Significance
Vascular plants contribute to most of the food and feed production on earth and deliver heating and construction materials in the form of wood. Yet, we know very little about how nonvascular plants acquired vascular tissues during evolution. Here, we provide molecular insights into how 2 evolutionarily conserved transcription factors were redeployed to control proliferation of vascular cells. This is an example of a developmental regulator for which the innovations leading to a vascular function are clearly correlated with the emergence of vascular plants. Thus, while cell division alone is likely not the only driver, it might have been an important evolutionary innovation enabling the emergence of vascular plants.
Keywords: plant evolution, plant vascular tissues, TMO5/LHW, tracheophytes, heterodimerization
Abstract
Vascular plants provide most of the biomass, food, and feed on earth, yet the molecular innovations that led to the evolution of their conductive tissues are unknown. Here, we reveal the evolutionary trajectory for the heterodimeric TMO5/LHW transcription factor complex, which is rate-limiting for vascular cell proliferation in Arabidopsis thaliana. Both regulators have origins predating vascular tissue emergence, and even terrestrialization. We further show that TMO5 evolved its modern function, including dimerization with LHW, at the origin of land plants. A second innovation in LHW, coinciding with vascular plant emergence, conditioned obligate heterodimerization and generated the critical function in vascular development in Arabidopsis. In summary, our results suggest that the division potential of vascular cells may have been an important factor contributing to the evolution of vascular plants.
Recent estimates suggest that plants contribute about 80% of all the biomass on earth (1). Most of the biomass is formed by vascular plants (tracheophytes) that can live for centuries and grow to large sizes. While nonvascular plants mostly rely on cell-to-cell symplastic transport, limiting movement of water and solutes to just a few cells, tracheophytes developed specialized vascular tissues with xylem and phloem cell types to facilitate transport over long distances (2). The acquisition of conducting tissues can therefore be regarded as a major evolutionary innovation. Although vascular plants are defined by the presence of vascular tissue with characteristic secondary cell-wall thickenings ensuring both structural support and transport over larger distances (3–6), earlier diverging land plants in the bryophyte group (mosses, liverworts, and hornworts) also contain water-conducting cells (WCC) and food-conducting cells, analogous to xylem and phloem cells (5, 7). Recent work in the moss Physcomitrella patens revealed that the differentiation of WCC hinges on NAC transcription factors orthologous to those that trigger xylem differentiation in the tracheophyte Arabidopsis thaliana (8). Thus, the transcriptional networks leading to a functional conducting tissue with secondary cell-wall depositions are largely conserved. Given that the auxin hormone response pathway, which has been shown to trigger vascular tissue formation in tracheophytes (9, 10), is also conserved in bryophytes (11–15), a key question is, what genetic innovation facilitated the establishment of elaborate vascular tissues in tracheophytes? A fundamental difference in the conducting tissues between bryophytes and tracheophytes is tissue dimensions. The core genetic network driving vascular cell division leading to an increase in girth in Arabidopsis involves a dimer of the TARGET OF MONOPTEROS 5 (TMO5) and LONESOME HIGHWAY (LHW) basic Helix–Loop–Helix (bHLH) transcription factors (16–22), representing a rate-limiting component in vascular cell proliferation. In this study, we show that proliferation of vascular cells may have been key to the evolution of elaborate vascular tissues.
Results
TMO5/LHW Acts as an Obligate Heterodimer in Arabidopsis Vascular Development.
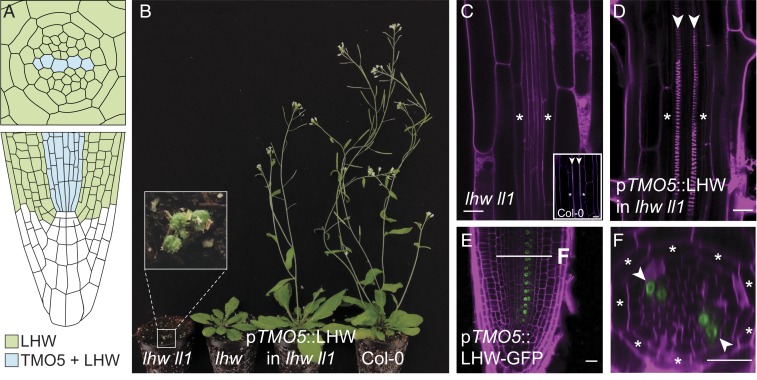
In Arabidopsis, both TMO5 and LHW have several paralogues, and higher-order mutants in either subclade strongly reduce the number of vascular cell files, leading to nearly indistinguishable phenotypes. Simultaneous misexpression of TMO5 and LHW triggers periclinal and radial cell divisions (PRD) in all root meristem cells (16, 17, 19–22). Thus, TMO5 and LHW are functionally interdependent. Within the root meristem, the activity of the TMO5/LHW dimer is limited to sites of common expression: TMO5 and homologs are expressed in young xylem cells, while LHW and homologs have broader expression domains (Fig. 1A) (17, 19, 20). Given that all LHW-clade genes have broader expression domains than TMO5-clade genes, it is conceivable that LHW-clade genes also act independently from TMO5. However, the strong lhw lhw-like1 (lhw ll1) double-mutant phenotype was almost fully rescued in overall growth (Fig. 1B) and root vascular development (Fig. 1 C and D) by expressing LHW only in TMO5-expressing cells (pTMO5::LHW in lhw ll1). Since pTMO5::LHW-GFP fluorescence was retained within the xylem cells (Fig. 1 E and F), suggesting that LHW protein might not move, it is unlikely that LHW has a vasculature-related function outside of the TMO5 domain. Thus, we can assume that the TMO5/LHW dimer likely acts as an obligate heterodimer in Arabidopsis. Given that TMO5 and LHW are phylogenetically distantly related bHLH proteins (23), this result urges the question of when and how the strict dependence on such an obligate heterodimer could have evolved.
Fig. 1.
Expression of LHW in the TMO5 promoter domain is sufficient to rescue the lhw ll1 double-mutant phenotype. (A) Schematic representation of the TMO5 and LHW expression domains in the Arabidopsis primary root (transverse cross-section in Upper and longitudinal section in Lower). Within the LHW domain (green), a subset of cells expresses both LHW and TMO5 (blue). (B–D) Complementation of the lhw ll1 double-mutant phenotype by a pTMO5::LHW transgene, as shown by flowering plant stature (B) and diarch pattern (D; surrounding endodermis layer marked by asterisks; xylem poles marked by arrowheads), compared to the lhw ll1 mutant (B and C). (E and F) Accumulation of LHW-GFP protein in the xylem domain of pTMO5::LHW-GFP root in longitudinal (E) and transverse (F) cross-section. Inset in C shows a control Col-0 root as reference. Asterisks mark surrounding endodermis and arrowheads mark xylem axis. (Scale bars, 20 µm.)
Deep Evolutionary Origin of TMO5 and LHW Genes.
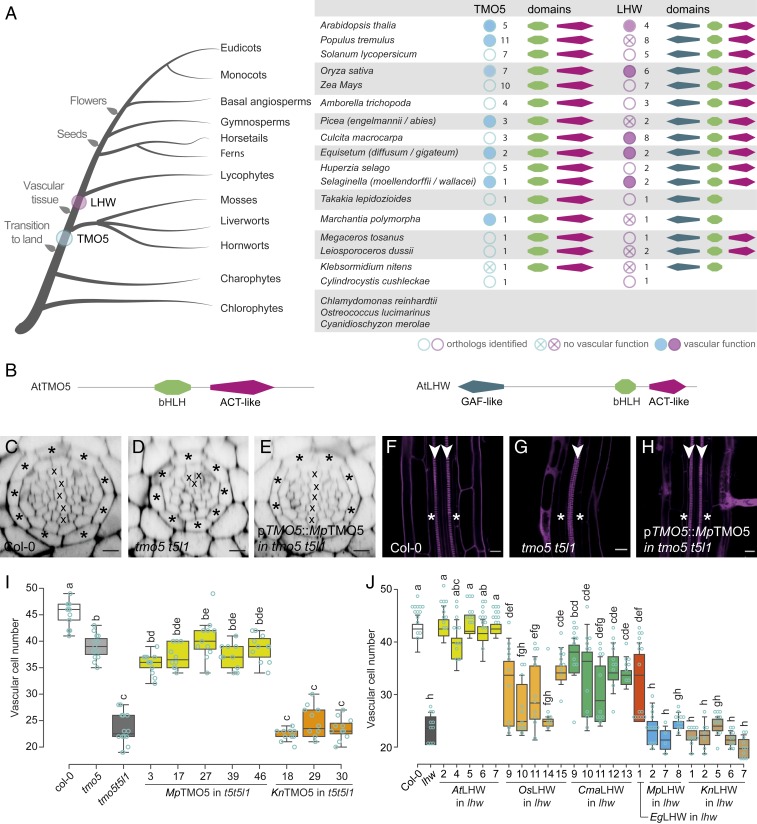
To explore whether the PRD-promoting function triggered by TMO5/LHW may have contributed to the evolution of vascular tissues, we applied a phylogenomic strategy for identifying orthologs in the green lineage, which represent homologs derived by speciation events assumed to have similar functions (24). We used full-length TMO5 and LHW Arabidopsis protein sequences as well as their homologs, TMO5-LIKE1 to 4 (T5L1 to T5L4) and LHW-LIKE1 to 3 (LL1 to LL3) (16), to search against transcriptome assemblies and annotated genome sequences of a representative group of 75 plants ranging from chlorophytes to angiosperms (see details in SI Appendix, Supplementary Material; SI Appendix, Figs. S1–S3 and Table S3). TMO5 and LHW orthologs were found to be present in all major plant lineages even including the charophytes, algal sisters to land plants (Fig. 2A; for individual gene identifiers and phylogenetic analyses with bootstrap values, see SI Appendix, Figs. S1 and S2). We used the closest bHLH family protein orthologs of TMO5 and LHW from several species as an outgroup to perform bootstrap replicas. The results indicated that all TMO5/LHW orthologs that we obtained are indeed grouped with AtTMO5/AtLHW (SI Appendix, Figs. S1 and S2). We further observed that most charophytes and bryophytes contain only a single copy of each gene, whereas numbers increased in the tracheophyte lineage (Fig. 2A). As bHLH family expansion occurred after charophytes split from the embryophytes (23), this suggests that TMO5 and LHW orthologs were among the first bHLH proteins established in the plant lineage. Thus, TMO5 and LHW genes did not appear with the establishment of vascular tissues during land plant evolution, but have a more ancient origin.
Fig. 2.
Acquisition of PRD function by extant TMO5 and LHW proteins. (A) Summary of TMO5 and LHW orthologs in representative species throughout the plant kingdom. Orthologs tested in complementation assays are marked by a blue or purple solid circle if complementation was observed and by a crossed circle if there was no complementation. The associated number of orthologs retrieved and the presence of specific protein domains (as mentioned in B) are indicated as well. (B) Protein domain architecture of AtTMO5 and AtLHW. (C–J) Interspecies complementation analysis of TMO5 (C–I) and LHW (J) orthologs. (C–H) Representative cross-section (C–E) and longitudinal sections (F–H) of a wild-type (C and F; Col-0), tmo5 t5l1 mutant (D and G), and a line carrying the pTMO5::MpTMO5 transgene in the tmo5 t5l1 background (E and H). Cell-file numbers in the vascular tissue (encircled by the endodermis layer; asterisks) were quantified in cross-sections and represented in box plots for TMO5 (I) and LHW (J) orthologs. Lowercase letters in I and J indicate significantly different groups as determined using a one-way ANOVA with post hoc Tukey HSD testing. Blue open circles in I and J represent data points. The presence of a diarch vascular architecture was used as another sign of complementation. Crosses in C–E mark xylem cell files; arrowheads (F–H) indicate xylem poles. (Scale bars in C–H, 20 µm.)
TMO5 Function Evolved Early and Is Highly Conserved.
Although TMO5 and LHW orthologs emerged early during plant evolution, it is plausible that the capacity to regulate vascular development is a derived property that evolved later. We therefore used a cross-species assay to investigate which TMO5 orthologs have the capacity to complement the Arabidopsis tmo5 t5l1 double mutant (henceforth “the capability of orthologs to restore PRD in Arabidopsis mutants” is referred to as “the PRD function”). Due to reduced PRD, this mutant shows a strongly reduced vascular bundle with only 1 protoxylem pole (monarch vascular pattern) (Fig. 2 D and G) instead of 2 protoxylem poles (diarch vascular pattern) in wild type (Fig. 2 C and F). We expressed the TMO5 orthologs (see SI Appendix, Tables S1 and S3, for nomenclature) from the native AtTMO5 promoter (Fig. 2 E, H, and I and SI Appendix, Figs. S4 and S5). We selected most of the orthologs from broadly used model systems in different clades to best represent the evolutionary tree and selected the closest ortholog from each species for all complementation experiments. All tracheophytic TMO5 orthologs that we tested (AtTMO5, PtTMO5, OsTMO5, PaTMO5, EdTMO5, and SmTMO5) complemented the mutant phenotype (SI Appendix, Fig. S4 and Table S1), revealing that the PRD function of this protein is conserved among the vascular plants that we analyzed. Surprisingly, however, the bryophytic MpTMO5 was likewise able to restore vascular development (Fig. 2 E, H, and I and SI Appendix, Figs. S4 and S5 and Table S1), suggesting a functional conservation of TMO5 extending beyond vascular plants to at least one bryophyte species. In contrast, the charophytic KnTMO5 could not complement (Fig. 2I and SI Appendix, Figs. S4 and S5 and Table S1). These data indicate that the land plant TMO5 orthologs tested are capable of performing the PRD function (Fig. 2A) and suggest that the protein function has remained highly conserved despite hundreds of millions of years of evolution.
Establishment of LHW Function Correlates with Emergence of Vascular Plants.
The high conservation in TMO5 function suggests that changes in TMO5 protein function are not connected to the appearance of vascular tissues. We next dissected the emergence of a PRD function for LHW orthologs in a similar complementation study using the lhw mutant and a broad range of LHW orthologs (Fig. 2J and SI Appendix, Figs. S4 and S5 and Table S1). Although only the AtLHW positive control showed a complete rescue of vascular bundle size; the vascular orthologs OsLHW, CmaLHW, EgLHW, and SwLHW could partly restore vascular cell numbers (Fig. 2J and SI Appendix, Figs. S4 and S5 and Table S1). In contrast, the nonvascular orthologs MpLHW, PhLHW, LduLHW, and KnLHW did not show any complementation (Fig. 2J and SI Appendix, Figs. S4 and S5 and Table S1). Although it is still possible that LHW orthologs from other nonvascular species might result in complementation, our results in 4 nonvascular plant species suggest that LHW ortholog function is less conserved than TMO5 function. Strikingly, the ability to mediate vascular cell proliferation is limited to tracheophyte lineages (Fig. 2A) and thus coincides with the emergence of vascular plants. This raises the intriguing possibility that the establishment of a functional TMO5/LHW heterodimer might have contributed to the emergence of elaborate vascular tissues.
TMO5/LHW Heterodimerization Is an Ancient Property.
We next asked if the PRD function of TMO5 and LHW orthologs correlates with their ability to form heterodimers. We first examined the interaction of AtLHW with TMO5 orthologs with a PRD function (AtTMO5 and MpTMO5) as well as KnTMO5, without PRD function, by Bimolecular Fluorescence complementation assay (BiFC). Surprisingly, all 3 TMO5 orthologs interacted with AtLHW (SI Appendix, Fig. S6 A and C), suggesting that the functional differences between KnTMO5 and MpTMO5 are not due to the differential capacity to interact with AtLHW. To investigate if heterodimerization capacity is conserved among the natural TMO5/LHW pairs, we next tested interactions within the same species. Similar to AtTMO5 and AtLHW, bryophytic MpTMO5 and MpLHW can heterodimerize, but this was not the case for the charophytic KnTMO5 and KnLHW (SI Appendix, Fig. S6 B, C, and F–H). Given that bHLH proteins require homotypic interactions with another bHLH protein (either a homo- or heterodimer) to bind DNA (25, 26), it is possible that KnTMO5/KnLHW proteins form homodimers instead. We therefore tested the homodimerization potential of all TMO5 and LHW orthologs and found that, indeed, all 3 TMO5 orthologs formed homodimers (note that no BiFC signal was observed for AtTMO5 homodimerization, but this interaction was confirmed by FRET-FLIM [Förster resonance energy transfer by fluorescence lifetime imaging]) (SI Appendix, Fig. S6 B and C). In contrast, none of the 3 LHW orthologs homodimerized (SI Appendix, Fig. S6B), suggesting that, at least in Klebsormidium nitens, LHW may have another bHLH interaction partner. In summary, these interaction studies indicate that the PRD function is not correlated with the capacity of forming TMO5/LHW heterodimers, as this ability already exists in the prevascular bryophyte Marchantia polymorpha. As such, it is possible that, in Marchantia, MpTMO5 and MpLHW regulate other developmental processes as heterodimers and that the later-evolved TMO5 and LHW were recruited for vascular development in tracheophytes.
A Facultative Interaction Predated Obligate Heterodimerization.
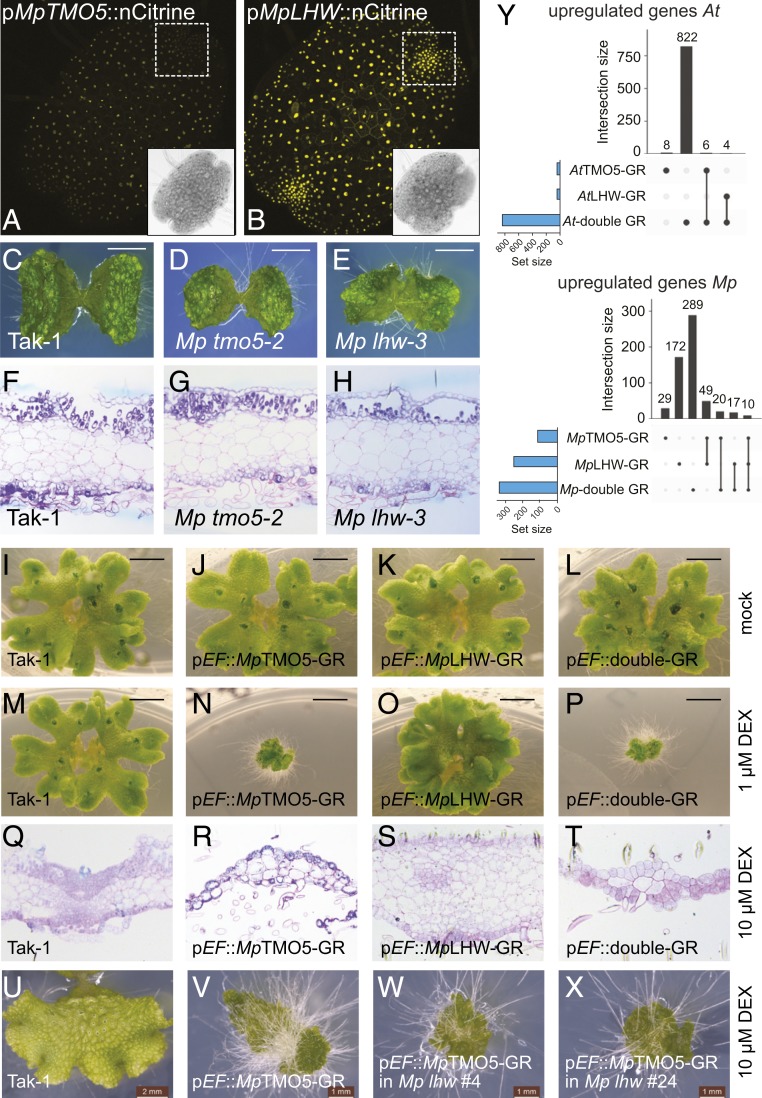
To understand the relation between TMO5 and LHW in a nonvascular plant, we turned to genetic analysis in M. polymorpha, a liverwort with single TMO5 and LHW orthologs and an exquisite set of genetic and genomic tools (27–29). We first analyzed pMpTMO5::nCitrine and pMpLHW::nCitrine expression domains and found both to be ubiquitously expressed in 1-d-old gemmalings, with pMpLHW::nCitrine showing higher expression levels and stronger fluorescence in meristematic notches (Fig. 3 A and B). Despite minor differences, MpTMO5 and MpLHW are thus coexpressed in most of their expression domains (SI Appendix, Fig. S7), consistent with heterodimerization potential. To explore MpTMO5 and MpLHW function, we generated 3 independent CRISPR/Cas9 mutants for each gene (SI Appendix, Fig. S8). Although general growth and horizontal expansion of thalli were reduced in both the Mptmo5 and Mplhw mutants (Fig. 3 C–E and SI Appendix, Fig. S8), at the cellular level, no obvious defects were observed in 10-d-old gemmalings (Fig. 3 F–H). In Marchantia, specialized rhizoids with pegged thickening of cell walls form bundles parallel to thalli and are responsible for water transport by capillary forces (30). We tested whether MpTMO5 and MpLHW are involved in the development of these pegged rhizoids. We observed no defect of pegged rhizoids in both Mptmo5 and Mplhw mutants (SI Appendix, Fig. S8C). Also, the band-shape thickening in the storage parenchymatous cells (morphologically similar to the thickening of xylem vessel cells) was not altered in these mutants (SI Appendix, Fig. S8C). To understand if MpTMO5 and MpLHW might be involved in cell-division patterning, we analyzed early gemma development. Division defects were occasionally observed in Mptmo5 mutants, which led to aberrant gemma shape (SI Appendix, Fig. S9). In Mplhw, however, no obvious division defects were observed compared to wild-type Tak-1 (SI Appendix, Fig. S9). These mutant phenotypes show that, although MpTMO5 might be involved in cell-division patterning during early gemma development, neither MpTMO5 nor MpLHW is critically required for normal development. This is in sharp contrast to Arabidopsis, where loss-of-function mutants are lethal (16). Furthermore, while there is some phenotypic resemblance, it is unclear if MpTMO5 and MpLHW functions are interdependent.
Fig. 3.
Marchantia TMO5 and LHW independently control gemmae and thallus development. (A and B) Expression of pMpTMO5::nCitrine (A) and pMpLHW::nCitrine (B) in 1-d-old gemmalings. Note that pMpLHW expression is ubiquitous in apical notches (marked by the dashed box in B), while pMpTMO5 expression is weak from the apical notch (dashed box in A). (C–H) Overall appearance (C–E) and histochemical cross-sections (F–H) of Tak-1 wild-type (C and F) Mp tmo5-2 mutant (D and G) and Mp lhw-3 mutant (E and H) gemmalings. (I–T) Overall appearance (I–P) and histochemical cross-sections (Q–T) of Tak-1 wild-type (I, M, and Q), pEF::MpTMO5-GR (J, N, and R), pEF::MpLHW-GR (K, O, and S), and pEF::double-GR (L, P, and T) thallus that were grown on mock medium (I–L) on 1 μM DEX for 21 d (M–P) or on 10 μM DEX (Q-T) for 7 d. Cross-sections (Q–T) were made near the meristematic notches. (U–X) Overall appearance of Tak-1 wild-type (U), pEF::TMO5-GR in Tak-1 background (V), and 2 independent Mp lhw mutants in the same pEF::TMO5-GR background (W and X), all grown on 10 μM DEX for 14 d. (Scale bars in C–E and U, 2 mm; in I–P, 5 mm; and in V–X, 1 mm.) (Y) Comparison of genome-wide transcriptional changes, as measured by RNA-seq upon induction (up-regulated genes only) of TMO5-GR, LHW-GR, or both (double GR) in Arabidopsis and Marchantia. UpSet plots show numbers of up-regulated expressed genes in each DEX-treated transgenic line compared to DEX-treated wild-type (Col-0 in Arabidopsis, Tak-1 in Marchantia). Total numbers of genes in each line are shown in blue bars (set size), while gene numbers that are either unique to each line or found in intersections (separated or connected black dots) are shown as black bars (intersection size).
Only simultaneous overexpression of AtTMO5 and AtLHW results in massive cell proliferation in Arabidopsis, while single misexpression of each gene induces very mild proliferation (16). Thus, to test functional interdependence, we next generated dexamethasone (DEX)-inducible single and double overexpression lines by driving MpTMO5-GR and MpLHW-GR from the strong pEF promoter (27). In both single overexpression lines, we observed strong morphological changes upon DEX treatment, resulting in distinctive phenotypes that differed between MpTMO5-GR and MpLHW-GR lines (Fig. 3 I–K and M–O). While induction of pEF::MpTMO5-GR resulted in dwarfed plants with increased rhizoid formation, the induction of pEF::MpLHW-GR caused a more compact thallus structure and absence of gemmae cups (Fig. 3 I–K and M–O). Interestingly, combined overexpression did not result in additional phenotypes compared to the single misexpression lines (Fig. 3 L and P). Moreover, the phenotypic changes in pEF::double-GR lines were not due to an increase of cell-division rate, as we did not observe a change in 5-ethynyl-2′-deoxyuridine staining after DEX treatment (SI Appendix, Fig. S10). Closer examination of tissue anatomy in the misexpression thalli revealed a strongly reduced number of cells in pEF::MpTMO5-GR, while pEF::MpLHW-GR induction caused a thickened thallus and cell proliferation (Fig. 3 Q–T). Hence MpTMO5 and MpLHW seem to induce distinct cellular phenotypes, consistent with a model in which they control development independently. To test this hypothesis directly, we generated CRISPR/Cas9 Mplhw loss-of-function mutations in the pEF::MpTMO5-GR background and asked if MpTMO5 misexpression phenotypes required MpLHW function. We observed the same phenotypic changes upon DEX treatment when the pEF::MpTMO5-GR construct was in a control Tak-1 or in a Mplhw mutant background (Fig. 3 U–X compared to Fig. 3 M–P; SI Appendix, Fig. S11), confirming that MpTMO5 does not need MpLHW to exert its function. These results hence support a model in which MpTMO5 and MpLHW function independently in M. polymorpha, in contrast to the obligate heterodimer complex found in A. thaliana.
To further support this hypothesis, we next determined if MpTMO5 and MpLHW regulate different sets of target genes by analyzing genome-wide transcriptional changes upon single or double overexpression in Marchantia. First, we generated a comparable dataset in Arabidopsis roots by similarly inducing either only AtTMO5, only AtLHW, or both together, followed by RNA sequencing (RNA-seq) analysis. As expected based on earlier transcriptome analysis (17, 22, 28, 29), AtTMO5 and AtLHW were clearly found to work cooperatively, as individual induction of AtTMO5 or AtLHW had a very limited transcriptional response (Fig. 3Y and SI Appendix, Table S4), while simultaneous induction of both proteins caused a large transcriptional response with over 800 genes differentially induced, including the known target genes LOG4 and AT4G38650 (blue bars in Fig. 3Y and SI Appendix, Table S4). We next performed the identical treatments on Marchantia 10-d-old gemmalings, followed by RNA-seq analysis. Confirming phenotypic data, and in stark contrast to the Arabidopsis data, single MpTMO5 or MpLHW induction resulted in, respectively, 108 and 248 differentially up-regulated genes, of which most are induced by only one of the transcription factors (blue bars in Fig. 3Y and SI Appendix, Table S4). Combined overexpression of MpTMO5- and MpLHW-induced transcript levels of almost 300 additional genes (blue bars in Fig. 3Y and SI Appendix, Table S4). When analyzing both up- and down-regulated genes together, a similar trend was observed (SI Appendix, Fig. S12 and Table S4). We further performed RNA-seq with Mp tmo5-2 and Mp lhw-1 to understand whether MpTMO5 and MpLHW might have a coregulation function on downstream targets. Consistent with the ectopic expression data, Mp tmo5-2 and Mp lhw-1 have a set of independently regulated genes and also a set of coregulated genes (SI Appendix, Fig. S12 and Table S4). In addition, we analyzed whether MpTMO5-regulated genes were mis-regulated in Mp lhw mutant and vice versa. However, we observed only very limited overlapping differentially expressed genes, indicating that MpTMO5 and MpLHW independently regulate different target genes (SI Appendix, Table S4). Thus, while each protein can act as a transcription factor on its own, regulating different sets of genes, there is a cooperative regulation of a set of additional genes. Importantly, we did not recover the orthologs of the known Arabidopsis TMO5/LHW target genes in Marchantia, and there was only a small overlap in induced genes from both species (SI Appendix, Table S4). This suggests that MpTMO5 and MpLHW do not share transcriptional targets with Arabidopsis TMO5/LHW, indicating a switch in target specificity during vascular plant evolution. Our data thus suggest that the TMO5/LHW function evolved from 2 proteins with distinct functions.
Subsequent Innovations Led to a PRD Function for TMO5/LHW.
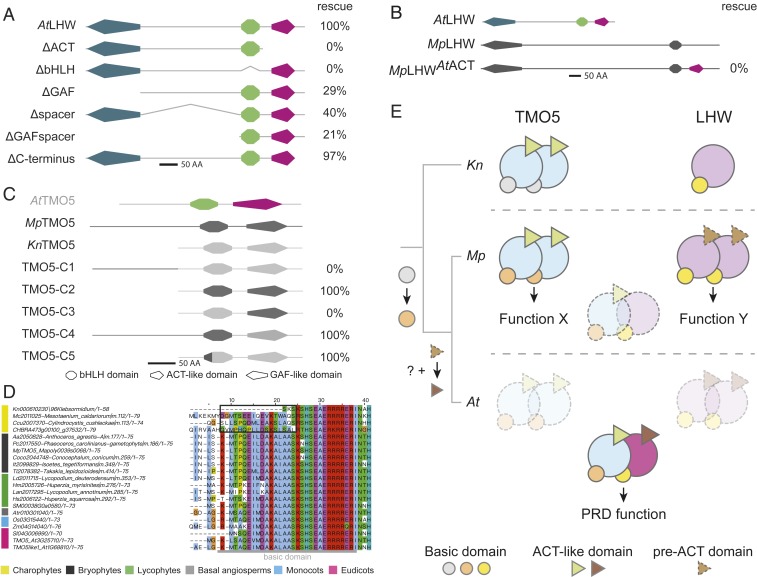
Clearly, modern TMO5/LHW function controlling PRD in vascular plants is not provided by gene absence or presence. Rather, properties of TMO5 and LHW proteins must underlie the acquisition of PRD activity. To identify the protein determinants conferring such activity, we analyzed protein domains in detail. In addition to the family-defining bHLH domain, an ACT-like domain was found in the C-terminal region of both AtTMO5 and AtLHW, even though the amino acid sequences of the ACT-like domain are largely different between AtTMO5 and AtLHW (Fig. 2 A and B and SI Appendix, Figs. S1, S2, and S13). ACT-like domains have been identified in other bHLH transcription factors, where they were shown to mediate protein–protein interactions (26, 31). In addition, AtLHW contains a GAF-like domain at the N terminus (Fig. 2B and SI Appendix, Fig. S2), which is often associated with small-molecule binding and sensory functions such as oxygen and light sensing (32). Interestingly, domain architectures of all TMO5 orthologs were identical in all species that we analyzed across green plants (Fig. 2 A and B and SI Appendix, Fig. S1), while domain architecture in LHW seems less conserved. Indeed, the presence of the ACT-like domain is variable in species of nonvascular clades and stabilizes in tracheophytes (Fig. 2 A and B and SI Appendix, Fig. S2). Given that all TMO5 orthologs tested can interact with AtLHW, but not vice versa (SI Appendix, Figs. S6 and S14), the domain architecture might be important for LHW function.
To first determine the importance of each domain for LHW function, we constructed domain deletions of AtLHW driven by its native promoter and analyzed the capacity to complement the lhw mutant (Fig. 4A and SI Appendix, Fig. S15A). LHW proteins with a deletion of the ACT-like domain (LHWΔACT) or the bHLH domain (LHWΔbHLH) failed to rescue the mutant. Deletion of the N-terminal GAF-like domain (LHWΔGAF) or the ∼300-amino-acid spacer between the GAF-like and bHLH domain (LHWΔspacer) could partially rescue the reduced vascular bundle size, resulting in a diarch phenotype in 29 and 40% of T1 roots, respectively (Fig. 4A and SI Appendix, Fig. S15A and Table S2). Interestingly, having only the bHLH and the ACT-like domain (LHWΔGAFspacer) was sufficient for partial rescue. Finally, removal of the 28-amino-acid C-terminal tail of LHW (LHWΔC terminus) did not affect the PRD function and fully rescued the mutant phenotype (Fig. 4A and SI Appendix, Fig. S15A and Table S2). We thus conclude that the bHLH and ACT-like domains are absolutely required for the PRD function of LHW, while the additional GAF domain and the spacer region are not. Given the importance of the ACT-like domain for a PRD function and considering the presence of this domain is conserved in the tracheophytes, we next tested if gain of the ACT-like domain was an important evolutionary step required for LHW to adopt a PRD function. A chimeric MpLHW protein containing the ACT-like domain of AtLHW (MpLHWAtACT) could not complement the Arabidopsis lhw mutant (Fig. 4B), illustrating that other, yet unknown, modifications contribute to acquiring PRD function.
Fig. 4.
Contributions of LHW and TMO5 domains to the PRD function. (A) Summary of domain deletions in Arabidopsis LHW and their ability to complement the Arabidopsis lhw mutant. The percentage (%) of rescue indicates the percentage of transgenic lines with restoration of diarch vascular pattern in roots. (B) Schematic representation of AtLHW, MpLHW, and the chimeric MpLHWAtACT. Protein domains derived from AtLHW are in light gray and MpLHW domains in dark gray. No complementation (0%) of the Arabidopsis lhw phenotype was observed with MpLHWAtACT. (C) Overview of domain swaps between MpTMO5 (dark gray) and KnTMO5 (light gray) and their ability to complement (% rescue) the diarch vascular pattern phenotype in the Arabidopsis tmo5 t5l1 mutant. The topology of Arabidopsis TMO5 is shown as a reference. (D) Multiple sequence alignment of TMO5 orthologs throughout the phylogenetic tree. The N-terminal region of the basic domain in charophyte (highlighted by the black square) has less homology to the region in land plants, indicating that the stabilization of this domain might be important for its function in land plants. The color bar on the left indicates different clades referred to in the legend below. All proteins in A, C, and D are drawn to scale (bar = 50 amino acids). (E) A model for TMO5/LHW evolution. Evolutionary scenario depicting the innovations in TMO5 and LHW proteins toward PRD function. The presumed ancestral TMO5 and LHW proteins (represented by orthologs in extant charophytes, including Klebsormidium), can form homodimers, but do not heterodimerize. At the origin of land plants, TMO5 (represented by the ortholog in the extant bryophyte M. polymorpha) gained its modern PRD function potential through mutations in its basic domain (orange circles). At the same time, TMO5 and LHW acquired the capacity to form heterodimers, yet control development independently from one another. A critical innovation to LHW in the ancestor of vascular plants (represented by the extant flowering plant Arabidopsis) favored obligate heterodimerization with TMO5 and completed the modern PRD function of this dimer. These changes, indicated by a different color of Arabidopsis LHW, included mutations in the ACT-like domain (brown triangle; the dashed triangle in Marchantia LHW represents the pre-ACT–like domain) together with another, yet unknown modification. Along with this switch to obligate heterodimerization, functions of each homodimer were lost.
The TMO5 domain architecture is conserved from charophytes to angiosperms, but KnTMO5 was not able to complement the Arabidopsis tmo5 t5l1 double mutant (Fig. 2I and SI Appendix, Figs. S4I and S5D). We therefore swapped domains of the complementing MpTMO5 into KnTMO5. Functionality of chimeric proteins was tested using complementation of the Arabidopsis t5t5l1 double mutant (Fig. 4C). Although exchange of the N-terminal fragment (TMO5-C1) was not sufficient to complement the mutant phenotype, swapping both the bHLH and ACT-like domain (TMO5-C2) did provide complementation (Fig. 4C), suggesting that modifications to the bHLH or the ACT-like domain might be key to acquiring the PRD function. Single domain swaps of the ACT-like domain (TMO5-C3) did not result in complementation. However, a swap of the bHLH domain and N terminus (TMO5-C4) was able to rescue the mutant phenotype. Given that the HLH region of the bHLH domain is highly conserved between AtTMO5, MpTMO5, and KnTMO5 (Fig. 4D), we investigated the differences in the basic region, which is required for DNA-binding specificity. Although AtTMO5 and MpTMO5 have similar basic regions, the first part is very different in KnTMO5 (Fig. 4D). Indeed, swapping only 16 amino acids upstream of the HLH domain, including the basic domain (TMO5-C5), was sufficient to create a PRD function for KnTMO5 (Fig. 4C and SI Appendix, Fig. S15B and Table S2). Moreover, multiple sequence alignments of the bHLH domain from 21 TMO5 orthologs (4 charophytic, 6 bryophytic, and 11 tracheophytic TMO5s) showed that the N-terminal part of the basic region is conserved in all land plants (Fig. 4D). Hence, we conclude that TMO5 is functionally conserved down to bryophytes and with a small modification to the basic region even up to charophytes. Given that TMO5 acts in a dedicated pathway associated with vascular plants, this is highly unexpected and establishes TMO5 as a highly conserved developmental regulator.
Discussion
Evolution of the TMO5/LHW Dimer and the Emergence of Vascular Plants.
Here, we propose a model for the evolution of the obligate TMO5/LHW heterodimer in vascular plants. In a first step with the emergence of land plants, point mutations in the basic region of the bHLH domain conferred the modern function to TMO5, which has been maintained in all subsequent lineages we tested (Fig. 4E). In a second step, conserving the presence of an ACT-like domain and other, yet unknown, modifications to LHW led to a preference for TMO5/LHW heterodimerization rather than homodimerization or interactions with other partners. This modification extended the range of target gene recognition and generated the function that is conserved among vascular plants (Fig. 4E). Nevertheless, it remains possible that evolution of target genes might also contribute to the TMO5-LHW regulatory network and emergence of vascular tissue. Intriguingly, the deep functional conservation of TMO5 suggests that modifications specifically to LHW were required to develop the PRD function of the TMO5/LHW complex. Importantly, the origin of these modifications to LHW that lead to an obligate TMO5/LHW heterodimer complex with PRD function is correlated with the appearance of tracheophytes. Although it is currently impossible to prove causality of this innovation to vascular tissue elaboration, it is worth highlighting that there is no such correlation for other pathways involved in vascular development, such as auxin signaling (11), CK signaling (33), or secondary cell-wall formation through VND transcriptional regulators (8). As such, we provide a unique example of a developmental regulator for which the innovations leading to a PRD function are correlated to the emergence of vascular plants. This leads to the intriguing hypothesis that the acquisition of cell-division capacity leading to a larger bundle of cells might have been a crucial innovation leading to vascular plant evolution. This is further supported by genetic analysis in Arabidopsis that revealed a large degree of self-organization in vascular tissue development, constrained by the number of cell files. Indeed, different doses of TMO5 and/or LHW lead to a range of vascular bundle cell-file numbers (16, 19, 20). Thus, while cell division alone is likely not the only driver, it conceivably is an important evolutionary innovation enabling the emergence of vascular plants.
Materials and Methods
A complete overview of the materials and methods used in this research can be found in SI Appendix. This includes information on the phylogeny analysis, complementation assays, cloning, protein–protein interaction studies and RNA-seq experiments.
Data Availability.
All RNA-seq data have been deposited in the National Center for Biotechnology Information as BioProject number PRJNA528622 (https://www.ncbi.nlm.nih.gov/bioproject/528622). All other data are available in the main text or SI Appendix.
Supplementary Material
Acknowledgments
We thank Veronique Storme and Helena Arents for help with statistical analysis. We acknowledge financial support from Netherlands Organization for Scientific Research Grants VIDI-864.13.001 (to B.D.R.) and VICI-865-14-001 (to D.W.); European Research Council Starting Grant 714055 (to B.D.R.); and European Molecular Biology Organization Long-Term Postdoctoral Fellowship ALTF 415–2016 (to H.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information database (accession no. PRJNA528622).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912470117/-/DCSupplemental.
References
- 1.Bar-On Y. M., Phillips R., Milo R., The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucas W. J., et al. , The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 55, 294–388 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Nieminen K., Blomster T., Helariutta Y., Mähönen A. P., Vascular cambium development. Arabidopsis Book 13, e0177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Rybel B., Mähönen A. P., Helariutta Y., Weijers D., Plant vascular development: From early specification to differentiation. Nat. Rev. Mol. Cell Biol. 17, 30–40 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Ohtani M., Akiyoshi N., Takenaka Y., Sano R., Demura T., Evolution of plant conducting cells: Perspectives from key regulators of vascular cell differentiation. J. Exp. Bot. 68, 17–26 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Fukuda H., Ohashi-Ito K., Vascular tissue development in plants. Curr. Top. Dev. Biol. 131, 141–160 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Ligrone R., Ducket J. G., Renzaglia K. S., Conducting tissues and phyletic relationships of bryophytes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 795–813 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu B., et al. , Contribution of NAC transcription factors to plant adaptation to land. Science 343, 1505–1508 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Sachs T., On determination of pattern of vascular tissues in peas. Ann. Bot. 32, 781–790 (1968). [Google Scholar]
- 10.Sachs T., The control of patterned differentiation of vascular tissues. Adv. Bot. Res. 9, 151–262 (1981). [Google Scholar]
- 11.Mutte S. K., et al. , Origin and evolution of the nuclear auxin response system. eLife 7, e33399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman J. L., et al. , Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Rensing S. A., et al. , The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Prigge M. J., Lavy M., Ashton N. W., Estelle M., Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr. Biol. 20, 1907–1912 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Kato H., et al. , Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. PLoS Genet. 11, e1005084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rybel B., et al. , A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437 (2013). [DOI] [PubMed] [Google Scholar]
- 17.De Rybel B., et al. , Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345, 1255215 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Schlereth A., et al. , MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Ohashi-Ito K., Bergmann D. C., Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 134, 2959–2968 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohashi-Ito K., Matsukawa M., Fukuda H., An atypical bHLH transcription factor regulates early xylem development downstream of auxin. Plant Cell Physiol. 54, 398–405 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Ohashi-Ito K., Oguchi M., Kojima M., Sakakibara H., Fukuda H., Auxin-associated initiation of vascular cell differentiation by LONESOME HIGHWAY. Development 140, 765–769 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Ohashi-Ito K., et al. , A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 24, 2053–2058 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Pires N., Dolan L., Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nehrt N. L., Clark W. T., Radivojac P., Hahn M. W., Testing the ortholog conjecture with comparative functional genomic data from mammals. PLOS Comput. Biol. 7, e1002073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murre C., Helix-loop-helix proteins and the advent of cellular diversity: 30 years of discovery. Genes Dev. 33, 6–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feller A., Machemer K., Braun E. L., Grotewold E., Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Althoff F., et al. , Comparison of the MpEF1α and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic Res. 23, 235–244 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Vera-Sirera F., et al. , A bHLH-based feedback loop restricts vascular cell proliferation in plants. Dev. Cell 35, 432–443 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Katayama H., et al. , A negative feedback loop controlling bHLH complexes is involved in vascular cell division and differentiation in the root apical meristem. Curr. Biol. 25, 3144–3150 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Shimamura M., Marchantia polymorpha: Taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol. 57, 230–256 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Feller A., Hernandez J. M., Grotewold E., An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J. Biol. Chem. 281, 28964–28974 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Aravind L., Ponting C. P., The GAF domain: An evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22, 458–459 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Pils B., Heyl A., Unraveling the evolution of cytokinin signaling. Plant Physiol. 151, 782–791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data have been deposited in the National Center for Biotechnology Information as BioProject number PRJNA528622 (https://www.ncbi.nlm.nih.gov/bioproject/528622). All other data are available in the main text or SI Appendix.