Abstract
Aims
To investigate the longitudinal exposure of English primary care patients to pharmacogenomic drugs to inform design of pre‐emptive testing.
Methods
Sixty‐three drugs were identified with dosing guidelines based on variants of 19 pharmacogenes in the Pharmacogenomics Knowledgebase on 01 September 2018. Prescribing of these pharmacogenomic drugs between 1993 and 2017 was summarised for a sample of 648 141 English patients aged 50–99 years on 01 January 2013, registered with Clinical Practice Research Datalink practices during 2011–12. Exposure of patients to pharmacogenomic drugs retrospectively (2, 10, 20 y) and prospectively (5 y) was described.
Results
During 2011–12, 58% of patients were prescribed at least 1 pharmacogenomic drug, increasing to 80% over the previous 20 years. Multiple exposure was common, with 47% patients prescribed ≥2 pharmacogenomic drugs and 7% prescribed ≥5 pharmacogenomic drugs over the next 5 years. The likelihood of exposure to pharmacogenomic drugs increased with age, with 89% patients ≥70 years prescribed at least 1 pharmacogenomic drug over the previous 20 years. Even among those aged 50–59 years, 71% were prescribed at least 1 pharmacogenomic drug over the previous 20 years. The pharmacogenomic drugs prescribed to the most patients were for pain relief, gastroprotection, psychiatric and cardiovascular conditions. Three pharmacogenes (CYP2D6, CYP2C19 and SLCO1B1) accounted for >95% pharmacogenomic drugs prescribed.
Conclusions
In primary care patients, exposure to pharmacogenomic drugs is extremely common, multiplicitous and has commenced by relatively early adulthood. A small number of pharmacogenes account for the majority of drugs prescribed. These findings could inform design of pre‐emptive pharmacogenomic testing for implementation in primary care.
Keywords: clinical pharmacology, general practice, pharmacogenomics
What is already known about this subject
Pharmacogenomic information can improve prescribing, but testing is currently limited in the NHS to specialist indications in small numbers of patients.
Pre‐emptive multigene pharmacogenomic testing could inform multiple prescribing decisions over a patient's lifetime.
The optimal design of pharmacogenomic testing for use in primary care is not known.
What this study adds
Within English primary care, multiple exposure to pharmacogenomic drugs is extremely common with 60% patients being prescribed ≥2 and 18% ≥5 pharmacogenomic drugs over 20 years.
Exposure to pharmacogenomic drugs has commenced by relatively early adulthood and increases with age.
Three pharmacogenes account for >95% pharmacogenomic drugs prescribed.
These findings could inform design of pre‐emptive pharmacogenomic testing for primary care.
1. INTRODUCTION
Pharmacogenomics is the study of how genes affect an individual's response to drugs and pharmacogenomic testing aims to provide information to improve the safety and effectiveness of drug treatment.1 Pharmacogenomic information can be considered actionable if it leads to changes in prescribing decisions, such as use of an alternative drug or dose. Medical centres in the USA now routinely offer genetic testing for common variants in genes associated with severe adverse drug reactions or poor treatment response.1, 2, 3, 4 However, in the UK pharmacogenomic testing is only available in a few specialist areas and has not yet been widely adopted in the National Health Service (NHS). Currently, British guidelines only mandate pretreatment pharmacogenomic testing for abacavir and https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5339 and then only in certain patient subgroups.5, 6, 7
The NHS England policy document Improving Outcomes through Personalised Medicine 8 and the 2016 annual report of the Chief Medical Officer (England)9 both highlight pharmacogenomics as an important opportunity to deliver precision medicine. Implementation of pharmacogenomics is 1 of the 3 main goals of the NHS genomic revolution, along with improving outcomes from cancer and rare diseases. The NHS Genomic Medicine Service is currently determining whether pharmacogenomic testing should be included in the 2019/20 National Genomic Test Directory.10 Pharmacogenomic testing is likely to take the form of a multi‐gene panel, which can offer better value than multiple single tests as costs and processing times fall.
Pharmacogenomic testing can be pre‐emptive before prescription, or reactive in response to treatment failure or an adverse drug reaction. Advantages of pre‐emptive testing include availability of information at the time of prescription, allowing this to be personalised to the patient. The use of a pre‐emptive multigene panel has the potential to provide a lifetime's worth of test results applicable to multiple drugs. In the UK, routine contact with primary care patients, such as NHS vascular risk checks, provide opportunities for pre‐emptive pharmacogenomic testing.11 The longitudinal electronic NHS primary care record, which follows patient journeys over many years, provides an excellent vehicle for aligning pharmacogenomic information with prescribing episodes. Clinical decision support within these electronic health records could be a key tool for the integration of pharmacogenomics into routine patient care.12
The Clinical Practice Research Datalink (CPRD) is a large database of anonymised longitudinal medical records from primary care in the UK.13 The aim of this study was to use CPRD data to investigate the longitudinal exposure of English primary care patients to drugs where choice or dosing was affected by knowledge of pharmacogenomics (pharmacogenomic drugs). Age and frequency of exposure to pharmacogenomic drugs and the most frequent drugs and pharmacogenes involved were determined to inform design of pre‐emptive multigene pharmacogenomic testing in primary care patients.
2. METHODS
2.1. Overview
Current and historical prescribing of pharmacogenomic drugs with dosing guidelines identified in the Pharmacogenomics Knowledgebase (PharmGKB) was summarised for a sample of primary care patients in England aged 50–99 years, actively registered throughout 2011–12 in 215 practices. Exposure of patients to pharmacogenomic drugs both retrospectively (over 2, 10 and 20 years) and prospectively (5 years) was described in subgroups divided by age.
2.2. Approval
This study is based on data from CPRD obtained under licence from the UK Medicines and Healthcare products Regulatory Agency. The data are provided by patients and collected by the NHS as part of their care and support. The interpretation and conclusions contained in this report are those of the authors alone. The protocol number (18_023_RA) was approved by the Independent Scientific Advisory Committee evaluation of joint protocols of research involving CPRD data in August 2018.
2.3. Population
The CPRD is a large, anonymised database of longitudinal medical records from primary care, which has been collecting data from participating UK general practices since 1987. It includes around 7% of the UK population and has been shown to be broadly representative with an age–sex structure similar to the whole population.13 However, since CPRD is based on practices using Vision software, historically this has resulted in a geographical bias within England, with practices concentrated in the South and relatively few in the in the North‐East and East of England.14 For this study, we selected 215 English practices, which had consented to external linkage to other datasets (about 75% of English practices) and were recording data deemed to be of research quality using internal CPRD metrics by 01 January 2012 and continued to collect such data continuously to at least 01 January 2015.13 All patients aged 50–99 years (n = 648 141) who were actively registered with these practices on 01 January 2013 and had been registered with the same practice for at least the previous 2 years were included. This provided the opportunity to analyse prescribing information back to age 30 years for many individuals. For all patients, we extracted all available primary care prescribing data between 1993 and 2017, and classified exposure to pharmacogenomics drugs by searching on drug substance/generic name. Since prescribing data in CPRD are only available when the patient is registered, we could only fully describe exposure for all 684 141 patients during 2011–12. For longer retrospective periods (10, 20 y) we had to restrict to (i) practices which were contributing to CPRD at that time, (ii) patients within these practices that were registered over the whole period. Similarly, our prospective description of prescribing to 2017 is only based on patients who were still active and alive in practices still contributing to CPRD. All prescribing data over the 20‐year retrospective and 5‐year prospective period were extracted. Figure 1 gives an overview of the number of practices and patients with available prescribing periods.
Figure 1.
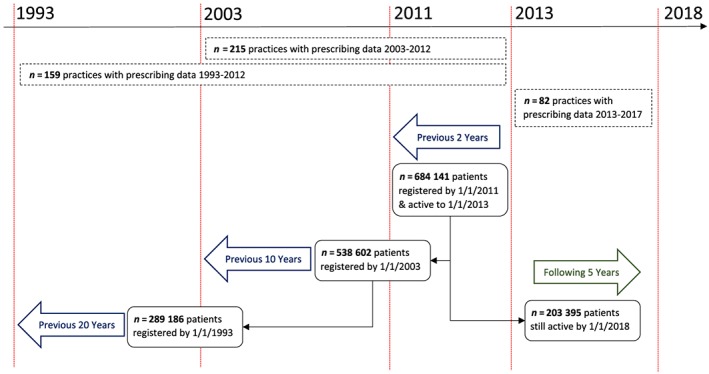
Overview of practices and patients included in available prescribing periods
2.4. Selection of pharmacogenomic drugs
Drugs included in the analysis were those with well‐established pharmacogenomic‐based drug/dosing guidelines in the PharmGKB,15, 16 which provides a repository of genotype‐based dosing recommendations published by the Clinical Pharmacogenetics Implementation Consortium, the Royal Dutch Pharmacogenetics Working Group, the Canadian Pharmacogenomics Network for Drug Safety and/or other professional societies. Sixty‐three drugs were identified as having dosing guidelines based on variants of 19 genes as of 01 September 2018 (Table 1).
Table 1.
Drugs with pharmacogenomic‐based dosing guidelines from Pharmacogenomics Knowledgebase listed by associated gene
| Gene | Drug with pharmacogenomic‐based dosing guideline |
|---|---|
| CTFR | ivacaftora |
| CYP2C19 | amitriptylinea, citaloprama , b, clomipraminea, clopidogrela , b, doxepina, escitaloprama , b, esomeprazoleb, imipraminea , b, lansoprazoleb, omeprazoleb, pantoprazoleb, sertralinea , b, trimipraminea, voriconazolea , b |
| CYP2C9 | acenocoumarolb, gliclazideb, glimepirideb, phenprocoumonb, phenytoina , b, warfarina , c |
| CYP2D6 | amitriptylinea , b, aripiprazoleb, atomoxetineb, clomipraminea , b, codeinea , b , c, desipraminea, doxepina , b, flecanideb, fluvoxaminea, haloperidolb, imipraminea , b, metoprololb, mirtazapineb, nortriptylinea , b, ondansterona, oxycodoneb, paroxetinea , b, propafenonea, risperidoneb, tamoxifena , b , c, tramadolb, trimipraminea, tropisetrona, venlafaxineb, zuclopenthixolb |
| CYP3A5 | tacrolimusa , b |
| CYP4F2 | warfarina |
| DPYD | capecitabinea , b, fluorouracila , b, tegafurb |
| F5 | Hormonal contraceptivesb |
| G6PD | rasburicasea |
| HLA‐A | carbamazepinea , b |
| HLA‐B | abacavira , b, allopurinola , d, carbamazepinea , b, oxcarbazepinea, phenytoina, ribavirinb |
| IFNL3 | Peginterferon alfa‐2aa, peginterferon alfa‐2ba, ribavirina |
| RARG | daunorubicinc, doxorubicinc |
| SLCO1B1 | simvastatina |
| SLC28A3 | daunorubicinc, doxorubicinc |
| TPMT | azathioprinea , b, cisplatinc, mercaptopurinea , b, tioguaninea , b |
| UGT1A1 | atazanavira, irinotecana , d |
| UGT1A6 | daunorubicinc, doxorubicinc |
| VKORC1 | acenocoumarolb, phenprocoumonb, warfarina , c |
Clinical Pharmacogenetics Implementation Consortium guideline,
Royal Dutch Pharmacogenetics Working Group guideline,
Canadian Pharmacogenomics Network for Drug Safety guideline,
other guideline
CTFR = cystic fibrosis transmembrane conductance regulator; CYP2C19 = cytochrome P450 2C19; CYP2C9 = cytochrome P450 2C9; CYP2D6 = cytochrome P450 2D6; CYP3A5 = cytochrome P450 3A5; CYP4F2 = cytochrome P450 4F2; DPYD = dihydropyrimidine dehydrogenase; F5 = coagulation factor V; G6PD = glucose‐6‐phosphate dehydrogenase; HLA‐A = human leucocyte antigen‐A; HLA‐B = human leucocyte antigen‐B; IFNL3 = interferon lambda 3; RARG = retinoic acid receptor γ; SLCO1B1 = solute carrier organic anion transporter family member 1B1; SLC28A3 = solute carrier family 28 member 3; TPMT = thiopurine S‐methyltransferase; UGT1A1 = UDP‐glucuronosyltransferase family 1 member A1; UGT1A6 = UDP‐glucuronosyltransferase family 1 member A6; VKORC1 = vitamin K epoxide reductase complex subunit 1 (Key: gene = protein)
The following drugs with pharmacogenomic‐based dosing guidelines are not licensed for prescription in the UK: phenprocoumon, tropisetron and desipramine.
2.5. Analysis
Exposure of patients to pharmacogenomic drugs over retrospective (over 2, 10 and 20 y) and prospective (5 y) available prescribing periods was determined and compared in subgroups divided by age on 01 January 2013. Use of a pharmacogenomic drug within an available prescribing period was defined as any prescription for a pharmacogenomic drug within that period, even where only a single prescription was recorded for a patient. Unless otherwise stated, any prescription for a pharmacogenomic drug within an available prescribing period was considered exposure, irrespective of whether that drug had already been prescribed to the patient prior to that time period. The pharmacogenomic drugs prescribed to the most patients and the genes most commonly implicated were identified. As exposure to pharmacogenomic drugs was not normally distributed, data were described using median (interquartile range).
2.6. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY.25
3. RESULTS
Among the 648 141 patients who were registered throughout 2011–12, complete prescribing data were further available retrospectively over 10 years (2003–12) for 538 602 patients (79%) and 20 years (1993–2012) for 289 186 patients (45%). Prescribing data were available prospectively for 203 395 patients over 5 years (2013–17; Figure 1).
3.1. Prevalence of exposure to pharmacogenomic drugs
Table 2 shows the exposure of patients to pharmacogenomic drugs over available prescribing periods. Even over a 2‐year period, 58% of patients were exposed to at least 1 pharmacogenomic drug (median per patient = 1, interquartile range = 0–2), and this rose to 80% among 289 185 patients with available data over the preceding 20 years (median = 2, interquartile range = 1–4). Over a 5‐year prospective period, 47% of patients were prescribed at least 2 pharmacogenomic drugs and 7% were prescribed at least 5 pharmacogenomic drugs. When all prescriptions were counted during the study, the proportion that were for pharmacogenomic drugs rose from 16% from 1993–2012 to 20% during 2013–2017. Figure S1 shows the distribution of the number of pharmacogenomic drugs in the different prescribing periods.
Table 2.
Exposure of patients to pharmacogenomic (PGx) drugs over available prescribing periods
| Available prescribing periods | Before 1 January 2013 | After 1 January 2013 | ||
|---|---|---|---|---|
| 2 years (2011–2012) | 10 years (2003–2012) | 20 years (1993–2012) | 5 years (2013–2017) | |
| Total practices with complete recording throughout period | 215 | 215 | 159 | 82 |
| Total patients registered throughout period | 684 141 | 538 602 | 289 186 | 203 395 |
| ‐ with ≥1 prescription for any drug, n (%) | 596 767 (87%) | 517 843 (96%) | 283 948 (98%) | 190 035 (93%) |
| ‐ with ≥1 prescription for any PGx drug, n (%) | 399 311 (58%) | 400 422 (74%) | 231 113 (80%) | 143 947 (71%) |
| PGx drugs | ||||
| ‐ mean per patient | 1.2 | 2.1 | 2.5 | 1.7 |
| ‐ median (interquartile range) | 1 (0–2) | 2 (0–3) | 2 (1–4) | 1 (0–3) |
| Number of patients with multiple PGx drugs | ||||
| ‐ with ≥2 PGx drugs, n (%) | 222 796 (33%) | 284 238 (53%) | 172 818 (60%) | 95 454 (47%) |
| ‐ with ≥5 PGx drugs, n (%) | 17 879 (3%) | 67 465 (13%) | 50 769 (18%) | 14 448 (7%) |
3.2. Prevalence of exposure to pharmacogenomic drugs by age
Figure 2a shows the percentage of patients prescribed pharmacogenomic drugs over available periods divided by age group. In patients aged 50–59 in 2013, 71% were exposed to at least 1 pharmacogenomic drug over the retrospective 20 year prescribing period and 12% were prescribed at least 5 pharmacogenomic drugs over the prospective 5 year period. Among patients aged 70 years and older, approximately 89% were prescribed at least 1 pharmacogenomic drug, and 25% were prescribed at least 5 over the preceding 20 years.
Figure 2.
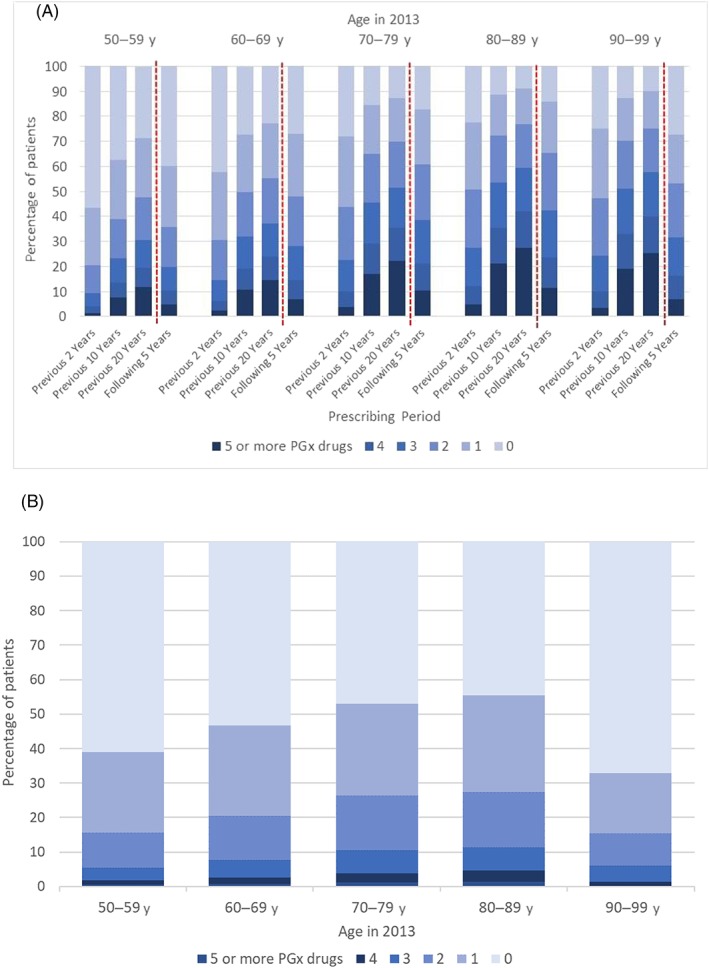
(A) Percentage of patients prescribed pharmacogenomic drugs over available prescribing periods by patient age in 2013. (B) Percentage of patients with a prescription for a pharmacogenomic drug in 2013–17 among those with no prescriptions for pharmacogenomic drugs during 2011–12, by patient age in 2013
Figure 2b shows incident prescribing of pharmacogenomic drugs between 2013–17 for 90 836 patients (45% of 203 395 patients with data for this period) who were not prescribed a pharmacogenomic drug during 2011–12. In this group, approximately 39% of patients aged 50–59 in 2013 were prescribed a new pharmacogenomic drug during 2013–17 after not receiving 1 during 2011–12. This increased across age groups to 55% of patients in the 80–89 year group. It should be noted that patients aged 90–99 years are likely to be dying at a faster rate than the other age groups and therefore may not have had the chance of being prescribed a pharmacogenomic drug, hence low rates in this group.
Figure 3 shows the proportion of women and men, by age group, prescribed pharmacogenomic drugs in each year over 20 years. There was a similar increase in men and women in the prescription of pharmacogenomic drugs in all age groups over the previous 20 years. The only exception was women in the 50–59‐year age group who had higher rates of pharmacogenomic drug prescribing in the mid‐1990s (when they were age 30–39 y) compared to the other age categories. On review, this was due to the prescription of hormonal contraceptives for systemic use which decreases in older ages. Figure 4 shows the cumulative exposure to pharmacogenomic drugs a 20‐year period, excluding those prescribed a pharmacogenomic drug in 1993 and therefore starting from an assumption of zero pharmacogenomic drugs in those included at the start of 1993. By 2003–4 around half of patients aged 70 and over had been exposed to a pharmacogenomic drug, for patients aged 50–59 this threshold was reached during 2008.
Figure 3.
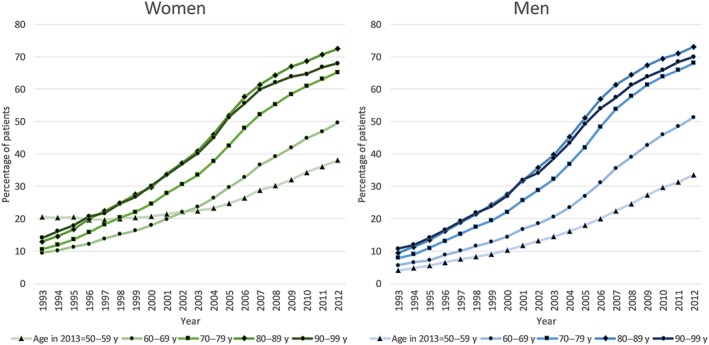
Annual percentage of patients prescribed at least 1 pharmacogenomic drug for different age groups in women and men over retrospective 20 year prescribing period (1993–2012)
Figure 4.
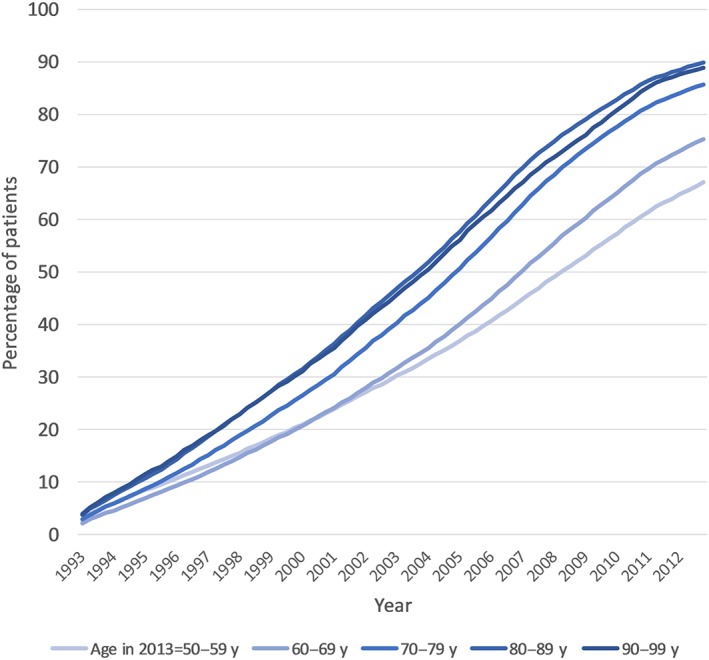
Cumulative percentage of patients prescribed at least 1 pharmacogenomic drugs over 20 years. As we did not have pre‐1993 prescribing data, we excluded patients who were prescribed a pharmacogenomic drug at any time in 1993 and then looked for first exposure afterwards.
3.3. Pharmacogenomic drugs with most frequent use
Table 3 shows the exposure of patients to individual pharmacogenomic drugs and their associated pharmacogenes. The pharmacogenomic drugs prescribed to the most patients were moderate strength opioids, proton‐pump inhibitors, statins, antidepressants, hormonal contraceptives, antiplatelets and anticoagulants. Information on drugs that were only prescribed in secondary care e.g. https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5343, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7063 and https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7069, was not available in the primary care dataset.
Table 3.
Exposure to individual pharmacogenomic (PGx) drugs licensed for prescription in the UK over available prescribing periods
| Available prescribing period | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| –2 years (2011–2012) n = 684 141 | −10 years (2003–2012) n = 538 602 | −20 years (1993–2012) n = 289 186 | +5 years (2013–2017) n = 203 395 | ||||||
| PGx drug | n | % | n | % | n | % | n | % | Corresponding pharmacogene(s) |
| Codeine | 136 997 | 20% | 221 623 | 41% | 139 784 | 47% | 66 463 | 33% | CYP2D6 |
| Omeprazole | 140 142 | 20% | 170 784 | 32% | 100 499 | 34% | 70 433 | 35% | CYP2C19 |
| Simvastatin | 156 472 | 23% | 168 050 | 31% | 94 681 | 32% | 44 472 | 22% | SLCO1B1 |
| Lansoprazole | 68 613 | 10% | 103 676 | 19% | 65 873 | 22% | 29 809 | 15% | CYP2C19 |
| Amitriptyline | 52 717 | 8% | 84 654 | 16% | 58 064 | 19% | 27 917 | 14% | CYP2D6, CYP2C19 |
| Tramadol | 45 784 | 7% | 79 492 | 15% | 46 585 | 16% | 19 074 | 9% | CYP2D6 |
| Citalopram | 39 295 | 6% | 58 446 | 11% | 34 553 | 12% | 15 061 | 7% | CYP2C19 |
| Warfarin | 26 479 | 4% | 28 395 | 5% | 17 951 | 6% | 9042 | 4% | CYP2C9, VKORC1, CYP4F2 |
| Paroxetine | 3850 | 1% | 8643 | 2% | 16 405 | 6% | 1263 | 0.6% | CYP2D6 |
| Clopidogrel | 20 030 | 3% | 28 392 | 5% | 16 232 | 5% | 11 607 | 6% | CYP2C19 |
| Hormonal contraceptives | 883 | 0% | 5935 | 1% | 15 909 | 5% | 143 | 0.1% | F5 |
| Sertraline | 13 176 | 2% | 17 878 | 3% | 13 733 | 5% | 9812 | 5% | CYP2C19 |
| Gliclazide | 20 569 | 3% | 20 184 | 4% | 11 951 | 4% | 7601 | 4% | CYP2C9 |
| Allopurinol | 15 322 | 2% | 15 520 | 3% | 9171 | 3% | 6409 | 3% | HLA‐B |
| Mirtazapine | 10 760 | 2% | 14 314 | 3% | 8120 | 3% | 5924 | 3% | CYP2D6 |
| Carbamazepine | 5214 | 1% | 9079 | 2% | 8090 | 3% | 1849 | 1% | HLA‐A, HLA‐B |
| Venlafaxine | 5425 | 1% | 10 472 | 2% | 7667 | 3% | 2238 | 1% | CYP2D6 |
| Esomeprazole | 5470 | 1% | 12 150 | 2% | 7155 | 2% | 3119 | 2% | CYP2C19 |
| Pantoprazole | 3706 | 1% | 8601 | 2% | 6644 | 2% | 1962 | 1% | CYP2C19 |
| Tamoxifen | 2909 | 0.4% | 6470 | 1% | 5145 | 2% | 1062 | 0.5% | CYP2D6 |
| Escitalopram | 3022 | 0.4% | 9390 | 2% | 4611 | 2% | 1066 | 0.5% | CYP2C19 |
| Fluorouracil | 4197 | 0.6% | 8129 | 2% | 4576 | 2% | 3764 | 2% | DPYD |
| Nortriptyline | 2767 | 0.4% | 4778 | 0.9% | 3859 | 1% | 1507 | 0.7% | CYP2D6 |
| Metoprolol | 3435 | 0.5% | 5040 | 0.9% | 3446 | 1% | 881 | 0.4% | CYP2D6 |
| Clomipramine | 867 | 0.1% | 1618 | 0.3% | 2680 | 0.9% | 291 | 0.1% | CYP2C19, CYP2D6 |
| Imipramine | 616 | 0.1% | 1423 | 0.3% | 2225 | 0.8% | 167 | 0.1% | CYP2C19, CYP2D6 |
| Oxycodone | 2786 | 0.4% | 4012 | 0.7% | 2108 | 0.7% | 1700 | 0.8% | CYP2D6 |
| Azathioprine | 1841 | 0.3% | 2500 | 0.5% | 1792 | 0.6% | 736 | 0.4% | TPMT |
| Phenytoin | 2139 | 0.3% | 2256 | 0.4% | 1741 | 0.6% | 589 | 0.3% | CYP2C9, HLA‐B |
| Trimipramine | 438 | 0.1% | 902 | 0.2% | 1710 | 0.6% | 116 | 0.1% | CYP2C19, CYP2D6 |
| Glimepiride | 2204 | 0.3% | 2676 | 0.5% | 1646 | 0.6% | 1103 | 0.5% | CYP2C9 |
| Flecainide | 1742 | 0.3% | 2196 | 0.4% | 1511 | 0.5% | 832 | 0.40% | CYP2D6 |
| Risperidone | 1958 | 0.3% | 2479 | 0.5% | 1409 | 0.5% | 665 | 0.3% | CYP2D6 |
| Haloperidol | 921 | 0.1% | 1466 | 0.3% | 1273 | 0.4% | 237 | 0.1% | CYP2D6 |
| Doxepin | 343 | 0.1% | 999 | 0.2% | 1251 | 0.4% | 101 | 0.1% | CYP2C19, CYP2D6 |
| Tacrolimus | 997 | 0.2% | 1662 | 0.3% | 887 | 0.3% | 713 | 0.4% | CYP3A5 |
| Ondansetron | 466 | 0.1% | 736 | 0.1% | 475 | 0.2% | 422 | 0.2% | CYP2D6 |
| Fluvoxamine | 58 | 0.0% | 131 | 0.0% | 433 | 0.2% | 27 | 0.0% | CYP2D6 |
| Aripiprazole | 604 | 0.1% | 579 | 0.1% | 269 | 0.1% | 315 | 0.2% | CYP2D6 |
Values are number (%) patients prescribed individual pharmacogenomic drugs licensed for prescription in the UK over available prescribing periods
Drugs are ranked from most to least commonly prescribed over the −20 year prescribing period. Drugs not included in the table were:
Rarely prescribed in primary care (number of prescriptions over 20 year period): propafenone (n = 186), mercaptopurine (n = 92), oxcarbazepine (n = 97), acenocoumarol (n = 106), zuclopenthixol (n = 108), voriconazole (n = 9), peginterferon alfa‐2a (n = 2), ribavirin (n = 4), tioguanine (n = 1), abacavir (n = 0), atazanavir (n = 0), atomoxetine (n = 1), capecitabine (n = 0).
Not prescribed at all in primary care during any available prescribing period: cisplatin, daunorubicin, doxorubicin, irinotecan, ivacaftor, peginterferon alfa‐2b, rasburicase, tegafur.
3.4. Pharmacogenes
The most relevant pharmacogenes for use in a primary care testing panel are CYP2C19, CYP2D6, SLCO1B1, CYP2C9, HLA‐B, VKORC1 and CYP4F2 (Table 4). Over 20 years (1993–2012), 57 and 56% of patients had prescriptions for drugs that had dosing guidelines based on CYP2D6 and CYP2C19 genotypes, respectively. Seven pharmacogenes (IFNL3, UGT1A1, CFTR, G6PD, RARG, SLC28A3, UGT1A6) were virtually never associated with a prescription in primary care. Figure 5 indicates the cumulative impact of testing the 5 pharmacogenes associated with the most prescriptions; the first 3 pharmacogenes (CYP2C19, CYP2D6 and SLCO1B1) account for virtually all pharmacogenomic drug prescribing (>95%) across all periods.
Table 4.
Exposure to drugs by related pharmacogenes over available prescribing periods
| Available prescribing period | –2 years (2011–2012) | −10 years (2003–2012) | −20 years (1993–2012) | +5 years (2013–2017) | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | n | % | n | % | n | % | n | % |
| CYP2D6 | 202 516 | 30% | 275 145 | 51% | 170 388 | 57% | 87 650 | 43% |
| CYP2C19 | 257 079 | 38% | 285 659 | 53% | 165 780 | 56% | 109 782 | 54% |
| SLCO1B1 | 156 472 | 23% | 168 050 | 31% | 94 681 | 32% | 44 472 | 22% |
| CYP2C9 | 50 250 | 7% | 51 496 | 10% | 31 897 | 11% | 17 717 | 9% |
| HLA‐B | 22 459 | 3% | 26 252 | 5% | 18 253 | 6% | 8773 | 4% |
| VKORC1 | 26 648 | 4% | 28 488 | 5% | 17 983 | 6% | 9094 | 4% |
| CYP4F2 | 26 479 | 4% | 28 395 | 5% | 17 951 | 6% | 9042 | 4% |
| F5 | 883 | 0.1% | 5935 | 1% | 15 909 | 5% | 143 | 0.1% |
| HLA‐A | 5214 | 0.8% | 9079 | 2% | 8090 | 3% | 1849 | 0.9% |
| DPYD | 4198 | 0.6% | 8132 | 2% | 4576 | 1.5% | 3764 | 2% |
| TPMT | 1936 | 0.3% | 2585 | 0.5% | 1829 | 0.6% | 795 | 0.4% |
| CYP3A5 | 997 | 0.2% | 1662 | 0.3% | 887 | 0.3% | 713 | 0.4% |
| IFNL3 | 3 | 0.0% | 5 | 0.0% | 4 | 0.0% | 1 | 0.0% |
| UGT1A1 | 0 | 0.0% | 2 | 0.0% | 0 | 0.0% | 1 | 0.0% |
| CFTR | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% |
| G6PD | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% |
| RARG | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 1 | 0.0% |
| SLC28A3 | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% |
| UGT1A6 | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% |
Values are number (%) of patients prescribed any drug with drug/dosing guidelines related to individual pharmacogenes over available prescribing periods. Pharmacogenes are ranked from most to least commonly associated with a prescribed drug over the −20 year prescribing period.
Figure 5.
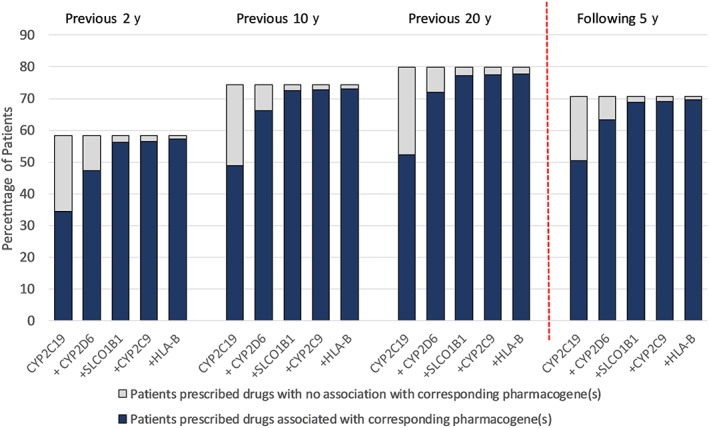
Cumulative impact of testing the 5 pharmacogenes associated with the most prescriptions over available prescribing periods. The overall height of the bars shows the percentage of patients in each prescribing period exposed to drugs affected by any pharmacogene. Blue bars show the cumulative effect of adding the 5 most common pharmacogenes successively from left to right. Grey bars indicate the proportion of patients prescribed drugs associated with pharmacogenes not yet considered (when moving left to right on the x‐axis). For each period, the first blue bar represents the % of patients prescribed any drug influenced by CYP2C19 only. The second blue bar is then % of patients with any drug that corresponds to CYP2C19 or CYP2D6 or both (and so on)
4. DISCUSSION
4.1. Main study findings
Our results demonstrate that exposure of primary care patients to pharmacogenomic drugs is extremely common. Eight‐in‐10 patients were exposed to at least 1 pharmacogenomic drug over a 20‐year period, with pharmacogenomic drugs comprising 16% of all drugs prescribed during that time. Exposure is commonly multiple, with 6/10 patients being prescribed 2 or more and almost 2/10 patients being prescribed 5 or more different pharmacogenomic drugs over the preceding 20 years. Prospectively, 5/10 patients were prescribed at least 2 pharmacogenomic drugs over 5 years. In patients who had not been prescribed a pharmacogenomic drug in 2011–12, 4/10 patients were prescribed at least 1 pharmacogenomic drug over the next 5 years.
Over 2/3 of patients aged 50–59 years were prescribed at least 1 pharmacogenomic drug, with an average of 2 pharmacogenomic drugs over the previous 20 years. This indicates that a pre‐emptive multigene panel done even at a relatively young age could have multiple applications, with the likelihood of exposure to pharmacogenomic drugs increasing with age across all available prescribing periods. Our data also highlight the pharmacogenomic drugs prescribed to the most patients in primary care and indicate that 3 pharmacogenes (CYP2C19, CYP2D6 and SLCO1B1) accounted for virtually all (>95%) of these common pharmacogenomic drugs across all periods.
4.2. Comparison with other studies
Our finding that primary care patients are commonly exposed to pharmacogenomic drugs is supported by recent studies from the USA. Schildcrout and colleagues investigated prescribing of 56 pharmacogenomic drugs in ~53 000 primary care patients at Vanderbilt University Medical Centre, with a median age of 54 years at the start of the study.17 Over a 5‐year period, 65% of individuals were exposed to at least 1 pharmacogenomic drug and 12% were exposed to ≥4. Our analysis showed similar results in a larger number of patients and over a longer period, whilst also providing relevance to the UK.
Samwald and colleagues investigated the prescribing of 61 pharmacogenomic drugs in ~73 million patients in the USA over a 4‐year period (2009–12).18 They also compared exposure by age group. Even in the youngest age group of 0–13 years, there was some exposure to pharmacogenomic drugs with 14% receiving at least 1 and 2% receiving at least 2 pharmacogenomic drugs. Similar to our study, they showed that exposure to pharmacogenomic drugs increased with age.
In the USA, in 2013, the 30 most commonly prescribed pharmacogenomic high‐risk drugs accounted for 738 million outpatient prescriptions.19 Samwald and colleagues showed that the most commonly prescribed pharmacogenomic drugs in their study were indicated for pain relief and cardiovascular conditions, similar to our findings.18 Additionally, the top 6 pharmacogenomic drugs (https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2955, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=553, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5488, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6853, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2949, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7150) in Schildcrout et al.'s study also featured high in our list.17 We found that 3 pharmacogenes (CYP2C19, CYP2D6 and SLCO1B1) accounted for virtually all (>95%) of these common pharmacogenomic drugs across all periods. As primary care patients are rarely exposed to more specialist pharmacogenomic drugs, this could inform design of focussed pharmacogenomic testing for implementation in primary care.
The most relevant pharmacogenes associated with the pharmacogenomic drugs in our study were CYP2C19, CYP2D6, SLCO1B1, CYP2C9, HLA‐B, VKORC1 and CYP4F2. Although we did not have genomic data, previous studies have shown that actionable variants in these genes are extremely common. In the USA, more than 97% of the population have at least 1 high‐risk diplotype from 12 pharmacogenes tested.19 The PREDICT programme at Vanderbilt University Medical Centre, reported that of 10 000 patients, 91% of individuals had at least 1 actionable genotype among 5 drug–gene interactions tested (CYP2C9/VKORC1–warfarin, CYP2C19–clopidogrel, SLCO1B1–simvastatin, CYP3A5–https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6784 and TPMT–thiopurines).20 Among 5400 Australian patients, 96% carried at least 1 actionable pharmacogenomic variant in CYP2D6, CYP2C1, CYP2C9 and VKORC1.21 The Mayo clinic found that 99% of patients (n = 1013) had an actionable pharmacogenomic variant in CYP2D6, CYP2C1, CYP2C9, VKORC1 and SLCO1B1.22 Bush and colleagues reported that of 5000 patients in the eMERGE‐PGx programme, 96% of all samples had ≥1 actionable pharmacogenomic variant within 92 pharmacogenes.23 The high prevalence of actionable pharmacogenomic variants is clearly of clinical importance given how common the prescribing of associated pharmacogenomic medications are in primary care patients.
4.3. Study strengths and limitations
To our knowledge, this is the first study to define the exposure of patients to pharmacogenomic drugs in a large UK primary care population over an extended period of time. Currently, there is no nationwide pre‐emptive pharmacogenomic testing programme in any healthcare system. Our findings will help to define the multigene panel needed for pre‐emptive testing in primary care patients.
While the age–sex structure of CPRD patients is broadly similar to that of the UK population,13 since CPRD is based on practices using Vision software, this has resulted in a geographical bias within England, with practices concentrated in the South and relatively few in the in the North‐East and East of England.14 Patients registered in practices located in the North or the Midlands had marginally higher rates of being prescribed a pharmacogenomic drug (82%) over the 20‐year period (1993–2012) than those patients in the South (78%). This difference was also seen in the 5‐year prospective period (2013–17) with higher rates in the North/Midlands (74%) than the South (70%). These small differences would suggest that the true national prescribing prevalence of pharmacogenomic drugs might be marginally underestimated, but does not affect our overall conclusions.
A strength of our study was that we used pharmacogenomic drugs with evidence‐based dosing guidelines in the PharmGKB. This indicates that pharmacogenomic testing in primary care patients could have real impact on prescribing decisions commonly made in clinical practice. The dosage of drugs used for different indications may affect the clinical significance of the drug–gene interaction e.g. https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1673 used as low dose for the treatment of cough vs higher doses for analgesia. However, in our data set, 99% of the 5 million prescriptions for codeine were in capsule or tablet form for use as an analgesic, indicating that pharmacogenomic guidelines would apply for the majority of prescribing decisions.
The aim of our study was to inform design of pre‐emptive multigene pharmacogenomic testing in primary care patients. As such, our analysis only included drugs initiated in or continued in primary care and did not include drugs for specialist indications, such as chemotherapeutic, antiretroviral and antiviral medications. Our results indicate that a relatively limited pharmacogenomic testing panel would cover the majority of prescribing decisions made in primary care (Figure 5). Whilst a broader panel may be required to cover specialist prescribing in secondary care, focussed reporting for primary care may assist in implementing guidelines into practice.
Within our study, we were able to include patients with up to 20 years of prescribing data retrospectively and 5 years of prescribing date prospectively. This provided insight into longitudinal prescribing to individuals and their multiple exposure to pharmacogenomic drugs over time. The main weakness of our approach is that by only including patients still registered at the beginning of 2013, we are only including survivors as, by definition, we did not include patients who have passed away prior to 2013. This means that the 90–99 year olds in 2013 in our sample are not truly representative of all 70–79 year olds in 1993. For our youngest age groups (50–59 y in 2013) loss to mortality will be less of an issue, but we will not be including 30–39‐year‐old patients from 1993 who subsequently de‐registered from their General Practice over the next 20 years. Despite this, we still showed that the 30–39 year olds who were continually registered over this period were commonly exposed to pharmacogenomic drugs. Information on younger age groups is needed to identify the first opportunity for pharmacogenomic testing that would capture lifetime exposure. We also attempted to look prospectively at prescribing over the following 5 years, but only included the survivors who have remained registered to practices that have continued to contribute to the CPRD over this period.
In our study, we assessed whether patients had had any exposure to pharmacogenomic drugs in defined available prescribing periods. This allowed us to identify pharmacogenomic drugs prescribed to the most patients, enabling us to infer where pharmacogenomic testing would have potential utility in primary care. However, pharmacogenomic exposure could be defined by only a single prescription for a drug and we did not evaluate duration of exposure or repeat exposures (e.g. to analgesic drugs). These factors would be important when considering the potential clinical utility of pharmacogenomic testing. We also defined exposure as being any prescription for a pharmacogenomic drug within an available prescribing period, irrespective of whether that drug had already been prescribed to the patient prior to that time period. This approach optimised numbers of patients available for analysis and generalisability to the whole population, however it did not assess incident prescribing. In a smaller subgroup of patients not taking pharmacogenomic medicines in 2011–2 we were able to show that patients of all ages frequently received new prescriptions for pharmacogenomic drugs (Figure 2b).
It would have been interesting to have had the genetic profiles of the patients. The lack of genomic information limited our ability to calculate the exact number of prescribing decisions that could have been influenced for each individual. However, previous studies have shown that most people have variants in pharmacogenes that affect metabolism of common drugs and therefore our analysis is likely to have broad relevance.19, 20, 21, 22, 23 The retrospective nature of the study is also a potential limitation as prescribing practice and exposure to drugs has clearly changed over the last 20 years. However, in England, the 100 most commonly prescribed drugs have been remarkably consistent over the last decade and the core list of most commonly prescribed drug groups has remained stable.24
As demonstrated in our study, a pre‐emptive multigene panel could impact on multiple prescribing decisions throughout a patient's life. However, 1 recognised barrier to the implementation of pharmacogenomics into healthcare services is the lack of evidence of clinical utility. Considering this, several initiatives are currently being carried out to assess the clinical value of pharmacogenomic testing. For example, the Ubiquitous Pharmacogenomics consortium is leading the PREPARE trial. This is a prospective, European randomised controlled trial that will implement pre‐emptive genotyping of a panel of 13 pharmacogenes consisting of 50 variants into routine care to guide drug and dose selection for 41 commonly prescribed drugs.25 The goal of the study is to show that pre‐emptive testing of patients for an entire panel of pharmacogenomic markers reduces the number of adverse drug reactions.
4.4. Implications for future research and practice
This study supports the development of a pre‐emptive multi‐gene panel, appropriate to primary care patients that could influence multiple prescribing decisions over the course of a patient's lifetime. Our findings could inform health economic modelling to establish the most cost‐effective time at which testing should be carried out in primary care patients. There are well established points within current NHS care, for example NHS health checks offered every 5 years to all 40–74 year olds, at which this pharmacogenomic test could be introduced.11
In conclusion, exposure of primary care patients to pharmacogenomic drugs is extremely common, multiplicitous and has commenced by relatively early adulthood. A small number of pharmacogenes account for the majority of drugs prescribed. These findings could inform design of a pre‐emptive pharmacogenomic testing panel and programme for implementation in primary care.
COMPETING INTERESTS
The authors have no competing interests to declare.
This study did not perform interventions with or administer substances to human subjects/patients and did not have a Principal Investigator.
CONTRIBUTORS
Emma Baker had the original idea for the study and all authors contributed to the study design. Iain Carey led the data analysis and all authors contributed to the interpretation of the data. James Kimpton wrote the first draft of the manuscript. All authors contributed to the revision of the manuscript related to its intellectual content. All authors approved the final version submitted for publication.
DATA AVAILABILITYS
This study is based on data from CPRD obtained under licence from the UK Medicines and Healthcare products Regulatory Agency. The data are provided by patients and collected by the NHS as part of their care and support. The interpretation and conclusions contained in this report are those of the authors alone. The protocol number (18_023_RA) was approved by the Independent Scientific Advisory Committee evaluation of joint protocols of research involving CPRD data in August 2018.
Supporting information
FIGURE S1
Distribution of number of pharmacogenomic drugs in different prescribing periods
Kimpton JE, Carey IM, Threapleton CJD, et al. Longitudinal exposure of English primary care patients to pharmacogenomic drugs: An analysis to inform design of pre‐emptive pharmacogenomic testing. Br J Clin Pharmacol. 2019;85:2734–2746. 10.1111/bcp.14100
REFERENCES
- 1. Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE. Pharmacogenomics and individualized medicine: translating science into practice. Clin Pharmacol Ther. 2012;92(4):467‐475. 10.1038/clpt.2012.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92(1):87‐95. 10.1038/clpt.2011.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson JA, Elsey AR, Clare‐Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands hospital personalized medicine program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14(7):723‐726. 10.2217/pgs.13.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right timedusing genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25‐33. 10.1016/j.mayocp.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. U.S. Food and Drug Administration . Table of Pharmacogenomic Biomarkers in Drug Labels. Available at https://www.fda.gov/drugs/science‐research‐drugs/table‐pharmacogenomic‐biomarkers‐drug‐labeling. Accessed February 2, 2019. 10.1111/j.1525-1594.2008.00596.x [DOI]
- 6. Joint Formulary Committee . British National Formulary (online). London: BMJ Group and Pharmaceutical Press. Available at http://www.medicinescomplete.com. Accessed April 10, 2019. [Google Scholar]
- 7. Mallal S, Phillips E, Carosi G, et al. HLA‐B*5701 screening for hypersensitivity to abacavir. And warfarin genetic dosage algorithm. N Engl J Med. 2008;358(6):568‐579. 10.1056/NEJMoa0706135 [DOI] [PubMed] [Google Scholar]
- 8. NHS England . Improving outcomes through personalised medicine. NHS Engl. 2016. https://www.england.nhs.uk/wp-content/uploads/2016/09/improving-outcomes-personalised-medicine.pdf. [Google Scholar]
- 9. Davies, SC . Annual Report of the Chief Medical Officer 2016: Generation Genome. Department of Health. 2017. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/631043/CMO_annual_report_generation_genome.pdf.
- 10. Raza S, Blackburn L, Moorthie S, et al. The Personalised Medicine Technology Landscape. PHG Foundation. 2018. 978‐1‐907198‐31‐1. https://www.phgfoundation.org/documents/phgf‐personalised‐medicine‐technology‐landscape‐report‐50918.pdf.
- 11. Vascular Programme (Department of Health) . Putting Prevention First. Vascular Checks: Risk Assessment and Management. London; 2008. http://webarchive.nationalarchives.gov.uk/20120503234407/http://www.dh.gov.uk/prod_consum_dh/groups/dh_digitalassets/@dh/@en/documents/digitalasset/dh_083823.pdf. [Google Scholar]
- 12. Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM. Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Heal Pharm. 2016;73(23):1967‐1976. 10.2146/ajhp160030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herrett E, Gallagher AM, Bhaskaran K, et al. Data resource profile: clinical practice research datalink (CPRD). Int J Epidemiol. 2015;44(3):827‐836. 10.1093/ije/dyv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kontopantelis E, Stevens RJ, Helms PJ, Edwards D, Doran T, Ashcroft DM. Spatial distribution of clinical computer systems in primary care in England in 2016 and implications for primary care electronic medical record databases: a cross‐sectional population study. BMJ Open. 2018;8(2):e020738 10.1136/bmjopen-2017-020738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whirl‐Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414‐417. 10.1038/clpt.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harding SD, Sharman JL, et al. The IUPHAR/BPS guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res. 2018;46:D1091‐D1106. 10.2217/pgs-2017-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schildcrout JS, Denny JC, Bowton E, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92(2):235‐242. 10.1038/clpt.2012.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samwald M, Xu H, Blagec K, et al. Incidence of exposure of patients in the United States to multiple drugs for which pharmacogenomic guidelines are available. PLoS One. 2016;11(10):e0164972 10.1371/journal.pone.0164972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical Pharmacogenetics implementation: current programs in five United States medical centers. Annu Rev Pharmacol Toxicol. 2014;(September;55(1):1‐18. 10.1146/annurev-pharmtox-010814-124835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Driest SL, Shi Y, Bowton E, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther. 2014;95(4):423‐431. 10.1038/clpt.2013.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mostafa S, Kirkpatrick CMJ, Byron K, Sheffield L. An analysis of allele, genotype and phenotype frequencies, actionable pharmacogenomic (PGx) variants and phenoconversion in 5408 Australian patients genotyped for CYP2D6, CYP2C19, CYP2C9 and VKORC1 genes. J Neural Transm. 2019;126(1):5‐18. 10.1007/s00702-018-1922-0 [DOI] [PubMed] [Google Scholar]
- 22. Ji Y, Skierka JM, Blommel JH, et al. Preemptive Pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable Pharmacogenomic genes using next‐generation DNA sequencing and a customized CYP2D6 genotyping Cascade. J Mol Diagnostics. 2016;18(3):438‐445. 10.1016/j.jmoldx.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bush WS, Crosslin DR, Owusu‐Obeng A, et al. Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100(2):160‐169. 10.1002/cpt.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Audi S, Burrage DR, Lonsdale DO, et al. The “top 100” drugs and classes in England: an updated “starter formulary” for trainee prescribers. Br J Clin Pharmacol. 2018;84(11):2562‐2571. 10.1111/bcp.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manson LE, van der Wouden CH, Swen JJ, Guchelaar H‐J. The ubiquitous pharmacogenomics consortium: making effective treatment optimization accessible to every European citizen. Pharmacogenomics. 2017;18(11):1041‐1045. 10.2217/pgs-2017-0093 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1
Distribution of number of pharmacogenomic drugs in different prescribing periods
Data Availability Statement
This study is based on data from CPRD obtained under licence from the UK Medicines and Healthcare products Regulatory Agency. The data are provided by patients and collected by the NHS as part of their care and support. The interpretation and conclusions contained in this report are those of the authors alone. The protocol number (18_023_RA) was approved by the Independent Scientific Advisory Committee evaluation of joint protocols of research involving CPRD data in August 2018.


