), which has 3 SCCB and 409 non-SCCB samples. Among the 274 differentially expressed CSPs genes identified in the UCLA-BLCA cohort, 72 were also differentially expressed in the TCGA-BLCA dataset (P < 0.05 in DESeq2 analysis). All of these 72 genes were similarly up-regulated or down-regulated in SCCB samples in both datasets (Fig. 3C and Dataset S7). Therefore, these differentially expressed CSPs are associated with phenotypes and are conserved across independent datasets.
Fig. 3.
Profiling CSP genes identifies CSPs associated with SCCB and the expression of PD-L1 in SCCB samples. Transcriptional analyses profile CSPs genes in samples from the UCLA-BLCA cohort. (A) A heatmap shows CSP genes differentially expressed in SCCB and non-SCCB samples from the UCLA-BLCA cohort and their expression levels in normal tissues. Data are shown by the z-score normalized by genes across samples (Left) or tissues types (Right) based on the TPM of each sample (Left) or the median TPM of a tissue type (Right). Each row represents a gene in both panels. In the Left, each column represents a sample from the UCLA-BLCA cohort. In the Right, each column represents a tissue type from GTEx database. Brain tissues and bladder tissues are highlighted. Red color indicates higher expression level (higher z-score); blue color indicates lower expression level (lower z-score). Scale bar indicates the z-score. (B) mRNA expression level of selected CSP genes in each group. The SCCB up-regulated or SCCB down-regulated CSPs have P < 0.05 in the DESeq2 analysis (suggesting significantly up-regulated or down-regulated in SCCB samples comparing to non-SCCB samples.) The similarly expressed CSPs have a P value >0.05 in the DEseq2 analysis. The TPM of each sample is shown. Each dot represents a sample with corresponding phenotype. The median and interquartile range are shown. Samples with a TPM less than 0.125 is not shown in the figure. (C) A plot of LFC in UCLA-BLCA and TCGA datasets showing the LFC of CSP genes that are differentially expressed in both the UCLA-BLCA dataset and the TCGA dataset. Each dot represents a CSP gene. CSPs have been prefiltered for P < 0.05 in both UCLA-BLCA and TCGA-BLCA datasets in DESeq2 analysis comparing SCCB versus non-SCCB samples. (D) Representative images of H&E and IHC using a Food and Drug Administration-approved PD-L1 antibody (Ventana SP-142) showed PD-L1 expression in non-SCCB and SCCB sample. (Scale bars, 100 μm.) (E) Quantification analysis of PD-L1 IHC staining in the UCLA-BLCA cohort. Data are shown by the percentage of PD-L1 staining positivity in tumor cells and TILs in non-SCCB and SCCB samples. Each dot represents a sample of given phenotype. The dot line shows 5% positivity. No statistical significance is observed between SCCB and non-SCCB samples in tumor cell or TILs (Student t test).
Evaluation of Therapeutic CSPs Suggested PD-1/PD-L1 Targeted Therapies as Treatment Options for SCCB.
To explore whether phenotype-associated CSPs have been used or investigated as potential immunotherapeutic targets, we reviewed the CSPs profile of SCCB in a publically available drug database (the drug gene interaction database) (45). We found 20 SCCB-down-regulated CSPs have been used as therapeutic antigens. SCCB up-regulated CSPs were not as frequently investigated since only 2 of them (NCAM1 and CD247) have therapeutic antibodies (Table 1). CSPs that are widely discussed as therapeutic targets—such as MS4A1, IGF1R, and TNF—were similarly expressed in SCCB and non-SCCB samples (Table 1). CSPs that have been approved for bladder cancer treatments, such as PD-L1 and PD-1, or have been shown as potential therapeutic targets for SCCB, such as DLL3, were also similarly expressed in SCCB and non-SCCB samples (Fig. 3B). Since PD-L1 protein expression is a treatment marker for PD-1 and PD-L1–targeted therapies in bladder cancer (46), we evaluated PD-L1 expression using a Food and Drug Administration-approved antibody (Ventana SP-142, Roche) (47) in our cohort. We found that PD-L1 exhibits a similar prevalence and staining intensity in tumor cells and tumor-infiltrating lymphocytes (TILs) in both SCCB and non-SCCB samples (Fig. 3 D and E). Although PD-L1 positivity of tumor cell positivity is only 0.5 to 3.5% in SCCB samples, 7 of 9 SCCB samples had greater than 5% positivity in TILs (SI Appendix, Table S1). Since PD-L1 positivity in TILs is the dominant factor associated with treatment outcome in bladder cancer patients (47), we predict potential responses to anti–PD-L1 treatment in SCCB patients.
Table 1.
A list of phenotype-associated CSP genes as potential therapeutic targets
| Phenotype | Gene name | LFC (SCCB vs. non-SCCB) | No. of therapeutic antibodies |
| SCCB up-regulated CSP genes | CD247 | 1.16 | 2 |
| NCAM1 | 1.28 | 1 | |
| SCCB down-regulated CSP genes | EGFR | −2.37 | 18 |
| ERBB2 | −1.88 | 14 | |
| ERBB3 | −1.28 | 10 | |
| APP | −1.04 | 6 | |
| ITGB1 | −1.04 | 5 | |
| ITGAV | −1.14 | 4 | |
| ITGA5 | −1.99 | 3 | |
| KDR | −0.57 | 3 | |
| PDGFRA | −0.85 | 3 | |
| ICAM1 | −0.98 | 2 | |
| ITGA2 | −2.02 | 2 | |
| NOTCH2 | −0.82 | 2 | |
| TACSTD2 | −3.04 | 2 | |
| TPBG | −0.72 | 2 | |
| BSG | −0.88 | 1 | |
| CD74 | −2.08 | 1 | |
| GPNMB | −1.41 | 1 | |
| ITGB5 | −0.65 | 1 | |
| ITGB6 | −1.83 | 1 | |
| MUC1 | −1.89 | 1 | |
| Similarly expressed CSP genes* | MS4A1 | −0.06 | 9 |
| IGF1R | −0.72 | 8 | |
| TNF | 0.52 | 7 | |
| TNFRSF10B | −0.45 | 6 | |
| CD19 | 0.25 | 4 |
Top 5 ranked by number of therapeutic antibodies
The Bladder-PARCB1 Cell Line Shares Similar CSP Profile with Clinical SCCB Samples.
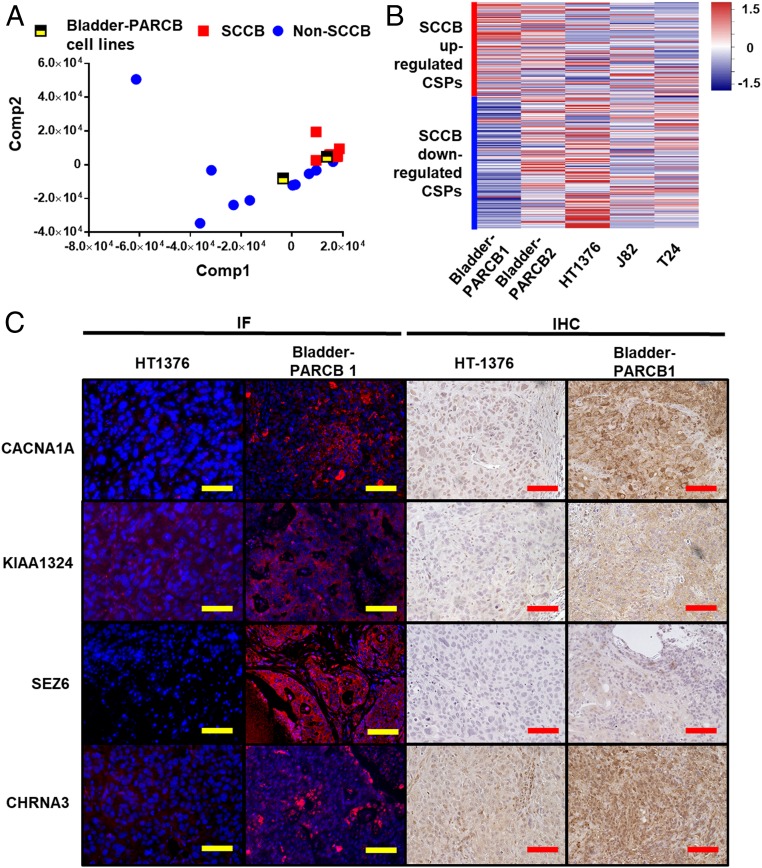
Cancer cell lines are useful tools for preclinical evaluation of CSPs as potential immunotherapy targets (48). To date, the bladder-PARCB cell lines are the only SCCB cell lines. We sought to evaluate whether they can be used as a representative tool for studying the SCCB surfacesome. We performed a partial least-square regression (PLSR), a supervised principal component analysis (PCA), by projecting CSP transcriptional profiles of bladder-PARCB cell lines to the UCLA-BLCA background separating SCCB and non-SCCB phenotypes. Interestingly, we found that the bladder-PARCB1 cell line clustered with SCCB samples while the bladder-PARCB2 cell line is closer to non-SCCB samples (Fig. 4A). We next compared the mRNA level of phenotype-associated CSP genes in the bladder-PARCB cell lines to those in urothelial carcinoma cell lines HT1376, J82, and T24. The bladder-PARCB1 cell line expresses higher levels of SCCB up-regulated CSP genes compared to urothelial carcinoma cell lines (Fig. 4B) (49). Thus, the bladder-PARCB1 cell line has a surfacesome more similar to SCCB than urothelial carcinoma cell lines do. We then validated the protein expression of SCCB up-regulated CSPs in tumors derived from the bladder-PARCB1 cell line and the HT1376 cell line using commercially available antibodies. Immunofluorescences (IF) and immunohistochemistry (IHC) showed that CSPs CHRNA3, CACNA1A, KIAA1324, and SEZ6 are expressed on the cell surface of tumors cells from the bladder-PARCB xenograft (Fig. 4C). We also identified higher expression levels in bladder-PARCB1 xenograft compared to in the HT1376 xenograft (Fig. 4C). To test whether the SCCB up-regulated CSPs are expressed in clinical SCCB samples, we performed IHC in our UCLA-BLCA cohort. We found that SCCB samples express NCAM1, while non-SCCB samples are negative or weakly positive for this marker; KIAA1324 expression is highly variable in both SCCB and non-SCCB samples; SEZ6 is expressed in a few tumor cells in SCCB samples, but is not expressed in non-SCCB samples (SI Appendix, Fig. S9). Our findings suggest that the PARCB model is a representative tool for investigating CSPs in the SCCB phenotype.
Fig. 4.
Bladder-PARCB cell lines share similar CSP profile with clinical SCCB samples. Bladder-PARCB cell lines express SCCB-associated CSPs at mRNA and protein levels. (A) PLSR projecting the CSP transcriptional profile of bladder-PARCB cell lines to the UCLA-BLCA cohort. The bladder-PARCB1 cell line is clustered with clinical SCCB samples. (B) Heatmap summarized the expression of phenotype-associated CSPs in bladder-PARCB cell lines and urothelial carcinoma cell lines NCI-HT1376, J82, and T24. Data are shown by the z-score normalized by genes across cell lines based on the TPM of genes in each cell line. Each row represents a gene. Red color indicates higher expression level (higher z-score); blue color indicates lower expression level (lower z-score). (C) Representative image of IF or IHC using antibody against CACNA1A, KIAA1324, CHRNA3, and SEZ6 in tumor derived from HT-1376 and PARCB cell lines. In the IF panel, blue color shows nuclear staining by DAPI, red color indicates positivity of CSPs. (Scale bars, 100 μm.)
Discussion
Current understanding of SCCB biology is largely limited by the lack of disease models and clinical cohorts. In the present study, we provided an integrated platform, including an SCCB disease model, SCCB cell lines, and a clinical cohort consisting of SCCB and non-SCCB bladder cancer patient samples to characterize this aggressive disease. Using the PARCB model, we demonstrated that both SCCB and non-SCCB phenotypes can be derived from urothelial cells with a defined genetic background, but can be distinguished by their transcriptomes. We further identified CSPs associated with the SCCB phenotype and showed that the PARCB model shared a CSP profile with clinical SCCB samples. Our platform improves our understanding of SCCB biology and provides a useful tool to further interrogate this disease.
SCCB is frequently observed alongside other bladder cancer phenotypes in clinical samples (10). However, whether SCCB originates from normal cells or is transdifferentiated from non-SCCB during cancer progression is not known. Using our PARCB model, we demonstrated that SCCB can be initiated from normal urothelial cells. However, we cannot rule out transdifferentiation as an alternative route of SCCB development. Small cell carcinoma can transdifferentiate from nonsmall cell carcinoma during anticancer treatments (50). For example, the neuroendocrine/small cell prostate cancer phenotypes can be induced by hormone-deprivation therapy (51). Tumors derived from bladder-PARCB single-cell clones show both small cell carcinoma and urothelial carcinoma phenotypes. This indicates that cells from 1 cancer phenotype can give rise to another phenotype, presumably through processes involving transdifferentiation. This is concordant with prior reports of therapy-induced transdifferentiation (52). Our model thus provides a resource for the investigation of this clinically impactful plasticity. The transdifferentiation of cancer cells can be dictated by epigenetic modification in neuroblastoma and nonsmall cell lung cancer (53, 54). Importantly, mutations on epigenetic modifiers KDM6A, ARID1A, CREBBP, EP300, and KMT2A/C/D are present in more than 70% of SCCB samples (11). We also found loss of epigenetic regulator EP300 mRNA in SCCB samples from the UCLA-BLCA cohort, suggesting dysregulation of the epigenetic network in SCCB. Emerging evidence shows that targeting the epigenetic regulation as a promising therapeutic strategy for blood, liver, and colorectal cancers (55). Therefore, our cell lines provided a useful tool to further investigate the epigenetic modification in dictating bladder cancer phenotypes and may lead to novel therapeutic strategies for aggressive bladder cancer.
The poor prognosis of SCCB indicates an unmet clinical need to develop new therapeutic strategies. Although PD-1/PD-L1–targeted immunotherapies (6) have been recently approved for metastatic bladder cancer, their efficacy in the subset of patients with SCCB remains unknown. Protein expression of PD-L1 in TILs predicts response to PD-1/PD-L1–targeted therapies in bladder cancer (46). We found that most SCCB samples had more than 5% PD-L1 positivity in TILs, suggesting a potential response to PD-1/PD-L1–targeted treatment. Moreover, PD-1/PD-L1–targeted therapies have antitumor effects in small cell lung cancer patients (56). Since small cell lung cancer and SCCB share histological features, such therapy may also have antitumor effects in SCCB patients. A recently initiated clinical trial (NCT03582475) in SCCB patients is evaluating pembrolizumab combined with chemotherapy. In addition, we nominated a list of CSPs that are associated with SCCB phenotype, including a set of CSPs that are highly expressed in brain tissues based on their mRNA expression. We showed that the expression of these CSPs are variable at both mRNA (Fig. 3A) and protein levels (SI Appendix, Fig. S9) in SCCB samples. Thus, further efforts are needed to comprehensively examine their protein expression levels to evaluate their potentials as therapeutic targets.
Bladder-PARCB cell lines are the only available human SCCB cell lines. They are representative cell models of SCCB that may grant us the ability to perform functional assays at the cellular level. We demonstrated that these cell lines recapitulate clinical SCCB features, such as the expression of NED markers as well as a similar metastatic behavior. This further extends the application of these cell lines to evaluate SCCB metastasis, which is the major cause of patient deaths (9). Notably, we showed that the bladder-PARCB1 cell line shares a CSP profile with clinical samples and express SCCB-associated CSPs. This will allow future preclinical works targeting these CSPs in this cell line. Moreover, our transcriptional analyses shown that there are differences in CSP profiles between bladder-PARCB1 and clinical SCCB samples. For example, GRIN2A and FAT3 are highly expressed in SCCB samples but is undetectable at the mRNA level in the bladder-PARCB1 cell line. This may be due to different genetic backgrounds of clinical SCCB samples and PARCB cell lines. Genetic alterations, such as E2F3 amplification and TERT promoter mutation, are frequently found in clinical SCCB samples (11). Although we observed up-regulation of TERT mRNA in bladder-PARCB1 cells compared to normal bladder epithelial cells, whether these genetic alterations may affect the CSP profile need to be further evaluated. We demonstrated that our epithelial transformation system is an efficient tool to test the effects of genetic events in tumorigenesis as well as tumor phenotype. Therefore, this system could be further used to explore the roles of other genetic alterations in the determination of CSP profile in urothelial cancers. Such study will contribute to identifying critical regulators of the SCCB surfacesome and developing novel targeted therapies.
Materials and Methods
Lentiviral Vector Cloning and Virus Packaging.
The PARCB factor vectors (FU-MYC-P2A-BCL2-CRW, FU-RB1-shRNA-TP53DN-CYW, and FU-myrAKT1-CGW) were cloned and lentiviruses were packaged by a previously described method (17). The FU-CRW backbone was used as a negative control (empty vector).
Isolation of Primary Urothelial Cells.
Donor tissues (including human urinary tract tissues and FFPE clinical samples) were provided in a de-identified manner and thus were exempt from Institutional Review Board approval. Surgical removed noncancerous human urinary tract tissues (renal pelvis, prostatic urethra, or bladder) were cut into 5-mm chunks and subjected to digestion (1 mg/mL type II collagenase [Fisher Scientific #17-101-015] and 1 mg/mL Dispase [Fisher Scientific, #17-105-041], with 1× Normalcin [Invivogen, #ant-nr-1] in DMEM with 10% FBS) at 37 °C for 12 h. The tissue was then filtered by a 40-µm cell strainer and the supernatant was collected. The cells in the supernatant were collected by centrifuge at 400 g for 5 min and were washed twice using PBS. The cell pellets were resuspended in DMEM with 10% FBS and cell number was counted.
FACS for CD49f+/EPCAM+ Cells.
Cells isolated from primary urothelial cells were stained using anti–CD49f-PE antibody (eBiosciences, #12-0495-82) and anti–EPCAM-APC (eBiosciences, #17-5791-82) antibody at a concentration of 5 μL per million cells. The EPCAM+/CD49f high and EPCAM+/CD49f low cell population was collected for epithelial transformation assay.
Epithelial Cell Transformation and Organoid Culturing.
The epithelial cell transformation and organoid method have been described in detail previously (17, 57). EPCAM+/CD49f high and EPCAM+/CD49f low primary urothelial cells, or primary bladder epithelial cells isolated from donors without urinary tract-related disease (American Type Culture Collection [ATCC], #PCS-420-010) were infected with lentivirus containing the PARCB factors and then mixed with Matrigel (Corning, #356234) to set up organoid culture. The organoids were cultured with bladder epithelial cell medium (ATCC, #PCS-440-030, #PCS-700-040) for 2 wk. Then, organoids were harvested by digesting with 1 mg/mL Dispase for 2 h. The organoids were washed 3 times with PBS to remove Dispase and resuspended in 30 µL Matrigel. The organoid–Matrigel mixture was implanted subcutaneously in immunodeficient NSG mice using a 27-gauge syringe. Xenografts were harvested 12 wk after implantation or when they reached 1-cm diameter, whichever came first.
Animal Protocol.
NSG mice were transferred from the Jackson Laboratories and housed and bred under the care of the Division of Laboratory Animal Medicine at UCLA. Subcutaneous injection of cells was performed according to protocols approved by UCLA’s Animal Research Committee.
Tissue Processing and Histology.
Tumor tissues were prefixed in 10% buffered formaldehyde for overnight at 4 °C, then moved into 70% ethanol for no less than 30 min. Tissues were then processed at the Translational Pathology Core Laboratory (TPCL), a College of American Pathologists/Clinical Laboratory Improvement Amendments-certified research facility in the UCLA Department of Pathology and Laboratory Medicine and a UCLA Jonsson Comprehensive Cancer Center Shared Facility. Paraffin-embedded tissues were sectioned into 4-µm sections and H&E staining was performed for every 5 sections. The histology was evaluated by experienced pathologists at UCLA.
Immunostaining, Immunoblotting, and Antibodies.
Immunostaining and immunoblotting were performed following methods that were previously described in detail (17). Antibodies targeting c-Myc (Abcam #ab32072, 1:500 dilution), p-AKT (Cell Signaling Technology, #9271, 1:500 dilution), BCL2 (Abcam, #ab32124, 1:500 dilution), P53 (Cell Signaling Technology, #2527, 1:500 dilution), RB (Abcam, #ab181616, 1:500 dilution), Ki67 (Abcam, #ab16667, 1:500 dilution), cleaved caspase 3 (Cell Signaling Technology, #9661, 1:500 dilution), GAPDH (Genetex, #GTX627408-01, 1:10,000 dilution), SYP (Santa Cruz, #sc-17750, 1:500 dilution for IHC, 1:2,000 dilution for immunoblotting), NCAM1/CD56 (Abcam, #ab75813, 1:500 dilution for IHC, 1:2,000 dilution for immunoblotting), and CHGA (Abcam, # ab15160, 1:100 dilution for IHC, 1:2,000 dilution for immunoblotting), KIAA1324 (Thermo Fisher, #PA5-72691, 1:200 dilution), SEZ6 (Thermo Fisher, #PA5-47683, 1:100 dilution), CHRNA3 (Thermo Fisher, #MA5-31685, 1:200 dilution) and CACNA1A (Thermo Fisher, #PA5-50634, 1:200 dilution) were used for immunostaining and immunoblotting.
For IHC, anti-mouse/rabbit biotinylated secondary antibodies were used to detect primary antibodies. ImmPACT DAB (Vector Laboratories, CAT# SK-4105) was used to visualize the staining.
For IF, Alexa 488 conjugated goat anti-mouse secondary antibody (Thermo Fisher A-11001) or Alexa 594 conjugated goat anti-rabbit secondary antibody (Thermo Fisher A-11012) were used to detect primary antibodies and to visualize the staining. A fluorescent microscope was used to capture the image and pseudo color were assigned using ImageJ. Brightness and contrast were adjusted using ImageJ to increase staining signal and reduce background. All pictures with the same staining were adjusted using the same parameters.
Cell Culture and the Establishment of Bladder-PARCB Cell Lines/Single-Cell Clones.
Primary bladder epithelial cells (ATCC, #PCS-420-010) and NCI-HT1376 cancer cell line were purchased from ATCC and maintained as recommended by ATCC.
The PARCB tumors from primary bladder epithelial cell lines (ATCC) were dissociated using 1 mg/mL type II collagenase (Fisher Scientific #17-101-015) and 1 mg/mL Dispase (Fisher Scientific, #17-105-041), with 1× Normalcin (Invivogen, #ant-nr-1) in DMEM with 10% FBS for 30 min. GFP+/RFP+/YFP+ cells were isolated using FACS and then cultured in stem cell culture medium (Advanced DMEM/F12 [Gibco #12634028] with 1× Glutamax [Gibco, #35050061], 1× B27 [Gibco, #17504044], 10 ng/mL human EGF [Peprotech #100-47], and 10 ng/mL human FGF-basic [Peprotech, #100-18B]). The cells were referred to as bladder-PARCB cell lines. The bladder-PARCB1 and PARCB2 cell lines have been cultured for more than 6 mo (1 or 2 passages per week) using stem cell culture medium in vitro. Both cell lines are propagated as organoids in tissue culture treated flasks. Single cells of cultured bladder-PARCB cell lines were dispensed into a 96-well plate using FACS (1 cell per plate). The plates were checked by microscopy to confirm the presence of only 1 cell in each well. Cells were cultured for 4 wk and the expanding cell clones were collected and cultured as single-cell clones. The single-cell clones were implanted subcutaneously into NSG mice to generate tumors.
Metastatic Assay of Bladder-PARCB Cells.
For metastatic assay, 5 × 106 bladder-PARCB1 cells in 200 μL PBS were injected into NSG mice through the tail-vein. The 18F-FDG PET scans were performed 16 wk after injection as described previously (58).
Collection of Clinical Samples.
FFPE urothelial cancer samples were collected from patients that had undergone surgery at the Ronald Reagan Medical Center (Los Angeles, CA). All samples were consented for and reviewed by certified genitourinary pathologists. The SCCB samples are defined by histology with typical small cell carcinoma histology including small round cell shape, large nuclear-to-cytoplasm ratio, and pyknotic round to oval nuclei. SCCB are further defined by >60% combined positivity of CHGA, ENO2, and SYP using IHC staining. Non-SCCB samples are defined by a muscle-invasive bladder cancer histology.
PD-L1 Staining and Scoring.
PD-L1 staining in the UCLA-BLCA cohort was performed using Ventana SP142 kit (Roche, #740-4859) following the manufacturer’s instruction. The slides were reviewed blindly and independently by 2 experienced pathologists. The discrepancies of results were resolved by re-evaluation of the slides. The prevalence of PD-L1 staining in tumor cells or TILs was shown as the percentage of positive area to total area, respectively. The staining intensity were scored from 1 to 3+. The intensity was then transformed to a quantitative scoring scale (intensity 1 = 1, 1+ = 1.5, 2 = 2, 2+ = 2.5, 3 = 3, 3+ = 3.5). A score range from A to B is then calculated as the average of A and B [i.e., 1 to 2+ would be calculated as (1 + 2.5)/2 = 1.75]. The percentages and intensity scores from the different pathologists were collected independently and the average value in each sample was calculated and shown.
Transcriptional Profiling of Urothelial Cancer Samples.
RNA was extracted from FFPE human urothelial cancer tissue using the Ambion RecoverAll kit (Thermofisher, #AM1975). RNA from cultured cells was extracted using Qiagen miRNeasy kit (Qiagen, #217004). Libraries were prepared using the Human FFPE RNA-seq kit (Nugen, #0341-32) according to the manufacture’s protocol. Libraries were pooled and sequenced at 150-bp paired-end using an Illumina HiSeq 3000. Raw RNA-seq files were mapped to the hg38 human genome using the TOIL pipeline (59), and transcripts were quantified using RNA-seq by expectation-maximization (RSEM). Differential expression analysis was performed using DESeq2 (60). Differential gene expression in SCCB and non-SCCB samples was identified by a P value cutoff of P < 0.05.
GSEA Analysis.
GSEA was performed using GSEA Desktop v3.0 (61, 62). The gene list from the DESeq2 analysis was filtered with a P value cutoff of P < 0.05. The genes were ranked using –log10(P value)*sign (log2 fold-change [LFC] SCCB vs. non-SCCB). GSEA was performed using the C5 all-gene ontology V6.2 dataset and preranked/classic setting. All positive- or negative-enriched gene sets (normalized P < 0.05) were ranked by normalized enrichment score. The key words related to a topic was determined by their frequencies in all gene sets followed by manual curation. Five categories including neuron (keywords: neuro, axon, synap), cell cycle (keywords: cell cycle, mitotic, mitosis, meiotic, epithelial cell proliferation, DNA replication, centrosome, spindle), cell death (keywords: cell death, apopto), cell adhesion (keywords: adhes, junction), and immune (keyword: immun, T cell, B cell) were analyzed. All gene sets that match any of these 5 categories were plotted.
Calculation of Pan-Small Cell Carcinoma Gene Signature Score.
The pan-small cell gene signature is derived from a previously published pan-small cell analysis (28). The genes were ranked by the PC1 in the varimax of PCA loading of RNA from normal lung, normal prostate, castration-resistance prostate cancer, neuroendocrine prostate cancer, lung adenocarcinoma, and small cell lung cancer clinical samples. The top 50 genes were used as the pan-small cell carcinoma gene signature to calculate the pan-small cell gene signature score in the UCLA-BLCA cohort. The log2 (normalized reads count) of each gene/sample was calculated. The z-score of each gene across all SCCB and non-SCCB samples were calculated. The sum of z-scores of the 50 genes was calculated in each sample and was used as the gene signature score for each sample. The gene-signature scores in non-SCCB samples were compared with those in SCCB samples by a Student t test and P < 0.05 is considered as statistically significant.
Identification of SCCB-specific CSP Genes.
CSP genes are determined by annotation of extracellular domain in Uniprot database. In total, 2,648 CSP genes were identified (Dataset S5). The CSP associated with a specific phenotype is determined by the P < 0.05 and LFC >0 (SCCB) or LFC < 0 (non-SCCB) in DESeq2 analysis. The RNA expression data of SCCB up-regulated or down-regulated CSP genes were transformed to transcripts per million (TPM) and then compared to tissue-expression profile from the GTEx dataset. Only genes that appear in both datasets are plotted. The GTEx data are obtained from the GTEx Portal on 29 July 2019. The RNA expression of urothelial carcinoma cell lines, NCI-HT1376, T24, and J82 were obtained from the Cancer Cell Line Encyclopedia database.
VIPER Analysis.
Aracne-AP, a computationally tractable version of the original Aracne framework, was used to reconstruct gene regulatory networks (63). The networks were built using the human neuroendocrine tumor interactome (regulonnet) and interactomes from TCGA RNA-seq datasets: BLCA, BRCA, CESC, COAD, ESCA, GMB, HNSC, KIRC, KIRP, LAML, LIHC, LUAD, LUSC, NET, OV, PAAD, PCPG, PRAD, READ, SARC, STAD, TGCT, THCA, THYM, and UCEC. Normalized enrichment scores were created using VIPER (30) with the SCCB to non-SCCB dichotomy, and using the built network as regulon. The P value of each regulator was calculated using a Student t test. P < 0.05 was considered as statistically significant.
PCA and PLSR Analysis.
PCA is an unsupervised learning approach that decomposes a matrix x into orthogonal principal components that maximize the variance explained in x. In our analysis data, the x matrix the RNA-seq expression matrix of the UCLA-BLCA datasets. The analysis was performed using the TPM of each sample.
PLSR is a supervised learning approach that decomposes a matrix x of predictors and dependent variables y into orthogonal components, while also maximizing the covariance between the 2 matrices. In our analysis data, the x matrix is the RNA-seq expression matrix of all CSP genes in the UCLA-BLCA dataset and the y matrix is a binary vector on the SCCB and non-SCCB samples classification. Projections onto this PLSR space were performed with the mRNA expression profile of the bladder-PARCB 1 and 2 cell lines. The analysis was performed using normalized counts from DESeq2 analysis.
RNA Expression Data of Urothelial Carcinoma Cell Lines.
The RNA expression of urothelial carcinoma cell lines, NCI-HT1376, T24, and J82 were obtained from the Cancer Cell Line Encyclopedia database (version CCLE_RNAseq_rsem_genes_tpm_20180929.txt.gz).
Statistics.
All statistical analyses were performed using Graphpad Prism 6. For the Student t test, P < 0.05 is considered as significant. For one-way ANOVA, the P value was subjected to Bonferroni correction.
Data Availability.
RNA-seq dataset was deposited at Gene Expression Omnibus (27), with accession number of GSE139822.
Supplementary Material
Acknowledgments
We thank the University of California, Los Angeles (UCLA) Tissue Procurement Core Laboratories for tissue preparation and the UCLA Technology Center for Genomics and Bioinformatics for performing RNA-seq analyses; the Cancer Cell Line Encyclopedia database for providing RNA-seq data of urothelial carcinoma cell lines (https://portals.broadinstitute.org/ccle); the GTEx supported by the Common Fund of the Office of the Director of the NIH, and by the National Cancer Institute (NCI), National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke for providing the normal tissue transcriptional profiles. The data used for the analyses described in this manuscript were obtained from the GTEx Portal (https://gtexportal.org/home/). The results shown in the manuscript are in whole or part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga). This work is supported by the following funding: A UCLA Broad Stem Cell Research Center postdoctoral fellowship (to L.W.); a Prostate Cancer Foundation Young Investigator award (to B.A.S.); NIH/NCI Grant K99/R00 Pathway to Independence Award K99CA218731 (to J.W.P.); a Gaba Broad Stem Cell Research Center Innovation Award (to A.I.C.); the Perkins Foundation (A.I.C.); NIH/NCI R01 Grant CA222877 (to T.G.G. and O.N.W.); UCLA Specialized Program of Research Excellence (SPORE) in Prostate Cancer NIH P50 CA092131 (to T.G.G. and O.N.W.); a grant from the Medical Research Grant Program of the W. M. Keck Foundation (to T.G.G. and O.N.W.); The Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research (T.G.G. and O.N.W.); and the Hal Gaba Fund for Prostate Cancer Research (T.G.G. and O.N.W.).
Footnotes
Competing interest statement: O.N.W. currently has consulting, equity, and/or board relationships with Trethera Corporation, Kronos Biosciences, Sofie Biosciences, and Allogene Therapeutics. T.G.G. currently has consulting and equity relationships with Trethera Corporation. The laboratory of T.G.G. has completed a research agreement with ImmunoActiva. None of these companies contributed to or directed any of the research reported in this article.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE139822).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915770117/-/DCSupplemental.
References
- 1.Sanli O., et al. , Bladder cancer. Nat. Rev. Dis. Primers 3, 17022 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Ghervan L., Zaharie A., Ene B., Elec F. I., Small-cell carcinoma of the urinary bladder: Where do we stand? Clujul Med. 90, 13–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choong N. W., Quevedo J. F., Kaur J. S., Small cell carcinoma of the urinary bladder. The Mayo Clinic experience. Cancer 103, 1172–1178 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Ghatalia P., Jung K., Litwin S., Bilusic M., Small cell carcinoma of the bladder: Comparison of survival in various treatment modalities at Fox Chase Cancer Center (FCCC) from 1995 to 2015. J. Clin. Oncol. 34, 465 (2016). [Google Scholar]
- 5.Morales A., Eidinger D., Bruce A. W., Intracavitary Bacillus Calmette-Guerin in the treatment of superficial bladder tumors. J. Urol. 116, 180–183 (1976). [DOI] [PubMed] [Google Scholar]
- 6.Ghatalia P., Zibelman M., Geynisman D. M., Plimack E., Approved checkpoint inhibitors in bladder cancer: Which drug should be used when? Ther. Adv. Med. Oncol. 10, 1758835918788310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koshkin V. S., et al. , Transcriptomic and protein analysis of small-cell bladder cancer (SCBC) identifies prognostic biomarkers and DLL3 as a relevant therapeutic target. Clin. Cancer Res. 25, 210–221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howlader N., et al., Eds., SEER cancer statistics review, 1975-2014. https://seer.cancer.gov/csr/1975_2014/. Accessed 19 September 2019.
- 9.Ries L. A. G., et al., Eds, “SEER Survival Monograph: Cancer Survival Among Adults: U.S. SEER Program, 1988-2001, Patient and Tumor Characteristics” (NIH Pub. No. 07-6215, National Cancer Institute, SEER Program, Bethesda, MD, (2007).
- 10.Abrahams N. A., Moran C., Reyes A. O., Siefker-Radtke A., Ayala A. G., Small cell carcinoma of the bladder: A contemporary clinicopathological study of 51 cases. Histopathology 46, 57–63 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Chang M. T., et al. , Small-cell carcinomas of the bladder and lung are characterized by a convergent but distinct pathogenesis. Clin. Cancer Res. 24, 1965–1973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson A. G., et al. ; TCGA Research Network , Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 174, 1033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McConkey D. J., Choi W., Dinney C. P., Genetic subtypes of invasive bladder cancer. Curr. Opin. Urol. 25, 449–458 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Choi W., et al. , Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 25, 152–165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jäger W., et al. , Patient-derived bladder cancer xenografts in the preclinical development of novel targeted therapies. Oncotarget 6, 21522–21532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balani S., Nguyen L. V., Eaves C. J., Modeling the process of human tumorigenesis. Nat. Commun. 8, 15422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park J. W., et al. , Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 362, 91–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X., Liu D., Fallon J. T., Zhong M., Distinct genetic alterations in small cell carcinoma from different anatomic sites. Exp. Hematol. Oncol. 4, 2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ismaili N., A rare bladder cancer—Small cell carcinoma: Review and update. Orphanet J. Rare Dis. 6, 75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouba E. J., Cheng L., Understanding the genetic landscape of small cell carcinoma of the urinary bladder and implications for diagnosis, prognosis, and treatment: A review. JAMA Oncol. 3, 1570–1578 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Swellam M., Abd-Elmaksoud N., Halim M. H., Khatab H., Khiry H., Incidence of Bcl-2 expression in bladder cancer: Relation to schistosomiasis. Clin. Biochem. 37, 798–802 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Golestani Eimani B., et al. , Expression and prognostic significance of bcl-2 and bax in the progression and clinical outcome of transitional bladder cell carcinoma. Cell J. 15, 356–363 (2014). [PMC free article] [PubMed] [Google Scholar]
- 23.van der Fels C. A. M., Rosati S., de Jong I. J., EpCAM expression in lymph node metastases of urothelial cell carcinoma of the bladder: A pilot study. Int. J. Mol. Sci. 18, E1802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu A. Y., et al. , Bladder expression of CD cell surface antigens and cell-type-specific transcriptomes. Cell Tissue Res. 348, 589–600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ismaili N., et al. , Small cell carcinoma of the urinary bladder: A case report and review of the literature. J. Cancer Res. Ther. 5, 133–136 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Amin M. B., Histological variants of urothelial carcinoma: Diagnostic, therapeutic and prognostic implications. Mod. Pathol. 22 (suppl. 2), S96–S118 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Liang et al., RNA-seq data of small cell carcinoma of the bladder (SCCB)/urothelial carcinoma (Non-SCCB) clinical samples, and bladder-PARCB cell lines. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139822. Deposited 1 November 2019.
- 28.Balanis N. G., et al. , Pan-cancer convergence to a small-cell neuroendocrine phenotype that shares susceptibilities with hematological malignancies. Cancer Cell 36, 17–34.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefebvre C., et al. , A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 6, 377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez M. J., et al. , Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 48, 838–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borromeo M. D., et al. , ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 16, 1259–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujino K., et al. , Insulinoma-associated protein 1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am. J. Pathol. 185, 3164–3177 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Goodison S., Urquidi V., Tarin D., CD44 cell adhesion molecules. Mol. Pathol. 52, 189–196 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., et al. , Aiolos promotes anchorage independence by silencing p66Shc transcription in cancer cells. Cancer Cell 25, 575–589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raju S., Joseph R., Sehgal S., Review of checkpoint immunotherapy for the management of non-small cell lung cancer. ImmunoTargets Ther. 7, 63–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J. K., et al. , Systemic surfaceome profiling identifies target antigens for immune-based therapy in subtypes of advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 115, E4473–E4482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singec I., et al. , Quantitative analysis of human pluripotency and neural specification by in-depth (Phospho)Proteomic profiling. Stem Cell Reports 7, 527–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shetty A., et al. , The neural cell adhesion molecule promotes maturation of the presynaptic endocytotic machinery by switching synaptic vesicle recycling from adaptor protein 3 (AP-3)- to AP-2-dependent mechanisms. J. Neurosci. 33, 16828–16845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green T., Heinemann S. F., Gusella J. F., Molecular neurobiology and genetics: Investigation of neural function and dysfunction. Neuron 20, 427–444 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Kiss B., et al. , Her2 alterations in muscle-invasive bladder cancer: Patient selection beyond protein expression for targeted therapy. Sci. Rep. 7, 42713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guancial E. A., et al. , FGFR3 expression in primary and metastatic urothelial carcinoma of the bladder. Cancer Med. 3, 835–844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlsson J., Wester K., De La Torre M., Malmström P. U., Gårdmark T., EGFR-expression in primary urinary bladder cancer and corresponding metastases and the relation to HER2-expression. On the possibility to target these receptors with radionuclides. Radiol. Oncol. 49, 50–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lonsdale J., et al. , Data from “The Genotype-Tissue Expression (GTEx) project.” GTEx portal. https://storage.googleapis.com/gtex_analysis_v8/rna_seq_data/GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9_gene_median_tpm.gct.gz. Accessed 29 July 2019.
- 44.Robertson A. G., et al. , Data from “Comprehensive molecular characterization of muscle-invasive bladder cancer.” GDC Data Portal. https://portal.gdc.cancer.gov/projects/TCGA-BLCA. Accessed 4 February 2019.
- 45.Cotto K. C., et al. , DGIdb 3.0: A redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 46, D1068–D1073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aggen D. H., Drake C. G., Biomarkers for immunotherapy in bladder cancer: A moving target. J. Immunother. Cancer 5, 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herbst R. S., et al. , Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerignoli F., et al. , In vitro immunotherapy potency assays using real-time cell analysis. PLoS One 13, e0193498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barretina J., et al. , Data from “The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.” CCLE portal. https://data.broadinstitute.org/ccle/CCLE_RNAseq_rsem_genes_tpm_20180929.txt.gz. Accessed 19 March 2019.
- 50.Grabowska M. M., Matusik R. J., Therapy-induced small-cell disease: From mouse to man and back. Nat. Rev. Urol. 15, 662–663 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Nadal R., Schweizer M., Kryvenko O. N., Epstein J. I., Eisenberger M. A., Small cell carcinoma of the prostate. Nat. Rev. Urol. 11, 213–219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morozumi K., et al. , Transdifferentiation of small cell carcinoma of the urinary bladder from urothelial carcinoma after transurethral resection of a bladder tumor, intravesical Bacillus Calmette-Guerin instillation, and chemotherapy: A case report. Case Rep. Oncol. 9, 786–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Groningen T., et al. , A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat. Commun. 10, 1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haley J. A., et al. , Altered transcriptional control networks with trans-differentiation of isogenic mutant-KRas NSCLC models. Front. Oncol. 4, 344 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ning B., Li W., Zhao W., Wang R., Targeting epigenetic regulations in cancer. Acta Biochim. Biophys. Sin. (Shanghai) 48, 97–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Majem M., Rudin C. M., Small-cell lung cancer in the era of immunotherapy. Transl. Lung Cancer Res. 6 (suppl. 1), S67–S70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldstein A. S., et al. , Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat. Protoc. 6, 656–667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salas J. R., et al. , 18F-FAC PET selectively images liver-infiltrating CD4 and CD8 T cells in a mouse model of autoimmune hepatitis. J. Nucl. Med. 59, 1616–1623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vivian J., et al. , Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35, 314–316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Lachmann A., Giorgi F. M., Lopez G., Califano A., ARACNe-AP: Gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics 32, 2233–2235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq dataset was deposited at Gene Expression Omnibus (27), with accession number of GSE139822.