Significance
The erm41 gene is considered the primary mechanism of intrinsic resistance to macrolides in Mycobacterium abscessus. Here we demonstrate that the hflX gene plays a significant and equally important role as erm41. We further describe an unusual mechanism of resistance to macrolide-lincosamide antibiotics mediated by the mycobacterial HflX that likely involves the dissociation of antibiotic-stalled ribosomes. An understanding of the various mechanisms employed by bacteria for resistance to an antibiotic is critical in predicting an effective therapeutic regimen against a pathogenic isolate, and can also inform the development of novel drugs.
Keywords: HflX, macrolides, Mycobacterium abscessus, erm41, ribosome
Abstract
Antibiotic resistance in bacteria is typically conferred by proteins that function as efflux pumps or enzymes that modify either the drug or the antibiotic target. Here we report an unusual mechanism of resistance to macrolide-lincosamide antibiotics mediated by mycobacterial HflX, a conserved ribosome-associated GTPase. We show that deletion of the hflX gene in the pathogenic Mycobacterium abscessus, as well as the nonpathogenic Mycobacterium smegmatis, results in hypersensitivity to the macrolide-lincosamide class of antibiotics. Importantly, the level of resistance provided by Mab_hflX is equivalent to that conferred by erm41, implying that hflX constitutes a significant resistance determinant in M. abscessus. We demonstrate that mycobacterial HflX associates with the 50S ribosomal subunits in vivo and can dissociate purified 70S ribosomes in vitro, independent of GTP hydrolysis. The absence of HflX in a ΔMs_hflX strain also results in a significant accumulation of 70S ribosomes upon erythromycin exposure. Finally, a deletion of either the N-terminal or the C-terminal domain of HflX abrogates ribosome splitting and concomitantly abolishes the ability of mutant proteins to mediate antibiotic tolerance. Together, our results suggest a mechanism of macrolide-lincosamide resistance in which the mycobacterial HflX dissociates antibiotic-stalled ribosomes and rescues the bound mRNA. Given the widespread presence of hflX genes, we anticipate this as a generalized mechanism of macrolide resistance used by several bacteria.
Mycobacterium abscessus has emerged as an important human pathogen during the last 10 y, causing superficial and deep-tissue infections after traumatic injury and/or surgery, as well as bronchopulmonary infections in patients with chronic lung damage, such as prior tuberculosis and cystic fibrosis, resulting in a persistent decline in pulmonary functions or acute respiratory failure (1). The major threat posed by this organism is its extremely low sensitivity to most FDA-approved antibiotics, making its infections incredibly difficult to treat (2, 3). The current treatment regimen against M. abscessus recommends a combination of an oral macrolide in conjunction with amikacin and 1 or more of the injectables (cefoxitin, imipenem, or tigecycline) for a period of several months (2, 4). The majority of these antibiotics target the ribosome, a 2.5-MDa ribonucleoprotein enzyme composed of a 50S and 30S subunit. The binding sites for most ribosome-targeting antibiotics are primarily concentrated at 3 locations within the ribosome: the decoding site on the 30S subunit, the peptidyl transferase center (PTC), and/or the nascent peptide exit tunnel (NPET) on the 50S subunit (5). Macrolide, lincosamide, and streptogramin B antibiotics are structurally distinct but are often considered together (MLSB antibiotics), as they have overlapping binding sites on the 50S subunit around the 23S rRNA nucleotide, A2058 (6). Macrolides are 14- to 16-member macrolactones and bind in the upper portion of the NPET between the PTC and the constriction formed by the proteins L4 and L22 (7, 8). Macrolide binding does not interfere with peptide bond formation per se, but hinders the passage of newly synthesized polypeptides, thereby interrupting translation elongation (9, 10). Lincosamides are smaller molecules that occupy the region between A2058 and the PTC in a way that overlaps with the aminoacyl moiety of the A-site tRNA, thereby preventing peptide bond formation (11).
Intrinsic resistance to macrolides is commonly attributed to 3 primary mechanisms: target modification, active efflux by ABC transporters and the Major Facilitator superfamily, and drug inactivation by esterases, lyases, and phosphorylases (12). Target modification at A2058 of the 23S rRNA by methylases confers cross-resistance to macrolide, lincosamide, and streptogramin B, commonly referred to as the MLSB phenotype, and is the most widespread mechanism of macrolide resistance (12, 13). More recently, the Antibiotic Resistance ATP binding cassette family F (ARE ABC-F) proteins have been shown to confer macrolide resistance by ribosome protection in several Gram-positive bacteria (14, 15). While some macrolide resistance genes are constitutively expressed, the majority are inducible by low doses of antibiotics through transcriptional or translational attenuation (16, 17). In mycobacteria, the most common mechanism of macrolide resistance involves mutations in the macrolide binding site on the 23S rRNA, as well as methylation of these residues by erm-encoded RNA methyltransferases (18, 19). The expression of mycobacterial erm genes is under the control of a transcriptional activator, WhiB7, which is in turn controlled by translational attenuation in the presence of subinhibitory concentrations of structurally unrelated antibiotics (20, 21). Deletion of whiB7 in Mycobacterium smegmatis, Mycobacterium tuberculosis, and M. abscessus results in multidrug sensitivity to MLSB and other ribosome-targeting antibiotics (22, 23).
Previously, we used genomewide transcriptomic profiling by RNAseq and identified ∼80 genes in the WhiB7 regulon of M. abscessus and M. smegmatis, one of which is MAB_3042c, a homolog of the universally conserved Translation Factor (TRAFAC) family of GTPases, HflX (22). Despite its widespread distribution, knockout strains of hflX are viable, and the precise biological function in most organisms is unclear (24, 25). The Escherichia coli HflX is known to be involved in splitting of stalled ribosomes generated during heat shock into free subunits, and the Staphylococcus aureus HflX was shown to disassemble hibernating 100S ribosomes (26, 27). Although binding of macrolides has been shown to interfere with the GTPase activity of E. coli HflX, the E. coli HflX has not been shown to be directly involved in antibiotic resistance (28). Recently, the hflX-r gene from Listeria monocytogenes was shown to confer macrolide resistance (29). We demonstrate here that Mab-HflX (MAB_3042c) and Ms-HflX (MSMEG_2736) are required for macrolide-lincosamide resistance in M. abscessus and M. smegmatis, conferring equivalent resistance as the erm genes, by an erm-independent pathway. We also demonstrate that Ms-HflX is a ribosome splitting factor, and disruption in its ability to dissociate ribosomes results in an inability to mediate antibiotic resistance. Our results suggest that a likely mechanism of mycobacterial HflX-mediated macrolide-lincosamide resistance involves dissociation of ribosomes stalled in the presence of these antibiotics.
Results
Deletion of the HflX Homolog Confers Hypersensitivity to Macrolide-Lincosamide Antibiotics in M. abscessus and M. smegmatis.
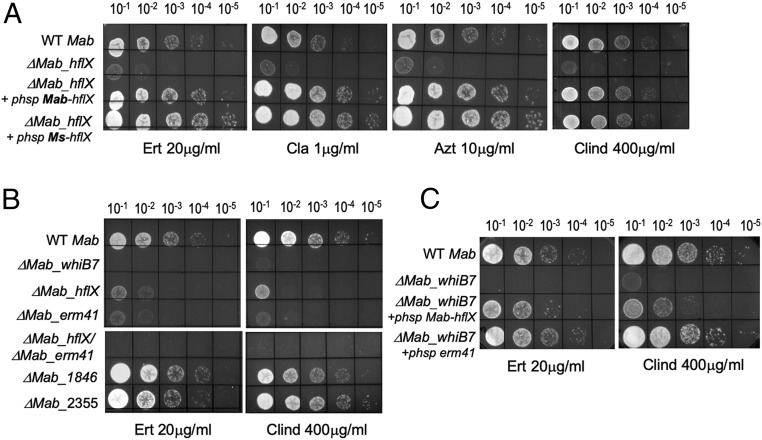
WhiB7, a transcriptional activator, is one of the earliest genes upregulated in response to ribosome-targeting antibiotics, and in turn activates the expression of ∼80 genes that comprise the WhiB7 regulon (22). A few genes in this regulon have known functions such as efflux pumps, the erm41 and eis2 genes; the roles of most genes are, however, unknown. One such gene induced in response to ribosome targeting antibiotics as part of the WhiB7 regulon is MAB_3042c, a homolog of the universally conserved ribosome binding protein, HflX (SI Appendix, Fig. S1). A comparison of the WhiB7 regulon of M. abscessus and M. smegmatis showed that MAB_3042c is also one of the few genes that is shared between the WhiB7 regulons of the 2 species; MSMEG_2736, the ortholog in M. smegmatis, displays ∼80% amino acid sequence identity to MAB_3042c (SI Appendix, Fig. S2) (22). Since HflX is known to bind the ribosome in several bacterial species studied, we explored whether MAB_3042c is involved in resistance to ribosome-targeting antibiotics (26, 30, 31). Isogenic deletions of MAB_3042c (Mab_hflX) in ATCC 19977 and MSMEG_2736 (Ms_hflX) in mc2155 were constructed using phage recombineering (22, 32). The resulting deletion strains ΔMab_hflX and ΔMs_hflX were hypersensitive to the macrolides erythromycin (ERT), clarithromycin (CLA), and azithromycin (AZT) and the lincosamide clindamycin (CLIND) (Fig. 1A, Table 1, and SI Appendix, Fig. S3 and Table S1); their sensitivity to several other ribosome-targeting antibiotics remained unchanged (SI Appendix, Fig. S4). Constitutive expression of hflX driven by Phsp60 from a chromosomally integrated copy in the respective mutant strains restored sensitivity of the mutant to wild-type levels; in fact, the complementing strains displayed increased tolerance to macrolides and clindamycin compared with wild-type bacteria (Fig. 1A and SI Appendix, Fig. S3). Moreover, overexpression of Ms_hflX in ΔMab_hflX also restored its antibiotic tolerance to wild-type levels, thereby suggesting a conserved function of Mab_hflX and Ms_hflX (Fig. 1A).
Fig. 1.
Deletion of M. abscessus hflX confers macrolide-lincosamide sensitivity. (A–C) Ten-fold serial dilutions of M. abscessus ATCC 19977, ΔMab_hflX, ΔMab_whiB7, ΔMab_erm41, ΔMab_hflX/ΔMab_erm41, ΔMab_1846, ΔMab_2355, and the indicated complementing strains were grown to A600 of 0.7 and spotted on Middlebrook 7H10 OADC containing 20 μg/mL erythromycin, 1 μg/mL clarithromycin, 10 μg/mL azithromycin, or 400 μg/mL clindamycin.
Table 1.
Survival of wild-type M. abscessus ATCC19977, ΔMab_HflX, ΔMab_HflX +phsp-HflX, ΔMab_erm41, and ΔMab_HflX/ΔMab_erm41 in a 2-fold dilution series of antibiotics in Middlebrook 7H9/OADC medium
| Antibiotic | Minimum Inhibitory Concentration (μg/mL) | |||||
| WT Mab | ΔMab_hflX | ΔMab_hflX + phsp-hflX | ΔMab_whiB7 | ΔMab_erm41 | ΔMab_hflX/ΔMab_erm41 | |
| Erythromycin | 2 | 0.25 | 2 | 0.625–0.125 | 0.25 | 0.125 |
| Clarithromycin | 1 | 0.125 | 1 | 0.0625 | 0.125 | 0.0625 |
| Azithromycin | 8 | 2 | 16 | 0.5 | 2 | 1.0 |
| Clindamycin | 200 | 50 | 200 | 12.5 | 25 | 25 |
The minimum concentration of antibiotic required to inhibit 99% of growth after 72 h is shown. Minimum inhibitory concentration values are representative of 3 independent assays.
Curiously, the M. abscessus WhiB7 regulon contains additional known and putative effectors of macrolide resistance; specifically, the full-length erm41 gene and homologs of ABC-F proteins, MAB_1846 and MAB_2355, transcription of which are induced on macrolide exposure. To evaluate their relative contribution to macrolide-lincosamide resistance, we constructed isogenic deletions in erm41, MAB_1846, and MAB_2355. Fig. 1B and Table 1 show that Δerm41 and ΔhflX were both equally hypersensitive to erythromycin and clindamycin; a ΔhflX/Δerm41 double-mutant and ΔwhiB7, however, displayed significantly increased sensitivity compared with either single mutant alone. Moreover, overexpression of either hflX or erm41 restored antibiotic sensitivity of the ΔwhiB7 mutant strain, suggesting that Erm41 and HflX act via independent pathways (Fig. 1C). ΔMAB_2355 and ΔMAB_1846 strains displayed mild hypersensitivity to erythromycin and clindamycin, respectively (Fig. 1B).
Mycobacterial HflX Dissociates 70S Ribosomes In Vitro, Independent of GTP Hydrolysis.
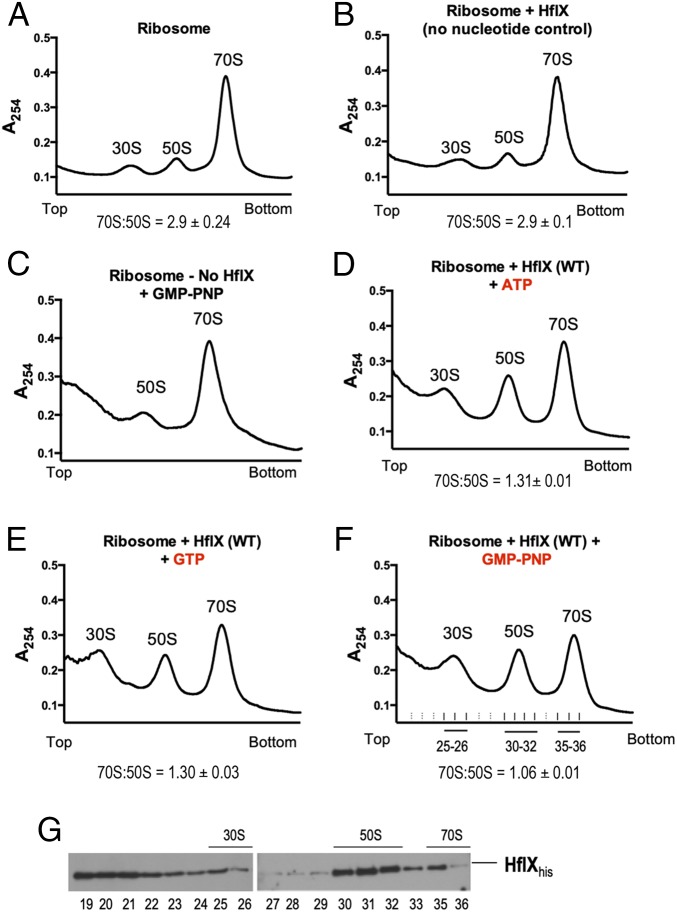
The E. coli hflX has previously been shown to be inducible under heat stress, and an E. coli ΔhflX mutant is highly heat sensitive. However, the viability of ΔMab_hflX and ΔMs_hflX remain unchanged on exposure to elevated temperatures (SI Appendix, Fig. S5 A and B). The E. coli HflX has also been shown to be involved in splitting of stalled ribosomes generated during heat shock into free subunits (26). To determine whether mycobacterial HflX is capable of similarly dissociating 70S ribosomes in vitro, we examined the effect of adding purified Ms-HflX to 70S ribosomes, using sucrose density gradient centrifugation (SDGC). Fig. 2 A–F shows that Ms-HflX indeed promoted dissociation of 70S ribosomes in the presence of either GTP or ATP, consistent with previous studies showing that E. coli HflX can bind to both ATP and GTP (31). No ribosome dissociation was observed in the absence of added nucleotides (Fig. 2B); moreover, maximum splitting efficiency was observed in the presence of the nonhydrolysable GTP analog GMP-PNP (Fig. 2F), which implied that although nucleotide binding is necessary for ribosome splitting, GTP hydrolysis is not. The distribution of HflX in ribosomal fractions was monitored by immunoblotting using anti-his antibodies that recognized his-tagged Ms-HflX. Although HflX could be detected in the top fractions as well as 70S fractions, an enrichment was observed in the 50S subunit fractions (Fig. 2G).
Fig. 2.
Nucleotide-dependent dissociation of 70S ribosomes by Ms-HflX in vitro. (A–F) Dissociation of 70S ribosomes (0.2 μM) was carried out in the presence of 3.0 μM Ms-HflX6his in HMA-7 buffer in the presence of 1 mM GTP, ATP, or GMP-PNP at 37 °C for 45 min and examined using a 5-mL analytical 10% to 40% SDGC. Reactions lacking either nucleotide or HflX were included as controls. Percentage area under the curve was calculated for 70S and 50S peaks, using PeakChart (v. 2.08, Brandel), and expressed as a ratio of 70S:50S. Data represent mean ± SD, n = 3. (G) The samples were collected using the Brandel Teledyne ISCO gradient fractionation system, methanol-chloroform precipitated, followed by immunoblotting with anti-his antibody to determine the presence of Ms-HflX6his in each fraction.
In Vivo Association of HflX with Ribosomes from Antibiotic-Treated Cells.
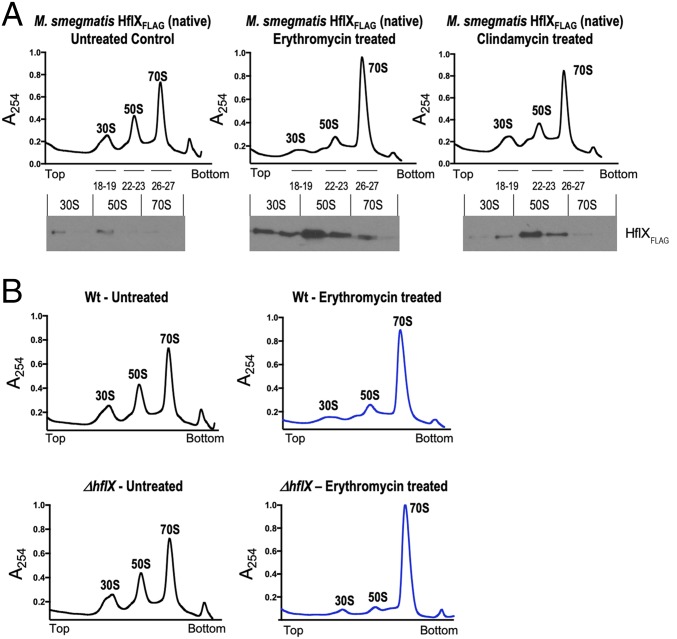
To determine whether HflX associates with ribosomal fractions in vivo, we constructed a strain in which Ms-hflX was C-terminally tagged with the 3X-FLAG epitope at its native chromosomal location. The distribution of endogenous HflX in ribosome fractions obtained from M. smegmatis treated with either ERT or CLIND, as well as an untreated control sample, was examined by SDGC coupled with immunoblotting. Fig. 3A shows that Ms-HflXFLAG was enriched in the 50S ribosomal fractions obtained from antibiotic-treated cells, but almost undetectable in corresponding fractions obtained from bacteria untreated with antibiotics.
Fig. 3.
(A) In vivo association of Ms-HflX with ribosomes. An M. smegmatis strain in which Ms-HflX was C-terminally tagged with the 3X-FLAG epitope at its native chromosomal location was grown to an OD of 0.7 and treated with either 20 μg/mL erythromycin or 16 μg/mL clindamycin for 1 h. Untreated cells were used as a control. A total of 50 pmoles crude ribosomes isolated from each sample were loaded on a 10-mL, 10% to 40% sucrose gradient, followed by ultracentrifugation in an SW41 rotor. Samples were collected from top to bottom on a Brandel fractionation system, and the distribution of endogenous Ms-HflXFLAG in ribosome fractions was analyzed by immunoblotting, using anti-FLAG antibody. (B) Ribosome profile of ΔMs_hflX compared with wild-type bacteria. Wild-type M. smegmatis and ΔMs-HflX strains were grown to an OD of 0.7 and treated with 20 μg/mL erythromycin for 1 h. Untreated cells were used as a control. Crude ribosomes were prepared from each sample, and equal quantities (50 pmoles) were loaded on 10-mL, 10% to 40% sucrose gradients. After ultracentrifugation, the samples were fractionated using the Brandel Teledyne gradient fractionation system, and results were normalized based on area under the curve (AUC). AUC values for WT-Untreated, WT-ERT treated, ΔhflX-untreated, and ΔhflX-ERT treated were 87.42, 89.66, 86.81, and 87.23, respectively. Polysome profile of erythromycin-treated samples are in blue, and untreated samples are shown in black.
We next analyzed the polysome profile of ribosomes isolated from wild-type and ΔMs_hflX strains when exposed to ERT, as well as in the absence of the drug, by SDGC. As seen in Fig. 3C, an increased accumulation of the 70S fraction was observed in the presence of ERT and likely represents ribosomes stalled in the presence of the drug. The quantity of 70S ribosome was, however, significantly (P < 0.05) greater in ERT-exposed ΔMs_hflX cells than that observed in ERT-exposed wild-type bacteria and could reflect a decrease in dissociation of antibiotic-stalled ribosomes in the absence of HflX (Fig. 3B and SI Appendix, Fig. S6).
Ribosome Splitting Function of HflX Correlates with Its Ability to Mediate Antibiotic Resistance.
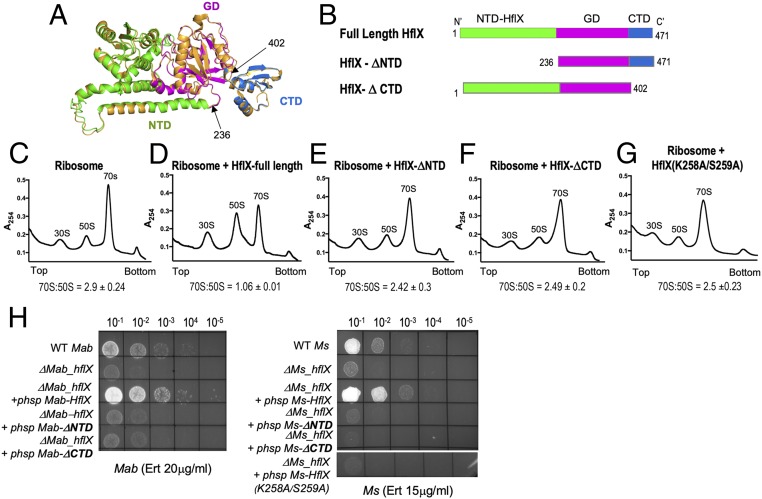
Bacterial and eukaryotic HflX are 3 domain proteins composed of a unique N-terminal HflX domain (NTD), a central GTPase domain (G-domain), and a C-terminal domain (CTD) (26, 33). Interaction with the large ribosomal subunit is a conserved feature of all HflX proteins and is thought to be facilitated by the N-terminal and C-terminal domains (30, 31, 34). Moreover, the ribosome splitting function of E. coli HflX has previously been shown to require both the N- and C-terminal domains (26). To determine whether the ribosome-splitting function of Ms-HflX influences its ability to confer macrolide resistance, we created deletions in the N- and C-terminal domains of Ms-HflX identified based on the structure of Ms-HflX modeled on ribosome-bound E. coli HflX (Fig. 4 A and B). The Ms-HflX mutants containing deletions of either the N-terminal (HflX236–471/ΔNTD) or the C-terminal (HflX1–402/ΔCTD) domains were then assayed for ribosome splitting activity in vitro and antibiotic resistance in vivo. Fig. 4 C–F shows that removal of either the NTD or the CTD of Ms-HflX resulted in a defect in their ability to dissociate 70S ribosomes. Preliminary analyses show that both mutant proteins retain their ability to bind ribosomes (SI Appendix, Fig. S7A). Importantly, both HflX-ΔNTD and HflX-ΔCTD from M. smegmatis and M. abscessus were functionally defective in their ability to complement the antibiotic sensitivity of their respective ΔhflX mutant strains (Fig. 4H, Table 2, and SI Appendix, Fig. S7 B and C). Finally, a point mutant of Ms_HflX (K258A/S259A) defective in ribosome splitting was also found to be defective in complementing the antibiotic sensitivity of ΔMs_hflX (Fig. 4 G and H). Together, these data indicate that the ability of mycobacterial HflX to mediate antibiotic resistance correlates directly with its ribosome splitting ability.
Fig. 4.
Ribosome splitting function of Ms-HflX correlates with its ability to confer antibiotic resistance. (A) A structural model of Ms-HflX guided by the structure of E. coli HflX was obtained using I-TASSER and overlaid on the structure of E. coli HflX (orange) using PyMOL (https://pymol.org/2/). The Ms-HflX model is color coded by domains, as shown on the Right. (B) Location of truncations are indicated. (C–G) Dissociation of 70S ribosomes (0.2 μM) was carried out in the presence of 3.0 μM of either full-length Ms-HflX6his, Ms-HflXΔNTD6his, Ms-HflXΔCTD6his or Ms-HflX(K258A/S259A)6his in HMA-7 buffer containing 1 mM GMP-PNP at 37 °C for 45 min and examined using a 5-mL analytical 10% to 40% SDGC and Brandel gradient fractionation. Percentage AUC was calculated for 70S and 50S peaks, using PeakChart (v. 2.08, Brandel), and expressed as a ratio of 70S:50S. Data represent mean ± SD, n = 3. (H) Wild-type, ΔhflX mutant, and complementing strains containing the respective HflX-ΔNTD, HflX-ΔCTD, and HflX(K258A/S259A) at either the Bxb1 attB site of ΔMs_hflX or the L5 attB site of ΔMab_hflX were assayed for growth on Middlebrook 7H10 containing indicated concentrations of antibiotics. Expression of HflX-ΔNTD and HflX-ΔCTD in the complementing strain was verified using real-time PCR (SI Appendix, Table S2).
Table 2.
Survival of wild-type M. abscessus ATCC19977, ΔMab_HflX, and complemented strains in a 2-fold dilution series of antibiotics in Middlebrook 7H9/OADC medium
| Antibiotic | Minimum Inhibitory Concentration (μg/mL) | |||
| WT Mab | ΔMab_hflX | ΔMab_hflX + phsp-hflX-ΔNTD | ΔMab_hflX + phsp-hflX-ΔCTD | |
| Erythromycin | 2 | 0.25 | 0.5 | 0.25 |
| Clarithromycin | 1 | 0.125 | 0.125 | 0.125 |
| Azithromycin | 8 | 2 | 2 | 2 |
| Clindamycin | 200 | 50 | 50 | 50 |
The minimum concentration of antibiotic required to inhibit 99% of growth after 72 h is shown. Minimum inhibitory concentration values are representative of 3 independent assays.
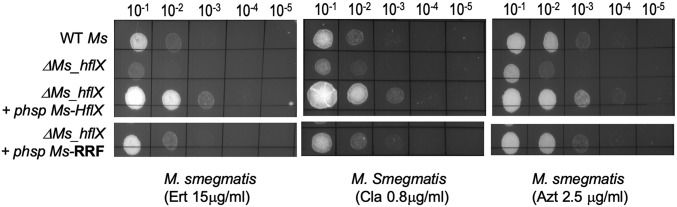
Macrolide Sensitivity of Mycobacterial ΔhflX Mutant Can Be Partially Restored by the Ribosome Recycling Factor.
The ribosome recycling factor (RRF) is required for dissociation of the posttermination complex during a normal translation cycle, thereby making the subunits available for a new round of translation. Although key differences exist between the requirements of HflX and RRF, both proteins are required for recycling ribosomes that are stalled under different physiological conditions (26). We therefore tested the ability of M. smegmatis RRF to complement the antibiotic resistance of ΔMs_hflX. As seen in Fig. 5 and SI Appendix, Table S1, constitutive expression of Ms-RRF partially restored antibiotic sensitivity of the mutant, supporting a role of ribosome splitting as a mechanism of antibiotic resistance. Partial complementation of HflX function by RRF in antibiotic resistance is also consistent with the notion that ribosomal substrates of HflX and RRF are likely to be fundamentally different (26).
Fig. 5.
Overexpression of the ribosome recycling factor (Ms-RRF) partially restores macrolide sensitivity of ΔMs_hflX. Complementing strains were created by integrating either Ms_RRF or Ms_hflX at the Bxb1 attB site of ΔMs_hflX. Expression of Ms-RRF in the complementing strain was verified using real-time PCR (SI Appendix, Table S2). Tenfold serial dilutions of wild-type M. smegmatis, ΔMs_hflX, and the complementing strains were grown to A600 of 0.7 and spotted on Middlebrook 7H10 ADC containing indicated concentrations of macrolides.
Discussion
The erm41 gene is considered to be the primary mechanism of intrinsic macrolide resistance in M. abscessus (18, 35). In the present study, we show that the M. abscessus hflX gene constitutes a significant effector of macrolide-lincosamide resistance and confers equivalent levels of resistance as erm41 via an erm-independent mechanism. We demonstrate that Ms-HflX strongly associates with ribosomal subunits in vivo in bacteria that are exposed to either ERT or CLIND, and an absence of HflX in the ΔMs_hflX deletion strain results in an increased population of 70S ribosomes on erythromycin exposure compared with wild-type bacteria. The mycobacterial HflX is also capable of dissociating 70S ribosomes in vitro independent of GTP hydrolysis, similar to that observed in E. coli HflX (26). These observations led us to hypothesize that mycobacterial HflX is likely involved in dissociation of ribosomes stalled in the presence of antibiotics similar to the recycling of prematurely stalled ribosomes by E. coli HflX during heat shock. Nevertheless, the cryoEM structure of E. coli HflX reveals that the NTD binds to the 50S ribosomal subunit and protrudes into the PTC, making extensive contact with ribosomal RNA, whereas the CTD interacts with the bL12 stalk base and occupies a position distant from the PTC (26). Owing to the conservation of mycobacterial HflX with that of E. coli, an alternate scenario is that the mycobacterial HflX-NTD occupies a similar position within the ribosome and occludes macrolide-lincosamide binding. SI Appendix, Fig. S8, however, reveals that the presence of HflX neither interferes with binding of 3H-ERT to ribosomes nor dissociates 3H-ERT that is already bound to ribosomes. Instead, the evaluation of HflX mutants demonstrating that mutants that are defective in their ability to restore antibiotic tolerance of ΔMs_hflX are also defective in ribosome splitting, as well as the ability of mycobacterial RRF to restore macrolide tolerance of ΔMs_hflX, together strengthen the importance of ribosome splitting in macrolide resistance (Figs. 4 and 5). Finally, the involvement of Ms-HflX-CTD, which is distant from the PTC, in conferring antibiotic tolerance reinforces the conclusion that antibiotic occlusion/ejection is unlikely to be the primary mechanism of HflX-mediated resistance to macrolides/lincosamides.
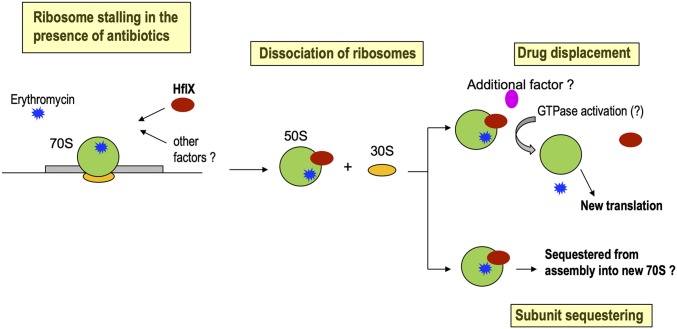
We therefore envisage a scenario in which binding of macrolides/lincosamides to ribosomes in the early stages of elongation results in accumulation of stalled, nonproductive ribosomes and a concomitant inhibition of translation. Association of HflX with antibiotic-stalled ribosomes presumably causes dissociation of the 50S and 30S subunits and rescue of the bound mRNA, which can then be translated by antibiotic-free ribosomes, thereby enabling survival in the presence of the drug. The postdissociated 50S subunit–HflX–antibiotic complex could either be sequestered from further rounds of translation due to the anti-association properties of HflX (26) or could serve as a substrate of accessory proteins that displace bound antibiotic/HflX. These possibilities are illustrated in Fig. 6. Moreover, it is likely that accessory proteins (if any) are also macrolide-lincosamide inducible and are included within the WhiB7 regulon. We speculate that MAB_1846 and MAB_2355, homologs of ABC-F proteins that displace ribosome-bound antibiotics, constitute strong candidates for accessory proteins that mediate recycling of the postdissociated 50S subunit–HflX–antibiotic complex. Although deletions in MAB_1846 or MAB_2355 alone do not significantly influence macrolide-lincosamide resistance, further investigations will be required to determine whether they act in concert with HflX, as well as to determine the identity of additional accessory proteins.
Fig. 6.
Model of mycobacterial HflX-mediated antibiotic resistance.
Our data in Fig. 3 shows preferential association of HflX with ribosomal fractions obtained from ERT/CLIND-treated cells; negligible levels of ribosome-bound HflX were observed in untreated cells. This difference could be attributed to a WhiB7-dependent increase in HflX expression in antibiotic-treated bacteria (SI Appendix, Fig. S9A). However, HflX levels in lysates of ERT-treated bacteria do not show an increase proportionate to the enrichment of HflX observed in ERT-treated ribosomes (∼10-fold; SI Appendix, Fig. S9). We therefore speculate that recruitment of HflX could be facilitated by structural changes within antibiotic-bound ribosomes that are recognized either directly or indirectly by HflX. However, we cannot exclude a basal level of HflX interaction with antibiotic-free ribosomes, which is suggested by the lethal effect of multicopy expression of HflX from a strong promoter (SI Appendix, Fig. S10). Further investigations will be needed to elucidate the characteristics of ribosomes that serve as substrates for HflX binding, as well as the role of accessory proteins (if any) in facilitating HflX binding to ribosome.
Two classes of ribosome-associated proteins have been implicated in antibiotic resistance: the Ribosome Protection Proteins that confer tetracycline resistance and the ARE ABC-F group of proteins that confer resistance to the macrolide-lincosamide-ketolide antibiotics. Both classes of proteins show homology to the translation factors, EF-G and EF-Tu, and actively trigger the release of the antibiotic bound to the ribosome (14, 36–39). Dissociation of ribosomes stalled in the presence of antibiotics and rescue of mRNA has, however, not been described as a mechanism of antibiotic resistance until very recently, when Duval et al. (29) demonstrated that deletion of Listeria monocytogenes hflX-r (lmo0762) confers sensitivity to erythromycin and lincomycin and results in accumulation of 70S ribosomes in the mutant strain on antibiotic exposure. Interestingly, L. monocytogenes encodes 2 HflX paralogs, only 1 of which is involved in antibiotic resistance. Such a duplication event is observed in other firmicutes, as well as proteobacteria; the actinomycetes, however, encode a single copy of hflX. Curiously, a phylogenetic analysis shows that mycobacterial and E. coli hflX are more closely related to lmo1296, the hflX paralog in L. monocytogenes incapable of conferring resistance to macrolides/lincomycin (SI Appendix, Fig. S11) (29). We also note that the E. coli HflX is unable to complement the antibiotic sensitivity of ΔMs_hflX and ΔMab_hflX mutants, suggesting either that E. coli HflX is not involved in macrolide-lincosamide resistance or that it confers antibiotic resistance only in its native host (SI Appendix, Fig. S12). Functional metagenomics from antibiotic-rich environments that identify hflX homologs as putative antibiotic resistance genes belong to both the HflX-r (Emergencia spp) and HflX (Simkania spp.) clades (40, 41). A possible explanation is that while all HflX proteins retain their ability to split 70S ribosomes into their subunits, their duplication allows each paralog to recognize ribosomes that are stalled under different physiological conditions. The ability of HflX to confer antibiotic tolerance may therefore be a generalized and widespread mechanism used by several bacteria, and not restricted only to species that contain the hflX-r paralog.
Materials and Methods
Referenced details of the materials and methods, including plasmids, strains, and oligonucleotides are provided in the SI Appendix. Proteins were purified using Ni-NTA chromatography. Antibiotic sensitivity assays were carried out either using broth dilution method or by spotting a 10-fold dilution series on plates containing the desired antibiotic concentrations. For analysis of polysome profiles on antibiotic exposure, bacteria were exposed to either ERT (20 µg/mL) or CLIND (16 µg/mL). Cells were lysed using the CryoMill (Retsch), and crude ribosomes were prepared as described (42), resuspended in HMA-8 buffer, layered on 10 mL of 10% to 40% sucrose gradients, and centrifuged using a Beckman SW 41 rotor followed by Brandel Teledyne fractionation. Ribosomes were purified from wild-type mc2155, ΔMs_hflX, and mc2155:Ms_hflX-FLAG strains as described (42). For in vitro splitting assays, purified ribosomes (0.2 μM) were incubated with 15-fold molar excess of full-length Ms-HflX, point mutants, or truncated mutants (Ms-HflX ΔNTD and ΔCTD) in a 50-µL total volume in HMA-7 buffer in the presence of 1 mM GTP, ATP, or GMP-PNP. The reactions were incubated at 37 °C for 45 min and layered on 5-mL 10% to 40% sucrose gradients.
Data Availability.
All data have been provided in the main article and SI Appendix.
Supplementary Material
Acknowledgments
We thank The Wadsworth Center’s Applied Genomics Technology Core and the Media Core for preparation of media and buffers. P.G. is supported by an NIH R21 grant (R21-AI146774), Cystic Fibrosis Foundation grant, and the Wadsworth Center.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.T.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906748117/-/DCSupplemental.
References
- 1.Lee M. R., et al. , Mycobacterium abscessus complex infections in humans. Emerg. Infect. Dis. 21, 1638–1646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith D. E., et al. ; ATS Mycobacterial Diseases Subcommittee; American Thoracic Society; Infectious Disease Society of America , An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175, 367–416 (2007). Correction in: Am. J. Respir. Crit. Care Med. 175, 744–745 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Nessar R., Cambau E., Reyrat J. M., Murray A., Gicquel B., Mycobacterium abscessus: A new antibiotic nightmare. J. Antimicrob. Chemother. 67, 810–818 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Floto R. A., et al. , US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis: Executive summary. Thorax 71, 88–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson D. N., The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 44, 393–433 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Poehlsgaard J., Douthwaite S., The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3, 870–881 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Kannan K., Mankin A. S., Macrolide antibiotics in the ribosome exit tunnel: Species-specific binding and action. Ann. N. Y. Acad. Sci. 1241, 33–47 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Ehrenberg M., Tenson T., A new beginning of the end of translation. Nat. Struct. Biol. 9, 85–87 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Schlünzen F., et al. , Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Berisio R., et al. , Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10, 366–370 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Tu D., Blaha G., Moore P. B., Steitz T. A., Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121, 257–270 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Roberts M. C., Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282, 147–159 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Leclercq R., Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34, 482–492 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Sharkey L. K., Edwards T. A., O’Neill A. J., ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio 7, e01975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross J. I., et al. , Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4, 1207–1214 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Min Y. H., Kwon A. R., Yoon E. J., Shim M. J., Choi E. C., Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob. Agents Chemother. 52, 1782–1789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hue K. K., Bechhofer D. H., Regulation of the macrolide-lincosamide-streptogramin B resistance gene ermD. J. Bacteriol. 174, 5860–5868 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nash K. A., Brown-Elliott B. A., Wallace R. J. Jr, A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53, 1367–1376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buriánková K., et al. , Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 48, 143–150 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burian J., Ramón-García S., Howes C. G., Thompson C. J., WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev. Anti Infect. Ther. 10, 1037–1047 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Burian J., Thompson C. J., Regulatory genes coordinating antibiotic-induced changes in promoter activity and early transcriptional termination of the mycobacterial intrinsic resistance gene whiB7. Mol. Microbiol. 107, 402–415 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Hurst-Hess K., Rudra P., Ghosh P., Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob. Agents Chemother. 61, e01347-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris R. P., et al. , Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 12200–12205 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdes S. Y., et al. , Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185, 5673–5684 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta D., Bandyopadhyay K., Datta A. B., Sardesai A. A., Parrack P., Properties of HflX, an enigmatic protein from Escherichia coli. J. Bacteriol. 191, 2307–2314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., et al. , HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat. Struct. Mol. Biol. 22, 906–913 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Basu A., Yap M. N., Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc. Natl. Acad. Sci. U.S.A. 114, E8165–E8173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coatham M. L., Brandon H. E., Fischer J. J., Schümmer T., Wieden H. J., The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome. Nucleic Acids Res. 44, 1952–1961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duval M., et al. , HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 115, 13359–13364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blombach F., et al. , An HflX-type GTPase from Sulfolobus solfataricus binds to the 50S ribosomal subunit in all nucleotide-bound states. J. Bacteriol. 193, 2861–2867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain N., et al. , E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides. Biochem. Biophys. Res. Commun. 379, 201–205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Kessel J. C., Hatfull G. F., Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Jain N., Vithani N., Rafay A., Prakash B., Identification and characterization of a hitherto unknown nucleotide-binding domain and an intricate interdomain regulation in HflX-a ribosome binding GTPase. Nucleic Acids Res. 41, 9557–9569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polkinghorne A., et al. , Chlamydophila pneumoniae HflX belongs to an uncharacterized family of conserved GTPases and associates with the Escherichia coli 50S large ribosomal subunit. Microbiology 154, 3537–3546 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Mougari F., et al. , Standardized interpretation of antibiotic susceptibility testing and resistance genotyping for Mycobacterium abscessus with regard to subspecies and erm41 sequevar. J. Antimicrob. Chemother. 71, 2208–2212 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Connell S. R., Tracz D. M., Nierhaus K. H., Taylor D. E., Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47, 3675–3681 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez-Pescador R., Brown J. T., Roberts M., Urdea M. S., Homology of the TetM with translational elongation factors: Implications for potential modes of tetM-conferred tetracycline resistance. Nucleic Acids Res. 16, 1218 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharkey L. K. R., O’Neill A. J., Antibiotic resistance ABC-F proteins: Bringing target protection into the limelight. ACS Infect. Dis. 4, 239–246 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Boël G., et al. , The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat. Struct. Mol. Biol. 21, 143–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau C. H., van Engelen K., Gordon S., Renaud J., Topp E., Novel antibiotic resistance determinants from agricultural soil exposed to antibiotics widely used in human medicine and animal farming. Appl. Environ. Microbiol. 83, e00989-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González-Plaza J. J., et al. , Functional repertoire of antibiotic resistance genes in antibiotic manufacturing effluents and receiving freshwater sediments. Front. Microbiol. 8, 2675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehta P., Woo P., Venkataraman K., Karzai A. W., Ribosome purification approaches for studying interactions of regulatory proteins and RNAs with the ribosome. Methods Mol. Biol. 905, 273–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data have been provided in the main article and SI Appendix.