Significance
Liver fibrosis interferes with normal organ function and supports tumor development in the liver. We uncovered a role of miRNAs in controlling liver fibrosis. In a comprehensive and systematic analysis we specify miRNA activities in targeting the fibrotic cellular microenvironment of the liver, in both mice and humans. We reveal and validate a complex network of 8 functionally connected miRNAs and 54 target genes to regulate structural, signaling, and remodeling components of the fibrotic extracellular matrix. We identify expression of this miRNA network to be controlled by the transcription factor Ppar. Thus, we expand the antifibrotic function of Ppar to controlling the synthesis of an antifibrotic miRNA network. This network may serve as a therapeutic target in antifibrotic therapies.
Keywords: microRNAs, fibrosis, hepatocellular carcinoma, PPARγ
Abstract
Liver fibrosis interferes with normal liver function and facilitates hepatocellular carcinoma (HCC) development, representing a major threat to human health. Here, we present a comprehensive perspective of microRNA (miRNA) function on targeting the fibrotic microenvironment. Starting from a murine HCC model, we identify a miRNA network composed of 8 miRNA hubs and 54 target genes. We show that let-7, miR-30, miR-29c, miR-335, and miR-338 (collectively termed antifibrotic microRNAs [AF-miRNAs]) down-regulate key structural, signaling, and remodeling components of the extracellular matrix. During fibrogenic transition, these miRNAs are transcriptionally regulated by the transcription factor Ppar and thus we identify a role of Ppar as regulator of a functionally related class of AF-miRNAs. The miRNA network is active in human HCC, breast, and lung carcinomas, as well as in 2 independent mouse liver fibrosis models. Therefore, we identify a miRNA:mRNA network that contributes to formation of fibrosis in tumorous and nontumorous organs of mice and humans.
Hepatocellular carcinoma (HCC) is the most frequent primary human liver malignancy. It represents the fifth most common cancer in men and the seventh in women (1). HCC is currently the third leading cause of cancer-related deaths worldwide (2).
To study molecular and cellular events underlying HCC formation, we have generated SRF- mice (3). These mice express in a mosaic, hepatocyte-specific fashion SRF-VP16, a constitutively active variant of serum response factor (SRF). SRF is a ubiquitously expressed transcription factor that regulates a wide range of biological processes (4). As a consequence of SRF-VP16 activity, SRF- mice develop hyperproliferative liver nodules that progress to lethal murine HCC (mHCC) (3). SRF-VP16–driven HCCs share several characteristics with human HCC (hHCC), including different features of the tumor microenvironment (3).
The tumor microenvironment is a complex composite of tumor and nontumor cells embedded within an extracellular matrix (ECM), which facilitates malignant tumor progression (5). HCC progression is a multistage process that typically arises in the context of liver fibrosis. Liver fibrosis is the consequence of an exaggerated wound-healing response to reoccurring or chronic liver injury and is characterized by excessive accumulation of ECM. The central event in liver fibrosis is the activation of hepatic stellate cells (HSCs). Activated HSCs (aHSCs) produce components of the ECM and growth factors, thus causing excessive ECM deposition, neoangiogenesis, and inflammation (6). These processes ultimately result in scarring and thickening of affected tissue, which interferes with normal liver function and facilitates HCC tumorigenesis.
Such a fibrotic microenvironment, caused by quantitative and qualitative changes in ECM depositions, is characterized by increased stiffness, which promotes tumorigenesis through elevated integrin signaling. This signaling leads to enhanced growth, survival, and proliferation of tumor cells (7). Furthermore, ECM deposition enhances HCC chemotherapy resistance and offers protection against immune cells (8).
Unfortunately, the sequence of molecular events leading to the formation of a fibrogenic microenvironment, including regulatory networks governing these events, is insufficiently understood.
Accumulating data support a regulatory role of microRNAs (miRNAs) in control of gene expression programs that underline different normal and pathologic processes, including cancer (9). miRNAs are 21- to 23-nucleotide-long RNAs that act as essential regulators of gene expression, directing degradation, destabilization, or translational repression of target mRNAs (10). In mammals, more than 60 of protein-coding genes are believed to be under the control of miRNAs (11). However, in the majority of cases which characterize the regulatory role of miRNAs, the magnitude of the described miRNA:mRNA regulation is mild. This seeming discrepancy of the extensive role that miRNAs have in different biological processes and the mild extent of their influence is explained by the capacity of individual miRNAs to target hundreds of different mRNAs simultaneously. If the miRNA targets are enriched in common pathways, then the sum of modest effects of individual miRNA:mRNA interactions can produce a stronger response than the direct interactions in isolation (12).
Although some miRNAs have already been implicated in the modulation of the fibrotic environment (13), our understanding of the extent of miRNA contribution to the control of fibrosis-related processes, especially in the context of HCC, is still very limited. Studies describing roles that miRNAs have in regulation of fibrosis and HCC typically have identified individual miRNA:target interactions. While these studies provide valuable insight into miRNA-directed regulation, such approaches ignore the complexity of miRNA signaling networks.
Furthermore, with the potential of a single miRNA to regulate hundreds of target mRNAs, miRNAs themselves have to be tightly and dynamically regulated. Specifically, mechanisms controlling miRNA expression, stability, and targeting efficiency may be exerted at all levels of miRNA biogenesis, processing, and functional maturation (14). These miRNA regulatory mechanisms are, currently, poorly defined in the context of fibrosis.
The complexity of regulation of miRNAs and miRNA-directed targeting leads to intricate networks of miRNAs and their target and regulatory genes. The nodes of these networks, which can be either miRNAs or mRNAs, are generally connected to many other nodes in these regulatory networks. Hubs, nodes in the network with an atypically high number of connections, are of special importance, as they represent sites of signaling convergence which can explain the network behavior and serve as potential targets for therapy and prediction of clinical outcome (15).
Therefore, in this study, we investigated and described 1) a miRNA:mRNA network that influences fibrotic microenvironment development in HCC with special emphasis on miRNA hubs which regulate a considerable number of genes in the network, 2) the conservation of the network in different fibrosis settings (e.g., across different fibrosis-facilitated carcinomas and different fibrosis models), and 3) the mechanism of regulation of miRNAs in the network.
As a starting point to investigate the role of miRNAs in regulating HCC-associated fibrosis we utilized the SRF- HCC model. Using transcriptome-wide experimental and bioinformatical tools, we identified a miRNA network that regulates different structural, signaling, and remodeling components of the ECM. The identified miRNA:mRNA network was also found to be dysregulated to different degrees in 2 murine fibrosis models, as well as 4 types of human fibrosis-facilitated carcinomas. We show that this complex network is composed of 8 miRNA hubs and 54 target genes which together regulate key components of the fibrotic microenvironment. Taken together, our findings indicate that the let-7 and miR-30 miRNA families, as well as miR-29c, miR-335, and miR-338, are important antifibrotic microRNAs (AF-miRNAs).
Additionally, we observed the dysregulation of the aforementioned miRNAs at the primary (pri-)miRNA level, suggesting transcriptional regulation of pri-miRNA synthesis. Upon investigation of 2 major transcriptional regulatory mechanisms (i.e., transcription factor-mediated regulation and genomic CpG methylation) we identified the transcription factor Ppar as a direct positive regulator of antifibrotic miRNA synthesis. Furthermore, we observed significant hypermethylation of let-7a, miR-335, and miR-338 gene promoters upon primary HSC (pHSC) activation, suggesting epigenetic mechanisms to contribute to the transcriptional control of these miRNAs.
Results
A Subset of miRNAs Targets ECM-Linked and Fibrosis-Associated Genes in mHCC.
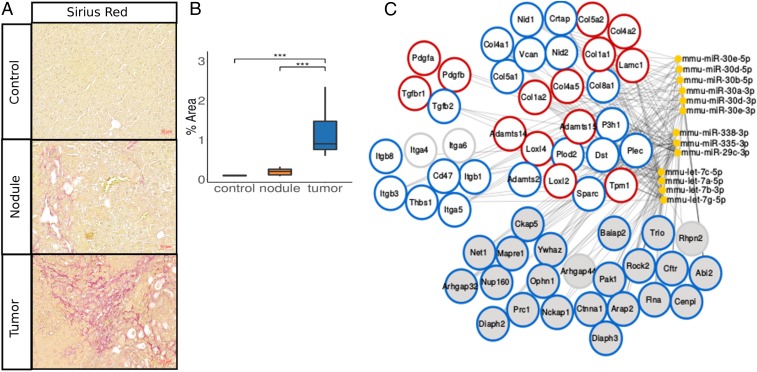
To study the role of miRNAs in the regulation of the fibrotic microenvironment during tumor progression we have used the SRF- mouse model of HCC formation. To evaluate the progression of fibrosis alongside tumor progression we performed Sirius Red and alpha-smooth muscle actin (Acta2) staining of SRF- precancerous nodular and HCC tissue, thereby highlighting fibrotic collagen depositions and HSC activation (16), respectively. This analysis revealed—in correlation with tumor progression—a gradual increase in both area covered by collagen (Fig. 1 A and B) and density of aHSCs (SI Appendix, Fig. S1 A and B).
Fig. 1.
A subset of miRNAs predicted to target ECM-linked and fibrosis-associated genes in mHCC. (A) Sirius Red staining of control, nodular, and tumor liver samples isolated from SRF- mice. (Scale bar, 50 m.) (B) Quantification of Sirius Red signal shown in A. (C) Network of miRNA:mRNA pairs, which encompasses predicted miRNA targeting of genes contributing to the ECM-related pathways highlighted in dark gray in SI Appendix, Fig. S1C. Genes are grouped in structural (upper right), signaling (upper left), and remodeling (middle right) components of the ECM, as well as genes related to integrin signaling (middle left) and Rho signaling (bottom). miRNAs mmu-miR-30e-5p, mmu-miR-30d-5p, mmu-miR-338-3p, mmu-miR-335-3p, mmu-miR-29c-3p, mmu-let-7a-5p, mmu-let-7c-5p, and mmu-let-7g-5p, which are predicted to target all ECM-related proteins of the network, were further experimentally characterized in the remainder of this study. Additionally, we chose to further characterize gene expression and predicted miRNA-mediated targeting of a subset of genes, which represent key structural, remodeling, and signaling components of the ECM. We highlighted these genes using red circles. Rims of gene nodes: red, ECM-related genes characterized further in this study and predicted to be targeted by the here-characterized miRNAs; blue, genes not characterized in this study but predicted to be targeted by the here-characterized miRNAs; and gray, genes not characterized in this study and predicted to be targeted by the here-noncharacterized miRNAs. Data are shown as median, first, and third quartiles (“box”) and 95% confidence interval of median (“whiskers”). ***p value 0.001.
To demonstrate that HSC activation occurs in the vicinity of SRF-VP16–expressing hepatocytes, we performed Egr1 immunostaining. As a target gene of SRF, Egr1 is not expressed in control samples, while its expression is high in SRF-VP16 nodular and tumor samples. Colocalization of Acta2 and Egr1 signals confirms that aHSCs are part of the tumor microenvironment, providing further support to the significance of fibrosis in SRF-VP16–driven tumor development (SI Appendix, Fig. S1A).
To investigate contributions of miRNAs to HCC formation and tumor microenvironment development, we performed small RNA-sequencing analysis (sRNA-seq) of nodular and tumor samples derived from livers of SRF- mice and corresponding controls. As the fibrotic microenvironment is more strongly developed in tumor tissue than in nodular tissue (Fig. 1 and SI Appendix, Fig. S1), in this study we have focused on miRNA changes in tumors.
To identify miRNA candidates with potential contributions to mHCC formation and microenvironment development, we applied 4 filtering criteria: 1) differential expression in tumors versus controls (padj≤ 0.05), 2) sufficient expression level (10 counts across all samples), 3) miRNA conservation (between mice and humans), and 4) similar expression pattern of at least one miRNA family member in hHCCs (10 of cases in The Cancer Genome Atlas’s [TCGA’s] cohort). Criteria 3) and 4) were applied to identify conserved miRNA expressions and functions with the final aim to compare the conclusions gained from our animal model to human patients. Upon applying these criteria, we found that 52 significantly down-regulated and 31 significantly up-regulated mouse miRNAs are dysregulated in at least 10 of human TCGA cases (1.5-fold).
Subsequent to identification of tumor-associated miRNAs, we performed screening of potential miRNA targets using the DIANA microT-CDS (17) and TargetScan (18) databases. To increase the accuracy of down-regulated miRNA target predictions, we performed RNA-sequencing (RNA-seq) analyses on largely overlapping samples as used for sRNA-seq (SI Appendix, Table S1) and correlated target mRNA candidates generated through bioinformatic analysis with the up-regulated genes found by RNA-seq. Similarly, targets of up-regulated miRNAs were identified.
To profile evolutionary conserved miRNA targeting, an analysis comparable to the above was performed on the TCGA datasets of hHCCs. Subsequently, miRNA:target mRNA pairs of both murine and human datasets were overlapped and only conserved pairs were further used in gene set enrichment (GSE) analysis, using KEGG (19) and Reactome (20) pathways. GSE analysis of down-regulated miRNAs showed strong overrepresentation of proteins involved in ECM function, integrin signaling, and Rho GTPase-related pathways (SI Appendix, Fig. S1C), indicating the importance of miRNAs in regulation of the fibrotic microenvironment in HCC development.
Breaking down these pathways into individual genes and their cognate regulatory miRNAs allowed us to identify a miRNA:target mRNA network, composed of a subset of miRNAs down-regulated in tumors. To identify miRNA hubs in the network, all miRNAs which regulate ECM-related genes were filtered for the number of target genes and network coverage. Only miRNAs which target at least 8 ECM-related genes in the network were retained. This miRNA network reveals that the miRNA families miR-30 and let-7, together with miR-335, miR-338, and miR-29c, control a majority of genes related to ECM function as well as integrin and Rho signaling and therefore represent miRNA hubs of the network. For simplicity, we refer to this subset of miRNAs as potential AF-miRNAs.
Collagens represent the most dominant structural proteins of the ECM (21). Most collagen family members in our network are regulated by the let-7 family and miR-29c, with a contribution of miR-335 and miR-338, while laminin gamma 1 (Lamc1) is targeted by miR-29c (Fig. 1C). ECM remodeling components, i.e., A Disintegrin and Metalloproteinases with a ThromboSpondin motif (Adamts) and LOX-like (Loxl) family members, are regulated by the let-7 family, miR-29c, and miR-338 (Fig. 1C). Components of the TGF- pathway (Tgfbr1 and Tgfbr2), the most potent positive regulator of fibrosis, are targeted by miR-335 and the let-7 family, while components of the PDGF pathway (Pdgfa and Pdgfb), responsible for induction of HSC proliferation, are targeted by miR-29c and miR-335. Integrin and Rho-GTPase signaling, which mediate signals from the ECM, are regulated by miR-30 family members. Rho-GTPase signaling is additionally regulated by miR-335 and miR-338 (Fig. 1C). Collectively, these results indicate that the identified AF-miRNAs are negative regulators of structural, remodeling, and signaling components of ECM organization.
AF-miRNAs Are Down-Regulated and Fibrosis-Associated Genes Are Up-Regulated in mHCC.
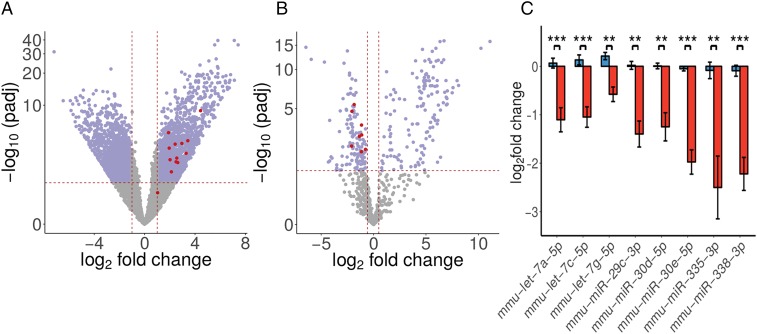
sRNA-seq performed on mouse SRF-VP16–driven HCC samples shows that the AF-miRNAs are significantly down-regulated in mHCC (Fig. 2 B and C).
Fig. 2.
AF-miRNAs are down-regulated and fibrosis-associated genes are up-regulated in murine HCC. (A) Volcano plot of genes identified in RNA-seq. Fibrosis-related genes characterized in this study are shown in red and significantly dysregulated genes (threshold 2-fold) in violet. (B) Volcano plot of miRNAs identified in sRNA-seq. AF-miRNAs characterized in this study are depicted in red (also listed in C), and significantly dysregulated miRNAs (threshold 1.5-fold) are depicted in violet. (C) sRNA-seq–derived, normalized read counts (-transformed) of AF-miRNAs in control (blue bars) and tumor (red bars) samples of SRF- mice. Data are shown as mean and SEM. ** value 0.01, *** value 0.001.
In this study, we focused on a subset of ECM-related genes which cover structural (Col1a1, Col1a2, Col4a2, Col4a5, Col5a2, and Lamc1), signaling (Pdgfa, Pdgfb, and Tgfbr1), and remodeling (Loxl2, Loxl4, Adamts14, Adamts15, and Tpm1) components of the ECM (Fig. 2A).
All aforementioned genes, except Col5a2, show significant up-regulation in the mouse SRF- model (SI Appendix, Fig. S2 A–C). Col5a2 shows clear, but statistically not significant ( value 0.058) up-regulation.
AF-miRNAs Are Down-Regulated and Fibrosis-Associated Genes Are Up-Regulated in the pHSC In Vitro Culture Fibrosis Model and in the In Vivo Murine Fibrosis Model.
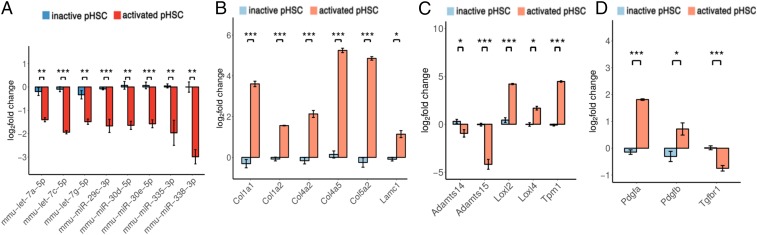
To probe the generality of our miRNA:mRNA network, we examined the expression of AF-miRNAs and fibrosis-associated target genes in 2 fibrosis models, pHSC in vitro culture and the carbon tetrachloride () in vivo mouse model.
pHSC activation, evident by loss of retinoid droplets and increased ECM production, occurs when cells are plated on standard plastic dishes (22). To ensure activation of pHSCs, cells were maintained in culture for 7 d (SI Appendix, Fig. S3). Using qPCR, we compared freshly isolated (inactive) pHSCs with pHSCs activated by prolonged culturing. The in vivo model was generated by prolonged administration of , which leads to hepatic fibrosis development (23).
All AF-miRNAs were found down-regulated in both fibrosis models (Fig. 3A and SI Appendix, Fig. S4A), while all measured structural ECM fibrosis-associated target genes were up-regulated in both models (Fig. 3B and SI Appendix, Fig. S4B). Similarly, all examined remodeling and signaling ECM components were up-regulated, except Tgfbr1 and Adamts, which were up-regulated in the model but down-regulated in the pHSC model (Fig. 3 C and D and SI Appendix, Fig. S4 C and D).
Fig. 3.
AF-miRNAs are down-regulated and fibrosis-associated genes are up-regulated in the pHSC fibrosis model. (A) Relative expression of mature miRNAs in inactive (freshly isolated) and activated (prolonged in vitro culturing) pHSCs. (B–D) Relative expression of fibrosis-associated structural (B), remodeling (C), and signaling (D) genes of the ECM in inactive and activated pHSCs. All samples are normalized to a randomly chosen control sample. Data are shown as mean and SEM. *p value 0.05, **p value 0.01, ***p value 0.001.
AF-miRNAs Target Structural, Signaling, and Remodeling Components of the ECM.
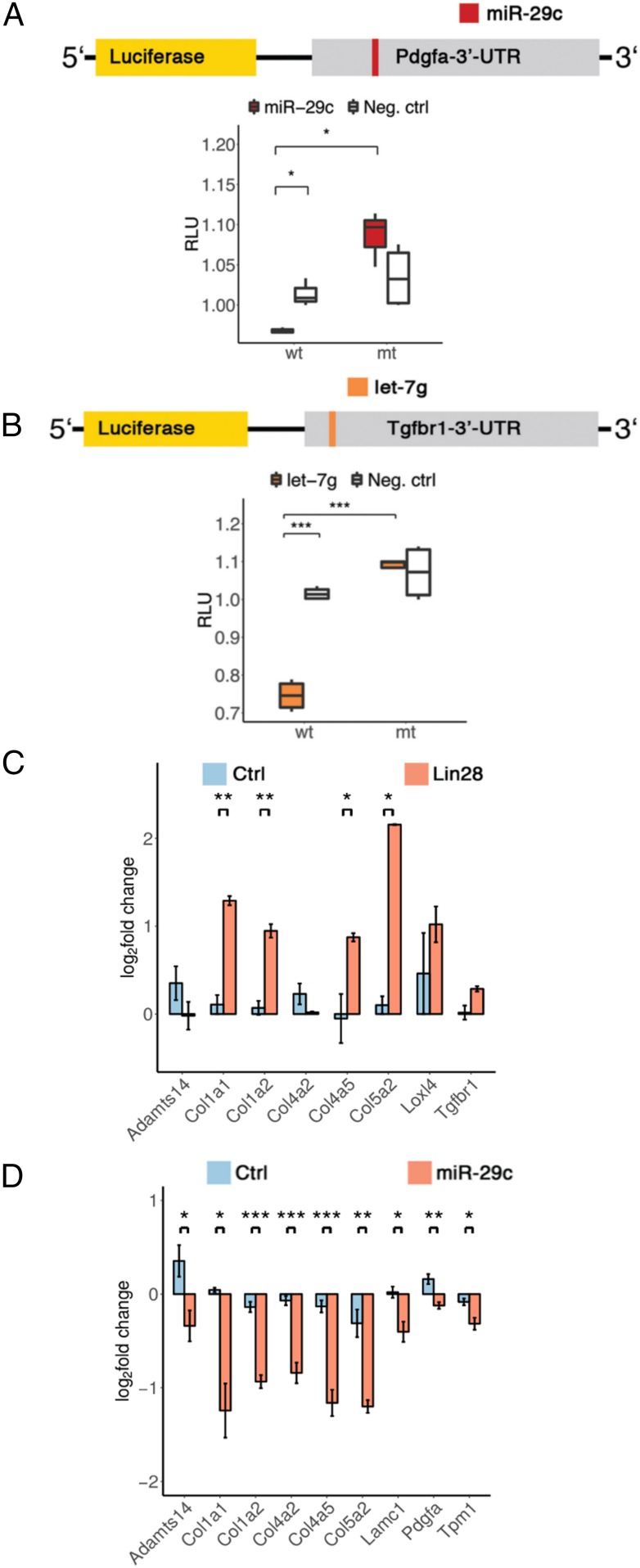
To experimentally validate the predicted AF-miRNA targeting of fibrosis-related genes, we performed luciferase assays and qPCR analyses (SI Appendix, Table S2). To modulate miRNA expression, we employed 3 strategies. First, we used miRNA mimics to overexpress miR-29c, miR-338, let-7a, let-7c, and let-7g. Second, we used miRNA inhibitors to inhibit miR-29c and let-7g. Third, we inhibited let-7a, let-7c, and let-7g expression by overexpressing Lin28a, which down-regulates the entire let-7 family (24).
For luciferase assays, we cloned 3’-untranslated regions (3’-UTRs) of Col1a1, Pdgfa, Tgfbr1, and Adamts15 genes downstream of the luciferase gene and assayed luciferase expression in NIH/3T3 cells upon miRNA mimic or inhibitor transfection. For validation, we mutated the miRNA binding sites and likewise assayed the mutant 3’-UTR constructs upon miRNA mimic or inhibitor transfection.
The luciferase reporter containing wild-type 3’-UTR of Col1a1 showed significant down-regulation upon let-7a, let-7c, let-7g, or miR-29c mimic expression compared to scrambled mimic, while the mutant 3’-UTR construct retained comparable levels of expression upon specific miRNA and scrambled mimic transfection (SI Appendix, Figs. S5A and S6A). In agreement, reverse effects were observed for Col1a1 luciferase assays performed with let-7g and miR-29c inhibitors (SI Appendix, Fig. S5J).
We observed a similar down-regulation of Pdgfa and Tgfbr1 constructs upon miR-29c and let-7a, let-7c, and let-7g mimics overexpression, respectively (Fig. 4 A and B and SI Appendix, Fig. S6C), as well as an up-regulation of the Tgfbr1 construct upon let-7g inhibitor transfection (SI Appendix, Fig. S5K). The Adamts15 luciferase construct showed a significant down-regulation upon miR-338, let-7a, let-7c, let-7g, and miR-29c mimic transfection (SI Appendix, Figs. S5B and S6B).
Fig. 4.
AF-miRNAs target structural, signaling, and remodeling components of the ECM. (A and B) Activities of wild-type and mutant (mutated miRNA site) 3’-UTR–luciferase constructs derived from (A) Pdgfa in NIH/3T3 cells transfected with miR-29c and scrambled miRNA mimic and (B) Tgfbr1 in NIH/3T3 cells transfected with let-7g and scrambled miRNA mimic. Let-7g– and miR-29c–transfected samples are colored in the plots according to the luciferase construct schematic. Samples transfected with scrambled miRNA mimic are shown in white (Neg. ctrl). (C and D) Relative expression of putative let-7 target genes associated with fibrosis in stable Lin28a-overexpressing NIH/3T3 cells (C) and putative miR-29c target genes associated with fibrosis in NIH/3T3 cells transfected with miR-29c mimics (D). (A and B) Data are shown as median, first, and third quartiles (“box”) and 95% confidence interval of median (“whiskers”). (C and D) Data are shown as mean and SEM. *p value 0.05, **p value 0.01, ***p value 0.001.
In qPCR analysis, we assessed the effect of the let-7 family on target gene expression in Lin28a-overexpressing NIH/3T3 cells (Fig. 4C). We validated Lin28a overexpression relative to control and confirmed its inhibitory effect on let-7a, let-7c, and let-7g (SI Appendix, Fig. S5 C and D). Lin28a-mediated inhibition of the let-7 family resulted in significant up-regulation of most collagens. While Tgfbr1 and Loxl4 showed a similar trend of up-regulation, Col4a2 and Adamts14 appeared to be weakly down-regulated (Fig. 4C).
Additionally, we investigated the effect of let-7c, let-7g, and miR-29c up-regulation (SI Appendix, Fig. S5 E–G) on target gene expression (Fig. 4D and SI Appendix, Fig. S5 H and I) upon miRNA mimic or scrambled mimic transfection. Up-regulation of all 3 miRNAs caused specific down-regulation of their target genes. While miR-29c and let-7c mediate significant inhibition of all their target genes with the exception of Col4a2 for let-7c (Fig. 4D and SI Appendix, Fig. S5I), the inhibitory effect of let-7g is more subtle (SI Appendix, Fig. S5H). In agreement with miRNA mimic experiments, inhibition of miR-29c and let-7g by miRNA inhibitor transfection in NIH/3T3 cells caused the up-regulation of most of their target genes (SI Appendix, Fig. S5 L and M).
AF-miRNAs Are Down-Regulated and Fibrosis-Associated Genes Are Up-Regulated in a Subset of Human HCCs.
Expression analysis of the AF-miRNAs in the hHCC dataset showed that the same miRNAs are down-regulated in at least 10 of TCGA cases with the exception of hsa-miR-30d-5p, which is down-regulated in only 3.2 of cases (SI Appendix, Fig. S7A).
The miRNAs hsa-let-7c-5p and hsa-miR-29c-3p show the highest frequency of down-regulation, 71 and 66, respectively. Hsa-miR-335-3p and hsa-miR-338-3p are down-regulated in a smaller number of cases, 30.1 and 10. Although hsa-miR-30d-5p is down-regulated in a very small fraction of tumors, hsa-miR-30e-5p, which shares the same targets as miR-30d, is down-regulated in 38 of cases (SI Appendix, Fig. S7A). In the human TCGA cohort, most of the fibrosis-associated genes show up-regulation (2-fold) in the majority of tumors with the exception of TGFBR1 and ADAMTS15, which show up-regulation in 11.8 and 7.5% of cases, respectively (SI Appendix, Fig. S7B).
A total of 29.7 of patients of the TCGA cohort have 25 (17 pairs) of AF-miRNA:fibrosis-associated gene pairs anticorrelated in the same patient (SI Appendix, Fig. S7 C and D). Across the cohort, 20 AF-miRNA:fibrosis-associated gene pairs are anticorrelated in at least 19.4 of patients, ranging from let-7c:COL4A2 which is anticorrelated in 57.8 of patients to miR-338:TGFBR1 which is anticorrelated in 2.2 (SI Appendix, Fig. S7E and Dataset S5).
The miRNA:mRNA Pairs Show Different Degrees of Association in Human Fibrosis-Facilitated Carcinomas.
To examine the recurrence of expression association of the AF-miRNAs and their target mRNAs in a wider range of human carcinomas, we implemented a multivariate linear regression approach (25).
The multivariate model assesses mRNA expression regulation, taking into consideration miRNA expression, changes in DNA copy number (CNV), and promoter methylation status of the respective gene. CNV and methylation data are used to assess the influence of miRNA-unrelated gene regulation. Using the multivariate model, we aimed to assess whether AF-miRNAs considerably contribute to fibrosis-associated gene regulation in vivo in different fibrosis-facilitated carcinomas.
The model was implemented across HCC, invasive breast carcinoma (BRCA), lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC), i.e., carcinoma types exhibiting fibrotic tumor microenvironments which advance tumor progression (5).
We first examined the recurrence of expression association of the miRNAs and their target mRNAs in individual carcinoma types. In hHCC, most ECM-related genes are consistently targeted by let-7g and miR-29c, while Rho GTPase-related genes are targeted by miR-30e (SI Appendix, Fig. S8B). Similarly, let-7g and miR-29c consistently inhibit the majority of ECM-related genes in BRCA and LUAD samples. However, in these carcinomas, miR-335 and miR-338 additionally contribute to targeting of ECM genes, while Rho GTPase-related genes are primarily modulated by miR-30d and miR-338 (SI Appendix, Figs. S8A and S9A). Furthermore, miR-335 regulates ECM- and integrin-related genes in LUAD and LUSC (SI Appendix, Fig. S9B).
Second, we analyzed the association recurrence across all examined carcinomas, defined by the association recurrence (REC) score. A negative REC score of miRNA:mRNA pairs across fibrosis-facilitated carcinomas indicates that miR-29c, let-7g, let-7a, and miR-335 consistently regulate different collagens (SI Appendix, Fig. S10). Adamts14 is regulated by miR-29c, let7a, and let-7g, while Adamts15 is targeted by let-7g and let-7c. Loxl2 and Loxl4 are modulated by miR-29c and let-7c and let-7a, respectively. Tgfbr1 is primarily regulated by miR-338, let-7a, and let-7c. The linear regression analysis, which assessed the extent of miRNA, DNA methylation, and CNV influence on mRNA expression, showed that identified miRNAs considerably contribute to the expression regulation of the fibrosis-associated genes.
To provide an overview of the characterized miRNA:mRNA interactions we compiled a table, which lists the interactions, the assays in which the interactions were validated, and the REC scores (SI Appendix, Table S3). While some validated miRNA:mRNA interactions have a positive REC score and thus the regression model does not provide support for regulation of the mRNA by the miRNA of these interaction pairs in the examined TCGA cohort, the same mRNAs are consistently regulated by other miRNAs of the network as indicated by a negative REC score.
Together, the AF-miRNAs are evidenced to commonly regulate ECM-related genes in human fibrosis-driven carcinomas, albeit to different extents in different carcinoma types.
Transcription Factor Ppar and DNA Methylation of miRNA-Encoding Gene Promoters Regulate Expression of AF-miRNAs.
To identify the mechanism of AF-miRNA regulation during pHSC activation we examined the expression of precursor molecules of AF-miRNAs. As miRNAs can be regulated at different stages of biogenesis, we quantified pri-miRNAs and precursor (pre-)miRNAs of AF-miRNAs using qPCR in inactive and activated pHSCs, as well as in the in vivo fibrosis mouse model.
We established that down-regulation of AF-miRNAs occurs at the pri-miRNA level in both fibrosis models, suggesting transcriptional regulation of pri-miRNA–encoding genes (Fig. 5B and SI Appendix, Fig. S11C). While we find all pri-miRNAs significantly down-regulated in the pHSC model, pri-miRNA down-regulation in the model is clear, albeit less pronounced. In agreement, we also observed pre-miRNA down-regulation in the pHSC model (SI Appendix, Fig. S11A).
Fig. 5.
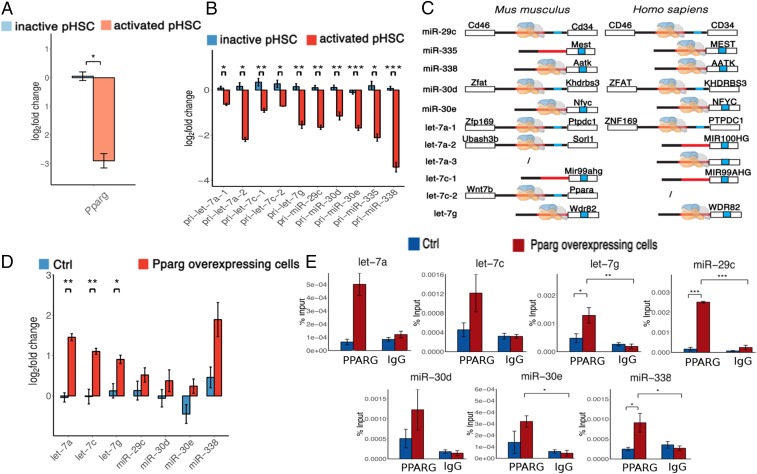
Transcription factor Ppar regulates expression of AF-miRNAs. (A) Relative expression of Pparg in the pHSC in vitro culture model. (B) Relative expression of AF-pri-miRNAs in the pHSC in vitro culture model. (C) Schematic representation of predicted Ppar binding to Mus musculus (Left) and Homo sapiens (Right) miRNA-encoding gene promoters. miRNA-encoding gene promoters are shown in red, whereas mature miRNAs are indicated in blue. miRNAs located in exons or introns of protein-coding genes share the promoter of the respective protein-coding genes. In the cases of miRNAs located in intergenic regions of the genome, the nearest neighboring genes are shown. Ppar is depicted on the individual miRNA-encoding gene promoter if it is predicted to bind to the respective promoter. (D) Relative expression of AF-miRNAs in a stable Ppar-overexpressing GRX hepatic stellate cell line. (E) ChIP analysis of Ppar binding to the promoters of AF-miRNAs in a Ppar-overexpressing GRX (red bars) and control GRX (blue bars) hepatic stellate cell line. (A–E) Data are shown as mean and SEM. *p value 0.05, **p value 0.01, ***p value 0.001.
To study how AF-miRNAs are regulated at the transcriptional level, we examined 2 major transcriptional regulation mechanisms: binding of potential transcription factors to the promoters of relevant miRNA-encoding genes and CpG methylation changes in promoters of miRNA-encoding genes upon HSC activation.
First, to identify miRNA promoters, we utilized publicly available Global Run-on sequencing (GRO-seq) data. As pri-miRNAs are rapidly cleaved to pre-miRNAs, mapping miRNA transcription start sites (TSSs) using conventional TSS mapping approaches is highly challenging (26). GRO-seq is a technique used to quantify nascent transcripts. GRO-seq data show sharp peaks around TSSs in both the sense and antisense directions and continuous signal of lower intensity throughout the entire transcript, allowing one to map TSSs of very transient transcripts (27). Nine mouse and 6 human GRO-seq datasets, which were retrieved from the GEO database, were used in miRNA TSS analysis. The source tissue/cells used to generate the retrieved datasets are primary fibroblasts, embryonic stem cells, embryonic fibroblasts, and liver tissue samples. Analysis of all datasets yielded consensus TSSs of miRNA-encoding genes which are present in the large majority of the datasets.
Locations of the identified miRNA promoters display high conservation in human and mouse genomes (Fig. 5C), indicating conservation of AF-miRNA regulation between mice and humans. To examine whether a majority of AF-miRNAs are regulated by common transcription factors, we used the FIMO tool of the MEME suite (28) to predict transcription factor binding to the mouse and human promoters of AF-miRNA–encoding genes.
To select final candidates of transcription factors, we applied 3 filtering criteria: 1) DNA binding prediction with 10 FDR, 2) binding to the majority of mouse and human miRNA promoters, and 3) differential expression (padj≤ 0.05) in inactive versus activated hepatic and pancreatic stellate cells (GEO datasets).
This approach identified Ppar as a potential transcription regulator of mouse and human let-7a, let-7g, miR-338, miR-29c, miR-30e, and miR-30d genes. Ppar was additionally identified as a potential regulator of mouse let-7c and human miR-335 (Fig. 5C). This prediction further indicates conservation of AF-miRNA regulation in mice and humans.
Next, we examined the expression of Pparg in our 2 fibrosis models using qPCR. We found that transcription factor Pparg is significantly down-regulated upon pHSC activation in both fibrosis models (Fig. 5A and SI Appendix, Fig. S11B).
We sought to confirm experimentally that Ppar binds to the identified miRNA-encoding gene promoters using chromatin immunoprecipitation (ChIP). Due to the high number of cells required to perform the procedure, we were unable to perform ChIP on pHSCs. Instead, as we hypothesize that reduced miRNA expression is a consequence of Pparg down-regulation, we generated a stable Ppar-overexpressing GRX hepatic stellate cell line. We used Ppar-overexpressing and corresponding control cells 1) to quantify mature miRNA expression upon transcription factor up-regulation and 2) to perform ChIP.
All miRNAs predicted to be regulated by Ppar show a definite trend of up-regulation upon Ppar overexpression (Fig. 5D). miR-335 could not be quantified due to low expression in the GRX cell line.
ChIP analysis showed high-affinity binding of Ppar to the promoters of AF-miRNA–encoding genes in cells overexpressing Ppar. Binding of Ppar was drastically lower in control GRX, demonstrating that expression levels of Ppar directly modulate its binding to miRNA-encoding gene promoters (Fig. 5E).
As genome CpG methylation impinges drastically on gene expression, we examined methylation changes in the promoters of miRNA-encoding genes in pHSCs. We observed significant hypermethylation of let-7a, miR-335, and miR-338 gene promoters upon pHSC activation, suggesting epigenetic mechanisms to contribute to transcriptional control of AF-miRNAs. Although not significant, let-7c-1 and miR-30e promoters were also hypermethylated upon pHSC activation (SI Appendix, Fig. S12).
To assess the effects of Ppar-mediated miRNA expression on fibrotic target genes, stable Ppar-overexpressing GRX cells were treated with the Ppar agonist 15-Deoxy-12,14-prostaglandin J2 (PGJ2) and simultaneously transfected with either inhibitors of miR-29c or inhibitors of let-7g. In the presence of scrambled inhibitor, the genes Col1a1, Col1a2, Col5a2, and Adamts14 showed significant down-regulation upon PGJ2 treatment, whereas the effect was less pronounced for Loxl2, Loxl4, and Tgfbr1 (SI Appendix, Fig. S13). Inhibition of miR-29c (SI Appendix, Fig. S13A) and let-7g (SI Appendix, Fig. S13B) in the presence of PGJ2 could partially reverse this effect and significantly increase expression of Col1a1, Col1a2, and Col5a2 as well as Tgfbr1, respectively. These data indicate that multiple fibrosis-associated genes of this network are—at least partially—regulated by Ppar-mediated expression of the AF-miRNAs miR-29c and let-7g.
In conclusion, we show that AF-miRNAs are significantly down-regulated in our mHCC model, while fibrosis-associated genes are up-regulated. Additionally, we show that AF-miRNAs are significantly down-regulated upon activation of pHSCs. treatment of mice, which induces fibrosis, leads to down-regulation of AF-miRNAs. Furthermore, using a linear regression model, we show that AF-miRNAs significantly contribute to the regulation of fibrosis-associated genes in human fibrosis-associated carcinomas, i.e., HCC, BRCA, LUAD, and LUSC. Also, modulation of AF-miRNA expression (let-7a, let-7c, let-7g, and miR-29c) causes anticorrelated expression of fibrosis-associated target genes. In vitro luciferase assays experimentally confirm predicted targeting of Col1a1 by miR-29c, let-7a, let-7c, and let-7g; Pdgfa by miR-29c; Tgfbr1 by let-7a, let-7c, and let-7g; and Adamts15 by miR-29c, miR-338, let-7a, let-7c, and let-7g. Therefore, all results outlined above support the conclusion that AF-miRNAs indeed act as antifibrotic miRNAs.
Based on this conclusion and our findings of AF-miRNA regulation, we propose the following model (Fig. 6): Upon HSC activation, Ppar expression decreases, consequently causing reduced transcriptional synthesis of antifibrotic pri-miRNAs. Reduced miRNA expression releases their inhibitory effect on fibrosis-associated target mRNAs, thereby increasing the abundance of profibrotic proteins.
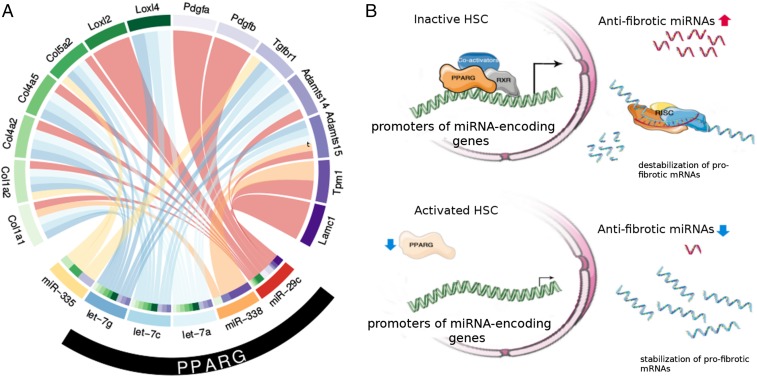
Fig. 6.
Schematic displays summarizing the regulation of AF-miRNAs and their target genes. (A) Circos plot summarizing the regulation of AF-miRNAs and their target genes. PPARG (black bar) is shown to up-regulate the here-validated miRNAs let-7g-5p, let-7c-5p, let-7a-5p, miR-338-3p, and miR-29c-3p. (B) Graphical model summarizing the transcriptional regulation of AF-miRNA–encoding genes by PPARG in HSCs. Reduced expression of PPARG upon activation of HSCs causes down-regulation of AF-miRNA expression. This permits elevated levels of profibrotic mRNAs, leading to the formation of a fibrotic ECM.
Discussion
We identified a network of functionally connected miRNA:mRNA pairs, which regulate cancer-associated fibrosis. This network regulates different structural, signaling, and remodeling ECM components, as well as ECM-linked integrin and Rho-GTPase signaling. We found key miRNA hubs of the network, i.e., miR-29c, miR-335, miR-338, let-7a, let-7c, let-7g, miR-30d, and miR-30e, to be down-regulated in the fibrosis-associated mHCC model, in 2 murine fibrosis models, and in 4 types of human carcinomas.
miR-29c is a known modulator of fibrotic environments in nasopharyngeal carcinoma, shown to reduce mRNA levels of different collagens and LAMC1 (29). We reexamined the role of miR-29c as regulator of fibrosis and experimentally validated Pdgfa, Tpm1, Adatms14, and Adamts15 as additional fibrosis-linked targets of this miRNA.
McDaniel et al. (30) found the let-7/Lin28 axis to be involved in human HSC activation upon alcoholic liver injury. Given that Lin28 is neither expressed in our mHCC (SI Appendix, Table S5, RNA-seq normalized reads) and pHSC culture model nor expressed in the RNA-seq dataset of human HSCs of Zhou et al. (31), we consider Lin28-mediated let-7 regulation in pHSCs and mHCC unlikely. Additionally to Col1a1 (30), we have experimentally validated 4 different collagen family members, Loxl4, and Tgfbr1 as let-7 targets.
To our knowledge, none of the other AF-miRNAs have previously been identified as regulators of fibrosis and none of the AF-miRNAs were previously identified as regulators of fibrotic microenvironments during HCC development.
Our study revealed miRNA-mediated targeting of relevant structural components of the ECM, namely laminin and collagens. Collagens represent the most abundant ECM component and collagen I deposition has been associated with increased incidence of tumor formation and metastasis (21). A study by Ramaswamy et al. (32) found a gene expression signature that distinguishes primary and metastatic adenocarcinomas and predicts the metastatic probability of tumors. A considerable proportion of the gene-expression signature described by Ramaswamy et al. (32) is composed of components of the tumor microenvironment, such as COL1A1 and COL1A2.
We show that, additionally to Col1a1 and Col1a2, AF-miRNAs also regulate other collagen subunits, such as Col5a2. Although collagen V is a minor constituent of the ECM compared to collagen I, collagen V is essential for fibrillogenesis, as its deletion leads to inability of collagen fibril assembly (33).
We also demonstrate that AF-miRNAs target components of the PDGF and TGF- signaling pathways. TGF- is considered the most potent fibrogenic cytokine. TGF- binds to type I receptor (Tgfbr1) and causes phosphorylation of downstream SMADs (34), thus inducing SMAD-mediated increased collagen I and III transcription (35). PDGF is a critical mitogen in the liver, which induces HSC proliferation (36).
Furthermore, we have shown that AF-miRNAs regulate remodeling components of the ECM. Tropomyosin 1 (Tpm1), an actin-binding protein, helps in orienting depositions of collagen and laminin in the ECM (37). Adamts proteases process procollagens and regulate collagen fibril deposition (38). We show that Adamts14 and Adamts15 are up-regulated in SRF--driven tumors and the mouse model, but down-regulated in the pHSC model, possibly indicating the necessity of higher Adamts abundance in tissues, where more extensive ECM remodeling may be required. Loxl proteins are primarily responsible for the regulation of collagen cross-linking. Loxl expression strongly correlates with tumor progression, metastasis, and consequently decreased patient survival (39).
Increased ECM stiffness, a consequence of elevated ECM deposition and remodeling, causes activation of MAPKs and Rho-GTPases via integrin signaling. Although we experimentally focus on miRNA-mediated ECM targeting, it is worth mentioning that the identified AF-miRNAs were also predicted to target integrin and Rho-GTPase signaling. These pathways are strong stimulators of tumor migration, invasion, and proliferation (40). Integrin 1 promotes cell invasion by sensitizing cancer cells to the changes in the ECM (41), while Rho-GTPases are indispensable in the regulation of cell migration and control of multiple aspects of M phase and progression of the cell cycle (40).
Mechanistically, we show that AF-miRNAs are regulated by the transcription factor Ppar. We show that Pparg expression, along with AF-miRNAs, is reduced in vivo and in vitro upon HSC activation and that overexpression of Pparg leads to increased AF-miRNA expression. Furthermore, we demonstrate direct Ppar binding to the AF-miRNA–encoding promoters.
Ppar heterodimerizes with the retinoid acid receptor (Rxr) and, upon DNA binding, recruits RNA polymerase and coactivators with histone acetyl transferase activity, causing remodeling of chromatin and enhancing transcription (42). Ppar:Rxr interacts with the peroxisome-proliferator response element (PPRE) in the promoter of its target genes. PPREs are typically found in various genes involved in lipid metabolism and energy homeostasis (43).
Additional to its role in the regulation of lipid metabolism, Ppar’s involvement in HSC activation has also been described. Ppar sustains HSC quiescence and promotes deactivation of HSCs (44). A study by Marra et al. (45) showed that PPAR inhibits HSC proliferation, migration, and chemokine expression, thus inhibiting fibrogenesis. Furthermore, HSC-specific interference of PPAR signaling aggravates liver damage and fibrosis induced by treatment (46). Studies conducted so far concluded that PPAR exerts its antifibrotic effects primarily by antagonizing TGF- signaling (38). Here, we propose an additional mechanism of Ppar-mediated regulation of fibrosis via stimulation of antifibrotic miRNA expression.
Transcription factors frequently regulate one or more classes of functionally related protein-coding genes. Ppar has been previously identified as a regulator of lipid-metabolism–related genes (43) and of fibrosis. Here, we find Ppar regulates a functionally connected class of miRNAs with antifibrotic properties. However, our discovery of antifibrotic miRNAs targeting key fibrosis-associated genes and Ppar-directed regulation of antifibrotic miRNAs describes an intricate coherent feed-forward loop, whereby miRNAs and transcription factors regulate common targets, thus significantly adding to the understanding of Ppar’s role in fibrosis regulation.
Activation of HSCs and down-regulation of miRNA expression occur in quite a short time (up to 7 d) in our experimental setup. Initial miRNA down-regulation correlates well with down-regulation of the positive regulator of AF-miRNA expression Pparg. However, we also identified hypermethylation of let-7a, let-7c-1, miR-335, miR-338, and miR-30e gene promoters upon pHSC activation. In our window of measurement, the identified methylation changes in all miRNA gene promoters are quite subtle. However, it is possible that initial changes in miRNA expression are mediated through transcription factor regulation, while more long-term changes are established through changes in the methylation of miRNA promoters.
In the last decade, several individual miRNA:target interactions have been characterized in fibrosis and HCC. However, such approaches ignore the complexity of miRNA signaling networks. Therefore, we use transcriptome-wide experimental and bioinformatical tools to identify miRNA hubs that influence HCC and its fibrotic microenvironment. We show a complex network of 8 miRNA hubs that target 54 ECM-related genes together regulate structural, signaling, and remodeling components of the fibrotic microenvironment.
Our findings indicate that the let-7 and miR-30 miRNA families, as well as miR-29c, miR-335, and miR-338, are important antifibrotic microRNAs. We show that these miRNAs are down-regulated in the fibrosis-associated mHCC model, in 2 murine fibrosis models, and in 4 types of human carcinomas.
Using a multivariate model, which assesses whether examined genes are indeed more likely to be targeted by miRNAs or by CpG methylation and CNV, we showed that AF-miRNAs considerably contribute to fibrosis-associated gene regulation in vivo in different human fibrosis-facilitated carcinomas and that changes in fibrosis-associated gene expression are more consistent with changes in miRNA expression than with changes in CNV or methylation.
The fact that a substantial part of the proteins in the described network is regulated through the activity of such a limited number of miRNAs emphasizes the relevance of miRNAs as powerful mediators of complex biological processes, in this case fibrosis.
A noteworthy aspect of our study is the identification of Ppar as a major transcription factor which regulates 7 of 8 miRNA hubs, therefore identifying Ppar as a regulator of a functionally connected class of miRNAs with antifibrotic properties.
Additionally, as all aforementioned miRNAs, with the exception of miR-335, are regulated—to a major extent—by Ppar, this transcription factor represents an attractive stimulatory target for antifibrotic therapy.
Materials and Methods
To profile the whole miRNome, sRNA-seq was performed on liver tumor and nodular tissue of SRF- mice alongside the corresponding controls. The library was generated using TruSeq Small RNA Library Prep Kits v2 (Illumina) according to the manufacturer’s protocol. Subsequent to identification of tumor-associated miRNAs, we performed screening of potential miRNA targets using the DIANA microT-CDS (17) and TargetScan (18) databases. To increase the accuracy of down-regulated miRNA target predictions, we performed RNA-seq analyses on largely overlapping samples as used for sRNA-seq.
To quantify mature miRNA, pre-miRNA, and pri-miRNA expression in RNA samples of fibrosis models, total RNA was reverse transcribed using the miScript II RT Kit (Qiagen) and the miScript SYBR Green PCR Kit (Qiagen) was used to quantify the RNA expressions.
To experimentally validate the functionality of predicted miRNA targeting, a luciferase gene reporter assay was used.
To validate PPAR binding to miRNA promoters, the ChIP protocol based on the procedure described by Daniel et al. (47) was used with some modifications.
Detailed description of all animal models, materials, and methods is contained in SI Appendix.
Data Availability.
sRNA-seq and RNA-seq FASTQ data are deposited in the NCBI Sequence Read Archive (SRA) under accession no. Bioproject: PRJNA522967.
The code for the bioinformatic analysis is available at https://ivanawinkler.github.io/mirna_paper/.
Requests for resources and reagents should be directed to and will be fulfilled by the corresponding author (A.N.).
Supplementary Material
Acknowledgments
We thank Elisa Izaurralde, who sadly passed away, for her continuous support. Brigitte Begher-Tibbe, Marion Bähr, and Heidemarie Riehle provided excellent technical support in immunostainings, methylation studies, and cell culture work, respectively. This work received financial support from the International Max Planck Research School (University Tübingen) (to I.W., C.B., S.W., O.K., and A.N.). Further support to A.N., I.W., and C.B. came from the Deutsche Konsortium für Translationale Krebsforschung (DKTK) Joint Funding Consortium “Noncoding mutations in cancer genomes.” Additional support to A.N. came from the Deutsche Forschungsgemeinschaft (DFG [German Research Foundation] - Project-ID 314905040 - TRR 209) and the German Cancer Aid (Project 109886). Further funding was provided by the Deutsches Bundesministerium für Bildung und Forschung (BMBF; LiSyM Project 031L0045, to J.G.H.). R.W. is supported by the DFG (SFB/TRR 57) and the Interdisciplinary Center for Clinical Research within the Faculty of Medicine at the Rheinisch-Westfälische Technische Hochschule (RWTH) Aachen (Project O3-1).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: sRNA-seq and RNA-seq FASTQ data are deposited in the NCBI Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra, under accession no. Bioproject: PRJNA522967. Processed sRNA-seq and RNA-seq (differential gene and miRNA expression table and normalized reads) are listed in SI Appendix. Output data of the linear regression analysis are also listed in SI Appendix. The code for the bioinformatic analysis has been deposited at https://ivanawinkler.github.io/mirna_paper/.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909145117/-/DCSupplemental.
References
- 1.El-Serag H. B., Hepatocellular carcinoma. N. Engl. J. Med. 365, 1118–1127 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Forner A., Reig M., Bruix J., Hepatocellular carcinoma. Lancet 391, 1301–1314 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Ohrnberger S., et al. , Dysregulated serum response factor triggers formation of hepatocellular carcinoma. Hepatology 61, 979–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olson E. N., Nordheim A., Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez-Gea V., Toffanin S., Friedman S. L., Llovet J. M., Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology 144, 512–527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez-Gea V., Friedman S. L., Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 6, 425–456 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R., Zeisberg M., Fibroblasts in cancer. Nat. Rev. Cancer 6, 392–401 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Heindryckx F., Gerwins P., Targeting the tumor stroma in hepatocellular carcinoma. World J. Hepatol. 7, 165–176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lujambio A., Lowe S. W., The microcosmos of cancer. Nature 482, 347–355 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel D. P., MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha M., Kim V. N., Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Bracken C. P., Scott H. S., Goodall G. J., A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 17, 719–732 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Lee Y. A., Wallace M. C., Friedman S. L., Pathobiology of liver fibrosis: A translational success story. Gut 64, 830–841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter J., Jung S., Keller S., Gregory R. I., Diederichs S., Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Anastasiadou E., Jacob L. S., Slack F. J., Non-coding RNA networks in cancer. Nat. Rev. Cancer 18, 5–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman S. L., Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 88, 125–172 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reczko M., Maragkakis M., Alexiou P., Grosse I., Hatzigeorgiou A. G., Functional microRNA targets in protein coding sequences. Bioinformatics 28, 771–776 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Friedman R. C., Farh K. K., Burge C. B., Bartel D. P., Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabregat A., et al. , Reactome graph database: Efficient access to complex pathway data. PLoS Comput. Biol. 14, e1005968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilkes D. M., Semenza G. L., Wirtz D., Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat. Rev. Cancer 14, 430–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiskirchen S., Tag C. G., Sauer-Lehnen S., Tacke F., Weiskirchen R., Isolation and culture of primary murine hepatic stellate cells. Methods Mol. Biol. 1627, 165–191 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Scholten D., Trebicka J., Liedtke C., Weiskirchen R., The carbon tetrachloride model in mice. Lab. Anim. 49(Suppl. 1), 4–11 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Newman M. A., Thomson J. M., Hammond S. M., Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen A., et al. , Analysis of microRNA-target interactions across diverse cancer types. Nat. Struct. Mol. Biol. 20, 1325–1332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q., et al. , Identification of active miRNA promoters from nuclear run-on RNA sequencing. Nucleic Acids Res. 45, e121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Core L. J., Waterfall J. J., Lis J. T., Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengupta S., et al. , MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 5874–5878 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDaniel K., et al. , The let-7/Lin28 axis regulates activation of hepatic stellate cells in alcoholic liver injury. J. Biol. Chem. 292, 11336–11347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou C., et al. , Long noncoding RNAs expressed in human hepatic stellate cells form networks with extracellular matrix proteins. Genome Med. 8, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramaswamy S., Ross K. N., Lander E. S., Golub T. R., A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Mouw J. K., Ou G., Weaver V. M., Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 15, 771–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derynck R., Zhang Y. E., Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425, 577–584 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Mann D. A., Marra F., Fibrogenic signalling in hepatic stellate cells. J. Hepatol. 52, 949–950 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Pellicoro A., Ramachandran P., Iredale J. P., Fallowfield J. A., Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 14, 181–194 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Hayes A. J., Benjamin M., Ralphs J. R., Role of actin stress fibres in the development of the intervertebral disc: Cytoskeletal control of extracellular matrix assembly. Dev. Dyn. 215, 179–189 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Bonnans C., Chou J., Werb Z., Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barker H. E., Cox T. R., Erler J. T., The rationale for targeting the LOX family in cancer. Nat. Rev. Cancer 12, 540–552 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Jaffe A. B., Hall A., Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Hamidi H., Ivaska J., Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters J. M., Shah Y. M., Gonzalez F. J., The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat. Rev. Cancer 12, 181–195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michalik L., Desvergne B., Wahli W., Peroxisome-proliferator-activated receptors and cancers: Complex stories. Nat. Rev. Cancer 4, 61–70 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Michalik L., Wahli W., Involvement of PPAR nuclear receptors in tissue injury and wound repair. J. Clin. Invest. 116, 598–606 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marra F., et al. , Ligands of peroxisome proliferator-activated receptor gamma modulate profibrogenic and proinflammatory actions in hepatic stellate cells. Gastroenterology 119, 466–478 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Tsuchida T., Friedman S. L., Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 14, 397–411 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Daniel B., Balint B. L., Nagy Z. S., Nagy L., Mapping the genomic binding sites of the activated retinoid X receptor in murine bone marrow-derived macrophages using chromatin immunoprecipitation sequencing. Methods Mol. Biol. 1204, 15–24 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
sRNA-seq and RNA-seq FASTQ data are deposited in the NCBI Sequence Read Archive (SRA) under accession no. Bioproject: PRJNA522967.
The code for the bioinformatic analysis is available at https://ivanawinkler.github.io/mirna_paper/.
Requests for resources and reagents should be directed to and will be fulfilled by the corresponding author (A.N.).