Significance
The 3-dimensional structure of Gga-AvBD11 (Gallus gallus avian β-defensin 11) is characterized by 2 packed β-defensin domains. This makes Gga-AvBD11 the type member of a structural family dubbed here avian-double-β-defensin (Av-DBD). Its high sequence conservation among bird species suggests essential roles in the avian egg. Our results present Gga-AvBD11 as an effector of innate immunity with a broad spectrum of activities against bacterial pathogens, Eimeria parasites, and avian influenza virus. In addition, it seems to modulate cell viability, proliferation, and invasiveness. Thus, Gga-AvBD11 is likely to assist in the proper development and defense of the avian embryo. Antibacterial, antiparasitic, and antiinvasive activities are mainly mediated by its N-terminal domain. Strikingly, antiviral activity requires the full-length protein at cytotoxic concentrations.
Keywords: defensin, avian egg, NMR structure, avian influenza virus, alarmin
Abstract
Out of the 14 avian β-defensins identified in the Gallus gallus genome, only 3 are present in the chicken egg, including the egg-specific avian β-defensin 11 (Gga-AvBD11). Given its specific localization and its established antibacterial activity, Gga-AvBD11 appears to play a protective role in embryonic development. Gga-AvBD11 is an atypical double-sized defensin, predicted to possess 2 motifs related to β-defensins and 6 disulfide bridges. The 3-dimensional NMR structure of the purified Gga-AvBD11 is a compact fold composed of 2 packed β-defensin domains. This fold is the archetype of a structural family, dubbed herein as avian-double-β-defensins (Av-DBD). We speculate that AvBD11 emanated from a monodomain gene ancestor and that similar events might have occurred in arthropods, leading to another structural family of less compact DBDs. We show that Gga-AvBD11 displays antimicrobial activities against gram-positive and gram-negative bacterial pathogens, the avian protozoan Eimeria tenella, and avian influenza virus. Gga-AvBD11 also shows cytotoxic and antiinvasive activities, suggesting that it may not only be involved in innate protection of the chicken embryo, but also in the (re)modeling of embryonic tissues. Finally, the contribution of either of the 2 Gga-AvBD11 domains to these biological activities was assessed, using chemically synthesized peptides. Our results point to a critical importance of the cationic N-terminal domain in mediating antibacterial, antiparasitic, and antiinvasive activities, with the C-terminal domain potentiating the 2 latter activities. Strikingly, antiviral activity in infected chicken cells, accompanied by marked cytotoxicity, requires the full-length protein.
Avian β-defensins 11 (AvBD11s) are atypical defensins found in birds that are essentially expressed in the oviduct (1). This localization suggests physiological roles related to avian reproduction and/or to the development of the avian embryo. AvBD11 from Gallus gallus (Gga-AvBD11, also known as VMO-2), which is also present in eggshell and in egg white, is one of the main components of the outer layer of hen egg vitelline membrane (VM) (2), the last protective barrier of the embryo (3–5). Consistent with its surmised role as an antimicrobial peptide, we have previously shown that purified Gga-AvBD11 displays antibacterial activities against gram-positive and gram-negative bacteria (6, 7). Next to its contribution to the egg’s innate immunity against pathogen infection, Gga-AvBD11 may also regulate embryo growth and the expansion of extraembryonic membranes. The VM outer layer assists in the growth of the extraembryonic vascularized yolk sac during embryogenesis, suggesting that Gga-AvBD11 may also have a significant role in angiogenesis and cell migration.
The sequence of Gga-AvBD11 contains 2 predicted β-defensin motifs (Fig. 1) (7) and represents the sole double-sized defensin (9.3 kDa) among all 14 AvBDs reported in the chicken species. Such atypical structural features raise questions regarding the independent or synergistic roles of the 2 predictive motifs. The presence of an AvBD11 protein was described in eggs of quail (8) and turkey (9), and similar sequences were also reported for the green lizard Anolis carolinensis [AcBD14 (10) and helofensin3-like protein (11)] and for the Komodo dragon (12), but not in other animal classes.
Fig. 1.
Amino acid sequences of Gga-AvBD11 and of the 2 derived peptides. Gga-AvBD11 precursor (accession NP_001001779) is a 104-amino acid protein encoded by 3 exons (residues overlapping the splice sites at exons 1 and 2 and exons 2 and 3 are given in green in the sequence). The signal peptide and the mature chain are highlighted in gray and purple, respectively. The position of cysteine residues and the spacing between cysteines are specified. The sequences of [1–40]Gga-AvBD11 and [41–82]Gga-AvBD11, delimited by the junction of exons 2 and 3 and further named N-ter and C-ter peptides, are highlighted in cyan and pink.
The benefit for the avian/reptile species of possessing a double-sized defensin is a fascinating question. The aim of the present work was to investigate the roles of Gga-AvBD11 and of its 2 isolated predicted β-defensin motifs (Fig. 1). The resolution of the Gga-AvBD11 structure by NMR revealed a compact 3-dimensional (3D) fold that we named avian-double-β-defensin (Av-DBD). The 3 molecules (Gga-AvBD11, N-terminal [N-ter] and C-terminal [C-ter] peptides) were tested for antimicrobial activities, for their immunomodulatory properties and their effect on cell viability and cell invasion. Our results clearly demonstrate that: 1) antibacterial effects are carried by the N-ter domain; 2) antiparasitic and antiinvasive activities are mainly mediated by the N-ter domain, but potentiated by the C-ter domain; and 3) antiviral (antiinfluenza) activity requires the full-length protein.
Results
AvBD11 Is Conserved in Birds.
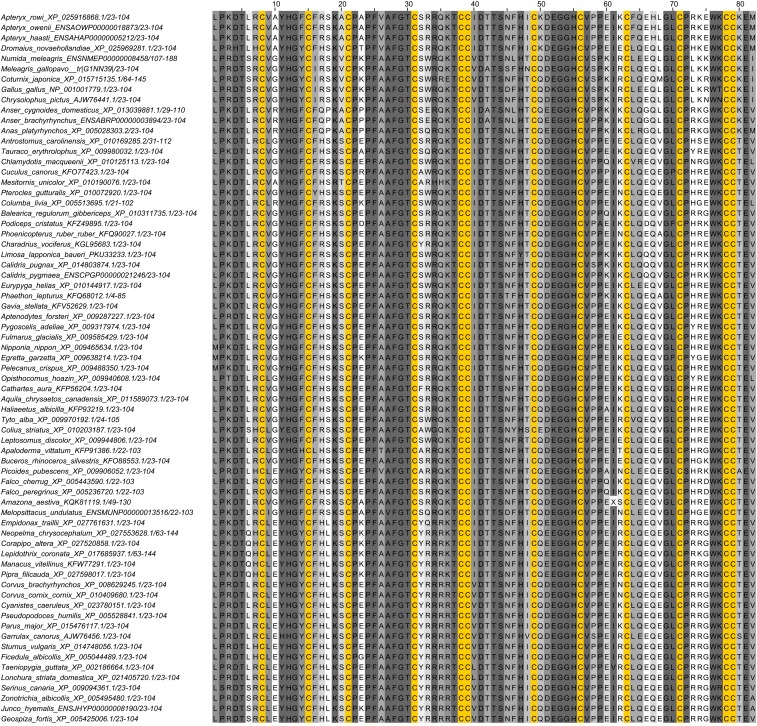
The AvBD11 gene was found in 69 avian species, which cover 32 orders out of the 40 orders of birds. The resulting AvBD11 proteins share high sequence identity with Gga-AvBD11 (Fig. 2). The strict conservation of the position of the 12 Cys residues suggests that the 3D structure of Gga-AvBD11 constitutes a structural archetype for all AvBD11s.
Fig. 2.
Multiple sequence alignment of AvBD11s. Protein sequences of AvBD11 of various bird species were retrieved from National Center for Biotechnology Information (NCBI) and Ensembl databases, aligned with Clustal, and drawn with JalView software (54). Sequences were ordered according to the classification of the International Ornithological Committee (IOC) World Bird List v9.2 (https://www.worldbirdnames.org/). Cysteine residues are highlighted in yellow. Other amino acids are colored according to the identity score, from light gray (low identity) to dark gray (high identity).
AvBD11 Sequence Contains Two Motifs Related to β-Defensins, with Different Physico-Chemical Properties.
In vertebrates, the 2 main groups of defensins, α- and β-defensins, differ in their disulfide pairing and the intercysteine spacing (13). In birds, only β-defensins have been identified (14). Mature Gga-AvBD11 is a cationic peptide (theoretical isoelectric point [pI]: 8.8) of 82 amino acids containing 12 cysteines involved in 6 disulfide bonds (7). The number of cysteines and the spacing between cysteines (CX6CX5CX9CX6CC and CX6CX6CX7CX6CC) led us to predict the presence of 2 β-defensin–related motifs, corresponding to exons 2 and 3, respectively (Fig. 1). Consistent with this, mass spectrometry (MS) fragmentation profile analysis readily revealed 2 fragmentation-resistant regions separated by a central fragmentation-sensitive area (SI Appendix, Figs. S1–S4). Using bioinformatics tools, we analyzed charge and hydrophobicity of each defined moiety: [1–40]Gga-AvBD11 is clearly cationic, while [41–82]Gga-AvBD11 is rather neutral (theoretical pI: 9.3 vs. 6.9; total net charge: +6 vs. 0, respectively). In addition, the C-ter domain is more hydrophilic than the N-ter domain.
Gga-AvBD11 Structure Is the Archetype of Avian-Double-β-Defensins.
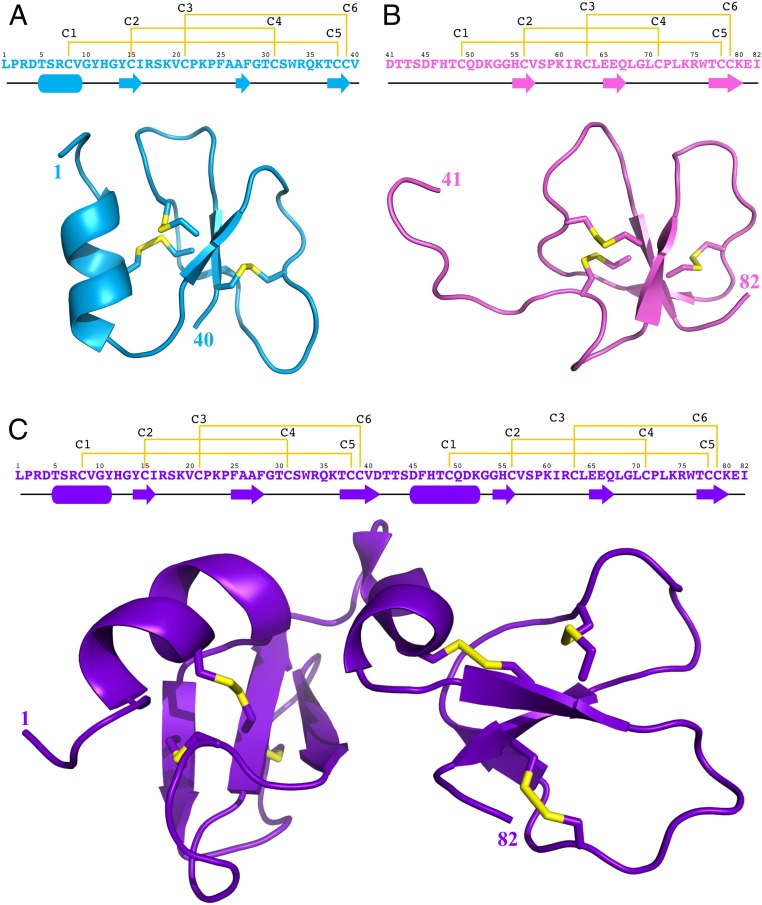
We determined by NMR the 3D structures of the entire protein [1–82]Gga-AvBD11 purified from hen eggs as well as the 2 peptide domains [1–40]Gga-AvBD11 and [41–82]Gga-AvBD11. The 2 peptides were produced by solid-phase peptide synthesis and oxidative folding under optimized conditions (SI Appendix, Figs. S5–S15). Their 3D structures were calculated by taking into account the restraints summarized in SI Appendix, Figs. S16–S18. Both N-ter and C-ter peptides fold into a typical β-defensin motif: a 3-stranded antiparallel β-sheet (residues 14–16, 27–28, 37–39 and 55–57, 65–67, and 77–80, respectively) stabilized by a C1–C5, C2–C4, and C3–C6 disulfide bridges array (Fig. 3 A and B). The [1–40]Gga-AvBD11 displays a short α-helix in its N-terminal part (residues 5 to 9). The 3D structure of the full-length protein reveals the presence of 2 β-defensin domains (Fig. 3C), with β2 and β3 strands slightly longer than in the N-ter peptide (β2:25 to 28, β3:37 to 41). The first helix is longer than in the N-ter peptide, and residues 45 to 52 that are unstructured in the C-ter peptide, form a second warped helix composed of a 310 helix turn (residues 45 to 48) followed by an α-helix turn (residues 49 to 52). Hydrophobic interactions involving Ala26, Phe28, Val40, Phe46, Gln67, and Leu68 stabilize a compact 3D fold (SI Appendix, Fig. S19). The NMR titration of the N-ter peptide with the C-ter peptide (SI Appendix, Fig. S20) does not indicate any interaction between the domains, which suggests an essential role of the covalent link between the 2 domains in the full-length protein.
Fig. 3.
Sequence, cysteines bonding pattern, and 3D structure of (A) N-ter peptide [1–40]Gga-AvBD11 in cyan, (B) C-ter peptide [41–82]Gga-AvBD11 in pink, and (C) the full-length [1–82]Gga-AvBD11 in purple. In each locant from Top to Bottom are represented the peptide sequence with the cysteines bonding pattern in yellow, then the elements of secondary structure, and, finally, the 3D structure with disulfide bridges shown in yellow (drawn with PYMOL) (55).
The Antibacterial Activities of Gga-AvBD11 Against Listeria monocytogenes and Salmonella Enteritidis Is Mediated by the N-ter Domain.
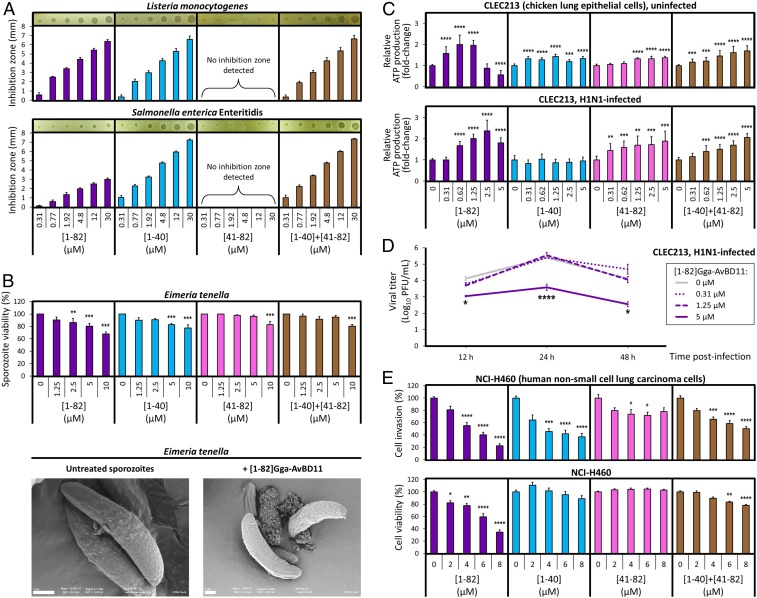
Antibacterial activities were tested against L. monocytogenes (gram-positive) and S. Enteritidis (gram-negative), 2 bacterial pathogens responsible for food poisoning. The N-ter peptide was active against both bacteria with minimal inhibition concentration values (0.14 μM and 0.31 μM, respectively) in the same range as those obtained with the entire Gga-AvBD11 (0.21 μM and 0.15 μM, respectively) (Fig. 4A and SI Appendix, Fig. S21). In contrast, the C-ter peptide showed no detectable activity against the 2 bacteria. Neither potentiation nor inhibition of the overall activity was observed when both peptides were used in combination. Altogether, these results suggest that the antibacterial activity of Gga-AvBD11 is carried by the cationic N-ter region.
Fig. 4.
Functional characterization of Gga-AvBD11 ([1–82]), its derived N-ter ([1–40]), and C-ter ([41–82]) peptides, and the combination [1–40]+[41–82]. (A) Antibacterial activities against L. monocytogenes and S. enterica sv. Enteritidis. Inhibition zones are shown as a function of molecule concentrations. Data are presented as means ± SEM of at least 4 independent experiments performed in duplicates. Representative pictures of inhibition zones are shown in Insets above the bar charts. (B) Antiparasitic activities against E. tenella sporozoites. (B, Top) Data represent means ± SEM of sporozoite viability for at least 3 independent experiments performed in duplicates. (B, Bottom) Scanning electron microscopy analysis of E. tenella sporozoites under control conditions or treated with Gga-AvBD11. (Scale bar, 1 µm.) (C) Relative ATP production as a measure for treatment/virus-induced cytotoxic or cytopathogenic effects in noninfected or H1N1 virus-infected CLEC213. Relative ATP production rates for treated and/or infected cells were calculated as fold changes with reference to mean control values (nontreated cells) set to 1. Shown are means ± SEM from a minimum of 2 independent experiments with 6 replicate samples. (D) H1N1 growth kinetics in Gga-AvBD11–treated CLEC213. CLEC213s were infected at an MOI of 0.01 and viral titers (PFU/mL) in cell culture supernatants at 12 h, 24 h, and 48 h pi were determined by conventional plaque assay on MDBK cells. Shown are means ± SEM from 1 experiment with duplicate samples. (E) Effects on invasion and viability of the human non–small-cell lung cancer cell line NCI-H460. Data are presented as the percentage of invasive cells or cell viability compared to those of control cells without any molecule added. Results are expressed as means ± SEM of at least 5 independent experiments performed at least in duplicates. (B–E) Statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Antiparasitic Activity Against E. tenella Sporozoites Is Mediated by the N-ter Peptide, but Most Pronounced for the Full-Length Gga-AvBD11.
Antiparasitic activity was determined by using a cell-free test inhibition assay on the infectious sporozoite forms of the protozoan E. tenella, the causative agent of cecal avian coccidiosis (15). Gga-AvBD11 showed a stronger antiparasitic activity than the N-ter peptide alone, which suggests amplification of the effect of the N-ter domain by the C-ter domain in the full-length molecule (Fig. 4B). However, the C-ter peptide proved to be inactive, except at the highest dose tested (10 µM), and neither synergy nor antagonism was observed when both peptides were used in combination. Sporozoites were further examined by scanning electron microscopy. Upon treatment with Gga-AvBD11, they underwent distinct structural alterations (Fig. 4B and SI Appendix, Fig. S22).
Gga-AvBD11 Displays Net Antiviral Activity at Concentrations Entailing Cytotoxicity.
Cytotoxic and antiviral activities were tested in a chicken lung epithelial cell (CLEC213) model using the commercial Viral ToxGlo assay (Fig. 4C). Gga-AvBD11 at 0.31 to 1.25 µM caused a significant increase in relative ATP production (as a measure for cell viability), while higher concentrations caused a modest (2.5 µM) or marked decrease (5 µM) in relative ATP production rates in noninfected cells. Consistently, treatment with 5.0 µM Gga-AvBD11 led to distinct morphological changes (cell shrinkage, rounded cell shape) and a dramatic loss of viable cells in the treated CLEC213 cultures (SI Appendix, Fig. S23). Similar to full-length Gga-AvBD11 at concentrations ≤1.25 µM, the N-ter and C-ter peptides (alone or in combination) caused a significant increase in relative ATP production in noninfected cells. However, unlike for Gga-AvBD11, no cytotoxicity was detected for the peptides. Antiviral activity was determined by assessing relative ATP production in CLEC213 infected with a cytopathic H1N1 avian influenza virus. Gga-AvBD11 treatment of infected cells resulted in a marked increase in relative ATP production rates, thus pointing to antiviral activity. At concentrations ≤1.25 µM, this increase was paralleled by a significant increase in relative ATP production in the noninfected cells (see above), indicating that the antiviral effects at these concentrations are probably delusive and rather related to proproliferative activities of Gga-AvBD11. However, at concentrations of 2.5 µM and 5.0 µM, relative ATP production in virus-infected cells remained on an elevated level (2.4- and 1.8-fold increase), indicating that the cytotoxic activity of Gga-AvBD11 at concentrations ≥2.5 µM is accompanied by net antiviral activity (outbalancing its cytotoxic effects). For the N-ter and C-ter peptides (alone or in combination), no such net antiviral activity was observed: significantly increased relative ATP production rates in virus-infected cells were paralleled by similar increases in the noninfected cells, pointing to proproliferative activities of the peptides in infected/noninfected cells as postulated for Gga-AvBD11 at concentrations ≤1.25 µM.
Finally, dose-dependent antiviral activity of the full-length protein was confirmed by determining the growth kinetics of the H1N1 virus in Gga-AvBD11–treated CLEC213 (Fig. 4D). While treatments with 0.31 µM and 1.25 µM had virtually no effect on viral replication, treatment with 5 µM Gga-AvBD11 led to a substantial reduction of viral titers at 12 h (12-fold), 24 h (59-fold), and 48 h (37-fold) postinfection (pi).
Gga-AvBD11 and Its N-ter and C-ter Peptides Affect Cell Viability and Invasion.
To explore the potential role of Gga-AvBD11 in regulating embryonic development and the expansion of the yolk sac membrane, we used National Cancer Institute (NCI)-H460 human cells derived from a human non–small-cell lung carcinoma. The use of this human cell line is motivated by the very limited availability of chicken cell lines with high proliferative and invasive capacities, and by the fact that the NCI-H460 cell line exhibiting high invasive capacities and an epithelium-like morphology is commonly used to assess pro- and antiinvasive agents in vitro (16, 17). The data from these assays are shown in Fig. 4E. Upon treatment of NCI-H460 cells with Gga-AvBD11, the number of cancer cells invading and passing through Matrigel significantly decreased at concentrations ≥4 µM. Gga-AvBD11 treatment also resulted in a dose-dependent, significant diminishment of cell viability. Still, at 4, 6, and 8 μM, the Gga-AvBD11 effect on invasiveness was even more dramatic, suggesting that this antiinvasive activity of Gga-AvBD11 is not solely linked to its action on cell viability. Treatment with the N-ter peptide induced a significant decrease in cell invasion ≥2 µM without affecting cell viability. The C-ter peptide exhibited no cytotoxicity and had only a modest effect on invasion. When the cells were treated with the 2 peptides in combination, distinct antiinvasive and cytotoxic activities were detected, but to a lesser extent than for Gga-AvBD11.
Gga-AvBD11 and Its Derivate Peptides Lack Proinflammatory Effects.
Using the transformed human bronchial epithelial cell line, BEAS-2B (CRL-9609) as a model, we explored proinflammatory treatment effects (SI Appendix, Fig. S25) by measuring interleukin-6 and -8 production in culture supernatants. We concluded that Gga-AvBD11, N-ter, and C-ter peptides lack proinflammatory properties in this particular cell model. It should be highlighted that a significant cytotoxic effect of Gga-AvBD11 was observed at the highest concentration tested (10 µM).
Discussion
The presence of an AvBD11 protein is predicted from the vast majority of avian genomes (18). Strikingly, however, neither a specific function nor a structural characterization of this protein has been established so far. Here, we focused on Gga-AvBD11 from G. gallus, as the archetype of a structural family. We propose to dub this family “avian-double-β-defensin” (Av-DBD), as we demonstrated that its compact fold is composed of 2 packed β-defensin domains (Fig. 3C and SI Appendix, Fig. S19). Indeed, the 3D structure of Gga-AvBD11 reported here exhibits 2 β-defensin domains, even though the intercysteine spacing is not canonical for the C-ter domain.*
Isolated N- and C-ter peptides display the typical 3-stranded antiparallel β-sheet of β-defensins (SI Appendix, Fig. S26), and their global folds are not drastically changed in the full-length protein (SI Appendix, Fig. S27), except for a slight lengthening of β2 and β3 strands and an additional helix at the beginning of the C-ter domain. The domains are packed together by a hydrophobic cluster leading to a compact structure (SI Appendix, Fig. S19). The residues involved in the cluster are highly conserved within the AvBD11 family (SI Appendix, Fig. S28). Phe28 constitutes the core of the cluster and is strictly unchanged. The surface of Gga-AvBD11 is globally positive. However, the C-ter domain exhibits a continuous negative area facing the cationic N-ter domain (SI Appendix, Fig. S29). This area involves residues globally conserved in the other AvBD11 sequences currently available (SI Appendix, Fig. S30). This preservation of hydrophobic and charged residues, in addition to the strict conservation of the position of Cys residues and the high degree of sequence identity in AvBD11s (Fig. 2), suggests that the 3D organization of Gga-AvBD11 is conserved in all birds. It should be mentioned that, to date, there is no reference for such a double-β-defensin in all structural databanks (SI Appendix, Fig. S31). It is noteworthy that other double-domain disulfide-rich proteins, e.g., double inhibitor cystine knot (ICK) peptides (22), are reported in the literature, yet with differences in the position of Cys residues, the disulfide pairing, and the 3D structure.
To explore which biological functions could be exerted by each domain of Gga-AvBD11, we scanned the Protein Data Bank for structural homologs. As expected, the N-ter domain (as part of the full-length protein or isolated peptide) fits with a large set of β-defensins, some α-defensins and snake crotamine. In contrast, the 3D structure of the C-ter peptide only matches platypus defensins, mouse mBD7, and crotamine. More surprisingly, it displays structural homologies with the C-ter domain of tick carboxypeptidase inhibitor (TCI) (SI Appendix, Fig. S32). Even if the 2 domains of the TCI structure (2k2x.pdb) both resemble β-defensin domains (23), the flexible linker between the 2 globular domains prevents DALI software (24) to ascertain any structural homologies with the compact 3D structure of the full-length Gga-AvBD11. Although the flexibility of this linker was reported to be important for the recognition mechanism and further interaction with carboxypeptidase A (25), Gga-AvBD11’s potential inhibitory activity against carboxypeptidase A was explored. Our results did not corroborate such an activity (SI Appendix, Fig. S33).
In order to identify other potential double-β-defensins (DBDs)—compact or not—in other species than birds, we extensively screened the sequence homologies related to Gga-AvBD11. We extracted a series of sequences from all branches of arthropods with the cysteine pattern of β-defensins (SI Appendix, Fig. S34), which led us to postulate a global organization into 2 β-defensin domains. However, the sequence identities with Gga-AvBD11 are drastically reduced (<34%) compared to bird sequences, and none of the residues involved in the packing of Gga-AvBD11 (SI Appendix, Fig. S19) is conserved in arthropods. This feature suggests a less compact structure (as in TCI) or at least a different packing between the 2 presumed β-defensin domains that could be predictive of divergent biological properties.
Our unsuccessful attempts to identify sequences similar to Gga-AvBD11 in vertebrates other than reptiles raise questions about the appearance of this family during evolution. It is now well accepted that 1) monodomain β-defensins found in vertebrates emerged from an ancestral cephalochordate big-defensin (26); 2) α-defensins, only found in mammals, are derived from β-defensins (26); and 3) cyclic θ-defensins, a restricted class uniquely found in old-world monkeys, are derived from α-defensins (27). Our current hypothesis predicts that AvBD11 originates from the duplication and fusion of an ancestral monodomain β-defensin gene, similar to what is reported for Gila monster helofensins (11). Because all our attempts to identify sequences related to double-β-defensins in other vertebrates than reptiles (mammals, fishes, amphibians), we cannot presume the existence of an ancestor gene, common to birds and arthropods. Probably, an independent duplication–fusion event might have occurred during the evolution of arthropods.
Genomic plasticity of defensin-encoding sequences is known to foster the adaptability of the innate immune system and to enable the acquisition of new functions, as exemplified by β-defensins of venomous vertebrates, which have evolved to target ion channels of their preys (28). In birds, 12 new AvBDs have been acquired through duplication events among galliform and passeriform birds (29). AvBD11 appeared in birds before the galliform–passeriform split that occurred around 100 million years ago (29), likely before the Paleognathae–Neognathae split (AvBD11 in ratite species) and it is highly conserved in the avian genomes currently available (Fig. 2). The analysis of previously published phylogenetic trees (18, 30) suggests that AvBD11 might have diverged from AvBD10. The appearance of such a double-domain protein during evolution could be driven either by its increased biological potency compared to a single-domain molecule, and/or by the necessity to acquire new functions carried only by the full-length protein. Because of the high concentration of Gga-AvBD11 in the vitelline membrane known to protect the embryo and to support the growth of the extraembryonic yolk sac, we assessed the antimicrobial activity of purified Gga-AvBD11 against various microbial pathogens and explored its biological effect on cell viability and invasion.
The 2 pathogenic bacteria L. monocytogenes and S. enterica were previously reported to be sensitive to Gga-AvBD11 (6, 7). Our present results clearly show that this antimicrobial activity is carried by the cationic N-ter domain. Cationicity is considered as one of the most critical determinants for antibacterial activities, triggering electrostatic interactions of peptides with the negatively charged membranes of bacteria (31). Hydrophobicity is also an essential parameter as it governs the peptide insertion into the lipid bilayer. The N-ter peptide respects these 2 key features, its cationic properties being shared by most AvBD11s, whereas the C-ter domain, less hydrophobic, exhibits variable charge properties (SI Appendix, Fig. S30). Consequently, the addition of the C-ter domain does not substantially affect the antibacterial potency of the N-ter domain (Fig. 4A and SI Appendix, Fig. S21).
Antiparasitic activities have been previously reported for other antimicrobial peptides such as cecropin, melittin, dermaseptin, cathelicidins (32, 33), and human α-defensins (34–36). The present work extends this activity to the β-defensin family. We demonstrated that Gga-AvBD11 and, to a lesser extent, also the N-ter peptide exhibit antiparasitic activities against infectious sporozoites of E. tenella, 1 of the 3 main Eimeria species responsible for avian coccidiosis (15). The exact mechanisms by which Gga-AvBD11 exerts its antiparasitic effects [aggregation (36), pore formation (35)] remains to be elucidated. However, it can be speculated that 1) the high cationicity of the full-length Gga-AvBD11 and the N-ter peptide might be crucial to trigger the first peptide/membrane interaction and that 2) the C-ter domain might increase the anti-Eimeria effect of the N-ter domain.
Testing of the cytotoxic and antiviral activities of Gga-AvBD11 and its N-ter and/or C-ter peptides in the CLEC213 infection model revealed that Gga-AvBD11 displays net antiviral (antiinfluenza) activity at concentrations that are cytotoxic in noninfected cells. Antiviral activity against Newcastle disease virus, an avian respiratory pathogen, has previously been reported for AvBD2 (37). However, to our knowledge, antiinfluenza activity of an avian β-defensin has so far never been reported. Strikingly, the concomitant cytotoxic and antiviral activities were only seen for the full-length molecule and not for the N-ter and C-ter peptides (alone or in combination). Distinct antiviral mechanisms have been shown for human α-, β-, and θ-defensins (38): virus neutralization through binding to viral envelope/capsid proteins or cellular cognate receptors, inhibition of viral fusion and postentry steps, and immunomodulatory activity. It is thus conceivable that the H1N1 virus inhibitory activity of Gga-AvBD11 may involve its binding to the viral hemagglutinin and/or neuraminidase glycoproteins, the viral entry receptor, or an innate immune receptor. One might speculate that Gga-AvBD11 binding to negatively charged α2,3 galactose-linked sialic acid moieties, the principal hemagglutinin receptor for avian influenza viruses, may restrict H1N1 virus infection by impeding viral adsorption and entry into CLEC213 cells. However, in this case, one would expect that the cationic N-ter peptide would display similar antiviral activity as the full-length molecule, which is clearly not the case—at least not in our experimental conditions. We hypothesize that the cytotoxic and antiviral activities of Gga-AvBD11 are mediated by its binding to a yet unidentified innate immune receptor, thereby leading to the activation of coalescing antiviral and proapoptotic signaling pathways. This notion is supported by our observation that 5 µM Gga-AvBD11 is a potent inducer of NF-κB activity in the HD11 chicken macrophage cell line (SI Appendix, Fig. S24). Interestingly, the proposed antiviral mechanism is in good agreement with a previous report describing AvBD13 as an activator of TLR4-mediated NF-κB signaling (39) and the concept of antimicrobial peptides serving as alarmins (40). Yet, it seems that the NF-κB–triggering activity of Gga-AvBD11 is restricted to the chicken (and potentially other avian/sauropsid species), as no such activity could be detected in the human cell lines used in this study.
Next to its role in protecting the embryo against pathogens, Gga-AvBD11 present in the vitelline membrane might participate in the growth of the extraembryonic yolk sac during egg incubation (roughly estimated to be at least 30 µM, based on purification yield of Gga-AvBD11 from VM). Indeed, as the vascularized yolk sac expands during embryonic development, the vitelline membrane undergoes major ultrastructural changes to finally rupture over the embryo (41). In the present work, irrespective of the epithelial cell line being used [a chicken lung epithelial cell line (SI Appendix, Fig. S23) as well as immortalized or transformed human epithelial cell lines, SI Appendix, Fig. S25)], we demonstrated that the full-length molecule exhibits cytotoxic effects. Interestingly, it was also shown to decrease cell invasiveness. These data are in agreement with results obtained with the human β-defensin 1 (hBD-1) on oral squamous cell carcinoma (OSCC) cell lines, prostate cancer cell lines, or bladder cancer cell lines (42–45). Generally, human β-defensins have been proposed to directly affect cancer cells by modulating their survival, proliferation, migration, and/or invasive properties (46), with hBD-1 being considered as a candidate tumor suppressor gene (43). Similar, but less pronounced effects were observed in the human cell lines by testing the N-ter peptide alone. The combined use of the N- and C-ter peptides was shown to affect cell viability, but to a lesser extent when compared to full-length Gga-AvBD11. This observation highlights the importance of the covalent link between the domains and/or the importance of the orientation of the 2 domains in the full-length molecule to warrant unmitigated activity/efficiency. Gga-AvBD11 is essentially concentrated in the outer layer of the vitelline membrane while embryonic development, yolk sac outgrowth, and expansion occur along the inner layer of the vitelline membrane, which has been reported to have growth promoting activity (47). The antiinvasive and cytotoxic activity of Gga-AvBD11 might thus be of major physiological importance to prevent anarchic and multidirectional cell growth of the embryo and of the yolk sac.
Besides these marked cytotoxic and antiinvasive effects, we did not detect any significant immunomodulatory activities of Gga-AvBD11 and the derived peptides in human cells, although such activities have been reported for a number of other defensins (46). As stated earlier, this points to a species- or class/clade-specific role of Gga-AvBD11 as an immunomodulatory antimicrobial peptide.
Conclusions
AvBD11 is an atypical defensin which is highly conserved among birds. The 3D structure of Gga-AvBD11 (from G. gallus) is reported here as the archetype of the structural family of avian-double-β-defensins (Av-DBD) characterized by 2 β-defensin domains packed via a hydrophobic cluster.
The conservation of AvBD11 within bird species and its abundant and principal expression in the oviduct are pointing to important biological activities related to avian reproduction and embryonic development. Our data allowed us to confirm the role of Gga-AvBD11 as an effector of innate immunity (most likely participating in the protection of the avian embryo) with a broad spectrum of antimicrobial activities: 1) N-ter domain-dependent antibacterial activity, 2) direct antiparasitic activity against the avian protozoan E. tenella, and 3) antiviral activity limiting influenza virus infection in avian cells—potentially by stimulating innate immunity. Moreover, from our experiments with human cancer cell lines as a model for studying cell proliferation and invasiveness, we concluded that Gga-AvBD11 might have an eminent role in driving and/or constraining the orientation and growth of embryonic and extraembryonic cell structures in the avian egg.
The appearance of such a multifunctional double-domain protein during evolution might have been driven by its superior effectiveness compared to a single-domain molecule and/or by the necessity to acquire new functions carried only by the full-length protein. Even though the C-ter domain has no impact on the antibacterial efficacy mediated by the cationic N-ter domain (as seen for the 2 bacterial pathogens used here), it was shown to potentiate/increase the anti-Eimeria, as well as the antiinvasive biological effects. Strikingly, other activities involving interactions with eukaryotic cells, notably cytotoxic and antiviral activities in mammalian/avian cells, seem to be restricted to the full-length Gga-AvBD11 protein.
In conclusion, we demonstrate here that Gga-AvBD11 is a protein with multiple activities, a Swiss knife capable of fulfilling a number of roles that are likely essential for the proper development of the avian embryo in the egg. We believe that Gga-AvBD11 still possesses hidden functions that merit further investigations. Studies on the contribution of Gga-AvBD11 in the formation of the protein fibers of the outer layer of the vitelline membrane (protein–protein interactions) but also in the growth of the yolk sac during embryonic development might be of particular interest to elucidate further its physiological role(s) and underlying mechanisms.
Methods
Three-Dimensional NMR Structures.
Synthetic [1–40]Gga-AvBD11, [41–82]Gga-AvBD11, and native Gga-AvBD11 purified from vitelline membranes as previously described (6) were dissolved in H2O:D2O at a concentration of 1.25 mM, 1.40 mM, and 1.08 mM, respectively. The pH was adjusted to 4.5.
Two-dimensional 1H-NOESY, 2D 1H-TOCSY, sofast-HMQC (15N natural abundance), and 13C-HSQC (13C natural abundance) were performed at 298K on an Avance III HD BRUKER 700-MHz spectrometer equipped with a cryoprobe. NMR data were processed using Bruker’s Topspin 3.2 and analyzed with CcpNmr (version 2.2.2) (48).
Structures were calculated using Crystallography and NMR System (CNS) through the automatic assignment software ARIA2 (version 2.3) (49) with NOE, hydrogen bonds and backbone dihedral angle restraints, and with 3 ambiguous disulfide bridges for [1–40]Gga-AvBD11 and [41–82]Gga-AvBD11 or 2 sets of 3 ambiguous disulfide bridges for Gga-AvBD11. Ten final structures were selected based on total energies and restraint violation statistics to represent each molecule and were deposited in the Protein Data Bank under the accession codes 6QES, 6QET, and 6QEU, respectively.
Radial diffusion assay (antibacterial assays).
Antibacterial assays were carried out on L. monocytogenes and S. enterica sv. Enteritidis American Type Culture Collection (ATCC) 13076 (International Center for Microbial Resources dedicated to Pathogenic Bacteria, CIRM-BP, INRA, Nouzilly), as previously described for the assessment of antibacterial activities of native AvBD11 (6, 7). Precultures and exponential-phase cultures were performed under agitation (180 rpm) at 37 °C either in brain heart infusion (BHI) or in tryptic soy broth (TSB), respectively, for L. monocytogenes and S. enterica sv. Enteritidis (ATCC 13076). Molten underlay medium (10 mM sodium phosphate buffer, pH 7.4, containing 0.03% TSB, 1% agarose type I low endosmosis, 0.02% Tween 20) was inoculated with exponentially growing bacteria at 3 × 105 colony-forming units (CFU)/mL and poured into a Petri dish. Wells were manually punched and filled with 5 µL of test peptides (native Gga-AvBD11 and N-ter and C-ter peptides, MSI-94) at different concentrations (30, 12, 4.8, 1.92, 0.768, and 0.3072 µM) that were then allowed to diffuse within the underlay for 3 h at 37 °C. Underlay gel was then covered with molten overlay (10 mM sodium phosphate buffer, pH 7.4, containing 6% TSB, 1% agarose type I low endosmosis) and incubated overnight at 37 °C. Antibacterial activities were revealed as clear inhibition zones surrounding the wells. Diameters of the inhibition zones were plotted against the logarithm of peptide concentrations and the linear regression equation was used to calculate the minimal inhibitory concentration (7, 12).
Antiparasitic assays.
Antiparasitic assays were performed against E. tenella sporozoites. Sporozoites from the yellow fluorescent protein-positive (YFP) E. tenella strain (50) were obtained by excystation of sporulated oocysts (51). Sporozoite viability was assessed after incubation in 10 mM PBS, pH 7.4 with or without antimicrobial peptides (1.25 to 10 µM) for 1 h at 41 °C, and staining with aqueous Evans’ blue (vol/vol). Live (green) and dead (red) parasites were counted by epifluorescence microscopy using a Zeiss Axiovert 200 microscope. Sporozoite viability is expressed as percentage of the live sporozoites incubated with PBS (considered as 100% viable). Statistical analysis was performed with Prism 5 software (GraphPad) using a 2-way ANOVA (Tukey–Kramer multiple group comparison).
Antiviral Activity Assay.
Cytotoxic and antiviral activities were tested using the Viral ToxGlo assay (Promega), a luminescence-based assay measuring ATP as a surrogate for cell viability (52). Antiviral activities against an H1N1 avian influenza virus (A/Mallard/Marquenterre/Z237/83) were assayed in a CLEC213 infection model (53). Viral ToxGlo assay-specific 50% tissue culture infectious dose (TCID50) was determined for the viral stock according to the manufacturer’s instructions. CLEC213s were seeded in 96-well plates (2.103 cells per well) and cultured in DMEM F12 media supplemented with 7.5% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin (Thermo Fisher Scientific) at 41 °C and 5% CO2. FBS was excluded from the media in subsequent treatment/infection experiments. To assess cytotoxicity, cells were washed with PBS and left untreated (controls) or treated with serial dilutions of Gga-AvBD11 and the N-ter or C-ter peptides (alone or in combination). To test for antiviral activity, protein/peptide dilutions were coincubated with H1N1 at 25× TCID50 (30 min at room temperature [RT]) and then overlaid onto the cells. At 48 h posttreatment/infection, the cells were washed and incubated with 100 µL of ATP detection reagent for 20 min at RT. ATP/luminescence values were measured using a GloMax-Multi Detection System (Promega). Relative ATP production rates for treated and/or infected cells were calculated as fold changes with reference to mean control values (nontreated cells) set to 1.
Dose-dependent antiviral activity of Gga-AvBD11 was tested by assessing H1N1 growth kinetics in nontreated/treated CLEC213. Briefly, CLEC213s were seeded in 6-well plates (8.105 cells per well) and cultured as described above. Gga-AvBD11 preparations (0 µM, 0.31 µM, 1.25 µM, or 5 µM) diluted in culture media containing 0.05 µg/mL L-1-p-tosylamino-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Worthington Biochemical Corporation) but no FBS were coincubated with H1N1 at a multiplicity of infection (MOI) of 0.01 (30 min at RT), and the mixtures were then overlaid onto the cells. At 12 h, 24 h, and 48 h pi, supernatant samples (200 µL) were collected and frozen at −80 °C. Viral titers in the samples expressed as PFU/mL were determined by conventional plaque assay on Madin-Darby canine kidney (MDCK) cells.
Statistical differences (nontreated vs. treated cells) were calculated by 1-way ANOVA, unpaired t test using GraphPad Prism v6.07 (GraphPad Software, Inc.).
Invasion and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) Cell Viability Assay.
Cell invasion and viability were analyzed using Matrigel and MTS assays, respectively, after 48 h of incubation with various concentrations of molecules (0, 2, 4, 6, or 8 µM). Cells used were the human non–small-cell lung cancer cell line NCI-H460 and the immortalized human bronchial epithelial cell line, BEAS-2B (CRL-9609) which were obtained from the ATCC (LGC Promochem).
Cell invasion was assessed using a modified Boyden chamber model. Two technical replicates for treated cells and 3 for control cells were analyzed in each independent experiment. The results were expressed as the percentage of invasive cells with reference to untreated control cell values. Results were expressed as mean ± SEM of 5 independent experiments.
Cellular viability was determined by MTS assay (CellTiter 96 AQueousOne Solution, Promega) (SI Appendix, Fig. S25). A Kruskal–Wallis test with Dunn’s test was used to compare results from cells treated with different concentrations of molecules with results from control cells. A Mann–Whitney U test was used to compare results of invasion and cytotoxicity obtained for 1 molecule tested at the same concentration for both assays. Statistical analyses were performed using GraphPad Prism version 6.0 for Mac OS X software.
Supplementary Material
Acknowledgments
This work was funded by the MUSE (Medicinal USe of Eggs) (2014-00094512) and Structure-Activity and Phylogenetic Relationships of avian beta-defensin 11 (SAPhyR-11) (2017-119983) project grants from the Région Centre-Val de Loire. We thank M. Chessé, J. Renault, and J.-C. Poirier (UMR0083 BOA, INRA, University of Tours, Nouzilly, France) for their technical support in the purification of Gga-AvBD11. We thank G. Gabant and C. Colas (mass spectrometry platforms of UPR4301, CBM, Orléans, France and FR2708, Orléans, France, respectively) for the MS analyses of synthetic peptides. We are grateful to P. Roingeard, J. Burlaud-Gaillard, and S. Georgault (Plateforme IBiSA de Microscopie Electronique, University of Tours, France) for the scanning electron microscopy analysis. We thank N. Winter, C. Schouler, and A. C. Lalmanach for providing access to their bacteriological laboratories and for scientific discussions; E. Kut and J. Cournet for technical assistance in the antiparasitic and/or antiviral assays; and L. Trapp-Fragnet, D. Garrido, and R. Guabiraba for providing data on viability/proliferation and NF-κB activation in Gga-AvBD11–treated chicken cells (all UMR1282, ISP, INRA, University of Tours, Nouzilly, France). We also thank A. Lebrun, C. Mathé, and D. Fouquenet for their technical assistance in the assessment of immunomodulatory, antiinvasive, and cytotoxic activities (CEPR UMR U1100, INSERM, University of Tours, Tours, France). Finally, we would also like to express our special gratitude to E. Kut (UMR1282, ISP, INRA, University of Tours, Nouzilly, France) for his considerable engagement in the realization of additional experiments.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of [1–40]Gga-AvBD11, [41–82]Gga-AvBD11, and [1–82]Gga-AvBD11 have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID codes 6QES, 6QET, and 6QEU).
*Depending on the cysteine spacing and the more or less lack of structural elements, several terms have emerged over the years to describe the different members of the enlarged β-defensin family: “defensin-like” (19), “defensin-related” (20), and even recently “intermediate defensin-like” (21). However, no term has yet been widely accepted and we prefer here to use the generic term “β-defensin.”
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912941117/-/DCSupplemental.
References
- 1.Xiao Y., et al. , A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: Implications for the origin and evolution of mammalian defensins. BMC Genomics 5, 56 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kido S., Morimoto A., Kim F., Doi Y., Isolation of a novel protein from the outer layer of the vitelline membrane. Biochem. J. 286, 17–22 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mann K., The chicken egg white proteome. Proteomics 7, 3558–3568 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Mann K., Proteomic analysis of the chicken egg vitelline membrane. Proteomics 8, 2322–2332 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Mann K., Macek B., Olsen J. V., Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics 6, 3801–3810 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Guyot N., et al. , Proteomic analysis of egg white heparin-binding proteins: Towards the identification of natural antibacterial molecules. Sci. Rep. 6, 27974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hervé-Grépinet V., et al. , Purification and characterization of avian beta-defensin 11, an antimicrobial peptide of the hen egg. Antimicrob. Agents Chemother. 54, 4401–4409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman M. A., Moriyama A., Iwasawa A., Yoshizaki N., Cuticle formation in quail eggs. Zool. Sci. 26, 496–499 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Mann K., Mann M., The proteome of the calcified layer organic matrix of Turkey (Meleagris gallopavo) eggshell. Proteome Sci. 11, 40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalla Valle L., Benato F., Maistro S., Quinzani S., Alibardi L., Bioinformatic and molecular characterization of beta-defensins-like peptides isolated from the green lizard Anolis carolinensis. Dev. Comp. Immunol. 36, 222–229 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Fry B. G., et al. , Novel venom proteins produced by differential domain-expression strategies in beaded lizards and gila monsters (genus Heloderma). Mol. Biol. Evol. 27, 395–407 (2010). [DOI] [PubMed] [Google Scholar]
- 12.van Hoek M. L., et al. , The Komodo dragon (Varanus komodoensis) genome and identification of innate immunity genes and clusters. BMC Genomics 20, 684 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selsted M. E., Ouellette A. J., Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557 (2005). [DOI] [PubMed] [Google Scholar]
- 14.van Dijk A., Veldhuizen E. J., Haagsman H. P., Avian defensins. Vet. Immunol. Immunopathol. 124, 1–18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalloul R. A., Lillehoj H. S., Poultry coccidiosis: Recent advancements in control measures and vaccine development. Expert Rev. Vaccines 5, 143–163 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Chang H. F., Cheng H. T., Chen H. Y., Yeung W. K., Cheng J. Y., Doxycycline inhibits electric field-induced migration of non-small cell lung cancer (NSCLC) cells. Sci. Rep. 9, 8094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsia T. C., et al. , Cantharidin impairs cell migration and invasion of human lung cancer NCI-H460 cells via UPA and MAPK signaling pathways. Anticancer Res. 36, 5989–5997 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y., et al. , Evolution of the avian β-defensin and cathelicidin genes. BMC Evol. Biol. 15, 188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres A. M., et al. , Defensin-like peptide-2 from platypus venom: Member of a class of peptides with a distinct structural fold. Biochem. J. 348, 649–656 (2000). [PMC free article] [PubMed] [Google Scholar]
- 20.Fox N. K., Brenner S. E., Chandonia J. M., SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 42, D304–D309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres A. M., et al. , Structure and antimicrobial activity of platypus ‘intermediate’ defensin-like peptide. FEBS Lett. 588, 1821–1826 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Maxwell M., Undheim E. A. B., Mobli M., Secreted cysteine-rich repeat proteins “SCREPs”: A novel multi-domain architecture. Front. Pharmacol. 9, 1333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arolas J. L., et al. , The three-dimensional structures of tick carboxypeptidase inhibitor in complex with A/B carboxypeptidases reveal a novel double-headed binding mode. J. Mol. Biol. 350, 489–498 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Holm L., Benchmarking fold detection by DaliLite v.5. Bioinformatics, btz536 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Pantoja-Uceda D., et al. , The NMR structure and dynamics of the two-domain tick carboxypeptidase inhibitor reveal flexibility in its free form and stiffness upon binding to human carboxypeptidase B. Biochemistry 47, 7066–7078 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Zhu S., Gao B., Evolutionary origin of β-defensins. Dev. Comp. Immunol. 39, 79–84 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Hazlett L., Wu M., Defensins in innate immunity. Cell Tissue Res. 343, 175–188 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Whittington C. M., Papenfuss A. T., Kuchel P. W., Belov K., Expression patterns of platypus defensin and related venom genes across a range of tissue types reveal the possibility of broader functions for OvDLPs than previously suspected. Toxicon 52, 559–565 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Hellgren O., Ekblom R., Evolution of a cluster of innate immune genes (beta-defensins) along the ancestral lines of chicken and zebra finch. Immunome Res. 6, 3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma D., et al. , Discovery and characterization of Coturnix chinensis avian β-defensin 10, with broad antibacterial activity. J. Pept. Sci. 18, 224–232 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Brogden K. A., Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Pretzel J., Mohring F., Rahlfs S., Becker K., Antiparasitic peptides. Adv. Biochem. Eng. Biotechnol. 135, 157–192 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Sierra J. M., Fusté E., Rabanal F., Vinuesa T., Viñas M., An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 17, 663–676 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Dabirian S., et al. , Human neutrophil peptide-1 (HNP-1): A new anti-leishmanial drug candidate. PLoS Negl. Trop. Dis. 7, e2491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madison M. N., et al. , Human defensin alpha-1 causes Trypanosoma cruzi membrane pore formation and induces DNA fragmentation, which leads to trypanosome destruction. Infect. Immun. 75, 4780–4791 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka T., et al. , Parasiticidal activity of human alpha-defensin-5 against Toxoplasma gondii. In Vitro Cell. Dev. Biol. Anim. 46, 560–565 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Liu C., et al. , Induction of avian β-defensin 2 is possibly mediated by the p38 MAPK signal pathway in chicken embryo fibroblasts after Newcastle disease virus infection. Front. Microbiol. 9, 751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holly M. K., Diaz K., Smith J. G., Defensins in viral infection and pathogenesis. Annu. Rev. Virol. 4, 369–391 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Yang Y., Jiang Y., Yin Q., Liang H., She R., Chicken intestine defensins activated murine peripheral blood mononuclear cells through the TLR4-NF-kappaB pathway. Vet. Immunol. Immunopathol. 133, 59–65 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Yang D., Han Z., Oppenheim J. J., Alarmins and immunity. Immunol. Rev. 280, 41–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen C., Ultrastructural changes in the avian vitelline membrane during embryonic development. J. Embryol. Exp. Morphol. 21, 467–484 (1969). [PubMed] [Google Scholar]
- 42.Avila E. E., Functions of antimicrobial peptides in vertebrates. Curr. Protein Pept. Sci. 18, 1098–1119 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Bullard R. S., et al. , Functional analysis of the host defense peptide human beta defensin-1: New insight into its potential role in cancer. Mol. Immunol. 45, 839–848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Q., et al. , Human beta-defensin-1 suppresses tumor migration and invasion and is an independent predictor for survival of oral squamous cell carcinoma patients. PLoS One 9, e91867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun C. Q., et al. , Human beta-defensin-1, a potential chromosome 8p tumor suppressor: Control of transcription and induction of apoptosis in renal cell carcinoma. Cancer Res. 66, 8542–8549 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Suarez-Carmona M., Hubert P., Delvenne P., Herfs M., Defensins: “Simple” antimicrobial peptides or broad-spectrum molecules? Cytokine Growth Factor Rev. 26, 361–370 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Haas H. J., Spratt N. T. Jr, Contributions to an analysis of the avian vitelline membrane’s potential to promote outgrowth of the yolk sac-serosal membrane. Anat. Rec. 184, 227–231 (1976). [DOI] [PubMed] [Google Scholar]
- 48.Vranken W. F., et al. , The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Rieping W., et al. , ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23, 381–382 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Gras S., et al. , Aminopeptidase N1 (EtAPN1), an M1 metalloprotease of the apicomplexan parasite Eimeria tenella, participates in parasite development. Eukaryot. Cell 13, 884–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rieux A., et al. , Eimeripain, a cathepsin B-like cysteine protease, expressed throughout sporulation of the apicomplexan parasite Eimeria tenella. PLoS One 7, e31914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noah J. W., et al. , A cell-based luminescence assay is effective for high-throughput screening of potential influenza antivirals. Antiviral Res. 73, 50–59 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Trapp S., et al. , Shortening the unstructured, interdomain region of the non-structural protein NS1 of an avian H1N1 influenza virus increases its replication and pathogenicity in chickens. J. Gen. Virol. 95, 1233–1243 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeLano W. L., The PyMOL Molecular Graphics System (Delano Scientific, San Carlos, 2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.