Significance
Ultrasmall nanoparticles, which have diameters <10 nm, are mobile, often cell active, and have marked therapeutic potential. Amorphous silica in this size range, at relatively low doses, was recently demonstrated to activate T cells in vitro prior to its dissolution into benign orthosilicic acid [Si(OH)4]. For both safety and therapeutic applications, a mechanism of action greatly facilitates translation. This work shows that ultrasmall amorphous silica nanoparticles directly ligate the T cell receptor (TCR) complex, notably the extracellular domains of CD3, and initiate signaling downstream. This surprising receptor selectivity, the well-known safety of the rapidly formed degradation product and the low cost of silica, bode well for use of these particles as affordable and accessible therapeutic TCR:CD3 agonists.
Keywords: T cell activation, ultrasmall silica nanoparticle, TCR:CD3 complex
Abstract
The impact of ultrasmall nanoparticles (<10-nm diameter) on the immune system is poorly understood. Recently, ultrasmall silica nanoparticles (USSN), which have gained increasing attention for therapeutic applications, were shown to stimulate T lymphocytes directly and at relatively low-exposure doses. Delineating underlying mechanisms and associated cell signaling will hasten therapeutic translation and is reported herein. Using competitive binding assays and molecular modeling, we established that the T cell receptor (TCR):CD3 complex is required for USSN-induced T cell activation, and that direct receptor complex–particle interactions are permitted both sterically and electrostatically. Activation is not limited to αβ TCR-bearing T cells since those with γδ TCR showed similar responses, implying that USSN mediate their effect by binding to extracellular domains of the flanking CD3 regions of the TCR complex. We confirmed that USSN initiated the signaling pathway immediately downstream of the TCR with rapid phosphorylation of both ζ-chain–associated protein 70 and linker for activation of T cells protein. However, T cell proliferation or IL-2 secretion were only triggered by USSN when costimulatory anti-CD28 or phorbate esters were present, demonstrating that the specific impact of USSN is in initiation of the primary, nuclear factor of activated T cells-pathway signaling from the TCR complex. Hence, we have established that USSN are partial agonists for the TCR complex because of induction of the primary T cell activation signal. Their ability to bind the TCR complex rapidly, and then to dissolve into benign orthosilicic acid, makes them an appealing option for therapies targeted at transient TCR:CD3 receptor binding.
Humans are exposed daily to high levels of amorphous silica particles. Ingestion probably accounts for the greatest exposure and studies show that an average adult in the United Kingdom ingests ∼35 mg of different particulate silicates per day via a combination of foods, tablets/capsules, and toothpaste (1, 2). Inhalation is another route of exposure as, not only are silicate aerosols ubiquitous in nature (3), inhalable silicates are common in manufactured sources, including toners, varnishes, and personal hygiene products (4). Indeed, systemic human exposure to silica particles will likely increase as amorphous silica finds use in drug and gene-therapy delivery systems, coatings for medical contrast agents, and cross-linking agents for tissue repair (5–8).
Ultrasmall silica nanoparticles (USSN), meaning those <10 nm in diameter, have received particular recent attention in view of biomedical applications (9). Ultrasmall particles are especially mobile, are in the dimensional range of biological macromolecules, and may well be cell-active in ways that are currently not well predicted or understood (10). Unintended environmental exposure is poorly described for particles of this size but, with silica for example, it is very likely that the gastrointestinal tract experiences their presence following the digestion of larger silica particles (2), while airborne exposure to USSN is also anticipated for some of the manmade sources described above (4). Of particular interest for USSN, however, is their therapeutic potential (9, 11). These particles, without any doping or surface modification, were recently shown to induce CD4+ and CD8+ T cell activation in vitro, regardless of the cells’ antigenic specificity (11). As such, USSN significantly increased the expression of the activation markers CD25 and CD69 on both CD4+ and CD8+ T cell subsets from primary cell cultures and induced the secretion of IFN-γ, indicating a Th1-type skewing. Unusually, this effect was not dependent on the presence of antigen-presenting cells (APCs), implying direct T cell–USSN interaction. Activation of T cells has immunotherapeutic potential for a broad range of conditions, from cancer to autoimmunity and transplantation tolerance (12, 13). Fresh, amorphous, USSN are inexpensive to synthesize and require no humanization compared to biological antibody-based agonistic approaches. However, a key challenge to translation of this material to an affordable and accessible treatment is an understanding of the mechanism of action, the cell signaling that results, and the precise activation status of the T cell after application of USSN. This study reports on these findings and paves the way for next-step therapeutic translation of USSN.
Results and Discussion
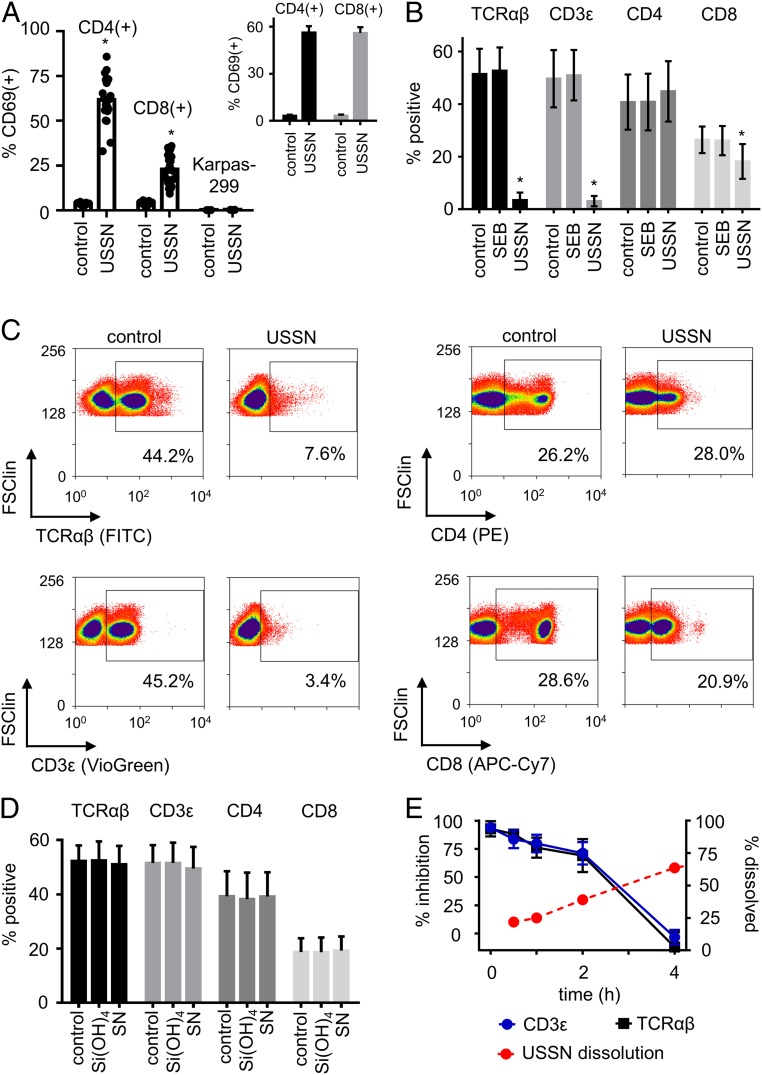
USSN had a median volume diameter (DV0.5) of 3.6 nm and gradually dissolved, as previously reported (11), when diluted in Hepes buffer or in our cell culture medium of RPMI and 10% FBS (SI Appendix, Fig. S1). We first confirmed that, as previously reported (11), these USSN, at 800 µM [Si], stimulated primary blood-derived human T cells, resulting in increased expression of the early T cell activation marker CD69, on CD4+ and CD8+ T cells in primary blood mononuclear cells (PBMC) from multiple different donors (Fig. 1A). Addition of 4% BSA did not affect baseline CD69 expression and, despite known interactions of this protein with larger silica species (14, 15), it did not inhibit the T cell stimulatory effect of USSN (Fig. 1 A, Inset vs. main image). While USSN also up-regulate CD69 on the Jurkat CD4 T cell line (11), similar activation was not evident for the Karpas-299 CD4 T cell line (Fig. 1A), which lacks a competent T cell receptor (TCR) complex (16). Since this implied that the TCR complex is required for USSN-induced stimulation, we undertook competitive binding studies to determine whether the particles and receptor complex directly interact. Fluorescent antibodies for the constant domain of the TCR αβ subunits and the CD3ε domain were almost entirely inhibited from binding to T cells when the mixed cell cultures were pretreated with USSN for 5 min (Fig. 1 B and C). In contrast, while competition for CD4 and CD8 antibody staining yielded some decrease in fluorescence intensity (Fig. 1C), the overall percentage of cells that were stained for these markers was barely affected (Fig. 1B), implying selectivity by USSN for the TCR complex. Neither dissolved silica [Si(OH)4] nor silica nanoparticles outside of the ultrasmall size range (DV0.5 = 14.1 ± 1.4-nm diameter) had any impact on antibody binding (Fig. 1D). Moreover, Staphylococcal enterotoxin B (SEB), a superantigen known to bind the variable region of the TCR, did not affect antibody binding to this receptor, suggesting that USSN selectivity is indeed for the constant region of the TCR complex.
Fig. 1.
T cell activation and TCR complex targeting by USSN. (A) CD69 expression on PBMC-residing CD4 and CD8 T cells, and on the Karpas-299 T cell line, with vehicle-only stimulation (control) or in response to 800 μM USSN. Cells were in RPMI media containing 10% FBS. Data represent the means ± SD for 23 different donors (CD4 and CD8 T cells) or for 3 independent replicates (Karpas-299). (Inset) CD69 expression on PBMC-residing CD4 and CD8 T cells with vehicle-only stimulation (control) or in response to 800 μM USSN in RPMI media containing 4% BSA. Data represent the means ± SD for 3 different donors. (*P < 0.05, denotes significance compared to the control, paired t-test.) (B) Percentage of cells positive for TCR, CD3, CD4, and CD8 within a T cell gate after treatment with vehicle, SEB, or 800 μM USSN for 5 min. Data represent the means ± SD for 4 donors (*P < 0.05, denotes significance compared to the control, paired t-test). (C) Representative flow cytometry plots showing the expression of TCR, CD3, CD4, and CD8 on cells within a T cell gate after treatment with vehicle or 800 μM USSN for 5 min. (D) Percentage of cells positive for TCR, CD3, CD4, and CD8 within a T cell gate after treatment for 10 min with either dissolved silica [Si(OH)4, at 800 μM] or with 800 μM silica nanoparticles (SN) just outside of the ultrasmall size range (DV0.5 = 14.1 ± 1.4-nm diameter). Data represent the means ± SD for 4 different donors. (E, Left y axis) Percent inhibition of TCR and CD3 within a T cell gate after treatment with 800 μM USSN for 0.1 to 4 h. Means ± SD for 4 donors. Right y axis shows the percent dissolution of 800 μM USSN in the media at 37 °C as abstracted from SI Appendix, Fig. S1C. Means ± SD for 3 replicates.
USSN dissolve over time when dispersed into tissue culture medium (SI Appendix, Fig. S1C), and we exploited this property to determine the longevity of the nanoparticle interaction with the TCR complex. When cells had been primed for 30 min with a single dose of USSN, the binding inhibition of TCR complex-targeted antibodies was less pronounced than at 5 min; by 4 h there was almost no inhibition at all (Fig. 1E). The loss of the inhibitory effect inversely mirrored the particle dissolution characteristics in the same culture medium (Fig. 1E).
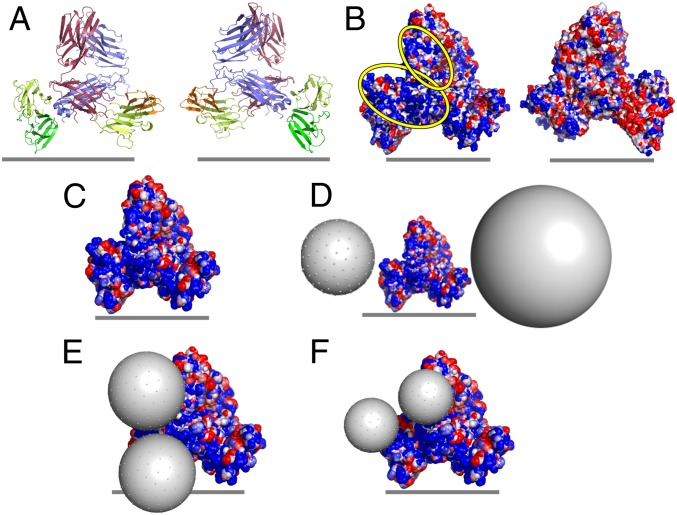
To probe the molecular feasibility of a direct interaction between USSN and the TCR complex, we created molecular models of the human TCR:CD3 complex and considered both size and electrostatic parameters in relation to the silica nanoparticles (Fig. 2). Prior studies have demonstrated a clear association between silica particle size and T cell stimulation because USSN-induced stimulatory responses markedly decreased when median hydrodynamic particle size increased from 3.6 to 5.1 nm to 7.8 nm, despite having very similar particle dissolution characteristics (11). In that same work, USSN surface charge was also shown to be negative (approximately −20 mV) at pH 7.1. Here, we show that electropositive patches, which could act as sites of interaction for the electronegative USSN, are present on both the human TCR and CD3 subunits (Fig. 2B and SI Appendix, Figs. S2 and S3), corroborating previous studies that modeled the electrostatic potential of the CD3 chains (17). These patches, along with the locations of the most strongly electropositive amino acids, were most prominent around TCRα and CD3γε, as well as at the interface between these 2 domains. Differently sized particles (pseudoatoms), representing USSN of varying hydrodynamic diameters, were manually positioned on the electropositive patches of the solvent accessible surface of the modeled human TCR:CD3 complex to assess the spatial feasibility of a direct interaction (Fig. 2 D–F). Steric interferences from the cell membrane hindered the 8-nm and 15-nm particles from achieving close proximity with certain electropositive patches of the TCR:CD3 complex (Fig. 2D). In contrast, these electropositive patches were much more accessible for the 5-nm and, as used in this work, 3.6-nm particles (Fig. 2 E and F and SI Appendix, Fig. S4).
Fig. 2.
The electrostatic potential of the TCR:CD3 complex surface and potential interfaces for USSN binding. (A) Ribbon representations of the modeled human TCR:CD3 complexes: red, TCRα chain; blue, TCRβ chain; orange, CD3δ chain; yellow, CD3ε chain; green, CD3γ chain. (B) Molecular surface representation of electrostatic potential (calculated by Bluues) for modeled human TCR:CD3 complexes. Electrostatic surfaces are shown over a range of ±2 kJ/mol q, with blue representing electropositive and red representing electronegative. Electropositive regions of interest are highlighted with yellow ovals. The Left and Right images in A and B represent front and back views of the receptor complex, related by a 180° rotation around the vertical axis, and the gray lines represent the approximate location of the cell membrane. (C) Electrostatic potential of the solvent-accessible surface of the human TCR:CD3 complex (colored as in B). (D–F) Theoretical representation of the potential interaction between the human TCR:CD3 complex and USSN, with USSN diameter = (D) 8 and 15 nm, (E) 5 nm, and (F) 3.6 nm.
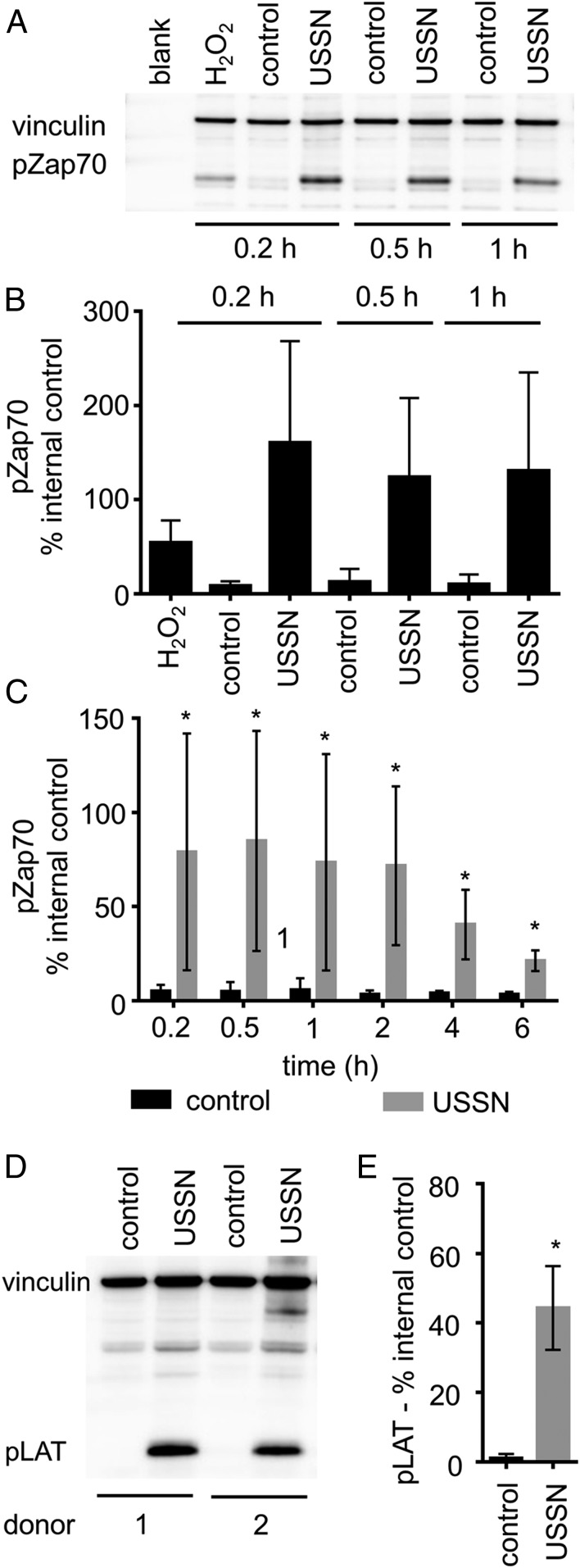
Overall, the data show that USSN, especially those less than 8 nm in diameter would, physically, be able to engage TCR complexes and, potentially, stimulate T cells. To confirm whether TCR complex engagement by USSN leads to T cell stimulation, we assessed whether the particles induced the phosphorylation of ζ-chain-associated protein 70 (Zap70) and linker for activation of T cells (LAT). These proteins become phosphorylated specifically after TCR ligation as members of the T cell activation signaling cascade (18). Jurkat cells, treated with USSN and incubated for up to 1 h, showed markedly increased levels of phosphorylated Zap70 (pZap70) in cell lysates compared to those from untreated cells and even compared to cell lysates from hydrogen peroxide-treated positive controls (Fig. 3 A and B). The effect was also evident in T cell-enriched primary human blood cells (Fig. 3C). Longer incubation periods showed that phosphorylation fell away between 2 and 6 h, probably due to particle dissolution (SI Appendix, Fig. S1C). The enriched T cell cultures also contained significant levels of phosphorylated LAT (pLAT) after USSN treatment (Fig. 3 D and E). Collectively, our data show that USSN initiate signaling downstream of the TCR following direct ligation of the receptor complex by the nanoparticles.
Fig. 3.
The effect of USSN on the phosphorylation of Zap70 and LAT in T cells. (A) A representative immunoblot of cultures stimulated for 0.2 to 1 h, showing the effect of 800 μM USSN on the phosphorylation of Zap70 in Jurkat cells. Blots were probed with antibodies to detect pZap70 (at Tyr319, protein ∼70 kDa) and vinculin (protein ∼128 kDa). (B) Percentage of pZap70 band volume compared to the vinculin band control at 0.2 to 1 h. Western blot data represent the means ± SD for 3 independent replicates. (C) Percentage of pZap70 band volume compared to the vinculin band control at 0.2 to 6 h in enriched T cell cultures. Western blot data represent the means ± SD for 6 different donors (*P < 0.05, denotes significance compared to the control, paired t-test). (D) A representative immunoblot of cultures stimulated for 0.2 h, showing the effect of 800 μM USSN on the phosphorylation of LAT in enriched T cells. Blots were probed with antibodies to detect pLAT (at Tyr191, protein ∼37 kDa) and vinculin (protein ∼128 kDa). (E) Percentage of pLAT band volume compared to the vinculin band control at 0.2 h in enriched T cell cultures. Western blot data represent the means ± SD for 6 different donors (*P < 0.05, denotes significance compared to the control, paired t-test).
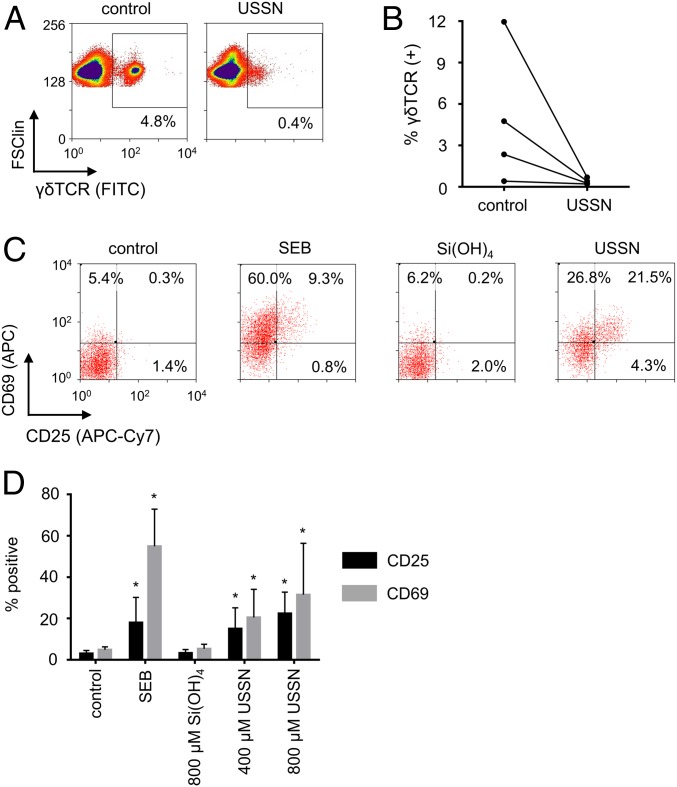
The cellular response of T cells bearing a γδ TCR was investigated next to understand better the selectivity of the interaction between USSN and the TCR:CD3 complex. γδ T cells, a T cell subset found abundantly in gut mucosa and less frequently in circulation, do not recognize MHC-presented antigen, but instead recognize soluble antigen, antigens presented by receptors such as CD1, and other receptors present on cells (19–23). Although the structure of the γδ TCR subunits differs significantly from that of the αβ TCR subunits (24), the USSN were also found to effectively inhibit cell surface γδ TCR antibody staining (Fig. 4 A and B). These results imply that the nanoparticles do not interact with the TCR subunits but instead engage the flanking CD3 subunits, a mechanism that is indeed known to lead to T cell activation (25). As with αβ T cells (11), USSN induced the expression of CD25 and CD69 on γδ T cells (Fig. 4 C and D).
Fig. 4.
The cellular impact of USSN on γδ T cells. (A) Representative flow cytometry plots showing the expression of γδ TCR on cells within a T cell gate after treatment with 800 μM USSN for 10 min. (B) Percentage of the cells positive for γδ TCR within a T cell gate after treatment with 800 μM USSN for 4 donors after 10 min. (C) Representative bivariate dot plots showing expression of CD25 and CD69 on γδ T cells with differing stimulants. (D) Percentage of PBMC-residing γδ T cells that are positive for CD25 and CD69 after 24 h with differing stimulants. Data shown represent the mean ± SD for 8 donors (*P < 0.05, denotes significance compared to the control, 2-tailed paired t-test).
The combined experimental and theoretical data demonstrate that USSN less than 8 nm in diameter directly ligate constant domains of the TCR:CD3 receptor complex, likely the CD3 subunits, and initiate signaling downstream of the TCR complex, leading to cell stimulation.
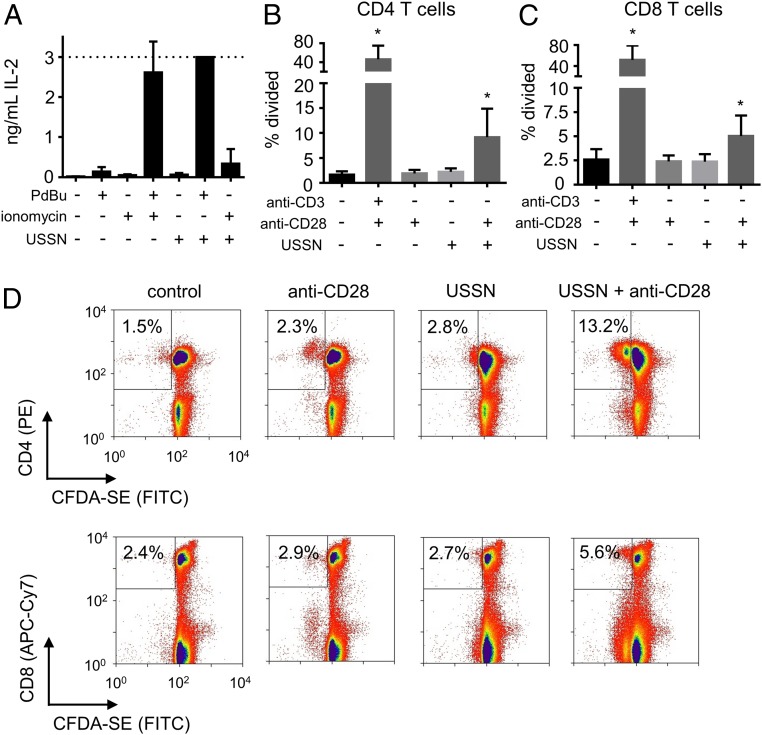
Previously, USSN were found incapable of inducing T cell expansion (11), a response that requires a secondary signal to occur in CD4 T cells, although not necessarily in CD8 T cells (26). Specifically, CD4 T cells require signaling through both the nuclear factor of activated T-cells and mitogen-activated protein kinase pathways to become fully activated (27, 28). These 2 pathways are induced by signaling through the TCR:CD3 complex and a costimulatory receptor (commonly CD28) (29). In vitro, calcium ionophores and phorbate esters are often used to induce these respective pathways (30, 31), and were employed here to help understand how signaling is induced by USSN. Given the long-term toxicity of these chemical inducers (e.g., at 72 h), the extent of activation was first assessed indirectly by measuring levels of the cytokine IL-2 in supernatants at 24 h. IL-2 is produced by fully activated T cells and is important in late-stage cell proliferation (32). Although, as anticipated, the chemical inducers had little effect individually, there was significant IL-2 secretion when cultures were treated with both the calcium ionophore ionomycin and the phorbate ester phorbol-12,13-dibutyrate (PdBu) (Fig. 5A). As such, IL-2 levels are a marker of collaboration between the 2 signaling pathways. USSN exposure alone did not induce IL-2 production, consistent with our previous work (11). However, IL-2 secretion significantly increased when the particles were treated in combination with PdBu but barely so with ionomycin. Thus, in agreement with the findings above, USSN-induced signaling occurs downstream of the TCR:CD3 complex while we additionally show that these particles do not induce secondary signaling. For confirmation, the proliferative response of CD4 T cells to USSN and an anti-CD28 costimulatory antibody was investigated. Clonal expansion was detected when cells were treated with USSN in combination with anti-CD28, but not without anti-CD28 (Fig. 5 B–D), further confirming that the particles bind to the TCR:CD3 complex and induce the primary, but not a secondary, T cell signal.
Fig. 5.
The effect of costimulation on USSN-induced IL-2 production and T cell proliferation. (A) PBMC supernatant IL-2 levels following 24 h culture with (+) or without (−) USSN (400 μM), PdBu (0.5 µg/mL), and ionomycin (0.2 µg/mL). Data represent the means ± SD for 4 independent replicates. (B and C) CD4 and CD8 T cell proliferation at 72 h following PBMC culture with (+) or without (−) USSN (400 μM), anti-CD3 (1 µg/mL) and anti-CD28 (1 µg/mL). Means ± SD for 8 different donors (*P < 0.05, denotes significance compared to the control, paired t-test). (D) Representative flow cytometry plots for the data in B and C with percentages representing fraction of CD4/8(+) cells which underwent rounds of division.
Conclusions
Uncoated inorganic nanoparticles were hitherto unknown to trigger primary TCR signaling, although their nonspecific adherence to T cell membranes is established (33). However, data presented here show that silica nanoparticles in the ultrasmall size range can now be considered a form of TCR agonist.
USSN markedly activate T cells, not only in complex cell culture media containing serum (Fig. 1A), but also at matched concentrations in a basic salt solution (11). They are directly active, therefore, and do not require conditioning by specific cell culture factors. The optimum USSN size for T cell activation and inhibition of TCR antibody binding is 3.6 to 5.1 nm (Fig. 1D) (11). Modeling studies provided a rationale for this, demonstrating electropositive sites on the complex’s TCR and CD3 subunits, which are spatially viable for particle binding (Fig. 2). However, since T cells with structurally dissimilar TCRs (i.e., γδ TCR and αβ TCR) showed comparable responses to USSN, it is most likely that the interaction is mediated by USSN binding at the flanking CD3 subunits of the TCR complex.
Given the demonstrable role of size and presumed role of charge in USSN effector function, it is plausible that other types of particulates in this size range will have similar T cell agonistic effects. However, this would need to be determined on a case-by-case basis because ultrasmall nanoparticles that are transiently stable against agglomeration or dissolution forces, when dispersed in complex aqueous systems, are probably a rarity (34–36). Indeed, silica nanoparticles are well known to exhibit anomalously low agglomeration rates as their size is decreased (37). This is caused by non-DLVO (Derjaguin, Landau, Vervey, and Overbeek) short-range steric repulsion arising from overlap of polysilicic acid chains (38). In addition, the T cell-stimulating effect of USSN was robust to high levels of the biologically ubiquitous protein albumin (Fig. 1A). Albumin is larger (longer) than—and its Stokes radius (3.48 nm) (39) similar to—the z-average size of the USSN used here, and thus classic protein adsorption or “corona” interactions with silica (40) were likely size-limited.
Other work has shown that metabolism of USSN is rapid and that the metabolite (orthosilicic acid) is benign (11, 41). As such, USSN offer the possibility of a safe and economical alternative to certain antibody-based, TCR:CD3 receptor-complex–targeted therapies. Although our current data do not allow extrapolation to in vivo oral or parenteral applications, they certainly pave the way for: 1) Extracorporeal treatment of T cells, in either mono or mixed cell cultures, for subsequent in vivo infusion; or 2) wound management where the environment is protein (albumin) rich and locally active γδ T cells drive the healing process (42–44).
Methods
USSN Dispersion Preparation.
Amorphous USSN dispersions with a median diameter of 3.3 to 3.8 nm were used throughout these investigations, and were prepared by diluting aliquots of alkaline sodium silicate solution (#338443; Sigma-Aldrich Chemical Co.) in ultrahigh purity water (18 MΩ cm) and adjusting pH with hydrochloric acid (HCl, 4 to 5 M). The dispersions were aged at room temperature for >12 h prior to use. Larger, amorphous silica nanoparticles were prepared using the same method, but by adding NaCl at a concentration of 154 mM to the dispersion immediately after pH adjustment to increase the rate of particle growth (45). Hydrodynamic particle size was determined using a Zetasizer NanoZS or NanoZSP (Malvern Instruments). Acquisition parameters for SiO2 (refractive index = 1.45, absorption = 0.01) in a water matrix (0.8872 cP, refractive index = 1.33) at room temperature were used and data were collected using a 173° backscatter angle.
USSN solubility in 50 mM Hepes buffer was determined by a molybdic acid dissolution assay (46). Silicic acid, not colloidal silica, and molybdic acid form a complex which absorbs at 400 nm. Next, 1 part diluted USSN dispersion (or SiF4 standard) and 2 parts molybdic acid solution [4.93 mM (NH4)6Mo7O24·4H2O in 0.15 M H2SO4] were mixed, incubated for 10 min, and absorbance at 400 to 405 nm was measured on a Labsystems Multiskan RC V1.5-0 plate reader (ThermoScientific) or a FLUOstar Omega 27 microplate reader (BMG Labtech).
USSN solubility in complex media was determined by passing particle dispersions through 3-kDa molecular mass cut-off centrifugal ultrafilters (Vivaspin-500, Sartorius Stedim Biotech; 13,000 rpm for 10 min), which have a nominal pore size of 1 to 2 nm (11, 47). Total Si analysis of the dispersion and the ultrafiltrate was performed by inductively coupled plasma optical emission spectroscopy at 251.611 nm on a Jobin Horiba Ultima 2C (Instrument SA) using a V-groove nebulizer and a flow rate of 0.83 mL/min. Samples were diluted with pH 10 NaOH solution prior to analysis. Calibration standards were prepared from SiF4 (1,000 or 10,000 mg/kg Si, Sigma-Aldrich Chemical Co.).
Cellular Studies.
Studies were approved by the UK Health Research Authority NHS Research Ethics Committee, REC reference 18/WM/0221.
Jurkat and Karpas 299 cells were acquired from Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH and maintained in RPMI-1640 media (#R0883, Sigma-Aldrich Co.) containing 10% FBS (#F9665, Sigma-Aldrich Co.), 0.3 g/L l-glutamine, 1% penicillin-streptomycin (also from Sigma-Aldrich Co).
PBMC were isolated using density gradient centrifugation from fresh single leukocyte cones (National Blood Service, Cambridge, UK). Cells were rested 2 h and stored in freezing medium (10% DMSO, 50% FBS, 40% RPMI 1640) at −80 °C or in N2(l). Cells were thawed, washed, and then rested 2 h in RPMI containing 10% FBS, 0.3 g/L l-glutamine, 1% penicillin-streptomycin, and 0.01 µg/mL DNase. T cell enrichment was conducted using a Pan T cell isolation kit (MACS Miltenyl Biotec, #130-096-535) according to the manufacturer’s specifications. T cells were then rested for a minimum of 2 h prior to use.
Cells suspended in RPMI containing FBS or BSA (Labtech, #PM-T1725) were exposed to USSN and controls. USSN, larger (but under 20 nm) silica nanoparticles and silicic acid were sterile-filtered (0.2 µm) prior to addition to growth media. SEB (Sigma Aldrich, #S4881), anti-CD3 (Biolegend, #300414), anti-CD28 (Biolegend, #302914), ionomycin (Sigma Aldrich), and PdBu (Sigma Aldrich) were employed as positive controls. Cells and supernatants were collected after the specified incubation period.
ELISA.
IL-2 levels in cell supernatants were assessed using an IL-2 (#DY202, R&D Systems) ELISA kits according to the manufacturer’s specifications.
Flow cytometry.
Flow cytometry was performed using standard methods, as previously described (11). In brief, a carboxyfluorescein diacetate, succinimidyl ester (CFDA-SE, 90%; Sigma-Aldrich Chemical Co.) dilution assay was used to track T cell proliferation. Prior to treatment, cells were labeled with CFDA-SE (at 106 cells/mL and with 0.1 μM CFDA-SE) for 7 min. Cellular stains and markers were used in accordance with the manufacturer’s suggested protocol, with minor changes. Briefly, at study termination a fluorescent antibody mixture was added to the cells in <100 μL PBS + 1% BSA and cultures were incubated for 20 min on ice. The 7-AAD viability stain was then added and the cells were incubated at room temperature for 10 min. Antibodies and stains used were CD3-VioGreen (Miltenyi Biotec, #130-096-910), CD3-PE (BD Pharmingen, #555340), CD4-PE (BD Pharmingen, #555347), CD25-FITC (BD Pharmingen, #555431), CD25-APC-H7 (BD Pharmingen, #560225), CD69-APC (BD Pharmingen, #555533), CD4-FITC (BD Pharmingen, #555346), CD8-APC-Cy7 (BD Pharmingen, #557834), CD8-APC (Miltenyi Biotec, #130-091-076), αβTCR-FITC (BD Pharmingen, #555547), γδTCR-FITC BD Pharmingen (#347903), and 7-AAD (Life technologies, #A1310; Biolegend, #420404). Cells were fixed with PBS + 2% paraformaldehyde after washing off unbound labels and were stored on ice. Flow cytometry analyses were conducted using a Beckman Coulter CYAN ADP flow cytometer, equipped with 3 solid-state lasers (405, 488, and 642 nm) and 11 detectors (in standard configuration). The machine was calibrated in accordance with the manufacturer’s protocols. Sample acquisition and analysis were conducted using Beckman Coulter Summit software. The total cell suspension or 400,000 events were analyzed. All samples were passed through 35-µM nylon mesh filters prior to analysis. Gating strategies are shown in SI Appendix, Fig. S5.
Western blot.
Western blot analyses were conducted on Jurkat cells and enriched T cell lysates (isolated from PBMC prior to treatment). The lysates and molecular weight standards (Bio-Rad, #1610376) were run on 4 to 12% bis-Tris gels (Invitrogen, #NP0321/0322) and transferred to the PVDF membrane (presoaked in methanol). After blocking for 1 h (in Thermo Scientific block buffer, #37571), the membrane was stained with primary antibody at 4 °C overnight, followed by staining with goat anti-rabbit IgG secondary antibody, HRP-conjugated (#A16096; 1:1,000 dilution) at room temperature for 0.5 h. The membrane was developed with Pierce ECL Plus Western blotting substrate (Thermo Scientific, #32132) and analyzed on a GeneGnome XRQ digital imager (Syngene) using the ECL Plus reagent settings. Band volume was determined through signal integration using GeneTools (Syngene) software. Primary antibodies used were vinculin (Thermo Scientific, #7000626; 1:1,000 dilution), pZap70 (Cell Signaling Technology, #2701; 1:1,000 dilution), pLAT (Cell Signaling Technology, #3584; 1:1,000 dilution). Precision Protein StrepTactin-HRP Conjugate (Bio-Rad, #1610380; 1:10,000 dilution) was used as a secondary antibody. Hydrogen peroxide was used as a positive control (48).
Theoretical Modeling.
Models of the human TCR:CD3 complex were generated from both solved structures and homology models. A homology model of murine CD3δ subunit was created using Modeler v9.17 (49). A BLAST search of the PDB using residues 22 to 105 of the murine CD3δ subunit (UniProt: P04235) resulted in 3 significant hits: 1XIW (17) (human CD3δ, 57% identity), 1XMW (50) (ovine CD3δ, 48% identity), and 1SY6 (51) (human CD3δ, 33% identity). The identity of 1SY6 was deemed to be too low for use as a modeling template. Sequences were aligned using structure-based (T-Coffee) (52) and sequence-based [BLAST (53) and MUSCLE (54)] approaches. Manual adjustment of alignments, based on the observed secondary structure of 1XIW and 1XMW, indicated that human CD3δ was the most appropriate template. Model accuracy was assessed using the molpdf, DOPE, and GA341 functions of Modeler. The stereochemistry of the 3 best-scoring models was assessed using MolProbity (55). The final model has 98.5% favored rotamers and a MolProbity score of 2.59 (42nd percentile).
The TCR:CD3 models were generated by superposition of the species-specific components with the published hybrid model (56) of the TCR:CD3 complex containing murine TCR (PDB ID code 3QJF) and human CD3 (PDB ID codes 1XIW and 1SY6) (51) components. The PDB files of proteins used to construct the final models were: Human TCR (PDB ID code 4P4K) (17), human CD3δε (PDB ID code 1XIW), human CD3εγ (PDB ID code 1SY6), murine TCR (PDB ID code 3QJF) (57), murine CD3δε (homology model plus PDB ID code 1XMW), and murine CD3εγ (PDB ID code 1JBJ) (58).
Electrostatic surfaces were generated based on generalized Born radii using the Bluues server (59). All structural manipulation and presentation were performed using the PyMOL Molecular Graphics System, v1.7.6.6 (Schrödinger). USSN representations are presented as pseudo atoms in which the atomic radius has been adjusted to represent that of the appropriate cluster.
Statistical Analysis.
Two-tailed Student t-test and a 2-way ANOVA, in which a P >0.05 was not considered significant, were used throughout.
Supplementary Material
Acknowledgments
These studies were supported by grants from the UK Medical Research Council (Grant MR/R005699/1) and the Natural Sciences and Engineering Research Council of Canada, as well as through sponsorship from HS Pharmaceuticals, LLC.
Footnotes
Competing interest statement: HS Pharmaceuticals, LLC sponsored B.V.’s PhD studentship through the Medical Research Council. The Medical Research Council has filed patent applications associated with the work and has agreed on an exclusive license with HS Pharmaceuticals LLC. B.V., R.E.H., and J.J.P. are among the inventors who will receive a share in financial return if the Medical Research Council receives income from this license.
This article is a PNAS Direct Submission.
Data deposition: Data is available on Open Science Framework Vis, B., https://osf.io/28wm4 (November 18, 2019), “Ultrasmall Silica Nanoparticles Directly Ligate the T Cell Receptor Complex.”
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911360117/-/DCSupplemental.
References
- 1.Lomer M. C., et al. , Dietary sources of inorganic microparticles and their intake in healthy subjects and patients with Crohn’s disease. Br. J. Nutr. 92, 947–955 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Peters R., et al. , Presence of nano-sized silica during in vitro digestion of foods containing silica as a food additive. ACS Nano 6, 2441–2451 (2012). [DOI] [PubMed] [Google Scholar]
- 3.McLaughlin J. K., Chow W. H., Levy L. S., Amorphous silica: A review of health effects from inhalation exposure with particular reference to cancer. J. Toxicol. Environ. Health 50, 553–566 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Shin J. H., et al. , Subacute inhalation toxicity study of synthetic amorphous silica nanoparticles in Sprague-Dawley rats. Inhal. Toxicol. 29, 567–576 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Bitar A., Ahmad N. M., Fessi H., Elaissari A., Silica-based nanoparticles for biomedical applications. Drug Discov. Today 17, 1147–1154 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Meddahi-Pellé A., et al. , Organ repair, hemostasis, and in vivo bonding of medical devices by aqueous solutions of nanoparticles. Angew. Chem. Int. Ed. Engl. 53, 6369–6373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaramuzzi K., et al. , Nanostructured SBA-15 silica: An effective protective vehicle to oral hepatitis B vaccine immunization. Nanomedicine 12, 2241–2250 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Skrastina D., et al. , Silica nanoparticles as the adjuvant for the immunisation of mice using hepatitis B core virus-like particles. PLoS One 9, e114006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zarschler K., et al. , Ultrasmall inorganic nanoparticles: State-of-the-art and perspectives for biomedical applications. Nanomedicine 12, 1663–1701 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Kopp M., Kollenda S., Epple M., Nanoparticle-protein interactions: Therapeutic approaches and supramolecular chemisty. Acc. Chem. Res. 50, 1383–1390 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Vis B., et al. , Non-functionalised ultrasmall silica nanoparticles directly and size-selectively activate T cells. ACS Nano 12, 10843–10854 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Kuhn C., Weiner H. L., Therapeutic anti-CD3 monoclonal antibodies: From bench to bedside. Immunotherapy 8, 889–906 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Buter J., et al. , Phase I/II study of low-dose intravenous OKT3 and subcutaneous interleukin-2 in metastatic cancer. Eur. J. Cancer 29A, 2108–2113 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Clemments A. M., Botella P., Landry C. C., Protein adorption from biofluids on silica nanoparticles: Corona analysis as a function of particle diameter and porosity. ACS Appl. Mater. Interfaces 7, 21682–21689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehman S. E., Mudunkotuwa I. A., Grassian V. H., Larsen S. C., Nano-bio interactions of porous and nonporous silica nanoparticles of varied surface chemistry: A structural, kinetic and thermodynamic study of protein adsorption from RPMI culture medium. Langmuir 32, 731–742 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Sandberg Y., et al. , Human T-cell lines with well-defined T-cell receptor gene rearrangements as controls for the BIOMED-2 multiplex polymerase chain reaction tubes. Leukemia 21, 230–237 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Arnett K. L., Harrison S. C., Wiley D. C., Crystal structure of a human CD3-ε/δ dimer in complex with a UCHT1 single-chain antibody fragment. Proc. Natl. Acad. Sci. U.S.A. 101, 16268–16273 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brownlie R. J., Zamoyska R., T cell receptor signalling networks: Branched, diversified and bounded. Nat. Rev. Immunol. 13, 257–269 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Dar A. A., Patil R. S., Chiplunkar S. V., Insights into the relationship between toll like receptors and gamma delta T cell responses. Front. Immunol. 5, 366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vantourout P., Hayday A., Six-of-the-best: Unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 13, 88–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiest D. L., “Development of γδ T cells, the special-force soldiers of the immune system” in T-Cell Development: Methods and Protocols, Bosselut R., Vacchio M. S., Eds. (Humana Press, 2016), pp. 23–32. [DOI] [PubMed] [Google Scholar]
- 22.Silva-Santos B., Serre K., Norell H., γδ T cells in cancer. Nat. Rev. Immunol. 15, 683–691 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Luoma A. M., Castro C. D., Adams E. J., γδ T cell surveillance via CD1 molecules. Trends Immunol. 35, 613–621 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allison T. J., Garboczi D. N., Structure of gammadelta T cell receptors and their recognition of non-peptide antigens. Mol. Immunol. 38, 1051–1061 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Trickett A., Kwan Y. L., T cell stimulation and expansion using anti-CD3/CD28 beads. J. Immunol. Methods 275, 251–255 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Wang B., Maile R., Greenwood R., Collins E. J., Frelinger J. A., Naive CD8+ T cells do not require costimulation for proliferation and differentiation into cytotoxic effector cells. J. Immunol. 164, 1216–1222 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Smith-Garvin J. E., Koretzky G. A., Jordan M. S., T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macian F., NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Macián F., et al. , Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109, 719–731 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Chatila T., Silverman L., Miller R., Geha R., Mechanisms of T cell activation by the calcium ionophore ionomycin. J. Immunol. 143, 1283–1289 (1989). [PubMed] [Google Scholar]
- 31.Bell J., T-cell turn-off. Nat. Rev. Immunol. 2, 460 (2002). [Google Scholar]
- 32.D’Souza W. N., Lefrançois L., IL-2 is not required for the initiation of CD8 T cell cycling but sustains expansion. J. Immunol. 171, 5727–5735 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Hewitt R. E., Vis B., Pele L. C., Faria N., Powell J. J., Imaging flow cytometry assays for quantifying pigment grade titanium dioxide particle internalization and interactions with immune cells in whole blood. Cytometry A 91, 1009–1020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira D. I. A., Lederer B., Powell J. J., A balanced salt solution that prevents agglomeration of nano iron oxo-hydroxides in serum-free cellular assays. Mater. Res. Express 2, 015403 (2015). [Google Scholar]
- 35.Murdock R. C., Braydich-Stolle L., Schrand A. M., Schlager J. J., Hussain S. M., Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol. Sci. 101, 239–253 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Moquin A., Neibert K. D., Maysinger D., Winnik F. M., Quantum dot agglomerates in biological media and their characterization by asymmetrical flow field-flow fractionation. Eur. J. Pharm. Biopharm. 89, 290–299 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Higashitani K., et al. , Orders of magnitude reduction of rapid coagulation rate with decreasing size of silica nanoparticles. Langmuir 33, 5046–5051 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi M., Juillerat F., Galletto P., Bowen P., Borkovec M., Aggregation and charging of colloidal silica particles: Effect of particle size. Langmuir 21, 5761–5769 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Axelsson I., Characterization of proteins and other macromolecules by agarose gel chromatography. J. Chromatogr. A 152, 21–32 (1978). [Google Scholar]
- 40.Kubiak-Ossowska K., Tokarczyk K., Jachimska B., Mulheran P. A., Bovine serum albumin at a silica surface explored by simulation and experiment. J. Phys. Chem. B 121, 3975–3986 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Jurkić L. M., Cepanec I., Pavelić S. K., Pavelić K., Biological and therapeutic effects of ortho-silicic acid and some ortho-silicic acid-releasing compounds: New perspectives for therapy. Nutr. Metab. (Lond.) 10, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toulon A., et al. , A role for human skin-resident T cells in wound healing. J. Exp. Med. 206, 743–750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Havran W. L., Jameson J. M., Epidermal T cells and wound healing. J. Immunol. 184, 5423–5428 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hewitt R. E., Powell J. J., Faria N., Bastos C., Vis B., “Compositions comprising nanosilica particles and their use in methods of activating T lymphocytes for therapy.” European Patent 3496728 (2018).
- 45.Belton D. J., Deschaume O., Perry C. C., An overview of the fundamentals of the chemistry of silica with relevance to biosilicification and technological advances. FEBS J. 279, 1710–1720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander G. B., Heston W., Iler R. K., The solubility of amorphous silica in water. J. Phys. Chem. 58, 453–455 (1954). [Google Scholar]
- 47.Guo L., Santschi P. H., “Ultrafiltration and its applications to sampling and characterisation of aquatic colloids” in Environmental Colloids and Particles: Behaviour, Separation and Characterization, Wilkinson K. J., Lead J. R., Eds. (IUPAC, 2007), vol. 10, pp. 159–221. [Google Scholar]
- 48.Griffith C. E., Zhang W., Wange R. L., ZAP-70-dependent and -independent activation of Erk in Jurkat T cells. Differences in signaling induced by H2o2 and Cd3 cross-linking. J. Biol. Chem. 273, 10771–10776 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Webb B., Sali A., “Protein structure modeling with MODELLER” in Protein Structure Prediction (Methods and Protocols), Kihara D., Ed. (Humana Press, 2014), vol. 1137, pp. 1–15. [DOI] [PubMed] [Google Scholar]
- 50.Sun Z.-Y. J., et al. , Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 16867–16872 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kjer-Nielsen L., et al. , Crystal structure of the human T cell receptor CD3 ε γ heterodimer complexed to the therapeutic mAb OKT3. Proc. Natl. Acad. Sci. U.S.A. 101, 7675–7680 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notredame C., Higgins D. G., Heringa J., T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 54.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natarajan A., et al. , Structural model of the extracellular assembly of the TCR-CD3 complex. Cell Rep. 14, 2833–2845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Newell E. W., et al. , Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k). J. Immunol. 186, 5823–5832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Z. J., Kim K. S., Wagner G., Reinherz E. L., Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 ε γ heterodimer. Cell 105, 913–923 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Walsh I., et al. , Bluues server: Electrostatic properties of wild-type and mutated protein structures. Bioinformatics 28, 2189–2190 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.