Significance
Abortive viral infections are defined as cells that have been infected with a virus but did not produce any progeny virus as a result of the infection. Recent single-cell studies have shown that abortive infection is observed frequently even during infection of susceptible and permissive cell types. Here, we recovered populations of susceptible and permissive cells that survived infection with herpes simplex virus at high multiplicity of infection. We found that these abortive cells maintain viral genomes in a quiescent state at least for 5 wk. These viral genomes can reactivate to produce new progeny viruses. These findings suggest that abortive HSV-1 infection leads to spontaneous latency-like infection in nonneuronal cells, challenging the current paradigm of herpesviruses latency.
Keywords: herpesviruses, latency, spontaneous reactivation
Abstract
Abortive viral infections are usually studied in populations of susceptible but nonpermissive cells. Single-cell studies of viral infections have demonstrated that even in susceptible and permissive cell populations, abortive infections can be detected in subpopulations of the infected cells. We have previously identified abortive infections in HeLa cells infected with herpes simplex virus 1 (HSV-1) at high multiplicity of infection (MOI). Here, we tested 4 additional human-derived nonneuronal cell lines (cancerous or immortalized) and found significant subpopulations that remain abortive. To characterize these abortive cells, we recovered cell populations that survived infection with HSV-1 at high MOI. The surviving cells retained proliferative potential and the ability to be reinfected. These recovered cell populations maintained the viral genomes in a quiescent state for at least 5 wk postinfection. Our results indicate that these viral genomes are maintained inside the nucleus, bound to cellular histones and occasionally reactivated to produce new progeny viruses. We conclude that abortive HSV-1 infection is a common feature during infection of nonneuronal cells and results in a latency-like state in the infected cells. Our findings question the longstanding paradigm that alphaherpesviruses can establish spontaneous latency only in neuronal cells and emphasize the stochastic nature of lytic versus latency decision of HSV-1 in nonneuronal cells.
One of the hallmarks of herpesviruses is their ability to cause lifelong latent infections. For each herpesvirus, latent infections are limited to specific subtypes of cells. While latency is considered a dormant state of the virus, recent studies identified lytic gene expression during latency (1–4). Latently infected individuals can asymptomatically shed progeny virus (5), questioning the quiescent state during latent infection.
In the case of herpes simplex virus 1 (HSV-1), spontaneous latency is established within sensory or sympathetic peripheral neurons. Even in these neuronal populations, specific subtypes vary in the establishment of latency and in their signals of reactivation (6, 7), suggesting that there is more than one mechanism to establish and maintain latency.
Many experimental models to study HSV latency were established, including in vivo animals, ex vivo animals, in vitro neuronal cells, and in vitro nonneuronal cells [recently reviewed by Suzich et al. (8)]. To enable quiescent HSV-1 infection in nonneuronal cells, the virus must be maintained in a nonreplicative state. To achieve quiescent state, viral infections were carried out either with mutant viral strains (mostly carrying mutations in immediate early genes), in the presence of antiviral drugs, or using special infection conditions (for example, high temperature) (9–14).
In both latent and lytic infections, the viral genomes are found in the nucleus of the infected cell. In an individual cell, only a limited number of viral HSV-1 genomes initiate expression and replication (15–17), suggesting that not all of the viral genomes that enter the nucleus during lytic infection are actively participating in the lytic cycle (18).
Single-cell studies of viral infections have suggested that up to 40% of the infected cells do not produce progeny viruses (19–21). Thus, abortive infection is a common outcome for many viral infections. Abortive infections could also result from infection of nonpermissive cells like HSV-1 in human monocytes (22). Here, we identify and characterize a population of HSV-1 permissive nonneuronal cells that recovered from acute infections.
Results and Discussion
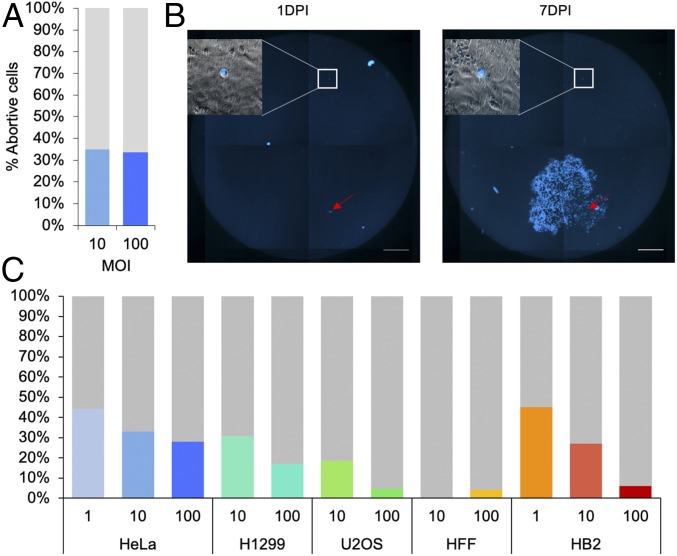
To identify a correlation between the number of HSV-1 genomes that initiate replication within individual cells to viral gene expression, we have previously infected GFP-expressing HeLa cells with a mix of 14 barcoded HSV-1 recombinants that carry the mCherry gene under the cytomegalovirus immediate early (CMV-IE) promoter at high multiplicity of infection (MOI). Three hours postinfection (hpi), individual infected cells were sorted onto a monolayer of uninfected highly permissive Vero cells (16). These experiments identified that high viral gene expression correlates with higher number of incoming viral genomes initiating replication. However, about 33% of infected cells, i.e., mCherry-positive cells, did not produce an infectious center plaque but remained fluorescent and without cytopathic effects for 6 d (Fig. 1A). This phenomenon was independent of the MOI the cells were infected with (either MOI 10 or 100). To ensure that the high percentage of abortive infections is not unique to HeLa cells we infected various cell lines, cancerous cell lines (HeLa, H1299, and U2OS) and noncancerous immortalized cell lines (HFF and HB2). All infections were carried out by HSV-1 OK22 that carries the mTurq2 gene under the CMV-IE promoter. Three hours postinfection, the infected cells were collected and redistributed in limiting concentrations on a monolayer of uninfected Vero cells. The infected cells were monitored by fluorescence (both cellular and viral encoded XFPs; see Methods for details) at 1, 2, and 7 d postinfection (dpi, Fig. 1B). We observed that in most cell lines, abortive HSV-1 infection is a common feature (Fig. 1C). The percentage of abortive infections is variable among the different cell lines independent of their origin (cancerous vs. noncancerous). Among the cancerous cells, U2OS had the lowest percentage of abortive cells, possibly as U2OS have several abnormalities in their antiviral intrinsic immunity factors and are considered to be highly permissive to HSV-1 infections. Interestingly, only very few abortively infected HFF were recovered after 1 week and only in MOI 100.
Fig. 1.
High rate of abortive cells following high MOI infection with HSV-1. (A) HeLa cells were infected with HSV-1 at MOI 10 and 100 as indicated. At 3 hpi, cells were sorted according to their fluorescence level. The cells were placed on monolayers of uninfected Vero cells and incubated for 6 d. At 6 dpi, the cells were scanned with a fluorescence microscope to detect infectious centers originating from the single cells. Out of the single cells studied, the percentage of cells that did not produce infectious centers (blue) compared to the cells that did produce infectious centers (gray) is shown. (B) A representative image of a well containing a monolayer of Vero cells on which infected HeLa cells were placed. The HeLa cells were infected at MOI 10, and 3 hpi were manually diluted and placed on the Vero cells, so that each well had only 1 to 5 infected cells. The cells were scanned in a fluorescence microscope 1 and 7 dpi to monitor the state of the sorted cells. An abortive single HeLa cell is visualized inside the square (magnification of the fluorescence image overlaid with bright field image is shown). An infectious center arising from a productive infected cell is marked with a red arrow. (Scale bar, 0.5 mm.) (C) The percentage of abortive cells detected 7 dpi (colored bars) out of the total infected cells (gray bars) for 5 cell types, HeLa, H1299, U2OS, HFF, and HB2, infected with HSV-1 at MOI 1, 10, or 100 (as indicated). For each cell type, experiments were performed as in B, and an average of at least 2 experiments per each condition is shown.
In most cell lines, MOI 100 resulted in fewer abortive cells detected 7 dpi compared to MOI 10. In HeLa cells the differences between MOI 100 and 10 were the smallest, and in contrast in HB2 cells the differences were the largest. In both cell lines, we also tested MOI 1. At MOI 1, ∼45% of cells were abortively infected in both cell lines (Fig. 1C).
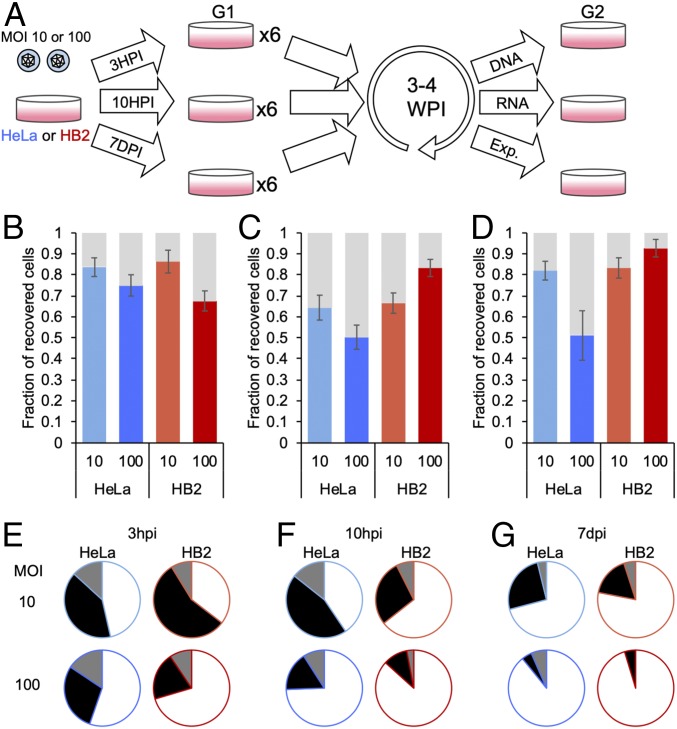
While abortive viral infections are well documented, the fate of abortively infected cells was less studied. To characterize these abortive cells, we attempted to recover cells that survived infection. We chose to continue with 2 cell lines, HeLa and HB2 cells (cancerous and noncancerous, respectively). We infected both cell types, and 3 hpi, we trypsinized the infected cells and diluted them into fresh wells. Three to four weeks later, the wells were split again and were examined by microscopy a few days later (Fig. 2A). In about 80% of wells, growing populations of cells were detected, independent of the cell type or MOI (Fig. 2B). These results indicate that abortive infection is a common outcome of viral infection even at high MOI and that abortively infected cells can be recovered. Further, these cells continue to propagate.
Fig. 2.
Recovered cell populations that survived acute HSV-1 infection. (A) Schematic illustration of the experimental system to recover cell populations after infection. HeLa and HB2 cells were infected at either MOI 10 or 100 with HSV-1. At either 3 hpi, 10 hpi, or 7 dpi, cells were trypsinized, diluted with medium, and spread into 6 new wells (G1). Cells were incubated for 3 to 4 wk, allowing the surviving population to regrow. After the incubation period, cells from each well were further divided into new wells (G2) and were subjected to further experiments. (B–D) HeLa and HB2 cells were infected at either MOI 10 or 100 with HSV-1 as indicated. At either 3 hpi (B), 10 hpi (C), or 7 dpi (D), cells were replated as described in A. Each G2 well was monitored for the presence of cells. The average fraction of wells containing surviving cells over all wells per experiment was calculated. SEMs among different experiments are represented by error bars. (E–G) The surviving cells were analyzed for the activity of the virus in these cells. The percentage of G1 wells arising from (E) 3 hpi, (F) 10 hpi, and (G) 7 dpi wells with all 3 G2 wells with either no active viral replication (white) or active viral replication (black) is presented. A third alternative in which only 1 or 2 of the 3 G2 wells showed active viral replication, and the other wells from the same G1 well had no active replication, was also detected (gray). The averages of more than 10 different-day experiments for each condition were calculated (B–G).
One possible explanation of the high rates of cells recovered is that the trypsin digest of the infected cells at 3 hpi interrupted the viral replication process and increased the rate of abortive infection. To verify that this is not the cause of the observed abortiveness phenomenon, we repeated the experiment with modifications in the experimental conditions. We either trypsinized the infected cells at 10 hpi and only then diluted them to fresh wells, or we only replaced medium at 10, 24, and 48 hpi (to reduce the viral progeny burden arising from productive infection in the plate) and redistributed the cells a week after infection (Fig. 2A). In both late trypsinization conditions at MOI 10, a comparable percentage of wells showed growing cell populations, indicating that abortive infection is not enhanced by the early trypsinization process (Fig. 2 C and D). At MOI 100, HeLa cells at late trypsinization conditions had a lower percentage of wells (∼50%) that contained abortive cell populations; HB2, on the other hand, had a higher percentage of wells containing recovered cell populations, as time of trypsinization was delayed (up to 92% at trypsinization at 7 dpi).
Each G1 well (G1, Fig. 2A) that was maintained for 3 to 4 wk was split into 3 new wells (G2, Fig. 2A), and incubated for a few days before microscopically examining the cells. Three possible outcomes were detected in the positive cell populations: all 3 G2 wells contained actively infected cells, all 3 wells contained cells without any sign of viral infection or a mix of the 2 phenotypes among the 3 wells (Fig. 2 E–G). In most infection conditions (MOI, cell types, and timing of first split), the majority of wells had no sign of infection. Events of 3 wells with mixed phenotypes were only a minority of the events. Both active infection and the mixed phenotypes were less frequent in HB2 cells and as the time of first trypsinization was delayed (Fig. 2 E–G).
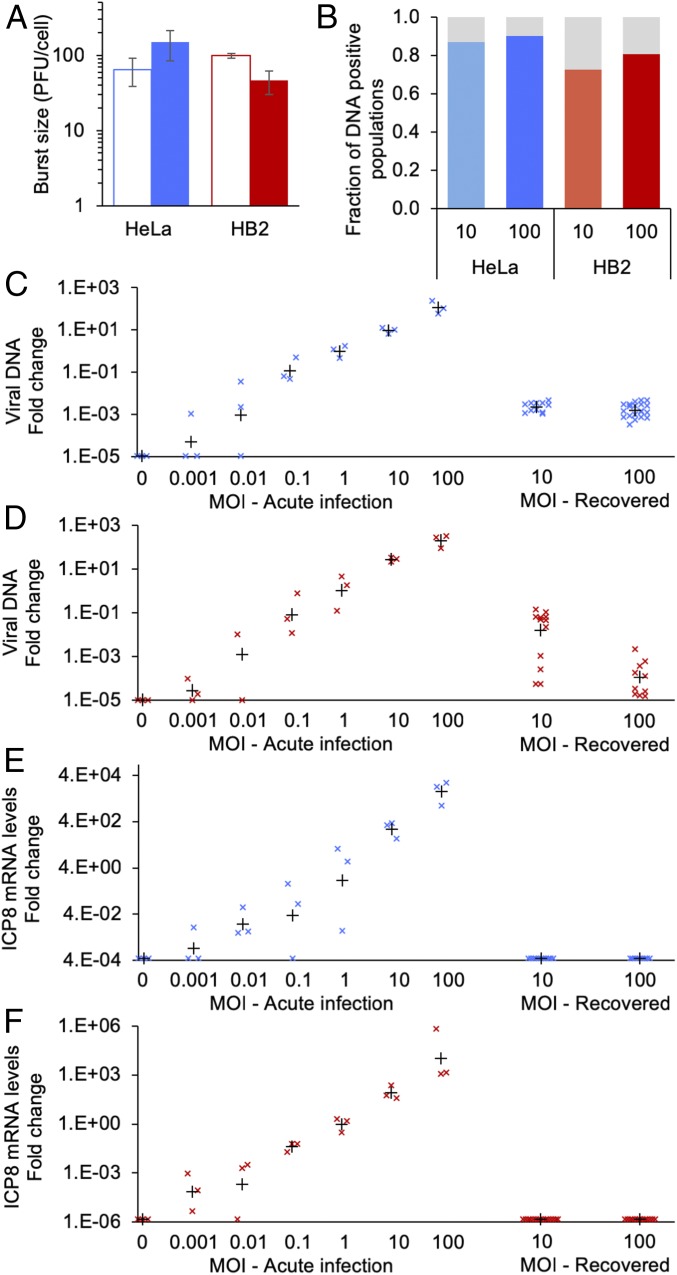
Obtaining populations of the cells that survived the infection allowed us to characterize the fate of the abortive cells. We focused on cells in which no sign of infection was detected in all 3 G2 wells (white color in pie charts, Fig. 2 E–G). To ensure that the recovered cells are not a result of genetically distinct subpopulation that evolved resistance to the virus, we reinfected the recovered population and compared the burst size to infection in naive cells. We found small differences between the naive and recovered cells both in HeLa and in HB2 cells (Fig. 3A). These differences suggest that the resistance to HSV-1 infection does not result from genetic mutations. Reinfection was carried out with HSV-1 that carries the EYFP gene (OK12), while the initial infection was done with a virus that carries the mTurq2 gene (OK22). All progeny viruses obtained from the reinfection expressed yellow fluorescence.
Fig. 3.
Recovered cell populations contain viral genomes. (A) Recovered populations from MOI 100 infection of HeLa or HB2 cells with no active viral replication (full bars) were reinfected with OK12, and viral burst size measured in plaque forming units (PFU) per cell at 24 hpi was compared to infection with OK12 on naive cells (empty bars). Three replicates were titered, and the average and SD are presented. (B) At least 20 samples of HeLa or HB2 recovered populations with no active viral replication were taken from MOI 10 and 100 (as indicated). At 4 to 6 wpi, the samples were lyzed and analyzed with qPCR with 2 viral primers and 1 host primer as control. The fraction of viral positive samples was taken out of the host positive samples. (C and D) Levels of viral DNA were analyzed by qPCR with primers for the UL3 region. Samples from HeLa (C) or HB2 (D) recovered cells are compared to samples of acute infection at different MOIs (as indicated) that were collected at 3 hpi. The levels of viral DNA (marked with “x”) were compared to host DNA and normalized to DNA levels at MOI 1. The mean relative DNA level for each group is marked by a black plus sign. (E and F) Levels of ICP8 mRNA from HeLa (E) or HB2 (F) recovered cells are compared to samples of acute infection at different MOIs (as indicated) that were collected at 3 hpi. The levels of ICP8 mRNA (marked with “x”) were compared to host mRNA and normalized to ICP8 mRNA levels at MOI 1. The mean relative ICP8 mRNA level for each group is marked by a black plus sign.
To test if the recovered cells (with no sign of infection) maintained viral DNA, we collected total DNA from 1 G2 well. In ∼90% of HeLa and ∼75% of HB2 cell populations recovered after 4 to 5 wk postinfection (wpi), viral DNA was detectable by qPCR (Fig. 3B). The levels of viral genomes recovered from HeLa cells were similar between MOI 10 and 100 and can be compared to levels of viral genomes of acute infection at MOI 0.01 (Fig. 3C). In recovered HB2 cells, viral DNA levels varied among the different samples, and in general, higher levels were observed at MOI 10 compared to 100 (Fig. 3D).
We collected total RNA from the parallel G2 well of these cell populations. For each condition (HeLa or HB2, MOI 10 or 100), at least 10 samples were analyzed. Quantitative RT-PCR was used to identify immediate early, early, and late messenger RNAs (mRNAs). No viral mRNAs were detected in all cell populations, for which their corresponding G2 wells were positive for viral DNA, suggesting a quiescent state of the viral genomes. For example, mRNA levels of the early protein ICP8 are not detected in HeLa nor in HB2 cells, compared to mRNA levels obtained from acute infection even at low MOI (Fig. 3 E and F). Similarly, latency-associated transcript (LAT) was also not detected in these cell populations. Recent work suggests that LAT expression in quiescent nonneuronal cells is lower compared to latent neuronal cells (23).
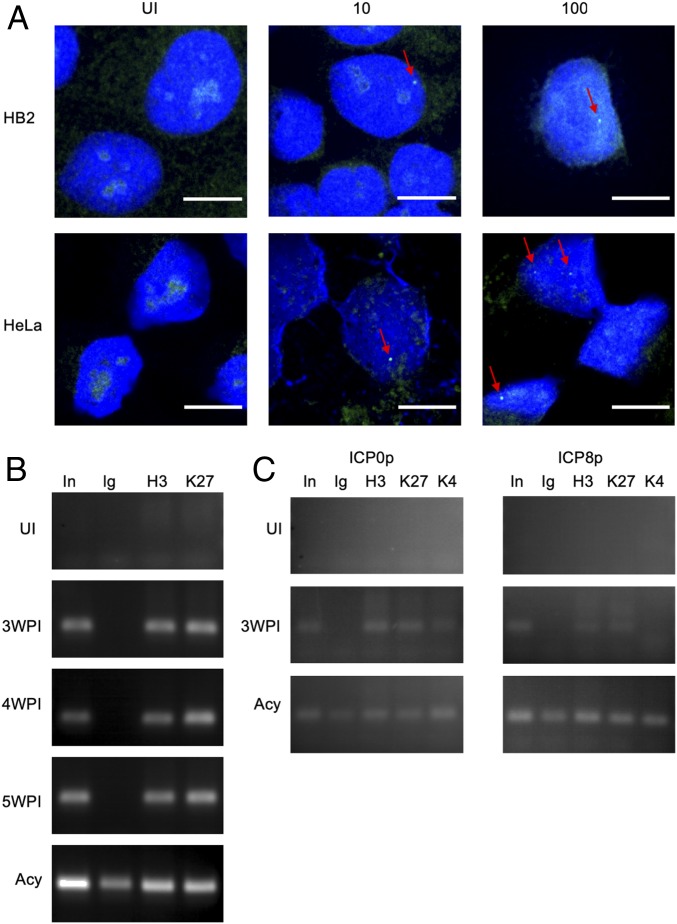
Quiescent HSV-1 genomes are found as episomes inside the host nuclei (24). To demonstrate that the viral DNA is located within the nucleus of the abortively infected cells, we performed a fluorescent in situ hybridization (FISH) assay. HeLa and HB2 cells were fixed at 4 wpi (HeLa) and 3 wpi (HB2) and hybridized with fluorescent probes. We found cells with one or more specific dense fluorescent spot within the nuclei (Fig. 4A). These spots were found only in cells that have been previously exposed to the virus. We note that in most of the recovered cells we have not been able to detect these spots. Similar fluorescent spots were characterized and described previously as viral DNA in latently infected mouse primary trigeminal ganglia sensory neurons (25). We therefore conclude that the observed fluorescent spots are most likely condensed viral genomes.
Fig. 4.
Quiescent viral genomes detected in recovered cell population. (A) FISH images of uninfected (UI) HB2 or HeLa cells and cells infected at MOI 10 or 100 recovered 4 wpi (HeLa) or 3 wpi (HB2). Red arrows indicate green fluorescent foci. Viral DNA is labeled with green probes, and DAPI is presented in blue. (Scale bar, 10 µm.) (B) Chromatin immunoprecipitation (ChIP) for UI cells, recovered HeLa cell populations (originally infected at MOI 100) at different time points postinfection as marked, and cells infected for 48 hpi in the presence of Acyclovir (Acy). PCR results of UL3 gene from Input (In) and pulldown samples with nonspecific IgG (Ig), Histone H3 antibody (H3), and Histone H3 tri methyl K27 (K27) are presented. (C) ChIP for UI cells, recovered HB2 cell populations (originally infected at MOI 100), and cells infected for 48 hpi in the presence of Acy. PCR results for promoter regions of ICP0 and ICP8 genes from In and pulldown samples with Ig, Histone H3, K27, and Histone H3 tri methyl K4 (K4) are presented.
At the quiescent condition, HSV-1 genomes are associated with host histones and are retained in a heterochromatin state (26–29). To test the conditions in which the viral genomes are found within the abortive cell populations, we performed a ChIP assay. We were able to identify that in recovered HeLa, viral genomes are associated with host histones (H3 Ab, Fig. 4B). Moreover, these genomes were associated with histones that were marked with a known silencing marker, histone 3 lysine 27 trimethyl (H3K27me3), for at least 5 wk (Fig. 4B). To further verify that viral genomes are in heterochromatin state, we performed the ChIP assay on HB2 cells at 3 wpi and tested specifically promoter regions of immediate early (ICP0) and early (ICP8) genes. We observed that in these regions the histones were not only marked by the H3K27me3 modification but were also missing the H3K4me3 activation marker (Fig. 4C). These results indicate that following abortive infection, the viral genomes are maintained quiescent within the cell nuclei at a heterochromatin state. The ChIP results corroborate the FISH findings, as both support that the viral genomes are maintained condensed within the nucleus of the recovered cells.
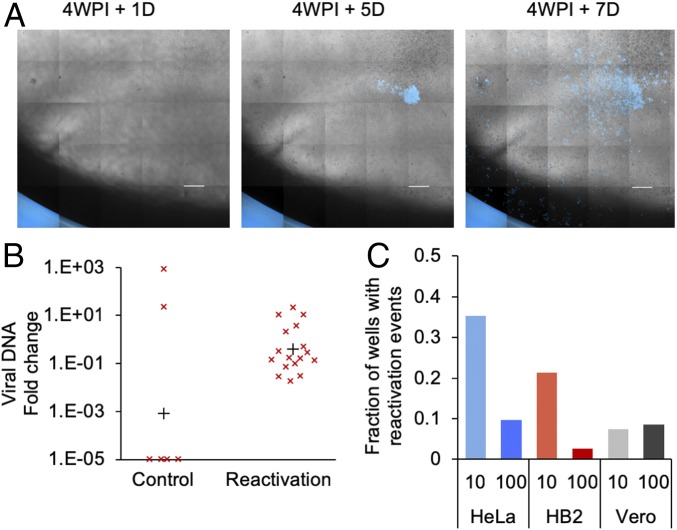
As mentioned above, some of the abortive cell populations that were monitored for 4 wpi showed active replication of the virus. As it is unlikely that cells can replicate during active viral infection or maintain active replication for 4 wk, we speculate that these events might be spontaneous reactivation. Indeed, continuous observation of abortive cell populations that had no sign of active replication for 4 wpi exhibited viral-derived fluorescence in plaques that emerged several days later (Fig. 5A). To verify that these are indeed reactivation events we picked the plaque regions under inverted fluorescent microscope as well as recovered cells that showed no active infection, as control, and tested the samples for progeny viruses and viral DNA. All reactivation samples contained progeny viruses and showed detectable amount of viral DNA, compared to the control (Fig. 5B). Among the abortive HeLa and HB2 cell populations that were observed several times after 4 wpi, we detected more reactivations in MOI 10 compared to 100 (Fig. 5C). Interestingly, in HB2 populations arising from the delayed (1 wk) trypsinization, we were unable to detect reactivation, similar to the very low rate of mixed infection phenotype observed in these cells (Fig. 2G). We speculate that the mixed infection phenotype is also a result of spontaneous reactivation occurring before the first microscopic examination of the cultures.
Fig. 5.
Recovered cells can spontaneously reactivate from quiescent state. (A) Representative images of a well containing a monolayer of recovered HeLa Cells. At 4 wpi + 1 d with no observed viral replication; at 4 wpi + 5 or 7 d a viral plaque (in blue) emerges. (Scale bar, 1 mm.) (B) Plaques observed on monolayer of recovered HB2 cells were picked under fluorescence microscopes, and DNA was collected from these samples (reactivation). As a control, random areas from recovered HB2 monolayers with no signs of reactivation were picked in a similar way. Relative levels of viral DNA from each sample (marked with “x”) were measured by qPCR and analyzed, as in Fig. 3D. The mean relative DNA level for each group is marked by a black plus sign. (C) Four weeks postinfection, HeLa, HB2, or Vero populations with no active viral replication, taken from MOI 10 and 100 (as indicated), were scanned at 2 time points: 12 h after the second split and 3 to 7 d later. The fraction of wells, out of all wells per plate, that showed no active infection at the first scan and an active infection at the second scan was calculated. Data from 2 experiments were accumulated.
To verify that our results are not unique to the specific cell types we tested, we repeated the reactivation experiment on Vero cells. Vero cells are commonly used in HSV-1 research. We obtained recovered population from Vero cells infected at MOI 10 or 100 and observed similar reactivation rates in both cases after 4 wk (Fig. 5C).
Taken together, our findings indicate that in many cell types, a subpopulation of HSV-1–infected cells are unable to complete productive infection (Fig. 1). These abortive cells maintain the HSV-1 genomes in a quiescent state for more than a month and can spontaneously reactivate. These features of the abortive infection resemble latent infection defined as “the persistence of a viral genome within tissue where, at any given time, there is a population of cells that lack detectable infectious virus, viral proteins, or viral lytic transcripts that are dormant but have the capability of being reactivated” (30). We therefore suggest that abortive cells can be used as a model for latent genomes.
In MOI 1, we observed the highest rates of abortive infection (Fig. 1C). We speculate that some of these abortive cells result from infection with defective viral genomes that are unable to complete the infection process. However, high rates of abortive infection were observed at high MOI; in this case, it is implausible that all entering genomes are defective and are unable to complement each other. Therefore, we suggest that abortiveness probably results from the cell condition prior to infection or the initial viral host interactions. As these abortive cells can be reinfected (Fig. 3A), it is unlikely that they result from different genetic backgrounds, but rather from a transient epigenetic state.
Our previously single-cell studies of HSV-1 infections suggest that preexisting cellular diversity contributes to the outcome of infection (16, 31). However, these preexisting conditions can explain only part of the diversity in the outcome. Recent studies on persistent retrovirus (HIV and human T cell leukemia virus [HTLV]) infections have suggested that stochastic autoregulated viral gene expression is an important factor in entering and maintaining latency (32–35). Our results that quiescent infection can occur not only in neuronal cells, but also within nonneuronal cells, suggest that HSV-1 may have a similar stochastic gene expression regulation that results in persistent infection.
The number of latent viral genomes in neuronal cells varies and can reach up to 1,000 genomes per cell (36). In our FISH experiments, we detected between 1 and 20 fluorescent spots per cell. However, in most recovered cells we could not detect any spots. This result together with the expected low expression of LAT RNA (23) may indicate that the LAT RNA levels are below our detection limits in these recovered cell populations. Further, in our DNA and ChIP samples, we did not observe an increase in the amount of viral DNA as time passes. Therefore, we speculate that there is no mechanism for active viral replication and genome maintenance in these abortive cells, but rather a dilution of the initial number of genomes between the replicating cells. In tissue cultures, cell replication is very rapid compared to most cells in vivo; thus, if such abortive cells are maintained in vivo, a longer time for diluting out the quiescent viral genomes is expected.
In most cases, infection of neuronal cells in vitro by HSV-1 leads to acute productive infection; as these neuronal cells are unable to replicate, it will be hard to recover populations of neuronal cells that survived the infection as was done here for nonneuronal cells. In most models of HSV-1 latency in neuronal cells in vitro some spontaneous reactivation can be observed. In our experiments, spontaneous reactivation is much more frequent (Fig. 5). We speculate that this high frequency could result from a leakier state of latency in nonneuronal cells. Alternatively, cell division may increase the likelihood of reactivation.
Our results introduce the possibility that the high frequency of herpes simplex shedding observed in clinical samples (5) may, in part, be due to these abortive infections. If in peripheral tissue some cells remain abortive in situ, they might reactivate more frequently or may contaminate PCR sampling for this tissue. These results may explain why PCR-positive shedding is more common than infectious virus shedding (37). In the future, reexamination of clinical samples is required to corroborate this hypothesis.
Materials and Methods
Complete descriptions of the methods used in this study are found in SI Appendix, Materials and Methods. Sequences of PCR primers and FISH probes are provided in SI Appendix, Table S1. FISH protocol was adapted from ref. 38 and visualized with fluorescent oligonucleotides described previously (39). ChIP protocol was adapted from ref. 40.
Data Availability.
All data are presented in the paper. For more information, please contact the corresponding author.
Supplementary Material
Acknowledgments
We thank Ariel Ionescu and Eran Perlson for valuable support and all O.K. and M.S. laboratory members for their comments. This work was supported by grants from the Israel Science foundation, grant 1387/14 (to O.K.) and grant 1134/16 (to M.S.) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910537117/-/DCSupplemental.
References
- 1.Ma J. Z., Russell T. A., Spelman T., Carbone F. R., Tscharke D. C., Lytic gene expression is frequent in HSV-1 latent infection and correlates with the engagement of a cell-intrinsic transcriptional response. PLoS Pathog. 10, e1004237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markus A., Lebenthal-Loinger I., Yang I. H., Kinchington P. R., Goldstein R. S., An in vitro model of latency and reactivation of varicella zoster virus in human stem cell-derived neurons. PLoS Pathog. 11, e1004885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell T. A., Tscharke D. C., Lytic promoters express protein during herpes simplex virus latency. PLoS Pathog. 12, e1005729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shnayder M., et al. , Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. MBio 9, e00013-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wald A., et al. , Frequent genital herpes simplex virus 2 shedding in immunocompetent women. Effect of acyclovir treatment. J. Clin. Invest. 99, 1092–1097 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrera J. R., Charron A. J., Leib D. A., Neuronal subtype determines herpes simplex virus 1 latency-associated-transcript promoter activity during latency. J. Virol. 92, e00430-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanez A. A., Harrell T., Sriranganathan H. J., Ives A. M., Bertke A. S., Neurotrophic factors NGF, GDNF and NTN selectively modulate HSV1 and HSV2 lytic infection and reactivation in primary adult sensory and autonomic neurons. Pathogens 6, E5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzich J. B., Cliffe A. R., Strength in diversity: Understanding the pathways to herpes simplex virus reactivation. Virology 522, 81–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris R. A., Preston C. M., Establishment of latency in vitro by the herpes simplex virus type 1 mutant in1814. J. Gen. Virol. 72, 907–913 (1991). [DOI] [PubMed] [Google Scholar]
- 10.McMahon R., Walsh D., Efficient quiescent infection of normal human diploid fibroblasts with wild-type herpes simplex virus type 1. J. Virol. 82, 10218–10230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill F. J., Prolongation of herpes simplex virus latency in cultured human cells by temperature elevation. J. Virol. 24, 41–46 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell J., Preston C. M., An in vitro latency system for herpes simplex virus type 2. J. Gen. Virol. 67, 397–403 (1986). [DOI] [PubMed] [Google Scholar]
- 13.Scheck A. C., Wigdahl B., De Clercq E., Rapp F., Prolonged herpes simplex virus latency in vitro after treatment of infected cells with acyclovir and human leukocyte interferon. Antimicrob. Agents Chemother. 29, 589–593 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiraki K., Rapp F., Establishment of herpes simplex virus latency in vitro with cycloheximide. J. Gen. Virol. 67, 2497–2500 (1986). [DOI] [PubMed] [Google Scholar]
- 15.Shapira L., Ralph M., Tomer E., Cohen S., Kobiler O., Histone deacetylase inhibitors reduce the number of herpes simplex virus-1 genomes initiating expression in individual cells. Front. Microbiol. 7, 1970 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen E. M., Kobiler O., Gene expression correlates with the number of herpes viral genomes initiating infection in single cells. PLoS Pathog. 12, e1006082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor M. P., Kobiler O., Enquist L. W., Alphaherpesvirus axon-to-cell spread involves limited virion transmission. Proc. Natl. Acad. Sci. U.S.A. 109, 17046–17051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobiler O., Weitzman M. D., Herpes simplex virus replication compartments: From naked release to recombining together. PLoS Pathog. 15, e1007714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Combe M., Garijo R., Geller R., Cuevas J. M., Sanjuán R., Single-cell analysis of RNA virus infection identifies multiple genetically diverse viral genomes within single infectious units. Cell Host Microbe 18, 424–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heldt F. S., Kupke S. Y., Dorl S., Reichl U., Frensing T., Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat. Commun. 6, 8938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y., Yongky A., Yin J., Growth of an RNA virus in single cells reveals a broad fitness distribution. Virology 385, 39–46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albers I., Kirchner H., Domke-Opitz I., Resistance of human blood monocytes to infection with herpes simplex virus. Virology 169, 466–469 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Harkness J. M., Kader M., DeLuca N. A., Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J. Virol. 88, 6847–6861 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rock D. L., Fraser N. W., Latent herpes simplex virus type 1 DNA contains two copies of the virion DNA joint region. J. Virol. 55, 849–852 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maroui M. A., et al. , Latency entry of herpes simplex virus 1 is determined by the interaction of its genome with the nuclear environment. PLoS Pathog. 12, e1005834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshmane S. L., Fraser N. W., During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63, 943–947 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q. Y., et al. , Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U.S.A. 102, 16055–16059 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cliffe A. R., Garber D. A., Knipe D. M., Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 83, 8182–8190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwiatkowski D. L., Thompson H. W., Bloom D. C., The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83, 8173–8181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloom D. C., Alphaherpesvirus latency: A dynamic state of transcription and reactivation. Adv. Virus Res. 94, 53–80 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Drayman N., et al. , Dynamic proteomics of herpes simplex virus infection. MBio 8, e01612-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen M. M. K., et al. , A post-transcriptional feedback mechanism for noise suppression and fate stabilization. Cell 173, 1609–1621.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahgoub M., et al. , Sporadic on/off switching of HTLV-1 Tax expression is crucial to maintain the whole population of virus-induced leukemic cells. Proc. Natl. Acad. Sci. U.S.A. 115, E1269–E1278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Razooky B. S., Pai A., Aull K., Rouzine I. M., Weinberger L. S., A hardwired HIV latency program. Cell 160, 990–1001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouzine I. M., Weinberger A. D., Weinberger L. S., An evolutionary role for HIV latency in enhancing viral transmission. Cell 160, 1002–1012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawtell N. M., Poon D. K., Tansky C. S., Thompson R. L., The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J. Virol. 72, 5343–5350 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koelle D. M., Wald A., Herpes simplex virus: The importance of asymptomatic shedding. J. Antimicrob Chemother. 45 (suppl. T3), 1–8 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Cremer M., et al. , Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol. Biol. 463, 205–239 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Tomer E., et al. , Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses. FASEB J. 33, 9388–9403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shamay M., Greenway M., Liao G., Ambinder R. F., Hayward S. D., De novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J. Biol. Chem. 285, 36377–36386 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented in the paper. For more information, please contact the corresponding author.