Significance
HLA alleles have previously been implicated with scleroderma risk, but, in this study, using a European American ancestral cohort and a newly recruited large cohort of African Americans, we comprehensively define the HLA alleles and amino acid residues associated with scleroderma. Scleroderma is characterized by mutually exclusive and specific autoantibodies. We demonstrated ancestry-predominant HLA alleles that were much more strongly associated with autoantibody subsets of scleroderma than with the overall risk of SSc. We bioinformatically predicted immunodominant peptides of self-antigens and demonstrated homology of these peptides with viral protein sequences from Mimiviridae and Phycodnaviridae families. Our findings suggest the hypothesis that scleroderma-specific autoantibodies may arise through molecular mimicry, driven by the interaction of specific viral antigens with corresponding HLA / heterodimers.
Keywords: scleroderma, HLA, autoantibodies, molecular mimicry, mimivirus
Abstract
Systemic sclerosis (SSc) is a clinically heterogeneous autoimmune disease characterized by mutually exclusive autoantibodies directed against distinct nuclear antigens. We examined HLA associations in SSc and its autoantibody subsets in a large, newly recruited African American (AA) cohort and among European Americans (EA). In the AA population, the African ancestry-predominant HLA-DRB1*08:04 and HLA-DRB1*11:02 alleles were associated with overall SSc risk, and the HLA-DRB1*08:04 allele was strongly associated with the severe antifibrillarin (AFA) antibody subset of SSc (odds ratio = 7.4). These African ancestry-predominant alleles may help explain the increased frequency and severity of SSc among the AA population. In the EA population, the HLA-DPB1*13:01 and HLA-DRB1*07:01 alleles were more strongly associated with antitopoisomerase (ATA) and anticentromere antibody-positive subsets of SSc, respectively, than with overall SSc risk, emphasizing the importance of HLA in defining autoantibody subtypes. The association of the HLA-DPB1*13:01 allele with the ATA+ subset of SSc in both AA and EA patients demonstrated a transancestry effect. A direct correlation between SSc prevalence and HLA-DPB1*13:01 allele frequency in multiple populations was observed (r = 0.98, P = 3 × 10−6). Conditional analysis in the autoantibody subsets of SSc revealed several associated amino acid residues, mostly in the peptide-binding groove of the class II HLA molecules. Using HLA / allelic heterodimers, we bioinformatically predicted immunodominant peptides of topoisomerase 1, fibrillarin, and centromere protein A and discovered that they are homologous to viral protein sequences from the Mimiviridae and Phycodnaviridae families. Taken together, these data suggest a possible link between HLA alleles, autoantibodies, and environmental triggers in the pathogenesis of SSc.
Systemic sclerosis (scleroderma, SSc) is a systemic autoimmune disease that is clinically heterogeneous and is characterized by progressive thickening of the skin and internal organs, leading to morbidity and mortality. A hallmark of SSc is the presence of circulating antinuclear antibodies (ANA), which are observed in 90 to 95% of patients (1). Anticentromere antibody (ACA), antitopoisomerase I antibody (ATA), anti-U3-ribonucleoprotein antibody (fibrillarin, AFA), and anti-RNA polymerase III antibody (ARA) are the common autoantibodies reported in SSc and are mutually exclusive and specific for SSc (2, 3). These autoantibodies are associated with distinct patterns of skin and internal organ involvement and are markers of prognosis and survival (4–6). Compared with European Americans (EA), African Americans (AA) have a higher prevalence of ATA and AFA and also tend to have a more severe phenotype comprising diffuse skin involvement and greater interstitial lung disease, leading to increased mortality (5, 7–10).
Genetic factors, along with environmental factors, contribute to the risk of SSc with HLA genes reported to have the strongest influence on SSc susceptibility, and these alleles have an even stronger effect within the SSc-specific autoantibody subsets (11–24). These HLA alleles encode variations in the antigen-binding grooves of the HLA molecules that determine their binding affinity for specific antigens presented to T helper cells (25). Aberrant self-peptide or foreign peptide presentation via class II HLA molecules on the antigen-presenting cells (APCs) leads to activation of autoreactive T helper cells that play a crucial role in activation of B cells, autoantibody formation, and autoimmunity induction. Thus, HLA alleles coding for a specific antigen-binding groove sequence on the APCs recognize a specific self-peptide causing activation of T helper cells and production of autoantibodies.
HLA-mediated presentation of foreign antigens can activate autoantigen-specific T cells either by presenting peptides derived from self-proteins or by presenting peptides that are homologous to self-antigens but derived from microbial proteins. This mechanism, whereby the amino acid sequence of a microbial peptide is homologous to the peptide sequence from a self-protein, thus causing activation of T helper cells and leading to B cell activation and autoantibody production against the self-protein, is called molecular mimicry (26). Molecular mimicry has been proposed in the pathogenesis of several autoimmune diseases, including multiple sclerosis, type 1 diabetes mellitus, spondyloarthropathies, Graves’ disease, systemic lupus erythematosus (SLE), and SSc (27–33). For instance, Epstein–Barr virus, a ubiquitous human DNA virus, exhibits molecular mimicry with common SLE self-antigens (30). Presence of viruses in SSc tissues and viral infections acting as a trigger for autoimmunity have been proposed as environmental risk factors in the pathogenesis of SSc (33–35).
Given the potential importance of the HLA region and the strong genetic risk it confers in SSc, we investigated the relationships between genetic HLA associations, autoantibodies, and autoantigens, and identified candidate foreign peptides with homology to autoantigens, which might promote SSc pathogenesis through molecular mimicry. AA SSc patients were obtained from the Genome Research in African American Scleroderma Patients (GRASP) cohort that was created to enroll a large number of AA patients with SSc for conducting systematic and comprehensive genetic studies (8, 36). Genotypes from 2 cohorts of AA and EA ancestries were used to impute HLA classical alleles of 3 class I and 5 class II genes, which were evaluated for association with SSc and common SSc-specific autoantibodies. In the AA SSc cohort, we identified 2 African ancestry-predominant alleles that could explain, at least in part, the increased SSc frequency and severity observed in AAs. Upon analyzing the autoantibody subsets of SSc, an even stronger and specific HLA allele association was observed in both the AA and EA cohorts. Pairs of HLA molecule alpha and beta chains (HLA / heterodimers) were used to bioinformatically predict immunodominant peptides derived from the specific self-antigens that bind these HLA molecules. Remarkably, these immunodominant peptides demonstrated significant sequence homology with peptides derived from viruses in the Mimiviridae and Phycodnaviridae families, suggesting potential molecular mimicry.
Results
SSc Patients and Population Stratification.
To identify HLA alleles associated with SSc, we examined 662 AA SSc patients and 946 AA controls enrolled in the GRASP cohort along with 723 EA SSc patients and 5,347 EA controls obtained from the database of Genotypes and Phenotypes (dbGaP) (SI Appendix, Table S1). The gender and autoantibody information for the AA and EA SSc patients is shown in SI Appendix, Table S2. Principal component (PC) analyses (PCAs) were performed, and the top 10 PCs were used as covariates to correct for population stratification in the association analyses for both the AA and EA populations individually (SI Appendix, Fig. S1).
Classical HLA Allele Imputation.
To assess imputation accuracy in the AA cohort, we compared the HLA*IMP:03 allele concordance with exome sequence-based types from 763 GRASP individuals, both SSc and controls, using the HLA*PRG:LA software (37). The concordance rates for class I and class II HLA alleles present at minor allele frequency of 1% were 96% or higher (SI Appendix, Table S3). The allele frequencies in both AA and EA SSc cases and controls for the HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, and HLA-DPB1 genes are presented in SI Appendix, Tables S4 and S5.
Classical HLA Allele Associations with SSc.
In 662 AA SSc cases and 946 controls, 2 independent class II HLA alleles were significantly associated with AA SSc after correcting for population stratification (Table 1 and SI Appendix, Fig. S2). In AA SSc, a predominantly African ancestral allele, HLA-DRB1*08:04 was the most strongly SSc-associated, with an odds ratio (OR) of 3.2. A conditional regression analysis accounting for the effect of HLA-DRB1*08:04 allele identified a second African-predominant allele, HLA-DRB1*11:02, which was independently associated with SSc, with an OR of 2.3 (Table 1).
Table 1.
Logistic regression and conditional analysis of HLA classical alleles in AA SSc
| Frequency (%)† | Unconditioned | Conditioned | ||||
| HLA allele | (SSc/Ctrls) | OR (95% CI) | P value | OR (95% CI) | P value | |
| All SSc vs. controls | HLA-DRB1*08:04 | 24.3/9.3 | 3.2 (2.4-4.2) | 3.26 × 10−16 | 3.2 (2.4-4.2) | 3.26 × 10−16‡ |
| SSc = 662; control = 946 | HLA-DQB1*03:19 | 18.4/8.8 | 2.4 (1.8-3.2) | 2.45 × 10−8 | ||
| HLA-DQB1*03:01 | 37/25.8 | 1.8 (1.4-2.2) | 1.41 × 10−6 | |||
| HLA-DRB1*07:01 | 11.5/20.0 | 0.6 (0.4-0.7) | 2.72 × 10−6 | |||
| HLA-DQA1*02:01 | 11.5/20.0 | 0.6 (0.4-0.7) | 3.18 × 10−6 | |||
| HLA-DRB1*11:02 | 13.6/7.1 | 2.2 (1.6-3.0) | 9.39 × 10−6 | 2.3 (1.6-3.2) | 1.84 × 10−6 | |
| HLA-DPA1*02:01 | 62.1/51.5 | 1.6 (1.3-1.9) | 3.20 × 10−5 | |||
| HLA-DPB1*13:01 | 16.9/9.7 | 1.9 (1.4-2.6) | 3.21 × 10−5 | |||
| AFA+ SSc vs controls | HLA-DRB1*08:04 | 42.6/9.3 | 7.4 (4.9-11.3) | 2.61 × 10−19 | 7.4 (4.9-11.3) | 2.61 × 10−19‡ |
| SSc = 129; control = 946 | HLA-DQB1*06:09 | 20.9/6.6 | 3.8 (2.3-6.3) | 1.37 × 10−6 | 4.1 (2.4-7.0) | 2.04 × 10−6 |
| HLA-DQB1*03:01 | 45.0/25.8 | 2.5 (1.7-3.6) | 9.16 × 10−6 | |||
| HLA-DQB1*03:19 | 22.5/8.8 | 3.0 (1.9-4.9) | 2.53 × 10−5 | |||
| HLA-DRB1*13:02 | 29.5/13.8 | 2.6 (1.7-4.0) | 3.70 × 10−5 | |||
| ATA+ SSc vs. controls | HLA-DPB1*13:01 | 30.6/9.7 | 4.3 (2.9-6.3) | 2.35 × 10−12 | 4.3 (2.9-6.3) | 2.35 × 10−12‡ |
| SSc = 183; control = 946 | HLA-DQB1*02:01 | 7.7/25.1 | 0.3 (0.2-0.5) | 1.10 × 10−8 | 0.3 (0.2-0.5) | 2.17 × 10−7 |
| HLA-DRB1*03:01 | 3.3/14.9 | 0.2 (0.1-0.5) | 4.92 × 10−7 | |||
| HLA-DQB1*03:01 | 43.7/25.8 | 2.3 (1.7-3.1) | 2.64 × 10−6 | |||
| HLA-DPA1*02:01 | 69.9/51.5 | 2.3 (1.6-3.2) | 2.73 × 10−6 | |||
| HLA-DRB1*08:04 | 21.3/9.3 | 2.8 (1.8-4.2) | 8.13 × 10−6 | |||
| HLA-DQA1*05:01 | 51.4/39.1 | 1.6 (1.2-2.2) | 2.90 × 10−3 | 2.1 (1.5-3.0) | 2.21 × 10−5 | |
| ARA+ SSc vs. controls | None significant | |||||
| SSc = 119; control = 946 | ||||||
| ACA+ SSc vs. controls | None significant | |||||
| SSc = 64; control = 946 | ||||||
Independent associations by conditional regression analyses are shown in bold.
Frequency of individuals with 1 or 2 alleles.
Unconditioned; common AA haplotype: HLA-DRB1*08:04/DQA1*05:01/DQB1*03:01.
In 723 EA SSc cases and 5,437 controls, 3 independent HLA classical alleles, HLA-DQB1*02:02, HLA-DPB1*13:01, and HLA-DRB1*11:04, were significantly associated with SSc (Table 2 and SI Appendix, Fig. S2). The most significantly associated allele, HLA-DQB1*02:02, was disease-protective, with an OR of 0.5, whereas HLA-DPB1*13:01 and HLA-DRB1*11:04 were disease risk alleles, with ORs of 2.6 and 2, respectively (Table 2). Conditioning on the final HLA model for each ancestral population, neither the classical HLA alleles nor the HLA region single nucleotide variants remained significant at the statistical threshold (SI Appendix, Figs. S3 and S4). None of the class I HLA alleles showed any statistically significant independent association with SSc in either the AA or EA cohort.
Table 2.
Logistic regression and conditional analysis of HLA classical alleles in EA SSc
| Frequency (%)† | Unconditioned | Conditioned | ||||
| HLA allele | (SSc/Ctrls) | OR (95% CI) | P value | OR (95% CI) | P value | |
| All SSc vs. controls | HLA-DQB1*02:02 | 10.2/18.0 | 0.5 (0.4-0.6) | 3.55 × 10−9 | 0.5 (0.4-0.6) | 3.55 × 10−9‡ |
| SSc = 723; Control = 5,437 | HLA-DRB1*07:01 | 15.1/23.7 | 0.5 (0.4-0.7) | 6.06 × 10−9 | ||
| HLA-DQA1*02:01 | 15.8/24.3 | 0.6 (0.4-0.7) | 1.04 × 10−8 | |||
| HLA-DPB1*13:01 | 8.3/3.3 | 2.6 (1.9-3.5) | 1.75 × 10−8 | 2.6 (1.9-3.6) | 1.04 × 10−8 | |
| HLA-DRB1*11:04 | 10.5/4.7 | 2.2 (1.7-2.9) | 9.25 × 10−8 | 2.0 (1.5-2.7) | 1.04 × 10−8 | |
| HLA-B*44:03 | 5.1/9.7 | 0.5 (0.3-0.7) | 4.72 × 10−6 | |||
| HLA-DRB1*01:01 | 23.7/17.3 | 1.5 (1.2-1.8) | 4.39 × 10−5 | |||
| AFA+ SSc vs. controls | Not tested | |||||
| SSc = 0; control = 5,437 | ||||||
| ATA+ SSc vs. controls | HLA-DPB1*13:01 | 32.2/3.3 | 13.7 (8.9-21.0) | 1.47 × 10−24 | 13.7 (8.9-21.0) | 1.47 × 10−24‡ |
| SSc = 115; control = 5,437 | HLA-DRB1*11:04 | 25.2/4.7 | 6.3 (3.9-10.0) | 8.62 × 10−12 | 6.5 (4.0-10.6) | 1.59 × 10−11 |
| HLA-DPA1*02:01 | 48.7/26.3 | 2.9 (2.0-4.2) | 8.70 × 10−8 | |||
| HLA-DPA1*01:03 | 54.8/32.6 | 2.7 (1.8-3.9) | 4.65 × 10−7 | |||
| ARA+ SSc vs. controls | None significant | |||||
| SSc = 123; control = 5,437 | ||||||
| ACA+ SSc vs. controls | HLA-DRB1*07:01 | 3.4/23.7 | 0.1 (0.05-0.2) | 4.79 × 10−20 | 0.1 (0.05-0.2) | 4.79 × 10−20‡ |
| SSc = 238; control = 5,437 | HLA-DQA1*02:01 | 4.6/14.5 | 0.1 (0.1-0.2) | 4.85 × 10−18 | ||
| HLA-DQB1*02:02 | 2.9/18.0 | 0.1 (0.1-0.3) | 2.44 × 10−14 | |||
| HLA-DQB1*05:01 | 42.4/22.3 | 2.3 (1.8-3.0) | 4.21 × 10−9 | 2.0 (1.5-2.6) | 2.93 × 10−6 | |
| HLA-DQA1*01:01 | 47.5/26.7 | 2.2 (1.7-2.9) | 7.08 × 10−9 | |||
| HLA-DRB1*01:01 | 34.5/17.3 | 2.2 (1.7-3.0) | 1.32 × 10−7 | |||
| HLA-DQA1*04:01 | 14.3/5.4 | 2.7 (1.8-4.0) | 4.18 × 10−6 | 2.7 (1.8-4.0) | 6.67 × 10−6 | |
| HLA-DQB1*03:03 | 2.1/8.9 | 0.2 (0.1-0.5) | 6.29 × 10−6 | |||
| HLA-DRB1*08:01 | 11.8/4.6 | 2.6 (1.7-3.9) | 4.97 × 10−5 | |||
Independent associations by conditional regression analyses are shown in bold.
†Frequency of individuals with 1 or 2 alleles.
‡Unconditioned; Common EA haplotypes: HLA-DRB1*11:04/DQA1*05:01/DQB1*03:01 and HLA-DRB1*07:01/DQA1*02:01/DQB1*02:02.
Classical HLA Allele Associations within SSc Autoantibody-Positive Subsets.
SSc has highly specific and mutually exclusive autoantibodies, and thus we hypothesized that the HLA allelic associations would be stronger within the SSc-specific autoantibody subsets. We tested this hypothesis by stratifying the SSc samples into subsets of SSc-specific autoantibody-positive patients and evaluating association of HLA alleles. In 129 AFA+ AA SSc patients, the OR for HLA-DRB1*08:04 increased from 3.2 (P = 3.26 × 10−16) in overall SSc to 7.4 (P = 2.61 × 10−19) in the AFA+ subset (Table 1). Although not detected in overall SSc, HLA-DQB1*06:09 was independently associated in the AFA+ SSc subset. In 183 ATA+ AA SSc, HLA-DPB1*13:01, HLA-DQB1*02:01, and HLA-DQA1*05:01 were independently associated with ATA+ SSc (Table 1). The association of HLA-DPB1*13:01 with ATA+ SSc was particularly strong (OR = 4.3) as compared to overall SSc (OR = 1.9). None of the HLA classical alleles were statistically significantly associated with SSc in 119 ARA+ AA SSc patients, nor in 64 ACA+ AA SSc.
AFA data were not reported for the EA SSc patients in dbGaP, so we were unable to evaluate the AFA+ subset in the EA SSc patients. In 115 ATA+ EA SSc patients, HLA-DPB1*13:01 and HLA-DRB1*11:04 were independently associated in the EA ATA+ SSc patients (Table 2). Also, as seen in AAs, the association of HLA-DPB1*13:01 in ATA+ SSc subset was much stronger (OR = 13.7, P = 1.47 × 10−24) than its association with overall SSc in EAs (OR = 2.6, P = 1.75 × 10−8; Table 2). Interestingly, as in the ARA+ SSc subset in AAs, none of the classical HLA alleles were statistically significantly associated with the ARA+ SSc subset in EAs. In 238 ACA+ EA SSc, HLA-DRB1*07:01, associated with SSc protection, was the most statistically significantly associated allele, with OR = 0.1 and P = 4.79 × 10−20 (Table 2 and SI Appendix, Fig. S2). HLA-DRB1*07:01, part of the HLA-DRB1*07:01/DQA1*02:01/DQB1*02:02 haplotype, is in strong linkage disequilibrium (LD) with HLA-DQB1*02:02 (r2 = 0.95), and likely explains the association of HLA-DQB1*02:02 with overall SSc.
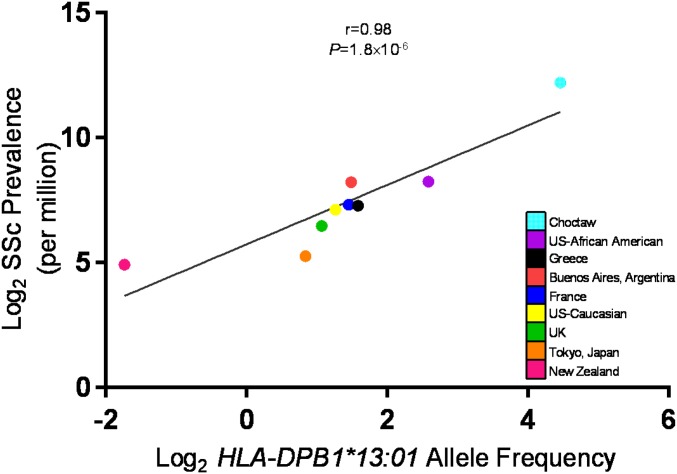
HLA-DPB1*13:01 and SSc.
HLA-DPB1*13:01 was identified as a strong risk allele in both the AA and EA ATA+ SSc; therefore, we examined whether HLA-DPB1*13:01 was enriched in the ATA+ SSc subset exclusively. In AAs, HLA-DPB1*13:01 was present in 11.7% of ATA− SSc and 9.7% of controls, which was statistically not different, whereas 30.6% of ATA+ SSc carried it. Similarly in EAs, HLA-DPB1*13:01 was present in 32.3% of ATA+ SSc and only 3.8% of ATA− SSc and 3.3% of controls. HLA-DPB1*13:01 was not only enriched in the ATA+ SSc subset but also had a higher frequency in the AA control population as compared to the EA control population. Given the fact that AAs have a higher incidence and prevalence of SSc, we next explored SSc prevalence and HLA-DPB1*13:01 allele frequency in several populations around the world. We observed a direct correlation between SSc prevalence in any given population and the HLA-DPB1*13:01 frequency, with a correlation coefficient of 0.98 and P = 1.8 10−6 (Fig. 1 and SI Appendix, Table S6). Even after removing the Choctaw population with the highest prevalence of SSc, the correlation coefficient remained 0.81 (SI Appendix, Fig. S5).
Fig. 1.
Population frequency of HLA-DPB1*13:01 allele and SSc prevalence.
Classical HLA Allele Associations within SSc Autoantibody-Negative Subsets.
On observing the enrichment of HLA-DPB1*13:01 in the ATA+ SSc subset, we systematically examined the autoantibody-negative subsets for association with the subset-specific independent HLA alleles. HLA-DRB1*08:04 was statistically significantly associated in both the AFA+ and AFA− subsets in the AA SSc. This was consistent with the strong association of HLA-DRB1*08:04 in overall AA SSc patients. The other independent HLA associations identified in the autoantibody-positive SSc subsets were not observed in the autoantibody-negative SSc subsets, highlighting the specificity of these associations with these SSc-specific autoantibodies (SI Appendix, Tables S7 and S8).
Amino Acid Residue Associations with SSc Autoantibody Subsets.
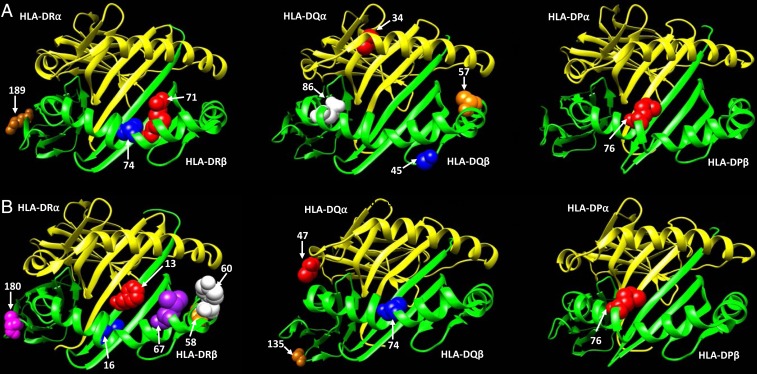
We performed amino acid association analysis for each of the class II HLA genes in the AA and EA SSc autoantibody subsets. In the AFA+ AA SSc subset, HLA-DRB1 amino acid (aa) positions 74 and 189, which are in tight LD, showed the strongest association, followed by aa position 71 (SI Appendix, Fig. S6A). In HLA-DQB1, aa positions 45 and 86 were independently associated with SSc risk in the AFA+ subset (SI Appendix, Fig. S6A). In the ATA+ AA SSc subset, HLA-DPB1 aa position 76, HLA-DQB1 aa positions 45 and 57, and HLA-DQA1 aa position 34 were independently contributing toward SSc risk (SI Appendix, Fig. S6A). Interestingly, aa position 45 on HLA-DQB1 was important for both the AFA+ and ATA+ SSc subsets in the AA population (SI Appendix, Fig. S6A). In the ACA+ EA SSc subset, HLA-DRB1 aa positions 60, 16, 13, and 180, HLA-DQB1 aa positions 135 and 74, and HLA-DQA1 aa position 47 were independently associated with SSc (SI Appendix, Fig. S6B). In the ATA+ EA SSc subset, HLA-DPB1 aa position 76 was strongly associated with SSc risk, similar to in the AA population. HLA-DRB1 aa positions 58 and 67 were also independently contributing toward SSc risk in the ATA+ subset in EAs (SI Appendix, Fig. S6B). We highlighted these independently associated aa residues in 3-dimensional (3D) ribbon models of HLA-DR, HLA-DQ, HLA-DQ, and HLA-DP with a direct view of the peptide-binding groove. All of the above-mentioned SSc-associated amino acids were part of the peptide-binding groove of class II HLA molecules, except for HLA-DR aa positions 180 and 189 and HLA-DQ aa position 135 (Fig. 2 and SI Appendix, Fig. S7).
Fig. 2.
Ribbon model of the HLA-DR, HLA-DQ, and HLA-DP proteins with independently associated amino acid residues, based on PDB ID codes 6atf, 1s9v, and 3lqz, respectively. (A) Scleroderma-associated aa positions in AAs; (B) Scleroderma-associated aa positions in EAs.
Classification and Regression Tree.
We used an established exploratory method (Classification and Regression Tree [CART]) as an alternative approach to identify interactions among the classical HLA alleles in the autoantibody subsets in the AA and EA populations. The alleles partitioning out higher in the decision tree suggest greater importance than the ones lower in the tree. In the ATA+ AA SSc subset, 30.6% carried HLA-DPB1*13:01, and, furthermore, 2 higher-order HLA allelic interactions were identified. HLA-DPB1*13:01−/HLA-DQB1*02:01+ was seen in 6% of patients, and HLA-DPB1*13:01−/HLA-DQB1*02:01−/HLA-DQA1*05:01+ was seen in 33.3% of patients (Fig. 3A and SI Appendix, Fig. S8A). In the AFA+ AA SSc subset, 42.6% carried HLA-DRB1*08:04; 14% carried HLA-DRB1*08:04−/HLA-DQB1*06:09+, and 5.4% carried HLA-DRB1*08:04−/HLA-DQB1*06:09−/HLA-DRB1*13:04+ (Fig. 3A and SI Appendix, Fig. S8B). Taken together, the susceptibility HLA alleles account for 64% of ATA+ SSc patients and 62% of AFA+ SSc patients in the AA population. Similar analysis performed in the EA ATA+ SSc subset identified HLA-DPB1*13:01+ (32.2%), HLA-DPB1*13:01−/HLA-DRB1*11:04+ (20.9%), and HLA-DPB1*13:01−/HLA-DRB1*11:04−/HLA-DQA*03:01+ (4.3%), accounting for 53% of patients with risk alleles (Fig. 3B and SI Appendix, Fig. S9A). Fifty-four percent of EA ACA+ SSc patients were accounted for by HLA-DRB1*07:01−/HLA-DQB1*05:01+ (42%) and HLA-DRB1*07:01−/HLA-DQB1*05:01−/HLA-DQA1*04:01+ (11.7%) (Fig. 3B and SI Appendix, Fig. S9B). The ORs from the CART analysis were comparable to the multivariate logistic regression analyses shown in Tables 1 and 2.
Fig. 3.
Pie charts of independently associated classical HLA alleles. Data from ATA+, AFA+, and ACA+ subsets in (A) AAs and (B) EAs.
HLA Molecule and Chain-Pair Associations with SSc Autoantibody Subsets.
The HLA alleles encode for class II HLA molecules that are composed of an alpha and a beta chain, and the resulting 3D structure defines the nature of the peptides that are effectively bound. We performed association analysis of the HLA haplotypes for HLA-DQA1/DQB1, HLA-DPA1/DPB1, and HLA-DRA1/DRB1 pairs within SSc autoantibody subsets to identify HLA / heterodimers. In the AAs, conditional regressional analysis identified 2 HLA / heterodimers independently associated with the AFA+ SSc subset and 3 HLA / heterodimers associated with the ATA+ SSc subset (Table 3). In the EAs, conditional regressional analysis identified 2 HLA / heterodimers associated with the ATA+ SSc subset and 2 / heterodimers associated with the ACA+ SSc subset (Table 3). HLA-DRA1*01:01/DRB1*07:01 was protective for ACA+ SSc subset in the EAs, and HLA-DQA1*05:01/DQB1*02:01 was protective for ATA+ SSc subset in the AAs. The other 7 independently associated HLA / heterodimers were all associated with increased SSc risk.
Table 3.
Logistic regression and conditional analysis of HLA / heterodimers in SSc autoantibody subsets
| SSc case group (n) | HLA / heterodimer | OR (95% CI) | P value |
| AA AFA+ SSc vs. controls | DRA1*01:01/DRB1*08:04 | 7.4 (4.9-11.3) | 2.6 × 10−19 |
| SSc = 129; control = 946 | DQA1*01:02/DQB1*06:09 | 4.6 (2.6-7.9) | 4.0 × 10−7† |
| AA ATA+ SSc vs. controls | DPA1*02:01/DPB1*13:01 | 4.8 (3.2-7.1) | 8.4 × 10−14 |
| SSc = 183; control = 946 | DQA1*05:01/DQB1*02:01 | 0.2 (0.1-0.5) | 5.3 × 10−6† |
| DQA1*05:01/DQB1*03:19 | 3.3 (2.0-5.5) | 1.6 × 10−5† | |
| EA ATA+ SSc vs. controls | DPA1*02:01/DPB1*13:01 | 15.7 (10.1-24.2) | 1.2 × 10−25 |
| SSc = 115; control = 5437 | DRA1*01:01/DRB1*11:04 | 6.4 (3.9-10.4) | 2.9 × 10−11† |
| EA ACA+ SSc vs. controls | DRA1*01:01/DRB1*07:01 | 0.1 (0.05-0.2) | 4.8 × 10−20 |
| SSc = 239; control = 5,437 | DQA1*01:01/DQB1* 05:01 | 2.0 (1.5-2.6) | 1.8 × 10−6† |
Significance upon conditioning on top associated α/β heterodimer(s).
Immunodominant Peptide Prediction.
We explored the possibility of whether nuclear self-antigens act as the source of peptides that are being recognized and presented by these HLA molecules. We utilized NetMHCIIpan 3.2 to identify class II HLA restricted peptides using the HLA / heterodimers and the correlated SSc self-antigens (topoisomerase I, fibrillarin, and centromere protein A or B [CENPA/CENPB]) (38). Peptide sequences that had a binding affinity of 500 nM to 2 associated HLA / heterodimers were selected. We bioinformatically identified immunodominant peptides on 3 regions of topoisomerase I that bind the multiple HLA risk / heterodimers in ATA+ SSc in the 2 ancestral populations (SI Appendix, Table S9). A similar search for immunodominant peptides in the AFA+ subset identified 5 regions on fibrillarin and, in the ACA+ subset, yielded 1 region on CENPA and 7 regions on CENPB that were bioinformatically predicted to bind the multiple HLA risk / heterodimers for the respective autoantibody subsets (SI Appendix, Tables S10, S11A, and S11B).
Molecular Mimicry.
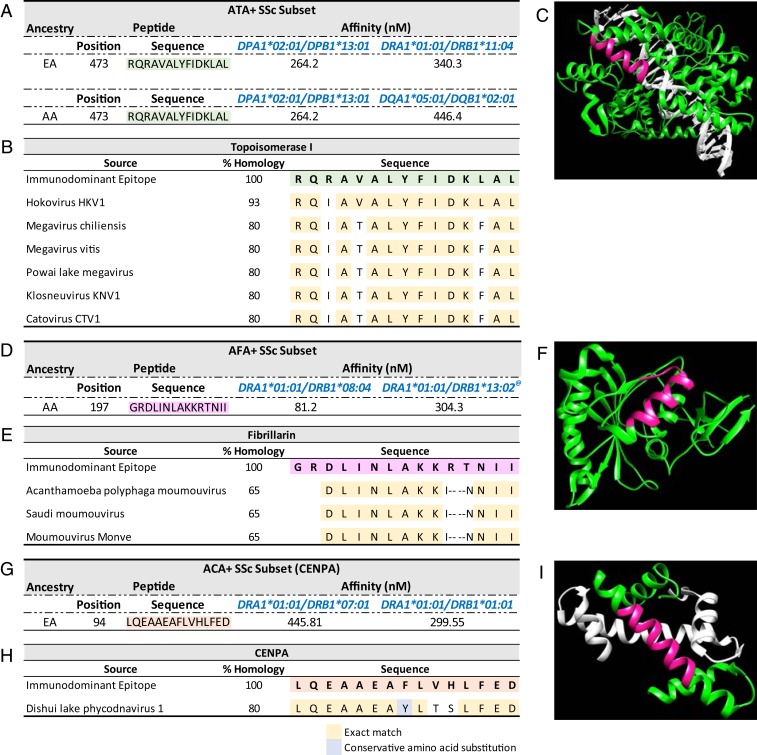
Next, we explored homology between the bioinformatically predicted immunodominant peptide sequences and microbial protein sequences to assess whether the autoantibodies observed in SSc may be induced by molecular mimicry. The bioinformatically predicted immunodominant peptide sequences from topoisomerase I (SI Appendix, Table S9) were compared for homology with microbial protein sequence databases. Several hundreds of homologous sequences were identified in fungi (E value 0.05) due to extensive similarities between human and fungi topoisomerase I proteins. There was no homology observed with bacterial sequences even at an E value of 1. Remarkably, only one sequence in topoisomerase I, “RQRAVALYFIDKLAL,” had high-quality matches in the viral database at an E value of 0.05. These homologous peptides were from viruses in the Mimiviridae family, part of the nucleocytoplasmic large DNA virus (NCLDV) clade, and had an extremely significant homology E value of 3.0 10−6 with Hokovirus (Fig. 4 and SI Appendix, Table S12). Given these findings, we examined the bioinformatically predicted immunodominant peptides from fibrillarin, CENPA, and CENPB for homology within the viral protein sequence database. On comparing several dozens of bioinformatically predicted immunodominant peptides, only one peptide sequence in fibrillarin, and another one in CENPA, had high-quality matches (E value 0.05) in the viral database (SI Appendix, Tables S10 and S11A). Fibrillarin sequence “GRDLINLAKKRTNII” and CENPA sequence “LQEAAEAFLVHLFED” were homologous to protein sequences from NCLDV in the Mimiviridae and Phycodnaviridae families with E values of 0.004 and 0.01, respectively (Fig. 4 and SI Appendix, Table S12). No high-quality matches were found for any of the CENPB sequences (SI Appendix, Table S11B).
Fig. 4.
Bioinformatically derived immunodominant peptides and homologous viral protein identification. (A) Predicted immunodominant peptides in topoisomerase I protein, (B) peptide sequences from microbial proteins homologous to topoisomerase I sequence, (C) 3D ribbon model of topoisomerase I with the identified immunodominant peptide in pink, (D) predicted immunodominant peptides in fibrillarin protein, (E) peptide sequences from microbial proteins homologous to fibrillarin sequence, (F) 3D ribbon model of fibrillarin protein with the identified immunodominant peptide in pink, (G) predicted immunodominant peptides in CENPA, (H) peptide sequences from microbial proteins homologous to CENPA sequence, and (I) 3D ribbon model of CENPA protein with the identified immunodominant peptide in pink. (These structures are based on PDB ID codes 1a35 for topoisomerase I, 2ipx for fibrillarin, and 3nqu for CENPA; in LD with DQA1*01:02/DQB1*06:09.)
These highly significant E values suggest that the homology was unlikely to occur by chance. To test this hypothesis even more rigorously, we contrasted the E values from randomly generated peptides to act as negative comparators. One hundred randomly generated 15-mer peptide sequences were compared to the viral sequence database for homology. None of these sequences matched any viruses from the Mimiviridae or Phycodnaviridae families (SI Appendix, Tables S13A and S13B). Additionally, an arbitrary 15-mer peptide selected from serum albumin residues 152 to 166, to act as a negative control for self-antigen peptides (39), did not show any homology with any viruses from the Mimiviridae or Phycodnaviridae families. The topoisomerase I immunodominant peptide “RQRAVALYFIDKLAL” is part of the catalytic domain of the topoisomerase I enzyme and is highlighted in pink on the 3D structure (Fig. 4C). The fibrillarin and CENPA immunodominant peptides are also highlighted on their respective protein structures (Fig. 4 F and I). Next, we compared the “RQRAVALYFIDKLAL” sequence in topoisomerase I, “GRDLINLAKKRTNII” sequence in fibrillarin, and “LQEAAEAFLVHLFED” sequence in CENPA for homology within the human protein database and discovered that these peptide sequences were unique to topoisomerase I, fibrillarin, and CENPA proteins, respectively, with E values of 0.05 (SI Appendix, Tables S14A, S14B, and S14C).
Bioinformatically Predicted Immunodominant Peptides in Other Ancestries.
We examined whether the bioinformatically predicted immunodominant peptides with homology to Mimiviridae and Phycodnaviridae viruses that were identified in the AA and EA SSc patients were also recognized by SSc-associated HLA alleles in SSc patients of other ancestries. SSc-associated HLA alleles in the ACA+ and ATA+ subsets in the Japanese, Chinese, Thai, Turkish, Iranian, Mexican, and Choctaw Indian populations were selected from published manuscripts (40–45). Upon examining the HLA risk alleles for the ATA+ subset in these populations, the “RQRAVALYFIDKLAL” sequence in topoisomerase I was predicted to bind with significant affinity to HLA-DRB1*15:02 in the Japanese and Thai populations, HLA-DRB1*08:02 in the Mexican population, HLA-DRB1*11:04 and HLA-DPB1*13:01 in the Turkish and Iranian populations, and HLA-DRB1*16:02 in the Choctaw Indian populations (SI Appendix, Table S15A) (40–44). Likewise, the “LQEAAEAFLVHLFED” sequence in CENPA was predicted to bind with significant affinity to HLA-DQB1*05:01 in the Japanese and Chinese populations and HLA-DQB1*03:01 in the Japanese population, which are the HLA risk alleles for the ACA+ subset in these populations (SI Appendix, Table S15B) (40, 45).
Discussion
This is the largest genetic study of AA SSc patients identifying African ancestry-predominant alleles, HLA-DRB1*08:04 and HLA-DRB1*11:02, that increase SSc risk. We demonstrate that an African ancestry HLA allele, HLA-DRB1*08:04, is associated with AFA that is common in the AA SSc patients and confers a risk of 7.4-fold. A previously unreported allele in SSc, HLA-DQB1*06:09, confers a risk of 4.1-fold in the AFA+ subset, independent of the effect of HLA-DRB1*08:04. We also report a very strong association of the HLA-DPB1*13:01 allele with the ATA+ subset of SSc that displays a transancestry effect. We show that the HLA-DRB1*07:01/DQA1*02:01/DQB1*02:02 haplotype confers an extremely protective effect in the ACA+ EA SSc subset with an OR of 0.1, and HLA-DRB1*07:01 has been reported to be protective for several other autoimmune diseases as well (46–50). Notably, there were no class I HLA alleles identified independently of class II HLA allele association, placing SSc firmly in the category of class II HLA disease. Lastly, the bioinformatically predicted immunodominant peptides on topoisomerase I, fibrillarin, and CENPA had significant homology to proteins from viruses in the Mimiviridae and Phycodnaviridae families, suggesting a potential environmental link in SSc pathogenesis.
An interesting observation in our study was the enrichment of the HLA alleles in SSc-specific autoantibody subsets that increased the risk severalfold. Supporting this hypothesis, we identified autoantibody subset-specific HLA alleles that, while not statistically significant in overall SSc, were significant in the autoantibody subsets with increased ORs. An exception to this was the strong association of HLA-DRB1*11:02 found in overall SSc in the AA population but not identified in any of the examined SSc-specific autoantibody subsets. On further analysis, it seems that the association of HLA-DRB1*11:02 was stronger in individuals with a speckled nuclear staining pattern, and its relevance to SSc will need to be further explored. The absence of any statistically significant association in the ACA+ SSc subset in the AAs was likely due to inadequate power because of the small sample size, since the frequency of ACA in the AA SSc patients is low (SI Appendix, Table S2). It is intriguing that the ARA+ SSc subset did not yield any statistically significant associations in either the AA or EA populations. This could possibly be due to the small sample size, leading to inadequate statistical power to detect an association. However, the ARA+ subset had a larger sample size than the ATA+ subset in EAs; thus it is possible that ARA+ SSc may not be one homogeneous entity. Instead, the ARA+ SSc subset could potentially represent a diverse collection of SSc phenotypes characterized by cancer association, presence of SSc renal crisis, or aggressive diffuse skin involvement. Perhaps further stratification of the ARA+ subset would yield statistically significant HLA associations. There is still a possibility that only non-HLA genes increase SSc risk in the ARA+ subset, but the HLA genes playing no role whatsoever in the pathogenesis of ARA+ SSc is unlikely.
HLA-DPB1*13:01 association with ATA+ SSc is very interesting, and the allele is present in a third of the ATA+ patients irrespective of African or European ancestry. This transancestry effect of HLA-DPB1*13:01 in AAs and EAs has previously been reported in the Choctaw native American SSc patients, who have a very uniform phenotype, with 95% ATA positivity and 65% HLA-DPB1*13:01 carrier frequency in the ATA+ subset (51, 52). The Choctaw native American SSc patients have not only the highest reported population carrier frequency of the HLA-DPB1*13:01 allele (45%) but also the highest reported prevalence of SSc (51, 52). In both the AA and EA populations, HLA-DPB1*13:01 frequency in the ATA− SSc subsets was similar to the controls and not statistically significant. This, along with the direct correlation of HLA-DPB1*13:01 frequency with SSc prevalence in various populations around the world, suggests a distinct role this allele may be playing in SSc pathogenesis. Interestingly, in the HLA-DP aa association analyses, aa position 76 isoleucine was the only statistically significant aa in both the AA and EA populations. It is possible that the HLA-DP aa position 76 isoleucine modifies the peptide-binding groove to recognize specific peptides that are presented by APCs to T helper cells, leading to an increase in SSc risk.
We identified several SSc-associated aa residues in all of the class II HLA genes, and most of them were part of the peptide-binding groove. Amino acid leucine, at position 74, in the peptide-binding groove was specific to HLA-DRB1*08:04 and was in perfect LD with serine at position 189 outside of the peptide-binding groove. These peptide-binding groove residue changes might be leading to APC recognition of specific peptides that, on presentation to T helper cells, lead to T helper cell activation and, in turn, B cell activation, ultimately resulting in autoimmunity. The residues outside the peptide-binding groove might play a role in altering the structure of the class II HLA molecule or modifying the interaction of the class II HLA molecule with the T cell receptor. Using CART analysis, we identified multiple HLA alleles for each of the SSc-specific autoantibody subsets (AFA+, ATA+, and ACA+) in both the AA and EA populations, which account for 53–64% of the SSc cases in each of these subsets.
Antibodies directed toward different nuclear or nucleolar self-antigens are seen in 95% of SSc patients (1). In this study, we explored the role of the class II HLA alleles and autoantigens and thus proposed self-antigens as a likely source of peptides that, once bound to the HLA molecules on APCs, are presented to T helper cells. We bioinformatically predicted immunodominant peptides that were recognized by multiple HLA risk alleles for each of the SSc-specific autoantibody subsets. The topoisomerase I peptide sequence “RQRAVALYFIDKLAL” that was bioinformatically predicted as an immunodominant peptide in both AA and EA ATA+ subsets is part of the catalytic domain of the molecule, unique to this protein and evolutionarily conserved. Hu et al. (53) have identified peripheral T cell lines from SSc patients recognizing the “RAVALYFIDKLA” peptide on topoisomerase I.
Molecular mimicry has been invoked previously as a potential mechanism driving autoimmunity in several diseases, including cytomegalovirus in multiple sclerosis and Epstein–Barr virus in lupus (27–30). We examined whether the bioinformatically predicted immunodominant peptide sequences identified in this study had homology to microbial protein sequences. Remarkably, “RQRAVALYFIDKLAL” sequence in topoisomerase I, “GRDLINLAKKRTNII” sequence in fibrillarin, and “LQEAAEAFLVHLFED” sequence in CENPA matched sequences from viruses of the Mimiviridae and Phycodnaviridae families that belong to the NCLDV clade, with an extremely high confidence level (54). Mimiviruses and phycodnaviruses are ubiquitous in aquatic environments, and humans are constantly being exposed to these viruses (55, 56). These viruses cannot infect human cells or replicate in them, but rather mimiviruses infect amoeba and phycodnaviruses infect algae (57, 58). Even though humans do not get infected with these viruses, phagocytosis by macrophages of virus-infected amoeba or algae can lead to processing of viral antigens and presentation via the class II HLA receptor to T helper cells (57). Activated T cells recognizing self-antigens with homology to viruses could arise from activation of quiescent T cells with receptors specific for host antigens resulting in autoreactive T cells. Alternatively, autoreactive T cells could arise by T cell receptor poly-specificity. Peptide recognition by T cell receptors is based on amino acid properties, and the binding motifs are degenerate, with only a small sequence needed for recognition. Similarities in peptides at critical residues that bind to the class II HLA molecules could lead to T cell cross-reactivity, ultimately leading to autoreactive T cells (59–62). Mimivirus and phycodnavirus peptides that have homology to topoisomerase I, fibrillarin, or CENPA could activate T helper cells, which, in turn, activate B cells with receptors that specifically recognize and target these nuclear antigens (topoisomerase I, fibrillarin, and CENPA, respectively). This could lead to the formation of ATA, AFA, and ACA observed in 74.8% of AA and 65.8% of EA SSc. Our data indicate that generation of a particular autoantibody has a strong relationship to class II HLA alleles, but the pathogenic potential of SSc-specific autoantibodies is unclear, and their presence could be an epiphenomenon. Constant exposure of the immune system to these nuclear antigens could lead to chronic autoimmunity. This raises an interesting hypothesis for a possible environmental link in SSc pathogenesis. An increased occurrence of antibodies against mimivirus collagen has been demonstrated in rheumatoid arthritis patients, along with antibodies against the mimivirus capsid protein L425 in a third of the rheumatoid arthritis patients (63). It is also possible that molecular mimicry to mimiviruses may just be an epiphenomenon not playing a direct role in SSc pathogenesis (64). Testing SSc samples for antibodies against mimivirus capsid protein would be an important step to demonstrate patient exposure to these viruses.
These HLA findings validate our understanding of SSc as an autoimmune disease and emphasize the relevance of class II HLA genes in SSc pathogenesis. The heterogeneity observed in SSc is best characterized by the robust HLA allelic associations demonstrated in the SSc-specific autoantibody subsets. These SSc-specific autoantibodies correlate not only with specific HLA alleles but also with distinct clinical phenotypes and disease outcomes (4–6). Stratifying SSc on the basis of autoantibodies and HLA alleles together for research and clinical trials may yield beneficial results. In the future, screening HLA-DPB1*13:01+ individuals for ANA and ATA could result in early identification and therapeutic intervention to block the development of SSc.
Materials and Methods
Patients and Controls.
This study included 662 AA SSc patients enrolled from 23 academic centers throughout the United States under the GRASP consortium with available genotype (Dataset S1) and serum SSc-specific autoantibody data (Dataset S2) (8, 36). The study was conducted in accordance with the Declaration of Helsinki, and participating centers secured local ethics committee approval prior to participant enrollment. All patients met the 1980 American College of Rheumatology (ACR) or the 2013 ACR/European League Against Rheumatism classification criteria for SSc or had at least 3 of the 5 features of CREST syndrome (calcinosis, Raynaud’s phenomenon, esophageal dysmotility, sclerodactyly, telangiectasias); 946 genetically similar unrelated controls were obtained from the Howard University Family Study, a population-based study of AA families and unrelated individuals (65). All cases and controls provided written informed consent. Genotype and phenotype data of 723 European ancestry SSc patients and 5,437 controls genotyped on the same platform were extracted from dbGaP (SI Appendix, Table S1). European ancestry SSc patients were a subset of those reported by Radstake et al. (14).
Autoantibody Testing.
Sera from the AA SSc patients were tested by a line immunoassay for SSc profile autoantibodies (Euroimmun Euroline profile kit). For the European ancestry SSc patients, reported autoantibody data were extracted from dbGaP accession phs000357.v1.p1.
Genotyping.
The AA SSc cases and controls were genotyped with the Illumina Infinium Multi-Ethnic Global Array kit. High-quality genotypes were imputed using the Michigan Imputation Server, and the required 6,114 markers were submitted to the HLA*IMP:03 server for HLA imputation. The European ancestry samples were genotyped on Illumina Human610-Quadv1_B chip (SI Appendix).
PCA.
For the 2 ancestral populations, PCA was used to evaluate the genetic similarity of the cases with the controls, to remove outliers, and to correct for residual dissimilarity separately (SI Appendix). Two-dimensional plots of the first 2 principal components of the cases and controls in each study are shown in SI Appendix, Fig. S1.
HLA Imputation.
We selected the HLA*IMP:03 tool to perform HLA imputation in the AA samples because it has a multiethnic reference panel of 10,561 individuals that includes 568 of African ancestry (SI Appendix) (66). We used available whole-exome sequence data for 763 of the AA samples to determine their HLA alleles using HLA*PRG:LA, allowing for comparison of HLA*IMP:03 imputed alleles with HLA*PRG:LA sequence-based alleles (37). For EA samples, SNP2HLA software and a mainly European ancestry reference of 5,225 individuals were used to impute classical HLA alleles and polymorphic HLA amino acids (67).
HLA Association and Conditional Analysis.
HLA alleles with frequency less than 0.01 were omitted, and a logistic regression association analysis was performed under a dominant model classically used for identifying HLA alleles associated with diseases (68) (SI Appendix). Regressions were corrected for genetic dissimilarity between the cases and the controls by including the top 10 PCs as covariates. To account for strong LD in the region, independent associations were identified by recursively including independent alleles as covariates. The total number of classical HLA alleles tested for association was 138 in both the populations, and there were 5 analyses conducted. Thus a Bonferroni’s multiple test corrected significance threshold of P 0.000072 was used for association analysis.
Amino Acid Analysis.
Amino acid associations with SSc were evaluated with a dominant model logistic regression analysis. Amino acids with frequency less than 0.01 were omitted. The P value threshold was set as P 0.000013 based on 800 amino acids tested across both population samples, multiplied by 5 sets of analysis.
The 3D Protein Modeling.
Protein Data Bank (PDB) ID codes 1a35, 2ipx, 3nqu, 6atf, 1s9v, and 3lqz were obtained for topoisomerase I, fibrillarin, CENPA, HLA-DR, HLA-DQ, and HLA-DP, respectively. UCSF Chimera was used to highlight individual aa positions (69).
CART Analysis.
CART analysis was performed to explore higher-order interactions among the HLA alleles using CART 6.0, Salford Systems (70).
Bioinformatic Prediction of Immunodominant Peptides.
An online computational tool, the NetMHCIIpan 3.2 server, was used to predict the binding of 15-mer peptide sequences within the protein of interest (topoisomerase I, fibrillarin, or CENPA/CENPB) to the SSc-associated major histocompatibility complex (MHC) class II / heterodimers within the respective autoantibody subsets. Peptides with a binding affinity of nM and observed in 2 of the SSc-associated HLA / heterodimers were prioritized as immunodominant peptides (38).
Molecular Mimicry.
The prioritized immunodominant peptide sequences were entered into the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool Standard Protein BLAST with the organism set to human (taxid:9606) to identify homologous sequences in other human proteins, and to fungi (taxid: 4751), bacteria (taxid: 2), and virus (taxid: 10239) to find homologous microbial sequences. Significant homology was defined by an E value of 0.05 (71).
Data Availability.
The AA genotype data are available as Dataset S1, and the corresponding phenotypic information is available as Dataset S2. The EA dataset is available from dbGaP (SI Appendix, Table S1).
Supplementary Material
Acknowledgments
This study was supported by research funding from the Scleroderma Research Foundation and the Intramural Research Programs of the National Human Genome Research Institute, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and the Center for Information Technology of NIH. Data were analyzed using the computational resources of the NIH high-performance computing Biowulf cluster (http://hpc.nih.gov). This work was supported, in part, by a Rheumatology Research Foundation Scientist Development award (P.G. and N.D.M.); NIH Grant T32-AR-048522 (N.D.M.); Chresanthe Staurulakis Memorial Discovery Fund and NIH Grant P30-AR-070254 (N.D.M., A.A.S., and F.M.W.); NIH Grant K01-AR-067280 (P.S.R.); NIH Grant P60-AR-062755 (P.S.R. and R.M.S.); and Nina Ireland Program for Lung Health (F.B.). We thank Dr. Daniella Schwartz for critical reading of the manuscript.
Footnotes
Competing interest statement: M.D.M. received consulting fees from Mitsubishi Tanabe, Astellas, Boehringer Ingelheim, Gerson Lehrman Group, Smart Analyst, and Guidepoint Global. R.T.D. received consulting fees from Eicos Sciences. D.K. received consulting fees from Actelion, Bristol-Myers Squibb, CSL Behring, Inventiva, EMD Merck-Serono, Sanofi-Aventis, GlaxoSmithKline, Corbus, Cytori, UCB, Bayer, Boehringer Ingelheim, and Genentech-Roche, and research support from Bayer, Bristol-Myers Squibb, and Pfizer, and owns stock or stock options in Eicos Sciences, Inc. (CiViBioPharma, Inc.). L.A.S. received consulting fees from Axon Pharma. H.G. received consulting fees from Pfizer, AbbVie, Actelion, and Horizon Pharmaceuticals. C.T.D. received research support from Gilead, Actelion, and Cytori. V.K.S. received research support from AbbVie. E.J.B. received consulting fees from Genentech. L.C. received consulting fees or served on the board of Reata, Bristol-Myers Squibb, Boehringer-Ingelheim, Eicos, and Mitsubishi Tanabe. S.L.B. and M.J.D. are coauthors on a 2015 research article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906593116/-/DCSupplemental.
References
- 1.Beck J. S., Anderson J. R., Gray K. G., Rowell N. R., Antinuclear and precipitating autoantibodies in progressive systemic sclerosis. Lancet 2, 1188–1190 (1963). [PubMed] [Google Scholar]
- 2.Reveille J. D., Solomon D. H., American College of Rheumatology Ad Hoc Committee of Immunologic Testing Guidelines , Evidence-based guidelines for the use of immunologic tests: Anticentromere, Scl-70, and nucleolar antibodies. Arthritis Rheum. 49, 399–412 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Bunn C. C., Black C. M., Systemic sclerosis: An autoantibody mosaic. Clin. Exp. Immunol. 117, 207–208 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho K. T., Reveille J. D., The clinical relevance of autoantibodies in scleroderma. Arthritis Res. Ther. 5, 80–93 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steen V. D., Autoantibodies in systemic sclerosis. Semin. Arthritis Rheum. 35, 35–42 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Hu P. Q., Fertig N., Medsger T. A. Jr, Wright T. M., Correlation of serum anti-DNA topoisomerase I antibody levels with disease severity and activity in systemic sclerosis. Arthritis Rheum. 48, 1363–1373 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Steen V., Domsic R. T., Lucas M., Fertig N., Medsger T. A. Jr, A clinical and serologic comparison of African American and Caucasian patients with systemic sclerosis. Arthritis Rheum. 64, 2986–2994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan N. D., et al. , Clinical and serological features of systemic sclerosis in a multicenter African American cohort: Analysis of the genome research in African American scleroderma patients clinical database. Medicine 96, e8980 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelber A. C., et al. , Race and association with disease manifestations and mortality in scleroderma: A 20-year experience at the Johns Hopkins Scleroderma Center and review of the literature. Medicine 92, 191–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan E., Furst D. E., Systemic sclerosis mortality in the United States: 1979-1998. Eur. J. Epidemiol. 20, 855–861 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Gourh P., et al. , Association of the PTPN22 R620W polymorphism with anti-topoisomerase I- and anticentromere antibody-positive systemic sclerosis. Arthritis Rheum. 54, 3945–3953 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Alkassab F., et al. , An allograft inflammatory factor 1 (AIF1) single nucleotide polymorphism (SNP) is associated with anticentromere antibody positive systemic sclerosis. Rheumatology 46, 1248–1251 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Agarwal S. K., et al. , Association of interleukin 23 receptor polymorphisms with anti-topoisomerase-I positivity and pulmonary hypertension in systemic sclerosis. J. Rheumatol. 36, 2715–2723 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radstake T. R., et al. , Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat. Genet. 42, 426–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rueda B., et al. , BANK1 functional variants are associated with susceptibility to diffuse systemic sclerosis in Caucasians. Ann. Rheum. Dis. 69, 700–705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gourh P., et al. , Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J. Autoimmun. 34, 155–162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourh P., et al. , Association of TNFSF4 (OX40L) polymorphisms with susceptibility to systemic sclerosis. Ann. Rheum. Dis. 69, 550–555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorlova O., et al. , Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PLoS Genet. 7, e1002178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnett F. C., et al. , Major histocompatibility complex (MHC) class II alleles, haplotypes and epitopes which confer susceptibility or protection in systemic sclerosis: Analyses in 1300 Caucasian, African-American and hispanic cases and 1000 controls. Ann. Rheum. Dis. 69, 822–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gladman D. D., et al. , Increased frequency of HLA-DR5 in scleroderma. Arthritis Rheum. 24, 854–856 (1981). [DOI] [PubMed] [Google Scholar]
- 21.Reveille J. D., et al. , Association of amino acid sequences in the HLA-DQB1 first domain with antitopoisomerase I autoantibody response in scleroderma (progressive systemic sclerosis). J. Clin. Investig. 90, 973–980 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McHugh N. J., et al. , Anti-centromere antibodies (ACA) in systemic sclerosis patients and their relatives: A serological and HLA study. Clin. Exp. Immunol. 96, 267–274 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuwana M., Kaburaki J., Medsger T. A. Jr, Wright T. M., An immunodominant epitope on DNA topoisomerase I is conformational in nature: Heterogeneity in its recognition by systemic sclerosis sera. Arthritis Rheum. 42, 1179–1188 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Ramos P. S., Silver R. M., Feghali-Bostwick C. A., Genetics of systemic sclerosis: Recent advances. Curr. Opin. Rheumatol. 27, 521–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neefjes J., Jongsma M. L., Paul P., Bakke O., Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Fujinami R. S., Oldstone M. B., Wroblewska Z., Frankel M. E., Koprowski H., Molecular mimicry in virus infection: Crossreaction of measles virus phosphoprotein or of herpes simplex virus protein with human intermediate filaments. Proc. Natl. Acad. Sci. U.S.A. 80, 2346–2350 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujinami R. S., Oldstone M. B., Amino acid homology between the encephalitogenic site of myelin basic protein and virus: Mechanism for autoimmunity. Science 230, 1043–1045 (1985). [DOI] [PubMed] [Google Scholar]
- 28.Wandinger K., et al. , Association between clinical disease activity and Epstein-Barr virus reactivation in MS. Neurology 55, 178–184 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Ebringer A., Baines M., Ptaszynska T., Spondyloarthritis, uveitis, HLA-B27 and Klebsiella. Immunol. Rev. 86:101–116 (1985). [DOI] [PubMed] [Google Scholar]
- 30.James J. A., et al. , Lupus and Epstein-Barr. Curr. Opin. Rheumatol. 24, 383–388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moroncini G., Mori S., Tonnini C., Gabrielli A., Role of viral infections in the etiopathogenesis of systemic sclerosis. Clin. Exp. Rheumatol. 31(2 Suppl 76), 3–7 (2013). [PubMed] [Google Scholar]
- 32.Kasturi K. N., Hatakeyama A., Spiera H., Bona C. A., Antifibrillarin autoantibodies present in systemic sclerosis and other connective tissue diseases interact with similar epitopes. J. Exp. Med. 181, 1027–1036 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maul G. G., et al. , Determination of an epitope of the diffuse systemic sclerosis marker antigen DNA topoisomerase I: Sequence similarity with retroviral p30gag protein suggests a possible cause for autoimmunity in systemic sclerosis. Proc. Natl. Acad. Sci. U.S.A. 86, 8492–8496 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamamdzic D., Harley R. A., Hazen-Martin D., LeRoy E. C., MCMV induces neointima in IFN-gammaR-/- mice: Intimal cell apoptosis and persistent proliferation of myofibroblasts. BMC Muscoskelet. Disord. 2, 3 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lunardi C., et al. , Systemic sclerosis immunoglobulin G autoantibodies bind the human cytomegalovirus late protein UL94 and induce apoptosis in human endothelial cells. Nat. Med. 6, 1183–1186 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Gourh P., et al. , Brief report: Whole-exome sequencing to identify rare variants and gene networks that increase susceptibility to scleroderma in African Americans. Arthritis Rheum. 70, 1654–1660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dilthey A. T., et al. , HLA*LA—HLA typing from linearly projected graph alignments. Bioinformatics. 35, 4394–4396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen K. K., et al. , Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogeboom C., et al. , Peptide motif analysis predicts lymphocytic choriomeningitis virus as trigger for multiple sclerosis. Mol. Immunol. 67, 625–635 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Furukawa H., et al. , Human leukocyte antigen and systemic sclerosis in Japanese: The sign of the four independent protective alleles, DRB1*13:02, DRB1*14:06, DQB1*03:01, and DPB1*02:01. PLoS One 11, e0154255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louthrenoo W., et al. , Association of HLA-DRB1*15:02 and DRB5*01:02 allele with the susceptibility to systemic sclerosis in Thai patients. Rheumatol. Int. 33, 2069–2077 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Reyna T. S., et al. , HLA class I and II blocks are associated to susceptibility, clinical subtypes and autoantibodies in Mexican systemic sclerosis (SSc) patients. PLoS One 10, e0126727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez-Serna D., et al. , Analysis of the genetic component of systemic sclerosis in Iranian and Turkish populations through a genome-wide association study. Rheumatology 58, 289–298 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Kuwana M., et al. , Association of human leukocyte antigen class II genes with autoantibody profiles, but not with disease susceptibility in Japanese patients with systemic sclerosis. Intern. Med. 38, 336–344 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Zhou X. D., et al. , Association of HLA-DQB1*0501 with scleroderma and its clinical features in Chinese population. Int. J. Immunopathol. Pharmacol. 26, 747–751 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noble J. A., Valdes A. M., Genetics of the HLA region in the prediction of type 1 diabetes. Curr. Diabetes Rep. 11, 533–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newman B., et al. , CARD15 and HLA DRB1 alleles influence susceptibility and disease localization in Crohn’s disease. Am. J. Gastroenterol. 99, 306–315 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Goyette P., et al. , High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet. 47, 172–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mero I. L., et al. , Oligoclonal band status in Scandinavian multiple sclerosis patients is associated with specific genetic risk alleles. PLoS One 8, e58352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paradowska-Gorycka A., et al. , Association of HLA-DRB1 alleles with susceptibility to mixed connective tissue disease in Polish patients. HLA 87, 13–18 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Tan F. K., et al. , Hla haplotypes and microsatellite polymorphisms in and around the major histocompatibility complex region in a Native American population with a high prevalence of scleroderma (systemic sclerosis). Tissue Antigens 53, 74–80 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Arnett F. C., et al. , Increased prevalence of systemic sclerosis in a Native American tribe in Oklahoma. Association with an Amerindian HLA haplotype. Arthritis Rheum. 39, 1362–1370 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Hu P. Q., Oppenheim J. J., Medsger T. A. Jr, Wright T. M., T cell lines from systemic sclerosis patients and healthy controls recognize multiple epitopes on DNA topoisomerase I. J. Autoimmun. 26, 258–267 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Iyer L. M., Balaji S., Koonin E. V., Aravind L., Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117, 156–184 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Boughalmi M., et al. , High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Environ. Microbiol. 15, 2000–2007 (2013). [DOI] [PubMed] [Google Scholar]
- 56.La Scola B., et al. , Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology 53:344–353 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Ghigo E., et al. , Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 4, e1000087 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H., et al. , The genome of a prasinoviruses-related freshwater virus reveals unusual diversity of phycodnaviruses. BMC Genomics 19, 49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wucherpfennig K. W., et al. , Polyspecificity of T cell and B cell receptor recognition. Semin. Immunol. 19, 216–224 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wucherpfennig K. W., et al. , Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. J. Exp. Med. 179, 279–290 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinigaglia F., Hammer J., Defining rules for the peptide-MHC class II interaction. Curr. Opin. Immunol. 6, 52–56 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Reay P. A., Kantor R. M., Davis M. M., Use of global amino acid replacements to define the requirements for MHC binding and T cell recognition of moth cytochrome c (93-103). J. Immunol. 152, 3946–3957 (1994). [PubMed] [Google Scholar]
- 63.Shah N., et al. , Exposure to mimivirus collagen promotes arthritis. J. Virol. 88, 838–845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albert L. I., Inman R. D., Molecular mimicry and autoimmunity. N. Engl. J. Med. 341, 2068–2074 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Adeyemo A., et al. , A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 5, e1000564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Motyer A., et al. , Practical use of methods for imputation of HLA alleles from SNP genotype data. 10.1101/091009. (9 December 2016). [DOI]
- 67.Jia X., et al. , Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomson G., Hla disease associations: Models for the study of complex human genetic disorders. Crit. Rev. Clin. Lab. Sci. 32, 183–219 (1995). [DOI] [PubMed] [Google Scholar]
- 69.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Zhang H., Bonney G., Use of classification trees for association studies. Genet. Epidemiol. 19, 323–332 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The AA genotype data are available as Dataset S1, and the corresponding phenotypic information is available as Dataset S2. The EA dataset is available from dbGaP (SI Appendix, Table S1).