Significance
Staphylococcus aureus is one of the most virulent bacterial pathogens and, in particular, has the richest repertoire of cytotoxins: A single bacterium can secrete 6 different β-barrel pore-forming toxins, with different cell type and species specificities. Each toxin engages specific receptors on target cells, but the role the receptor plays in the pore-formation process is poorly understood. Here, we determine the crystal structures of a very potent S. aureus leukocidin (LukGH) in complex with its receptor (CD11b) from a sensitive (human) and an insensitive (murine) host, and track the receptor involvement in different steps on the pore-formation pathway. These results advance the knowledge of receptor-mediated leukocidin pore formation and open ways for antileukocidin and anti-S. aureus approaches.
Keywords: host–pathogen interaction, pore forming toxins, receptor recognition, leukocidin, integrin
Abstract
Host–pathogen interactions are central to understanding microbial pathogenesis. The staphylococcal pore-forming cytotoxins hijack important immune molecules but little is known about the underlying molecular mechanisms of cytotoxin–receptor interaction and host specificity. Here we report the structures of a staphylococcal pore-forming cytotoxin, leukocidin GH (LukGH), in complex with its receptor (the α-I domain of complement receptor 3, CD11b-I), both for the human and murine homologs. We observe 2 binding interfaces, on the LukG and the LukH protomers, and show that human CD11b-I induces LukGH oligomerization in solution. LukGH binds murine CD11b-I weakly and is inactive toward murine neutrophils. Using a LukGH variant engineered to bind mouse CD11b-I, we demonstrate that cytolytic activity does not only require binding but also receptor-dependent oligomerization. Our studies provide an unprecedented insight into bicomponent leukocidin–host receptor interaction, enabling the development of antitoxin approaches and improved animal models to explore these approaches.
The pathogen Staphylococcus aureus is a versatile human pathogen with the unique ability to cause a wide range of diseases, such as skin and soft tissue infections, sepsis, or pneumonia, attributed to its immense diversity of host-targeting virulence factors (1). The secreted leukocidins, a family of bicomponent pore-forming toxins, are believed to be at the core of S. aureus immune evasion by lysing phagocytic cells, mainly neutrophil granulocytes but also monocytes and macrophages (2, 3). S. aureus produces up to 5 different leukocidins—γ-hemolysins HlgAB and HlgCB, LukSF-PV (PVL), LukED, and LukGH (also called LukAB) (2)—with their cell type and species specificity driven by binding to different proteinaceous receptors on the surface of the immune cells (2, 3). Following receptor binding, the toxins oligomerize to form a lytic, octameric, β-barrel pore on the cell membrane. Although the steps involved in the leukocidin structural changes occurring during the pore formation are at least partly understood, less is known about the role of the receptors in this process (4, 5).
The cellular receptors of all bicomponent toxins, except LukGH, are transmembrane-spanning G protein-coupled receptors (2, 3, 6). LukGH, however, binds to the extracellular α-I domain of the αM/β2 integrin (CD11b/CD18, macrophage-1 antigen, or complement receptor 3) (7). CD11b/CD18 is a member of the CD18 integrin family and is expressed on professional phagocytic cells (8) with a central role in the immune system, binding more than 40 protein ligands, including human fibrinogen and the complement fragment iC3b (9–11). Both the α- and β-subunits contain large ectodomains, one transmembrane domain each, and short cytoplasmic domains, which enable communication with the extracellular environment. The 2 ectodomains, supported by their upper and lower legs, come together to form the integrin head, which comprises the α-I domain, the canonical ligand binding site in the integrins. Integrin activation, the so-called “inside-out signaling,” results in an allosteric switch in the CD11b/CD18 ectodomain from a resting, bent state to the extended form, with the corresponding activation of the α-I domain (conversion to open form, see below) and ligand recruitment (12).
The human α-I domain (CD11b-I) was expressed recombinantly, independently of the other integrin subunits (13), and to date 13 crystal structures of CD11b-I in complex with natural ligands, antagonists, antibodies, or alone, have been solved (13–20). However, despite the critical role of CD11b-I in the immune system of different mammals (21), all available crystal structures were obtained with the human CD11b-I (huCD11b-I). Two different conformations have been observed: The so-called inactive (closed or low affinity) and active (open or high affinity) forms of CD11b-I. The latter involves the rearrangement of the metal coordinating residues at the metal ion-dependent adhesion site (MIDAS), to allow a carboxylate group from the ligand to complete the metal coordination, and a 10-Å downward shift of the C-terminal α7-helix (12, 14).
LukGH is expressed in human infections and appears to be the most potent S. aureus leukocidin based on in vitro and ex vivo data (22–25). It is however inactive or displays limited activity in the established S. aureus in vivo models, such as mouse and rabbit, which hinders the study of its role in S. aureus pathogenesis (7, 26). The variation in the CD11b-I sequences between different species was used to explain the LukGH species specificity, and activity was shown to correlate with binding to CD11b-I: That is, no binding to mouse CD11b-I (moCD11b-I) and very low activity toward murine polymorphonuclear neutrophils (PMNs) in vitro (7, 26). We have recently been able to improve LukGH cytotoxicity (∼10- to 15-fold) toward rabbit cells by increasing binding to the rabbit CD11b-I (rbCD11b-I) receptor, using alanine scanning and targeted mutagenesis to map the cytotoxin–receptor interaction (27). However, high-resolution structural data for the LukGH–CD11b-I interaction would allow rational design of LukGH variants with activity toward different species and provide mechanistic insights into receptor-mediated pore formation.
Here, we report the crystal structure of S. aureus pore-forming cytotoxin LukGH in complex with the CD11b-I domain of its integrin receptor CD11b/CD18. We use both the human and the mouse receptors for crystal and solution structural analysis to ascertain and characterize the 2 main requirements for activity: Binding and oligomerization. We find that the same receptor molecule is involved in binding and oligomerization via interactions with the LukH and LukG subunits from different LukGH dimers, respectively. We discuss the roles of receptor cell surface expression, activation, and clustering on LukGH activity and the molecular drivers of LukGH species specificity.
Results
Receptor Binding Is Necessary, but Not Sufficient for LukGH Cytotoxicity.
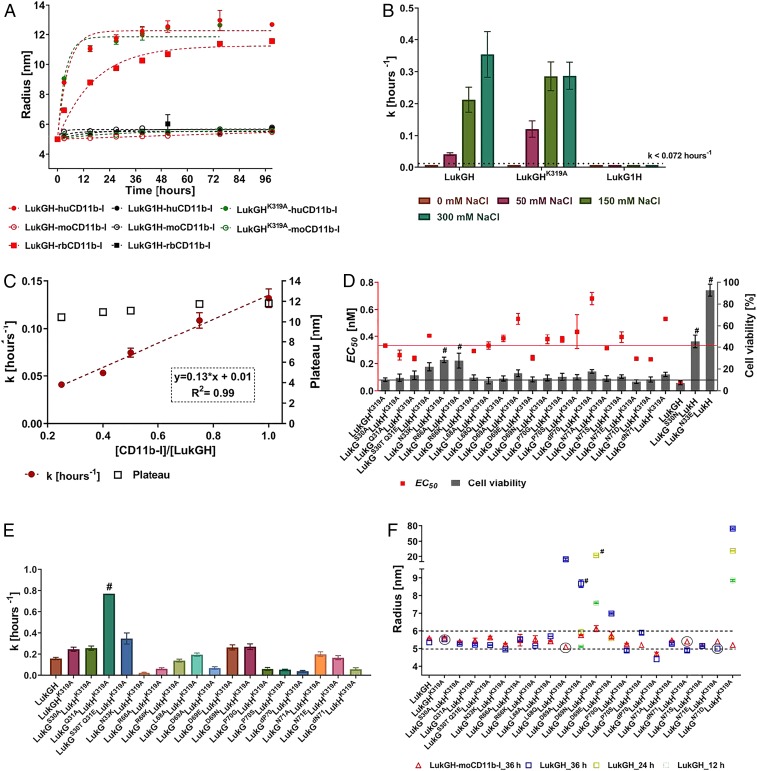
Using both human and rabbit cells and recombinant receptor molecules we have previously confirmed that LukGH binding to CD11b-I indeed correlated with its cytotoxic activity toward PMNs in these 2 species, and have identified mutations in LukGH that either decrease or enhance binding and activity (27). The most prominent change was seen with LukGHD312K [residue numbers correspond to the mature protein, i.e., after signal peptide cleavage (28)], a variant with increased affinity toward rbCD11b-I paralleled by 10- to 15-fold increased cytotoxicity toward rabbit PMNs (27). Mouse PMNs are resistant to LukGH at concentrations up to 30 µM, and the toxin binds the mouse receptor very weakly (Kd ∼1 µM) (Fig. 1 A–C) (7). We found 2 mutations in LukH, R294A and K319A (previously shown to decrease binding toward the human and rabbit receptor) (27), that significantly increase binding to moCD11b-I (Kd of 63 nM for LukGHK319A, similar to LukGHD312K with rbCD11b-I) (Fig. 1B) (27). However, these mutants display no activity toward mouse PMNs, at concentrations up to 20 µM, which are over 3 orders of magnitude greater than the EC50 (half maximal effective concentration) values of LukGH for rabbit and human PMNs (Fig. 1 B and C). Thus, the receptor binding–cytolytic activity correlation observed for human and rabbit does not apply to the mouse system and we hypothesized that another step in the pore-forming process, beyond receptor binding, is responsible.
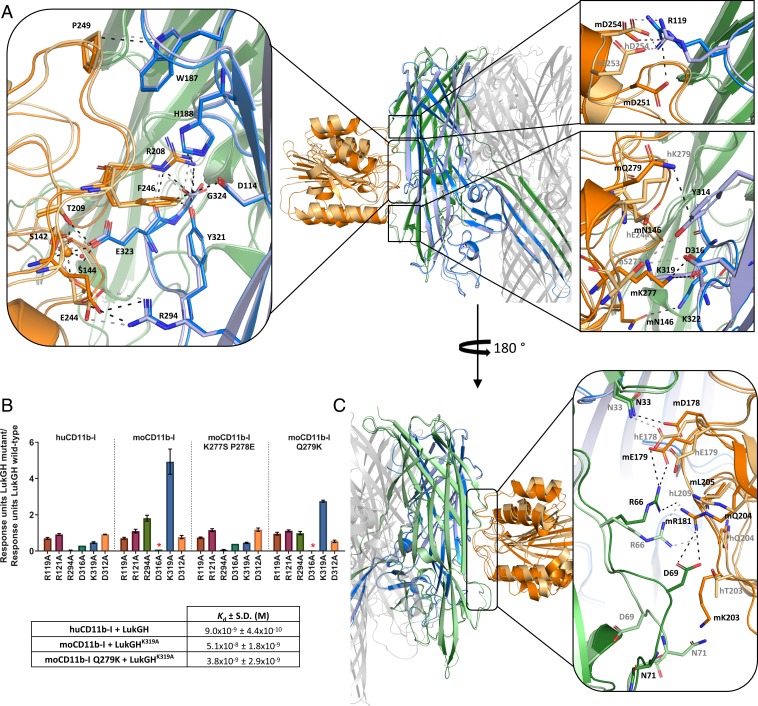
Fig. 1.
Binding and activity of LukGH wild-type and mutants to CD11b-I and crystal structure of LukGH-CD11b-I. (A) Steady-state analysis of LukGH wild-type binding to moCD11b-I. The steady state Kd is shown in the Inset. (B) Binding of LukGH to hu- or moCD11b-I expressed as response units (mean of 2 to 10 independent experiments ±SEM) and Kd (mean of 2 to 10 independent experiments ±SD). EC50 values of LukGH mutants toward differentiated HL-60 cells or mouse PMNs assessed in a luminescent cell viability assay measuring cellular ATP content (mean of 2 to 8 independent experiments ±SEM). For variants that had limited or no cytotoxicity (could not kill >75% of cells at the highest toxin concentration used), EC50 is not shown. (C) Cytotoxicity of LukGH, LukGHK319A, and LukED toward mouse PMNs assessed in a luminescent cell viability assay measuring cellular ATP content at cytotoxin concentrations of 30 µM, 20 µM, and 100 nM, respectively (mean of 3 independent experiments ±SEM). (D) Front and top view of LukGHK319A–moCD11b-I crystal structure. Dark blue and light green cartoons represent LukH and LukG from dimer 1 and dark green and light blue cartoon represent LukG and LukH from dimer 2, respectively. moCD11b-I is shown as an orange cartoon. Other dimers forming the octamer pore and bound CD11b-I molecules, are shown as a gray cartoon. Red spheres represent bound DMSO molecules from one asymmetric unit (dark red sphere represents DMSO 2). Comparison of moCD11b-I secondary structure (E) and MIDAS residues (F) from LukGHK319A–moCD11b-I structure (orange ribbon) with the active (1IDO, light pink ribbon) and inactive (1JLM, light gray ribbon) form of huCD11b-I. C-terminal α-helix is shown as light pink cartoon (1IDO) and gray cartoon (1JLM). Structures are aligned on moCD11b-I and MIDAS residues in E and F, respectively. The metal ions from the moCD11b-I structure and the inactive form of CD11b-I (1JLM) are shown as orange and gray spheres, respectively.
Structural Insight into LukGH–CD11b-I Interaction and Specificity.
Several attempts to crystallize huCD11b-I in complex with LukGH, including different LukGH constructs—that is, LukGH wild-type, LukGH with an impaired oligomerization interface (LukG1H) (28), and LukGH lacking the unstructured N terminus of LukH (33 and 41 amino acids)—were unsuccessful. However, we managed to crystallize moCD11b-I in complex with the full-length LukGHK319A mutant (with increased affinity to moCD11b-I), with crystals diffracting to 2.29-Å resolution (SI Appendix, Table S1). Subsequently, crystals were also obtained for the human variant (huCD11b-I) in complex with full-length wild-type LukGH, but anisotropically diffracted to lower resolution (2.75 Å along a* and b*, and 4.79 Å along c*) (SI Appendix, Table S1). Both crystal structures revealed one LukG and one LukH molecule bound to the CD11b-I in the asymmetric unit, with a total binding surface area of 701 and 340 Å2 for LukHK319A/moCD11b-I and LukG/moCD11b-I, and 695 and 246 Å2 for LukH/huCD11b-I and LukG/huCD11b-I interfaces, respectively. The LukG and LukH protomers from the asymmetric unit do not belong to the same LukGH dimer, as found in solution (28, 29), but to 2 different adjacent dimers (Fig. 1D). In the crystal structure, these LukG and LukH protomers assembled into a 4-fold rotationally (C4) symmetrical octameric pore composed of 4 LukG-LukH–CD11b-I heterotrimers, similar to that previously reported for LukGH alone (PDB ID code 4TW1) (28).
Except for the N termini and a few loops, essentially the entire LukH and LukG subunits are visible in the electron density map. For the moCD11b-I and huCD11b-I more than 20 amino acids of the C-terminal α-helix (α7) are not visible in the electron density maps (Fig. 1E), as also seen in the structure of the huCD11b-I–C3d complex (PDB ID code 4M76) (19), which is lacking the last 11 amino acids. moCD11b-I assumes an α/β Rossmann fold (Fig. 1E), similar to published huCD11b-I structures (78% identity between mo and huCD11b-I) (7). The RMSDCα between the CD11b-I domains in the complexes and the previously published conformations of huCD11b-I: Active (PDB ID code 1IDO) (13) and inactive (PDB ID code 1JLM) (14) are 0.62 and 1.63 Å over 169 superimposed Cα atoms for moCD11b-I, and 0.70 and 1.27 Å over 168 superimposed Cα atoms for huCD11b-I, respectively (Fig. 1E). The tendency of the α7-helix for downward shift and the coordination sphere of Mg2+ in the MIDAS site indicate that both human and mouse CD11b-I exist in the active conformation when bound to LukGH (Fig. 1 E and F).
Main Interaction Site of CD11b-I–LukGH and Its Conservation.
The main interaction site of LukGH with CD11b-I is located in the cap domain of LukH, in agreement with previous binding and mutagenesis data (27, 28, 30) (Fig. 2A). Interestingly, structural superposition of the LukGHK319A–moCD11b-I and LukGH–huCD11b-I structures over 706 Cα atoms resulted in RMSDCα of 1.11 Å. The largest structural difference is due to a shift in the position of the CD11b-I main chain (mean displacement of 1.9 Å, rotation by approximately 7° about a hinge nearly parallel to the pore axis and a maximal Cα-shift of 5 Å when the superposition is performed on LukH alone; RMSDCα of 0.41 Å over 270 Cα atoms), caused by different interactions at the edges of the binding epitope (vide infra) (Fig. 2 A, Center).
Fig. 2.
Binding epitope of LukGH-CD11b-I. (A) Binding epitopes of LukH–CD11b-I with detailed views of the specific interactions involved in CD11b-I binding in boxes, aligned on LukH. LukG, LukH, and CD11b-I from the LukGHK319A–moCD11b-I structure are shown in green, blue, and orange, respectively. The same protein components from the LukGH–huCD11b-I structure are shown in pale green, pale blue, and pale orange. Hydrogen bonds, salt bridges, the coordinate covalent bonds of Mg2+, as well as some other selected close contacts are shown as dashed lines colored black (for the moCD11b-I complex) or gray (for the huCD11b-I complex). (Left) Conserved interactions; (Right Upper and Lower) nonconserved interactions between the human and the mouse complexes. (B) Binding of LukGH mutants to CD11b-I variants relative to LukGH wild-type (mean of 3 independent experiments ±SEM, except for LukGHD316A with one experiment). Asterisks represent samples where no binding was detected (RU < 0.05 nm). Inset table shows Kd of selected LukGH and CD11b-I variants (mean of 2 to 3 independent experiments ±SD). (C) Binding epitopes of LukG-CD11b-I with a detailed view of the specific interactions involved in CD11b-I binding in the box, aligned on LukH. Color coding as in A.
The core of the interface is well conserved between the mouse and human structures (Fig. 2 A, Left and SI Appendix, Table S2). Central to these interactions is the MIDAS site, where the LukH residue E323 completes the octahedral coordination sphere around the metal ion, together with the conserved CD11b-I residues (S144, S142, and T209) and 2 water molecules, as seen for the active conformation of huCD11b-I (Figs. 1F and 2 A, Left, and SI Appendix, Fig. S1) (13, 14). The importance of this interaction is supported by the lack of receptor binding and cytolytic activity of the LukGHE323A variant (27, 30) and by the fact that Mg2+ substitution at the MIDAS site by Ca2+ impairs LukGH binding (SI Appendix, Table S3). Additional interactions involve the salt bridges between the side chains of E244 (CD11b-I) and R294 (LukH) and the side chain of R208 (CD11b) and the C-terminal carboxyl group of G324 (LukH). Polar contacts between R208 (CD11b-I) and H188 and Y321 (LukH), van der Waals contacts between F246 (CD11b-I) and D114, H188, and Y321 (LukH), and a hydrophobic interaction between P249 (CD11b-I) and W187 (LukH) are also observed (Fig. 2 A, Left). Mutagenesis studies at these positions in LukH confirm their involvement in binding and activity (27). While residues E244, R208 and F246 are conserved between human, rhesus macaque, pig, rabbit, and mouse CD11b-I variants, residues R208 and F246 are replaced by Q and Y, respectively, in guinea pig (SI Appendix, Fig. S2A).
The LukH–CD11b-I complex is stabilized by several salt bridges and polar interactions (SI Appendix, Table S2), explaining why the LukGH affinity for CD11b-I decreases with increasing the ionic strength, even though protein stability is not affected (SI Appendix, Fig. S2B and Table S4). At the extremities of the interface (Fig. 2 A, Right), the LukH–CD11b-I interactions vary in the 2 species. The main driver is the S277 huCD11b-I residue, which is K in the mouse variant. K277 forms a salt bridge with D316 (LukH) in the LukGHK319A–moCD11b-I complex (not present for the human complex) (Fig. 2 A, Right Lower). It appears that reduction of the size and removal of the positive charge (K319A) is needed to prevent steric clashes and electrostatic repulsion between K277 and K319, explaining the increased binding of the LukGHK319A variant to moCD11b-I. Instead, S277 from huCD11b-I forms hydrogen bonds with the side chains of LukH residues Y314 and D316, which brings the main chain of huCD11b-I closer to LukH (Fig. 2 A, Right Lower).
Since S277 is conserved between different species, except for mouse, we performed “humanizing mutations” (i.e., we introduced a K277S P278E double mutation in moCD11b-I) to confirm the above hypothesis. The LukGH variants with mutations in the region involved in the interaction with K277 (LukGHR294A, LukGHK319A, LukGHD316A) showed a similar binding pattern for moCD11b-I K277S P278E and huCD11b-I, with decreased binding affinity for LukGHR294A and LukGHK319A, while those with mutations remote from this interaction site (LukGHR119A, LukGHR121A, LukGHD312A) were not distinguished by the CD11b-I variant (Fig. 2B). The decreased binding affinity of LukGHK319A for huCD11b-I is probably due to loss of a salt bridge between K319 and E244 (CD11b-I). Additionally, we “humanized” the neighboring Q279 in moCD11b-I (moCD11b-I Q279K), which makes a N–H···π interaction with the aromatic side chain of LukH Y314 in the mouse but not in the human complex (the corresponding K279 residue is oriented away from the interface). The moCD11b-I Q279K variant had significantly increased affinity toward LukGHK319A compared to moCD11b-I K277S P278E (Fig. 2B and SI Appendix, Fig. S2C). Additional interactions are present in the moCD11b-I complex only, including the salt bridge at the top of the interface (D251 [moCD11b-I]–R119 [LukH]) (Fig. 2 A, Right Upper) and a hydrogen bond at the bottom (N146 [moCD11b-I]–K322 [LukH]) (Fig. 2 A, Right Lower).
While the LukH residues forming the salt bridges in human and mouse complexes are mostly conserved, except for LukH R119 and K319, only 2 positions from CD11b-I involved in salt bridge formation are conserved between human, rabbit, mouse, pig, rhesus macaque, and guinea pig (SI Appendix, Fig. S2A and Table S2). The conservation of the CD11b-I residues involved in the binding epitope in the mouse and human CD11b-I complex structure between different species reveals the highest similarity between human and rhesus macaque (89% identity), which correlates with activity toward macaque PMNs (26) and the highest divergence between human and guinea pig (63% identity).
Second Binding Interface between LukG and CD11b-I.
Unexpectedly, besides the LukH–CD11b-I epitope, we observed a second binding region between CD11b-I and LukG from an adjacent dimer of the LukGH octamer (Figs. 1D and 2C and SI Appendix, Supplementary Table S5). This interface is partly conserved between the mouse and human complexes: For example, the hydrogen bond between R66 (LukG) guanidinium group and L205 (CD11b-I) main-chain carbonyl group. However, most of the residues contacting the 2 loops in LukG in this interface differ between the 2 species: For example, N33 (LukG) side chain makes a hydrogen bond with the carboxyl group of D178 in moCD11b-I and E178 in huCD11b-I. Particularly interesting is the interaction of moCD11b-I with the loop 68-72 in LukG: Due to steric hindrance by K203 in moCD11b-I, the loop is flipped by up to ∼180° compared to the uncomplexed structures (PDB ID codes 5K59 and 4TW1) and to the complex with the human receptor (SI Appendix, Fig. S3), which in turn flips the side chain of LukG D69, allowing formation of a salt bridge with R181 in moCD11b-I (Fig. 2 C, Right). The flip is presumably kinetically unfavorable, as D69 loses hydrogen bonds with 3 residues from the adjacent β-sheet. The difference electron density map suggests flexibility of this loop and the presence of some other minor alternate conformations, which we have not been able to model satisfactorily (SI Appendix, Fig. S3). Such disorder is not observed in the complex with huCD11b-I, which has T at position 203 and does not appear to interact with the LukG 68-72 loop.
All LukG residues involved in the second binding interface are variable in the currently available LukG sequences (75 to 80% conservation level), in contrast to the main interface, where more than half of the residues are fully conserved (the remaining show 76 to 99% conservation) (SI Appendix, Tables S2 and S5).
CD11b-I Promotes LukGH Oligomerization in the Absence of a Cell Surface.
The ability of CD11b-I to bind at the oligomerization interface indicates that the receptor alone (in absence of a cell surface) may promote oligomerization. To further investigate this, we developed a noninvasive oligomerization assay using dynamic light scattering (DLS), by mixing LukGH with CD11b-I in a 1:1 molar ratio and monitoring the molecular size of the mixture, expressed as radius, over time. The hydrodynamic radia of LukGH and CD11b-I alone are ∼5 nm and ∼2 nm, and do not change for up to 36 to 48 h (SI Appendix, Fig. S4A). When the 2 components were mixed, we observed a time-dependent increase in radius from ∼5.5 nm to ∼11 to 12 nm over several hours, after which a plateau was reached (Fig. 3A). We assign the lower radius (∼5.5 nm) to the LukGH–CD11b-I complex, based on data with an oligomerization-deficient variant, the LukG1H dimer, which binds huCD11b-I, but is lacking cytolytic activity (28), and shows no change in size when mixed with CD11b-I (Fig. 3A). The higher (∼11 nm) radius corresponds to the final oligomerization product, a relatively stable structure that does not aggregate in the time frame of the experiment (up to 96 h), which is, most probably, an assembly similar to the octameric pore found in the crystal. We observed oligomerization of LukGH in the presence of human and rabbit CD11b-I, but not mouse CD11b-I, which parallels the activity data (Fig. 3A). Moreover, the LukGHK319A variant, which shows strong binding to moCD11b-I is still unable to oligomerize in the presence of moCD11b-I, explaining its lack of cytolytic activity.
Fig. 3.
Oligomerization of LukGH in solution, binding and activity of LukGH oligomerization variants. (A) Change of LukGH, LukG1H, and LukGHK319A (at 5 mg/mL) plus hu-, rb-, or moCD11b-I (at 2.5 mg/mL) cumulant radius, over time, measured in 25 mM Hepes, pH 7.5, 1 mM MgCl2, 150 mM NaCl (mean of 1 to 2 replicates ±SEM). The dotted lines represent fitting of the data to a one-phase association model with fixed y0 = 5 at x0 = 0 h (GraphPad Prism). (B) Oligomerization rate constant (k) and plateau for LukGH, LukGHK319A, and LukG1H (at 5 mg/mL) plus huCD11bI (2.5 mg/mL) in 25 mM Hepes, pH 7.5, 1 mM MgCl2, 0 to 300 mM NaCl (mean of 1 to 2 replicates ±SEM). Data were fitted as in A giving R2 > 0.93. (C) Oligomerization rate constant (k) of LukGH (4.5 mg/mL) plus increasing amounts of huCD11b-I (2.3 mg/mL) in 25 mM Hepes, pH 7.5, 1 mM MgCl2, 150 mM NaCl (mean of 2 replicates ±SEM). Linear regression fit (GraphPad Prism) is shown in red with equation in Inset. (D) Activity of LukGH mutants toward differentiated HL-60 cells expressed as EC50 and percent cell viability at maximal toxin concentration (100 nM) (mean of 2 independent experiments ±SEM). Red and black line represent EC50 value and percent cell viability of LukGHK319A mutant, respectively. Variants that had limited or no cytotoxicity (could not kill >75% of cells at the highest toxin concentration used) are marked with “#.” (E) Oligomerization rate constant (k) of LukG oligomerization mutants coexpressed with LukHK319A (at 4.5 mg/mL) plus huCD11b-I (2.3 mg/mL) (mean of 2 replicates ±SEM). Data were fitted as in A, in all cases, except for LukGQ31A LukHK319A (#, ambiguous fit), yielding R2 > 0.94. (F) Cumulant radius of LukGH_variant–moCD11b-I complexes (at 4.5 mg/mL for LukGH and 2.3 mg/mL for CD11b-I) and individual LukGH variants at 36 h of incubation in 25 mM Hepes, pH 7.5, 1 mM MgCl2,150 mM NaCl (mean of 1 [circled] or 2 replicates ±SEM). In case the sample shows increased radius at time 36 h, earlier time points are shown (24 and 12 h). Dotted lines represent ±10% change from a 5.5-nm radius. Samples with sum of squares error >10 are marked with “#.”
When the oligomerization rate was approximated to a first-order rate constant, we observe that huCD11b-I–induced oligomerization of LukGH is ∼3× faster than that induced by rbCD11b-I at physiological NaCl concentrations (150 mM) (Fig. 3B and SI Appendix, Fig. S4B). There is, however, a marked dependence of oligomerization rate on NaCl concentration (i.e., it increases with increasing NaCl concentration from 0 to 150 mM), with some variations at higher salt concentrations for different receptors (Fig. 3B and SI Appendix, Fig. S4B).
In order to investigate the stoichiometry requirements for CD11b-I–mediated oligomerization of LukGH, we measured the oligomerization efficiency and rate at different CD11b-I to LukGH ratios. The oligomerization appears complete at ratios as low as 1:4 (one CD11b-I molecule per LukGH octamer), with the oligomerization rate increasing almost linearly with increasing the ratio to 1:1 (4 CD11b-I molecules per LukGH octamer), indicative of a catalytic role of CD11b-I in this process (Fig. 3C).
Using site-directed mutagenesis, as described in the SI Appendix, Supplementary Results and Discussion, we could clearly confirm the involvement of LukG residues N33, R66, D69, P70, and N71 in both oligomerization and activity with the human system (Fig. 3 D and E), and for R66 and D69 also with rabbit cells (SI Appendix, Fig. S5A), in agreement with structural data. The most striking loss of activity was seen when LukG N33 was mutated to the negatively charged E, presumably due to repulsion at the second interface (LukG N33 interacts with E178 in huCD11b-I) (Figs. 2 C, Right and 3D). Importantly, all of the tested variants showed no change in binding to huCD11b-I, confirming that loss of activity was not due to decreased binding affinity (SI Appendix, Fig. S5B).
None of the oligomerization site mutants, coexpressed with LukHK319A showed any activity toward mouse PMNs up to cytotoxin concentrations of 800 to 1,000 nM, no improved affinity toward moCD11b-I, and no increase in radius in presence of moCD11b-I, when tested by DLS (Fig. 3F and SI Appendix, Fig. S5 C and D).
Fab Binding to the LukG Subunit of LukGH–huCD11b-I Prevents Its Cell Membrane Independent Oligomerization.
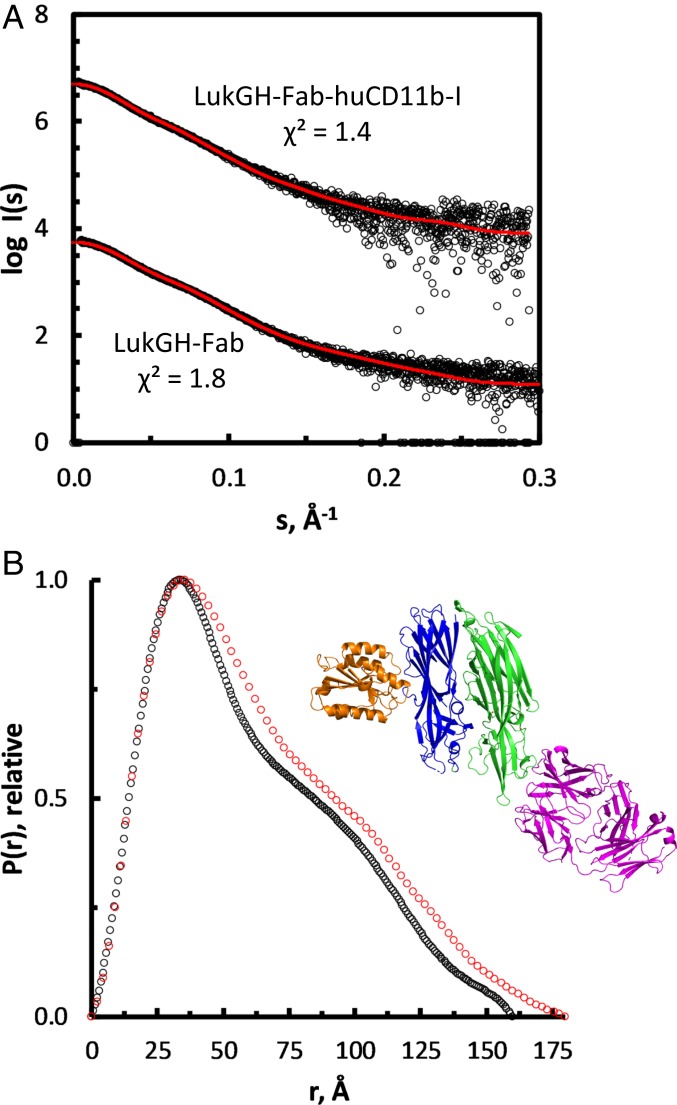
In order to gain insight into the structural organization of the LukGH–receptor complex in solution, we used solution small-angle X-ray scattering (SAXS) for LukGH and the LukGH–huCD11b-I complex in the presence of the Fab fragment of a LukGH neutralizing antibody (Fig. 4 and SI Appendix, Table S6). The Fab was used to stabilize the dimer and to allow the elution of the complex from the size-exclusion chromatography column to ensure the sample’s monodispersity. First, the complex of the Fab fragment with LukGH was analyzed in the absence of the receptor, and compared to the crystal structure of the complex we have previously determined (29). The computed distance distribution p(r) (Fig. 4B) indicates that the molecule is a multidomain (distinctive bumps) and an elongated particle [skewed p(r) shifted to shorter distances]. Furthermore, the overall structural parameters derived from SAXS (molecular mass, radius of gyration [RG], and maximum dimension [Dmax]) (SI Appendix, Table S6) are fully compatible with a monomeric construct and strongly support that the binding of the Fab fragment prevents the oligomerization of LukGH. Moreover, the experimental data are in good agreement with the theoretical curve calculated from a structural model derived from the available crystal structure (PDB ID code 5K59) with a discrepancy χ2 = 1.8 (Fig. 4A).
Fig. 4.
SAXS analysis of complex formation. (A) Scattering data as logI(s) vs. s plot compared to the theoretical scattering of the respective models. These comprise the interfaces as retrieved from the crystal structures; χ2 values are indicated. Curves are shifted along the y axis for better visualization. (B) Distance distribution profile of LukGH-Fab (black) and LukGH–Fab–huCD11b-I (red). The Inset shows the expected complex formation as cartoon representation, with the LukG, LukH, huCD11b-I, and Fab subunits in green, blue, orange, and purple cartoons, respectively.
Next, we analyzed the LukGH–Fab complex bound to huCD11b-I. Noticeable increases observed for the overall parameters (RG, from about 4.8 to about 5.1 nm, Dmax, from 16 to 18 nm) and an increase by about 20 kDa in the molecular mass are in line with the stable 1:1 complex formation (Fig. 4 and SI Appendix, Table S6) corresponding to LukGH–Fab complex bound to one huCD11b-I. No concentration-dependent alterations in the SAXS data are observed, indicating that the receptor is tightly bound to LukGH also in the presence of the Fab fragment. Moreover, the experimental data are in very good agreement (χ2 = 1.4) (Fig. 4A) with the scattering curve computed from a model combining the LukG:Fab interface (PDB ID code 5K59) and the LukH:huCD11b-I interface (crystal structure described here) (Fig. 4B). To further improve the fit, the program CORAL was used. Here, the missing amino acids (44 N-terminal residues of LukH and 22 C-terminal residues of CD11b-I) were modeled as dummy residues. With this approach, a χ2 value of 1.0 was achieved (SI Appendix, Fig. S6). Comparison of 20 individual runs suggests that the N-terminal of LukH is rigid and elongated.
Binding of the αLukGH-mAb#5.H1H2 Fab to the rim region of the LukG protomer (Fig. 5A) (29) in the LukGH dimer did not prevent binding of CD11b-I to LukGH (via the LukH protomer) in solution, but prevented oligomerization, as predicted from the crystal structure (αLukGH-mAb#5.H1H2 binds to the oligomerization interface) (29) and confirmed by DLS measurements in the presence of the Fab (Fig. 5B). However, on the cell surface, when LukGH is bound to the receptor, the αLukGH-mAb#5.H1H2 epitope is no longer accessible (Fig. 5A) and no αLukGH-mAb#5.H1H2 binding to cell-bound LukGH was detected (29).
Fig. 5.
Interaction of LukGH with Fab of αLukGH-mAb#5.H1H2 and activity in presence of LM2/1 and CBRM1/5. (A) Model of the LukGH–huCD11b-I octamer interacting with the Fab fragment of αLukGH-mAb#5.H1H2 (PDB ID code 5K59). The Fab is shown as purple surface, CD11b-I domain as orange cartoon, LukH1 and LukG1 forming Dimer 1 as dark blue and green cartoons, respectively, and LukH2 from adjacent dimer as light blue cartoon. The other LukG and LukH protomers and CD11b-I are shown in gray. Residues involved in binding of CBRM1/5 are shown as black spheres. (B) Change of the cumulant radius for LukGH plus huCD11b-I and/or αLukGH-mAb#5.H1H2 Fab measured in 25 mM Hepes, pH 7.5, 1 mM MgCl2, 150 mM NaCl at 1 mg/mL (mean of 1 to 2 replicates ±SEM). The red solid line represents fit of the data to a one-phase association model with fixed y0 = 5 at x0 = 0 h (GraphPad Prism). (C and D) Activity of LukGH toward LPS activated human PMNs, in presence and absence of 10 µg/mL LM2/1 (C) and CBRM1/5 (D) antibodies, assessed in a luminescent cell viability assay measuring cellular ATP content at different LukGH concentrations (mean of 3 replicates ±SEM).
We have also determined the effect of anti–CD11b-I antibodies with known epitopes (SI Appendix, Fig. S2A) on the activity of LukGH on lipopolysaccharide (LPS)-activated human PMNs. In the presence of the LM2/1 antibody, whose epitope is in the proximity of the LukGH binding epitope (SI Appendix, Fig. S2A), we observed an inhibition of LukGH activity (Fig. 5C), in agreement with a previous report (7). In contrast, the CBRM1/5 antibody, which recognizes a conformational epitope present only on the active CD11b-I form (31), enhances LukGH activity (Fig. 5D), presumably due to an allosteric activation. This is particularly interesting since an opposite effect (i.e., inhibition of binding) was observed with other CD11b ligands, ICAM-1 and fibrinogen, in the presence of CBRM1/5 (31).
Discussion
LukGH is a unique member of the bicomponent cytotoxin family, as it dimerizes in solution before receptor and target cell binding (28, 30). This feature has been proposed to be responsible for the very high cytotoxic activity of LukGH, which also correlates well with receptor up-regulation and activation on target cells (25, 28). At “high” receptor densities, on activated PMNs, the activity of LukGH is up to 3 orders of magnitude higher than on resting PMNs (25). Here, we provide the molecular basis for this correlation. A single receptor molecule is able to bind 2 adjacent dimers in the octamer, and implicitly a single LukGH dimer can bind 2 receptor molecules, via separate LukH and LukG interfaces. In addition, LukGH binds to the active form of the I-domain of CD11b, as all of the other bona fide CD11b ligands.
Using a combination of X-ray crystallography and SAXS, we were able to capture 2 intermediates in the receptor-mediated LukGH pore-formation pathway. The LukGH dimer–CD11b-I complex, stabilized by an oligomerization inhibitory Fab fragment, was analyzed by SAXS. This ternary complex involves interactions between CD11b-I and the cap domain of the LukH subunit, close to the LukGH oligomerization site, as also indicated by previous data generated with site-directed mutagenesis (27). Since there are no major structural changes in the LukGH dimer compared to the unligated form, this is presumably one of the first intermediates in the pathway. The second intermediate is the fully formed LukGH octamer complexed with 4 CD11b-I molecules, which in addition to the LukH interface (the binding interface), involves interactions with 2 LukG loops from a neighboring LukGH dimer (across the oligomerization interface). This is likely one of the final intermediates before insertion of the pore into the target cell membrane, although it is possible that not all 4 sites need to be occupied for pore formation to occur (see below). Interestingly, while the LukGH dimer–CD11b-I binding interface has 6 salt bridges, none is present in the oligomerization interface, at least with the human receptor. Accordingly, the ionic strength requirements for the 2 processes also appear to follow different trends (i.e., increase in ionic strength favors oligomerization but impairs binding). This corroborates the electrostatic nature of the LukGH dimer–CD11b-I interaction, and hydrophobic nature of the oligomerization interface, and may indicate different preferences for diverse microenvironments.
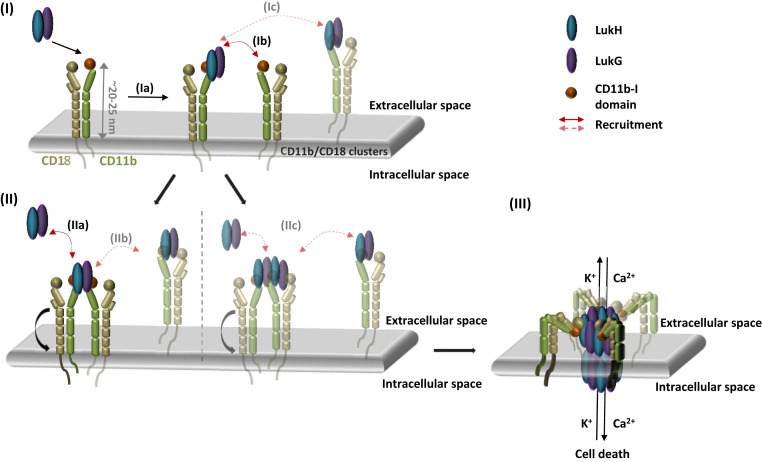
Based on all of the structural, mutagenesis, antibody inhibition, and cytotoxicity enhancement data presented here, we propose a mechanism of pore formation by LukGH on activated PMNs (Fig. 6). LukGH binds to its integrin receptor, CD11b/CD18, in an extended conformation, induced as a result of inside–out signaling following activation (12). This agrees with the potentiation of LukGH activity on LPS-stimulated PMNs by CBRM1/5, an anti–CD11b-I antibody that targets an epitope shielded in the bent integrin (Fig. 5 A and D). According to this model, initially LukGH binds an active CD11b-I domain on the cell surface, via its LukH subunit, presumably with concomitant recruitment of an adjacent CD11b-I domain (which may already have an occupied LukH site) via the LukG subunit (Fig. 6). Homodimerization of integrin α-domains, triggered by interactions between the homologous transmembrane domains, has been reported for the activated form of integrin αIIbβ3 (32). Recruitment of 2 additional LukGH dimers to form the octamer may not necessarily involve other I-domains (Fig. 6), since octameric pore formation is thought to be a highly cooperative process (as shown for S. aureus γ-hemolysin) (33). Moreover, DLS oligomerization data in solution suggest that there is no effect of additional receptor domains on oligomerization efficacy and only a small increase in oligomerization rate from 2 to 4 CD11b-I equivalents per LukGH octamer is observed.
Fig. 6.
Proposed model of LukGH–CD11b-I interaction and pore formation. (I) Binding of LukGH to CD11b-I via the LukH protomer (Ia) results in recruitment of a second integrin molecule via the LukG protomer (Ib) or alternatively, recruitment of a second integrin molecule with bound LukGH dimer (Ic). (II) After recruitment of the second integrin, via the LukG protomer, further LukGH dimer molecules are bound either as soluble LukGH dimers (IIa) or LukGH dimers bound to integrins (IIb). In the alternative version, 2 LukGH dimers bound to the 2 integrins (IIc) recruit further LukGH dimers in the same way as in IIa and IIb. (III) Bending of the integrin and insertion of the octameric pore containing 2 to 4 bound integrins into the membrane.
β-Barrel pore formation is a 2-step process, and the final step of insertion into the membrane is thought to occur after complete oligomerization of the cap domain (4). In the extended form of the integrin, the I-domain is ∼20 nm from the cell surface, so the integrin would have to bend to allow for the insertion of the pore (Fig. 6). Alternatively, the receptor may dissociate before pore insertion, via an unidentified mechanism, similar to the proposed receptor dissociation after pore formation/oligomerization in the case of another bicomponent leucocidin, LukSF (5). Ligand binding to the active I-domain of CD11b of the bent CD11b/CD18 integrin is also not unprecedented: ICAM-1 binding was shown to have antiinflammatory effects (34). Another aspect is the orientation of the LukGH pore relative to the cell surface when LukGH binds the receptor, as the alignment of CD11b-I on the available ectodomain crystal structures (PDB ID codes 3K71, 5ES4, 4NEH, 3K6S) does not result in a LukGH pore oriented perpendicular to the cell membrane (SI Appendix, Fig. S7). In principle, the region linking the I-domain with the rest of the α-chain in integrins is flexible (35, 36) and may allow the rotation of the LukGH pore toward the membrane. An intriguing possibility is that LukGH pores are able to kill adjacent cells, or that such a mechanism is used for LukGH-dependent bacterial escape from intracellular compartments (37).
Being able to specifically engage activated CD11b on the PMN surface is not the only advantage of the bivalent toxin–receptor interaction. CD11b/CD18 is known to bind a variety of endogenous ligands (SI Appendix, Table S7), with affinities in the high nanomolar range, some with epitopes overlapping with LukGH binding [e.g., C3d (19), iC3b (9), or human fibrinogen (10)] (SI Appendix, Fig. S2A). The concentration of these ligands varies with tissue type, but is particularly high in the blood (e.g., 1.5 to 4.0 mg/mL for fibrinogen). It is not yet clear whether LukGH is active in S. aureus bacterial sepsis, but the avid binding of LukGH to the CD11b receptors certainly provides a competitive advantage over the monovalent endogenous ligands. Following the same principle, the anthrax toxin protective antigen (PA) binds to its von Willebrand type I domain receptor, to the MIDAS site, via the PA_IV domain, but forms additional interactions using a neighboring domain (PA_II), leading to an ∼1,000-fold higher affinity compared to a typical integrin–ligand complex (38). Moreover, it was shown that the protonation of a histidine residue on the receptor, at the edge of this additional binding pocket, controls the pH-dependent dissociation from PA_II and subsequent pore formation, reminiscent of the CD11b-I interaction with LukG, where reduced interactions appear to favor oligomerization (38).
CD11b/CD18 and the other β2 integrins (CD11a, CD11c, and CD11d) play important roles in immune defense mechanisms, at the same time regulating immune responses (39). Whereas reduction or lack of β2 integrins leads to higher susceptibility to infection and impaired inflammatory responses, increased expression or activation of integrins has been linked to autoimmune diseases, such as systemic lupus erythematosus, rheumatoid arthritis, multiple sclerosis, as well as inflammation-aggravated conditions, such as stroke (39). The extremely high specificity and avidity of LukGH toward activated CD11b, and the availability of structural information for the interaction, make LukGH a suitable candidate for engineering potential therapeutic candidates, with or without functional pores, targeting integrins in inflammatory diseases. The caveats of using a nonhuman therapeutic protein, particularly for chronic indications, typically arise from the short half-life and formation of antidrug antibodies. However, these could be potentially circumvented for LukGH by exploiting its own ability to blunt the adaptive immunity via dendritic cell targeting (40) and by making use of its numerous and diverse natural sequence variants (41).
Materials and Methods
Production of Recombinant LukGH Variants.
LukGH variants were produced recombinantly in Escherichia coli, as described previously (27, 28), based on the wild-type sequence of the community-associated methicillin resistant S. aureus (CA-MRSA) USA300 (ST8) TCH1516 strain. Protein concentration was calculated based on the UV absorbance at 280 nm using the extinction coefficient (ε280 = 112 000 M−1 cm−1) calculated with ProtParam tool (ExPASy Server) (42) based on the LukGH protein sequence. Protein purity was determined by SDS/PAGE gels, stability by differential scanning fluorimetry, and the secondary structure by circular dichroism, as described in SI Appendix, Supplementary Material and Methods.
Production of moCD11b-I Variants and Expression and Purification of Recombinant huCD11b-I, rbCD11b-I, and moCD11b-I.
The I-domains (amino acids 127 to 321) of huCD11b, rbCD11b, and moCD11b (huCD11b-I, rbCD11b-I, and moCD11b-I) were cloned into pET24a (Novagen) vector at NdeI/XhoI (NdeI/BamHI for rbCD11b-I) sites and purified and biotinylated, as described in SI Appendix, Supplementary Material and Methods. Due to the lack of tryptophan in the amino acid sequence of hu-, mo-, and rbCD11b-I, protein concentration was determined based on the UV absorbance at 205 nm using the extinction coefficients [ε205(huCD11b-I) = 797 420 M−1 cm−1, ε205(rbCD11b-I) = 790 170 M−1 cm−1, ε205(moCD11b-I) = 794 570 M−1 cm−1] calculated with “A205 protein/peptide concentration webserver” (43). Protein purity and monomer content were assessed by nonreducing SDS/PAGE gel, stability by differential scanning fluorimetry and the secondary structure by circular dichroism, as described in SI Appendix, Supplementary Material and Methods.
Bio-Layer Interferometry.
Binding of LukGH (wild-type and mutants) to huCD11b-I, rbCD11b-I, or moCD11b-I (wild-type and mutants) was evaluated by Bio-Layer Interferometry (fortèBio Octet Red96 instrument, Pall Life Sciences), as described previously (27). In brief, biotinylated CD11b-I (2 to 4 μg/mL) was immobilized on streptavidin sensors (fortèBio, Pall Life Sciences). The association of LukGH (50 nM or 100 nM in assay buffer [PBS plus 1% BSA and 1 mM MgCl2 or CaCl2, or 25 mM Hepes, pH 7.5 plus 1% BSA plus 1 mM MgCl2 and NaCl [150 to 1,000 mM]) to the immobilized receptor and dissociation in assay buffer were monitored for 5 min each. Response units (RU) and where possible (for monophasic binding curve) equilibrium dissociation constants (Kd), were determined using the Data Analysis 7 software (fortéBio, Pall Life Sciences) by simultaneously fitting the association and dissociation curves to a 1:1 binding model. The steady-state Kd values were determined for LukGH wild-type binding to moCD11b-I and rbCD11b-I by measuring binding at multiple LukGH concentrations (100 to 2,200 nM and 20 to 400 nM, respectively) and fitting the data to a steady-state equilibrium model (Forte-Bio Analysis Software, v7).
Purification of LukGH–huCD11b-I–Fab and LukGH–Fab Complexes and SAXS Analysis.
LukGH and huCD11b-I, purified as described above, and the Fab of αLukGH-mAb#5.H1H2 (29) expressed in Chinese Hamster Ovary cells and purified by LC-κ affinity chromatography (CaptureSelect, Thermo Scientific), were mixed in 1:1:1.5 molar ratio, respectively. For the LukGH–Fab complex, LukGH and Fab were mixed in 1:1.5 molar ratio. Both complexes were concentrated and purified by size-exclusion chromatography as described in SI Appendix, Supplementary Material and Methods. Synchrotron radiation X-ray scattering data were collected at the EMBL P12 beamline of the storage ring PETRA III (DESY, Hamburg, Germany) (44) for both complexes (LukGH–Fab and LukGH–Fab–huCD11b-I) from a dilution series to examine concentration-dependent alterations, as described in SI Appendix, Supplementary Material and Methods. The indirect inverse Fourier transform of the SAXS data and the corresponding probable real space-scattering pair distance distribution [P(r) versus r profile] were calculated using GNOM (45), from which the Rg and Dmax were determined. The P(r) versus r profile was also used for volume and subsequent molecular weight estimates of the complexes, as described in Hajizadeh et al. (46). CRYSOL (47) was used to calculate the scattering profiles from the atomic coordinates of available crystal structures: The Fab fragment bound to LukGH, as deposited in PDB ID code 5K59 and for the LukGH–Fab–huCD11b-I complex, the interface as described in this work, was projected onto the former complex. The SAXS data (as summarized in SI Appendix, Table S6) and models are deposited in the Small Angle Scattering Biological Data Bank (www.sasbdb.org) with the following accession codes: SASDF45 (48) and SASDF55 (49) for the LukGH–Fab–huCD11b-I and the LukGH–Fab complexes, respectively.
DLS and Oligomerization Assay.
The increase in cumulant radius of LukGH after addition of CD11b-I from different species was followed by DLS using a Wyatt DynaPro DLS Plate Reader II instrument at 25 °C, at preset time points. The samples were prepared as described in SI Appendix, Supplementary Material and Methods. The cumulant radii of the samples, determined as described in SI Appendix, Supplementary Material and Methods, were plotted against incubation time and fitted to a “one-phase association” function in Prism 6 (GraphPad) with a fixed starting radius (y0 = 5 nm).
Cytotoxicity Assay.
Cell-based assays were performed using either differentiated HL-60 cells or human, rabbit, or mouse PMNs, and cytolytic activity of LukGH (wild-type and variants) was assessed as described previously (24, 25), as detailed in SI Appendix, Supplementary Material and Methods. Cell viability was determined with a Cell Titer-Glo Luminescent Cell Viability Assay Kit (Promega) according to the manufacturer’s instructions. Percent viability was calculated relative to mock-treated cells (100% viability). Data were analyzed by nonlinear regression using Prism 6 (GraphPad) and toxin activity is given as EC50 value (half-maximal effective concentration).
To determine LukGH competition with the LM2/1 (a-huCD11b [Mac-1a], eBioscience) and CBRM1/5 (Anti-HumanCD11b, Clone: CBRM1/5, eBioscience) antibodies, LPS-treated human PMNs [as described in Trstenjak et al. (27)], at a concentration of 2.5 × 104 cells per well in the assay medium, were preincubated with corresponding antibody (10 µg/mL) or assay buffer for 30 min prior to incubation with the toxin.
Protein Crystallization.
LukGHK319A with moCD11b-I or LukGH wild-type with huCD11b-I complexes were purified as described in SI Appendix, Supplementary Material and Methods. The purified complexes were concentrated to a final concentration of 5.0 mg/mL and 5.2 mg/mL for LukGHK319A–moCD11b-I and LukGH–huCD11b-I, respectively, and characterized by DLS and reducing SDS/PAGE gel. Diffraction quality crystals were obtained using hanging-drop vapor diffusion at 20 °C, in a drop containing 1-µL complex in 1-µL reservoir solution (25 to 30% [vol/vol] Jeffamine-600, 5 to 10% [vol/vol] DMSO) or 1-µL complex in 0.5-µL reservoir solution (30% [vol/vol] Jeffamine-600, 10% [vol/vol] DMSO), for LukGHK319A–moCD11b-I and LukGH–huCD11b-I, respectively. The crystals were harvested from the crystallization drop using a nylon loop and frozen directly in liquid nitrogen without addition of a cryoprotectant.
Diffraction Data Collection, Structure Determination, Refinement, and Interpretation.
Diffraction data were collected at 100 K at the European Synchrotron Radiation Facility at beamline ID30A-1 (MASSIF-1; wavelength 0.966 Å) for the LukGHK319A–moCD11b-I complex and at beamline ID29 (wavelength 1.072 Å) for the LukGH–huCD11b-I complex. Both datasets were processed using the XDS program package (50). Due to significant anisotropic diffraction, the LukGH–huCD11b-I dataset was corrected and merged using the STARANISO Server (51) incorporating the programs autoPROC (52), POINTLESS (53), and AIMLESS (54).
The LukGHK319A–moCD11b-I structure was solved by molecular replacement in Phaser (55) using LukG and LukH structures from their complex with a Fab fragment [PDB ID code 5K59, chains A and C (29)] as independent search models. After initial model building of LukGH in Coot (56) and 10 cycles of restrained refinement in REFMAC5 (57, 58), additional electron density corresponding to the moCD11b-I domain could be identified clearly and the missing component built in Coot and Buccaneer (59, 60). The structure of LukGH–huCD11b-I was solved by molecular replacement in Phaser (55) by searching sequentially with the LukGH dimer from the refined LukGHK319A–moCD11b-I structure and then with the modified huCD11b-I domain (PDB ID code 1IDO) (13) lacking the C-terminal α7-helix (residues 303 to 315). Both structures of the complexes were finalized by model building and refinement in Coot and Phenix (61). Due to the anisotropic low-resolution diffraction data, the LukGH–huCD11b-I structure was refined by applying additional dihedral-angle restraints derived from the refined LukGHK319A–moCD11b-I structure as a reference model. The data collection, refinement, and validation statistics are shown in SI Appendix, Table S1. The molecular interfaces and oligomeric states were analyzed in PISA (62) and the structures were superposed in the program LSQKAB (63) as a part of the CCP4 program suite (64). The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 6RHV (LukGH_K319A–moCD11b-I) (65) and 6RHW (LukGH–huCD11b-I) (66).
Supplementary Material
Acknowledgments
We thank M. Bowler and D. de Sanctis at the European Synchrotron Radiation Facility, Grenoble, France for providing assistance in using beamlines ID30A-1 (MASSIF-1) and ID29, respectively, and other European Synchrotron Radiation Facility staff members for their excellent support; I. Grishkovskaya and G. Mlynek for initial crystal screening; J. Zmajkovic for cloning the CD11b-I variants; S. Maier for technical assistance; and D. Mantus and L. Stulik for critical reading of the manuscript. The dynamic light scattering, circular dichroism, and differential scanning fluorimetry measurements were performed at the Vienna Biocenter Core Facilities Protein Technologies Facility (https://www.viennabiocenter.org/facilities/). K.D.-C.’s research was supported by the Christian Doppler Laboratory for Knowledge-based Structural Biology and Biotechnology, Federal Ministry of Economy, Family and Youth through the initiative “Laura Bassi Centres of Expertise,” funding the Centre of Optimized Structural Studies, 253275, COST action BM1405–Non-globular proteins, and by the University of Vienna. M.A.G. received support from iNEXT, Grant 653706, funded by the Horizon 2020 programme of the European Commission.
Footnotes
Competing interest statement: A.B. and H.R. are employees of X4 Pharmaceuticals GmbH, the legal successor of Arsanis Biosciences GmbH, which has developed an antileukocidin-GH antibody.
This article is a PNAS Direct Submission. V.J.T. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6RHV and 6RHW) and the Small Angle Scattering Biological Data Bank, https://www.sasbdb.org/ (accession nos. SASDF55 and SASDF45).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913690116/-/DCSupplemental.
References
- 1.Tong S. Y. C., Davis J. S., Eichenberger E., Holland T. L., Fowler V. G. Jr, Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonzo F. 3rd, Torres V. J., The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78, 199–230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spaan A. N., van Strijp J. A. G., Torres V. J., Leukocidins: Staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 15, 435–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita D., et al. , Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins. Nat. Commun. 5, 4897 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Haapasalo K., et al. , Staphylococcus aureus toxin LukSF dissociates from its membrane receptor target to enable renewed ligand sequestration. FASEB J. 33, 3807–3824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tromp A. T., et al. , Human CD45 is an F-component-specific receptor for the staphylococcal toxin Panton-Valentine leukocidin. Nat. Microbiol. 3, 708–717 (2018). [DOI] [PubMed] [Google Scholar]
- 7.DuMont A. L., et al. , Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proc. Natl. Acad. Sci. U.S.A. 110, 10794–10799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho M. K., Springer T. A., Biosynthesis and assembly of the α and β subunits of Mac-1, a macrophage glycoprotein associated with complement receptor function. J. Biol. Chem. 258, 2766–2769 (1983). [PubMed] [Google Scholar]
- 9.Zhang L., Plow E. F., Amino acid sequences within the α subunit of integrin αMβ2 (Mac-1) critical for specific recognition of C3bi. Biochemistry 38, 8064–8071 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Yakubenko V. P., Lishko V. K., Lam S. C. T., Ugarova T. P., A molecular basis for integrin alphaMbeta 2 ligand binding promiscuity. J. Biol. Chem. 277, 48635–48642 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Podolnikova N. P., Podolnikov A. V., Haas T. A., Lishko V. K., Ugarova T. P., Ligand recognition specificity of leukocyte integrin αMβ2 (Mac-1, CD11b/CD18) and its functional consequences. Biochemistry 54, 1408–1420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnaout M. A., Biology and structure of leukocyte β2 integrins and their role in inflammation. F1000Res 5, F1000 Faculty Rev-2433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J. O., Rieu P., Arnaout M. A., Liddington R., Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18). Cell 80, 631–638 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Lee J. O., Bankston L. A., Arnaout M. A., Liddington R. C., Two conformations of the integrin A-domain (I-domain): A pathway for activation? Structure 3, 1333–1340 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Baldwin E. T., et al. , Cation binding to the integrin CD11b I domain and activation model assessment. Structure 6, 923–935 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Xiong J. P., Li R., Essafi M., Stehle T., Arnaout M. A., An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain. J. Biol. Chem. 275, 38762–38767 (2000). [DOI] [PubMed] [Google Scholar]
- 17.McCleverty C. J., Liddington R. C., Engineered allosteric mutants of the integrin alphaMbeta2 I domain: Structural and functional studies. Biochem. J. 372, 121–127 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahalingam B., et al. , Stable coordination of the inhibitory Ca2+ ion at the metal ion-dependent adhesion site in integrin CD11b/CD18 by an antibody-derived ligand aspartate: Implications for integrin regulation and structure-based drug design. J. Immunol. 187, 6393–6401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bajic G., Yatime L., Sim R. B., Vorup-Jensen T., Andersen G. R., Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3. Proc. Natl. Acad. Sci. U.S.A. 110, 16426–16431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen M. R., et al. , Structural basis for simvastatin competitive antagonism of complement receptor 3. J. Biol. Chem. 291, 16963–16976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada Y., Ye X., Simon S., The integrins. Genome Biol. 8, 215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ventura C. L., et al. , Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5, e11634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumont A. L., et al. , Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol. Microbiol. 79, 814–825 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouha H., et al. , Disarming Staphylococcus aureus from destroying human cells by simultaneously neutralizing six cytotoxins with two human monoclonal antibodies. Virulence 9, 231–247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janesch P., et al. , Selective sensitization of human neutrophils to LukGH mediated cytotoxicity by Staphylococcus aureus and IL-8. J. Infect. 74, 473–483 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Malachowa N., et al. , Staphylococcus aureus leukotoxin GH promotes inflammation. J. Infect. Dis. 206, 1185–1193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trstenjak N., et al. , Adaptation of the Staphylococcus aureus leukocidin LukGH for the rabbit host by protein engineering. Biochem. J. 476, 275–292 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Badarau A., et al. , Structure-function analysis of heterodimer formation, oligomerization, and receptor binding of the Staphylococcus aureus bi-component toxin LukGH. J. Biol. Chem. 290, 142–156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badarau A., et al. , Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies. MAbs 8, 1347–1360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DuMont A. L., et al. , Identification of a crucial residue required for Staphylococcus aureus LukAB cytotoxicity and receptor recognition. Infect. Immun. 82, 1268–1276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond M. S., Springer T. A., A subpopulation of Mac-1 (CD11b/CD18) molecules mediates neutrophil adhesion to ICAM-1 and fibrinogen. J. Cell Biol. 120, 545–556 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R., et al. , Activation of integrin alphaIIbbeta3 by modulation of transmembrane helix associations. Science 300, 795–798 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Nguyen V. T., Kamio Y., Higuchi H., Single-molecule imaging of cooperative assembly of γ-hemolysin on erythrocyte membranes. EMBO J. 22, 4968–4979 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Z., et al. , Neutrophil recruitment limited by high-affinity bent β2 integrin binding ligand in cis. Nat. Commun. 7, 12658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie C., et al. , Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J. 29, 666–679 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen M., Springer T. A., Leukocyte integrin αLβ2 headpiece structures: The αI domain, the pocket for the internal ligand, and concerted movements of its loops. Proc. Natl. Acad. Sci. U.S.A. 113, 2940–2945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DuMont A. L., et al. , Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect. Immun. 81, 1830–1841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao M., Schulten K., Onset of anthrax toxin pore formation. Biophys. J. 90, 3267–3279 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schittenhelm L., Hilkens C. M., Morrison V. L., β2 Integrins as regulators of dendritic cell, monocyte, and macrophage function. Front. Immunol. 8, 1866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berends E. T. M., et al. , Staphylococcus aureus impairs the function of and kills human dendritic cells via the LukAB toxin. MBio 10, e01918-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badarau A., Trstenjak N., Nagy E., “Structure and function of the two-component cytotoxins of Staphylococcus aureus–Learnings for designing novel therapeutics” in Protein Reviews, Atassi M. Z., Ed. (Springer Singapore, 2017), vol. 18, pp. 15–35. [DOI] [PubMed] [Google Scholar]
- 42.Gasteiger E., et al. , “Protein identification and analysis tools on the ExPASy Server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Humana Press, 2005), pp. 571–607. [Google Scholar]
- 43.Anthis N. J., Clore G. M., Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 22, 851–858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanchet C. E., et al. , Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 48, 431–443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svergun D. I., Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992). [Google Scholar]
- 46.Hajizadeh N. R., Franke D., Jeffries C. M., Svergun D. I., Consensus Bayesian assessment of protein molecular mass from solution X-ray scattering data. Sci. Rep. 8, 7204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svergun D., Barberato C., Koch M. H. J., CRYSOL—A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995). [Google Scholar]
- 48.Graewert M., SASDF45 – Leukocidin/Integrin alpha-M complex (LukGH-huCD11b-I) in the presence of a neutralizing antibody Fab fragment. Small Angle Scattering Biological Data Bank. https://www.sasbdb.org/data/SASDF45/. Deposited 3 July 2019.
- 49.Graewert M., SASDF55 – Leukocidin (LukGH) from Staphylococcus aureus in complex with a neutralizing antibody. Small Angle Scattering Biological Data Bank. https://www.sasbdb.org/data/SASDF55/. Deposited 3 July 2019.
- 50.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tickle I. J., et al. , STARANISO (Global Phasing Ltd., Cambridge, United Kingdom, 2018). [Google Scholar]
- 52.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cowtan K., The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Cowtan K., Fitting molecular fragments into electron density. Acta Crystallogr. D Biol. Crystallogr. 64, 83–89 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krissinel E., Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43, W314–W319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kabsch W., A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 (1976). [Google Scholar]
- 64.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trstenjak N., Milić D., Djinović-Carugo K., Badarau A., Crystal structure of mouse CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH (LukH K319A mutant). Protein Data Bank. https://www.rcsb.org/structure/6RHV. Deposited 23 April 2019.
- 66.Trstenjak N., Milić D., Djinović-Carugo K., Badarau A., Crystal structure of human CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH. Protein Data Bank. https://www.rcsb.org/structure/6RHW. Deposited 23 April 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.