Abstract
Because of their replication mode and segmented dsRNA genome, homologous recombination is assumed to be rare in the rotaviruses. We analyzed 23,627 complete rotavirus genome sequences available in the NCBI Virus Variation database, and found 109 instances of homologous recombination, at least eleven of which prevailed across multiple sequenced isolates. In one case, recombination may have generated a novel rotavirus VP1 lineage. We also found strong evidence for intergenotypic recombination in which more than one sequence strongly supported the same event, particularly between different genotypes of segment 9, which encodes the glycoprotein, VP7. The recombined regions of many putative recombinants showed amino acid substitutions differentiating them from their major and minor parents. This finding suggests that these recombination events were not overly deleterious, since presumably these recombinants proliferated long enough to acquire adaptive mutations in their recombined regions. Protein structural predictions indicated that, despite the sometimes substantial amino acid replacements resulting from recombination, the overall protein structures remained relatively unaffected. Notably, recombination junctions appear to occur nonrandomly with hot spots corresponding to secondary RNA structures, a pattern seen consistently across segments. In total, we found strong evidence for recombination in nine of eleven rotavirus A segments. Only segments 7 (NSP3) and 11 (NSP5) did not show strong evidence of recombination. Collectively, the results of our computational analyses suggest that, contrary to the prevailing sentiment, recombination may be a significant driver of rotavirus evolution and may influence circulating strain diversity.
Keywords: dsRNA, genetic diversity, Reoviridae, segmented genome, RDP4
1. Introduction
The nonenveloped dsRNA rotaviruses of the family Reoviridae are a common cause of acute gastroenteritis in young individuals of many bird and mammal species (Desselberger 2014). The rotavirus genome consists of eleven segments, each coding for a single protein with the exception of segment 11, which encodes two proteins, NSP5 and NSP6 (Desselberger 2014). Six of the proteins are structural proteins (VP1-4, VP6, and VP7), and the remainder are nonstructural proteins (NSP1-6). The infectious virion is a triple-layered particle consisting of an outer capsid protein, VP7, a spike protein, VP4, an inner capsid protein, VP6, and a core protein, VP2. The RNA polymerase (VP1) and the capping enzyme (VP3) are attached to the inner capsid protein. For the virus to be infectious (at least when not infecting as an extracellular vesicle), the VP4 spike protein must be cleaved by a protease, which results in the proteins VP5* and VP8* (Arias et al. 1996). Because they comprise the outer layer of the virion, VP7 and VP4 are capable of eliciting neutralizing antibodies, and are used to define G (glycoprotein) and P (protease sensitive) serotypes, respectively (Matthijnssens et al. 2008a; Nair et al. 2017). Consequently, VP7 and VP4 are likely to be under strong selection for diversification to mediate cell entry or escape host immune responses (McDonald et al. 2009; Kirkwood 2010; Patton 2012).
Based on sequence identity and antigenic properties of VP6, ten different rotavirus groups (A–J) have been identified, with rotavirus A being the most common cause of human infections (Matthijnssens et al. 2012; Mihalov-Kovacs et al. 2015; Banyai et al. 2017). A genome classification system based on established nucleotide percent cut-off values has been developed for rotavirus A (Matthijnssens et al. 2008a, 2011). In the classification system, the segments VP7–VP4–VP6–VP1–VP2–VP3–NSP1–NSP2–NSP3–NSP4–NSP5/6 are represented by the indicators Gx–P[x]–Ix–Rx–Cx–Mx–Ax–Nx–Tx–Ex–Hx (x = Arabic numbers starting from one), respectively (Matthijnssens et al. 2008a, 2011). To date, between twenty and fifty-one different genotypes have been identified for each segment, including fifty-one different VP4 genotypes (P[1]–P[51]) and thirty-six different VP7 genotypes (G1–G36), both at 80 per cent nucleotide identity cut-off values (Steger et al. 2019).
The propensity of rotavirus for coinfection and outcrossing with other rotavirus strains makes it a difficult pathogen to control and surveil, even with current vaccines (Rahman et al. 2007; Matthijnssens et al. 2008a, 2009; Kirkwood 2010; Ghosh and Kobayashi 2011; Sadiq et al. 2018). Understanding rotaviral diversity expansion, genetic exchange between strains (especially between the clinically significant type I and type II genogroups), and evolutionary dynamics resulting from coinfections have important implications for disease control (Rahman et al. 2007; Matthijnssens et al. 2008a, 2009; Kirkwood 2010; Ghosh and Kobayashi 2011; Sadiq et al. 2018). Rotavirus A genomes have high mutation rates (Matthijnssens et al. 2010; Donker and Kirkwood 2012; Sadiq et al. 2018), undergo frequent reassortment (Ramig and Ward 1991; Ramig 1997; Ghosh and Kobayashi 2011; McDonald et al. 2016), and the perception is that these two processes are the primary drivers of rotavirus evolution (Doro et al. 2015; Sadiq et al. 2018). Genome rearrangements may also contribute to rotavirus diversity, but are not believed to be a major factor in rotavirus evolution (Desselberger 1996). Homologous recombination, however, is thought to be especially rare in rotaviruses due to their segmented dsRNA genomes and their polymerase’s transcription and replication mechanisms (Ramig 1997; McDonald et al. 2016; Varsani et al. 2018). Unlike +ssRNA (Lukashev 2005) viruses and DNA viruses (Pérez-Losada et al. 2015), dsRNA viruses cannot easily undergo intragenic recombination because their genomes are not replicated in the cytoplasm by host polymerases, but rather within nucleocapsids by viral RNA-dependent RNA polymerase (Ramig 1997; Patton et al. 2007). Genome encapsidation should significantly reduce the opportunities for template switching, the presumptive main mechanism of intramolecular recombination (Lai 1992; Pérez-Losada et al. 2015).
Despite the expectation that recombination should be rare in rotavirus A, there are nevertheless numerous reports of recombination among rotaviruses in the literature (Suzuki et al. 1998; Parra et al. 2004; Phan et al. 2007a,b; Cao et al. 2008; Martinez-Laso et al. 2009; Donker, Boniface, and Kirkwood 2011; Jere et al. 2011; Esona et al. 2017; Jing et al. 2018). However, a comprehensive survey of 797 rotavirus A genomes failed to find any instances of the same recombination event in multiple samples (Woods 2015). This result may be due to poorly fit recombinants that failed to increase in frequency in the population such that they would be resampled (Woods 2015). This study had less sequences available at the time and used different analytical techniques than we did. The main implications of this study are that recombination among rotavirus A is rare, usually disadvantageous, and not a significant factor in rotavirus evolution.
Since the number of publicly available rotavirus A whole segment genomes is now over 23,600, it is worth revisiting these conclusions to see if they are still valid. To this end, we used bioinformatics tools to identify possible instances of recombination among all available complete rotavirus A genome sequences available in the NCBI Virus Variation database as of May 2019. We found strong evidence for recombination events among all rotavirus A segments with the exception of NSP3 and NSP5. In several cases, the recombinants were fixed in the population such that several hundred sampled strains showed remnants of this same event. These reports suggest that rotavirus recombination occurs more frequently than is generally appreciated, and can significantly influence rotavirus A evolution.
2. Methods
2.1 Sequence acquisition and metadata curation
We downloaded all complete rotavirus genomes from NCBI’s Virus Variation Resource as of May 2019 (n = 23,627) (Hatcher et al. 2017). Laboratory strains were removed from the dataset. Genomes that appeared to contain substantial insertions were excluded as well. Avian and mammalian strains were analyzed separately as recombination analyses between more divergent genomes can sometimes confound the results. No well-supported events were identified among the avian strains. For each rotavirus genome, separate fasta files were downloaded for each of the eleven segments. Metadata including host, country of isolation, collection date, and genotype (genotype cut-off values as defined by Matthijnssens et al. (2008b)) were recorded. The genotype cut-off values allowed for the differentiation of events that occurred between two different genotypes—intergenotypic—from those which occurred between the same genotype—intragenotypic.
2.2 Recombination detection and phylogenetic analysis
All eleven segments of each complete genome were separately aligned using MUSCLE v3.8.31 (Edgar 2004) after removing any low quality sequence (e.g., ‘Ns’). Putative recombinants were identified using RDP4 Beta 4.97, a program, which employs multiple recombination detection methods to minimize the possibility of false positives (Martin et al. 2015). All genomes were analyzed, with a P-value cut-off < 10E-4, using 3Seq, Chimæra, SiScan, MaxChi, Bootscan, Geneconv, and RDP as implemented in RDP4 (Martin et al. 2015). We eliminated any strains that did not show a putative recombination event predicted by at least six of the above listed programs. We then ran separate phylogenic analyses on the ‘major parent’ and ‘minor parent’ sequences of putative recombinants in BEAST v1.10.4 using the general time reversible (GTR) + Γ + I substitution model (Suchard et al. 2018). ‘Parent’ in this case does not refer to the actual progenitors of the recombinant strain, but rather those members of the populations whose genome sequences most closely resemble that of the recombinant. Significant phylogenetic incongruities with high posterior probabilities between the ‘major parent’ and ‘minor parent’ sequences were interpreted as convincing evidence for recombination.
2.3 Phylogenetic analysis of VP1 and VP3
Segments 1 (VP1) and 3 (VP3) each showed evidence for recombination events resulting in a novel lineage, wherein many isolates reflected the same recombination event. To analyze these events more thoroughly, we split alignments of a subset of environmental isolates to include flagged clades, minor parent clades, and major parent clades along with outgroup clades to improve accuracy of tip dating. We generated minor and major parent phylogenies using BEAST v1.10.4 (Suchard et al. 2018). We used tip dating to calibrate molecular clocks and generate time-scaled phylogenies. The analyses were run under an uncorrelated relaxed clock model using a time-aware Gaussian Markov random-field Bayesian skyride tree prior (Minin et al. 2008). The alignments were run using a GTR + Γ + I substitution model and partitioned by codon position. Log files in Tracer v1.7.1 (Rambaut et al. 2018) were analyzed to confirm sufficient effective sample size values, and trees were annotated using a 20 per cent burn in. The alignments were run for three chains with a 200,000,000 Markov chain Monte Carlo chain length, analyzed on Tracer v1.7.1 (Rambaut et al. 2018), and combined using LogCombiner v1.10.4 (Drummond and Rambaut 2007). The best tree was visualized using FigTree v1.4.4 (Rambaut 2019) with the nodes labeled with posterior probabilities.
2.4 RNA secondary structure analysis
To test the hypothesis that recombination junctions were associated with RNA secondary structure, we generated consensus secondary RNA structures of different segments and genotypes using RNAalifold in the ViennaRNA package, version 2.4.11 (Bernhart et al. 2008; Lorenz et al. 2011). The consensus structures were visualized and mountain plots were generated to identify conserved structures that may correspond to breakpoint locations. We made separate consensus alignments in two ways, by using the same genotype and by combining genotypes. Due to the high sequence variability of the observed recombinants, particularly in segments 4 (VP4) and 9 (VP7), we note that the consensus structures may vary substantially depending on the sequences used in the alignments. Segments 7 (NSP3), 10 (NSP4), and 11 (NSP5) were not analyzed due to only having one or no recombination events. Consensus structures could not reliably be made for segment 2 (VP2). While the VP2 protein is relatively conserved across genotypes, it contains insertions particularly in the C1 genotype, yet shows recombination across C1 and C2 genotypes.
2.5 Protein structure and antigenic epitope predictions
To evaluate whether recombination events resulted in substantial (deleterious) protein structure changes, we employed LOMETS2 (Local Meta-Threading-Server) I-TASSER (Iterative Threading Assembly Refinement) (Zhang 2008; Roy et al. 2010; Yang and Zhang 2015) to predict secondary and tertiary protein structures. I-TASSER generates a confidence (C) score for estimating the quality of the protein models. To determine if any of the putative recombinants possessed recombined regions containing epitopes, we analyzed the amino acid sequences of all VP4, VP6, and VP7 recombinants and their major and minor parents. We used the Immune Epitope Database (IEDB) (Vita et al. 2019) and SVMTriP (Yao et al. 2012) to predict conserved epitopes.
3. Results
3.1 Strong evidence for homologous recombination in rotavirus A
We identified 109 putative recombination events (identified by 6/7 RDP4 programs; Table 1, Supplementary Table S1). Of these, sixty-seven recombination events were strongly supported, meaning they were detected by 7/7 RDP4 programs with a P-value cut-off < 10E-9 (Table 1). Most recombination events detected were observed in sequences uploaded to the Virus Variation database since the last large scale analysis (Woods 2015) so differences between these results and prior studies may simply reflect the recent increase in available genome sequences.
Table 1.
Recombination events identified among all mammalian rotavirus A genome sequences downloaded from NCBI Virus Variation database in May 2019 (Hatcher et al. 2017).
| VP1 | VP2 | VP3 | VP4 | VP6 | VP7 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of sequences analyzed | 1,710 | 1,600 | 1,905 | 1,990 | 2,176 | 3,887 | 1,962 | 2,186 | 1,881 | 2,430 | 1,900 |
| Putative recombination eventsa | 15 | 13 | 16 | 11 | 11 | 24 | 11 | 4 | 0 | 3 | 1 |
| Strongly supported eventsb | 7 | 8 | 15 | 7 | 6 | 14 | 7 | 1 | 0 | 1 | 1 |
| Recombination frequencyc | 4.1E-3 | 5.0E-3 | 7.9E-3 | 3.5E-3 | 2.8E-3 | 3.6E-3 | 3.6E-3 | 4.6E-4 | – | 4.1E-4 | 5.3E-4 |
| Intergenotypic recombination ratiod | 2/15 | 8/13 | 4/16 | 6/11 | 7/11 | 22/24 | 3/11 | 2/4 | – | 2/3 | 1/1 |
6/7 programs implemented in RDP4 identified putative recombination event (see Section 2).
Events identified by 7/7 programs implemented in RDP4 or events where more than one environmental isolate showed the same event (see Section 2).
Strongly supported events (row 3) divided by number of sequences analyzed (row 1).
Number of intergenotypic events out of all putative recombination events (row 2).
3.2 Putative recombination events observed in multiple environmental isolates
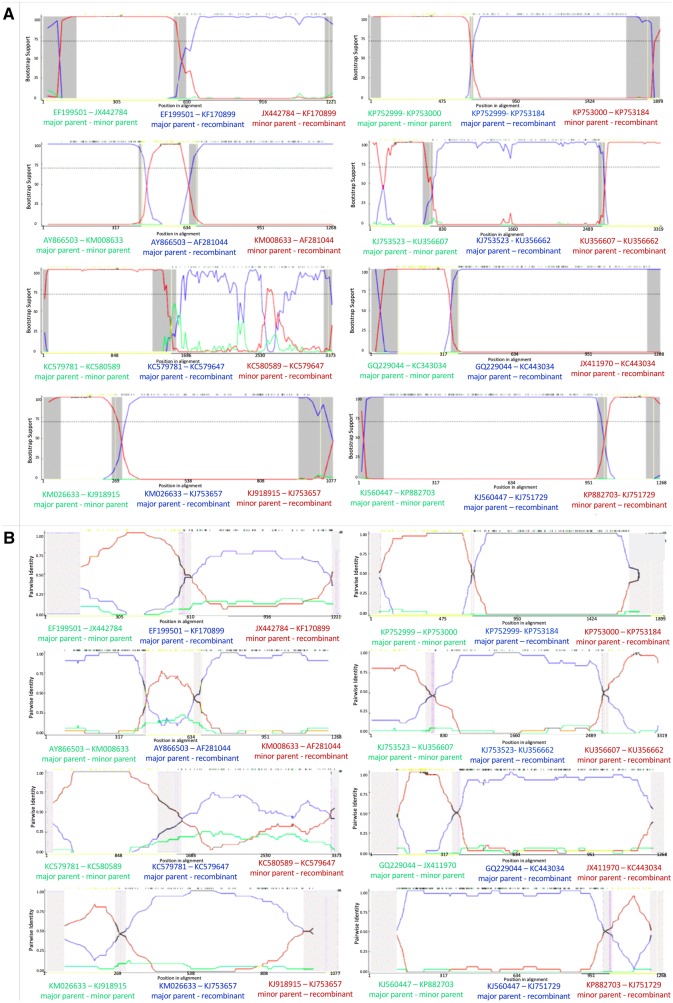
Eleven of the recombination events identified in Table 1 were observed in more than one environmental isolate (Fig. 1, Table 2). The observation of multiple sequenced strains with the same recombination event is strong evidence that the observed event was not spurious, and was not a consequence of improper analytical technique or experimental error. Assuming the events are not spurious, there are only two ways that multiple sequenced isolates will show the same recombinant genotype. Either multiple recombination events with the same exact breakpoints occurred at approximately the same time, or the event happened once and descendants of the recombined genotype were subsequently isolated from additional infected hosts. The latter scenario is more parsimonious, and suggests that the new genotype was not reproductively impaired. Indeed, some recombined genotypes may be more fit than their predecessors, but this outcome would need to be experimentally validated with infectivity assays.
Figure 1.
Bootscan (A) and RDP (B) analyses of putative recombinants where multiple environmental samples supported the event. From top left to bottom left: a G6-G6 event in VP7, a G9-G1 event, an R1-R1 event in VP1, and an N1-N1 event in NSP2. From top right to bottom right: an A8-A8 event in NSP1, an R2-R2 event in VP1, a G2-G1 event in VP7, and a G3-G1 event.
Table 2.
Recombination events observed in multiple independent environmental isolates (i.e., isolates from different patients and/or sequenced by different laboratories).
| Recombined segment | Genotype(s) involved | Number of independent isolates | Uncorrected and Corrected MCs | Breakpointsa | Accession numbers |
|---|---|---|---|---|---|
| Segment 1 (VP1) | R1 | 2 | 4.787E-18 and 1.349E-10 | 35–1,448 (1,290–1,498) | KC579647 |
| KC580176 | |||||
| Segment 1 (VP1) | R2 | Flagged in 285 samples | 1.305E-16 and 3.332E-09 | (541–711)–(1327–1445) | KU199270b |
| Segment 1 (VP1) | R2 | 2 | 1.756E-20 and 4.951E-13 | 656 (541–711)–2,661 (2,600–2,735) | KU356662 |
| KU356640 | |||||
| Segment 3 (VP3) | M2 | Flagged in 551 samples | 1.78E-12 and 4.982E-05 | 1,258 (872–1389)–1,798 (1,662–1,917) | KX655453 |
| Segment 3 (VP3) | M1 | Flagged in 107 samples | 2.491E-20 and 6.946E-13 | 2,158 (2,129–2,174)–2,531 (undetermined) | KJ919553 |
| KJ919517 | |||||
| KJ919551 | |||||
| KJ753665 | |||||
| JQ069727 | |||||
| Segment 7 (NSP1) | A8 (porcine) | 3 | 3.01E-37 and 8.622E-30 | (1,434–25)–(573–596) | KP753174 |
| KJ753184 | |||||
| KP752951 | |||||
| Segment 8 (NSP2) | N1 | 3 | 3.689E-13 and 4.054E-06 | 485 (455–499)–889 (868–914) | KJ753657 |
| KM026663 | |||||
| KM026664 | |||||
| Segment 9 (VP7) | G6 (bovine) | 2 | 6.240E-12 and 2.780E-05 | 1,048 (1,032–128)–481 (448–506) | HM591496 |
| KF170899 | |||||
| Segment 9 (VP7) | G1 and G2 | 2 | 5.507E-38 and 1.664E-30 | 55 (994–60)–291 (262–296) | KC443034 |
| MG181727 | |||||
| Segment 9 (VP7) | G1 and G3 | 2 | 1.4E-23 and 4.230E-16 | 857 (833–859)–1,019 (991–51) | KJ751729 |
| KP752817 | |||||
| Segment 9 (VP7) | G1 and G9 | 2 | 5.292E-19 and 1.599E-11 | 362 (346–366)–589 (558–605) | AF281044 |
| GQ433992 |
99% confidence intervals.
See Fig. 2.
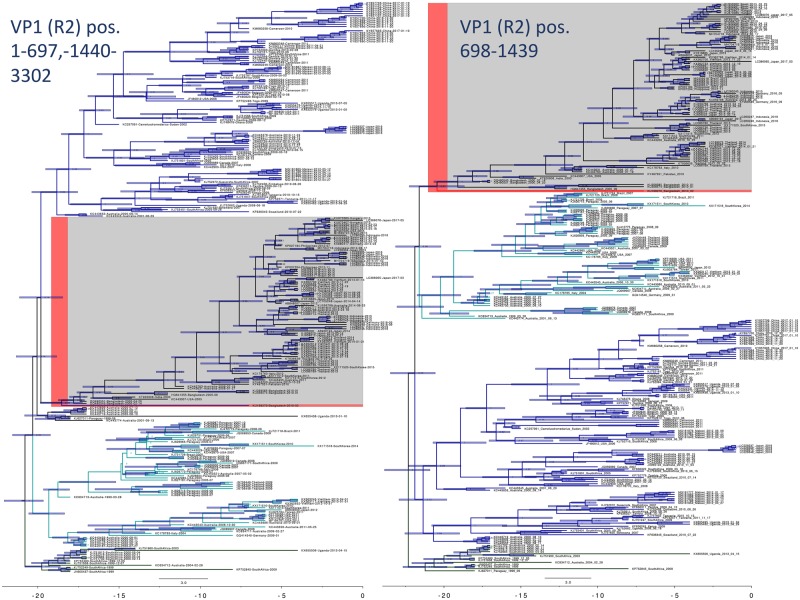
3.3 Segment 1 intragenotypic recombination resulting in a new lineage
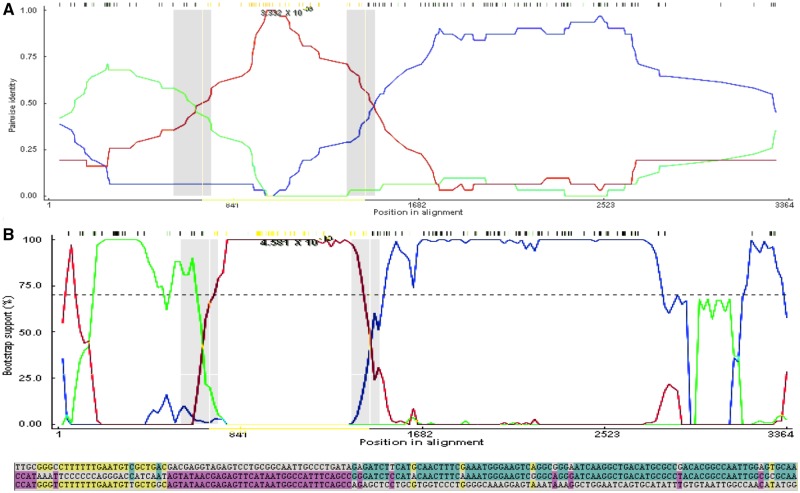
Segment 1 (VP1; ∼3,302 bp) showed evidence of a recombination event within the R2 clade that was fixed in the population, and resulted in a new lineage (highlighted clade in Figs 2 and 3, Table 2). The multiple comparison (MC) uncorrected and corrected probabilities were 1.305E-16 and 3.332E-09, respectively. The sequence most closely related to the recombined sequence was KU199270, a human isolate from Bangladesh in 2010 (Aida et al. 2016). Phylogenetic analysis using tip calibration suggests that the recombination event occurred no later than 2000–5 (node Cis), so if this is a true recombination event, the 2010 sequence is not the original recombinant. The recombinant region is 100 per cent similar to an isolate also from Bangladesh in 2010 (KU248372) (Aida et al. 2016), which also has a putative recombinant sequence in another region of its genome. The breakpoint regions (99% CI: (541–711)—(1,327–1,445)) may represent a potential hotspot for segment 1 recombination as multiple recombination events show breakpoints in this area (Table 2). Phylogenetic analysis of the alignment of 250 representative sequences containing only the putative recombinant region compared with the rest of the segment 1 sequence showed a consistent subclade shift within the R2 clade. The recombination events resulted in the incorporation of the following amino acid substitutions in the recombinant strains when compared to the major parent strain: 227 (K→E), 293 (D → N), 297 (K → R), 305 (N → K), and 350 (K → E).
Figure 2.
Bayesian phylogenetic analysis of a possible recombination event in VP1 that became fixed in the population. The strain closest to the recombination event is highlighted in red. Major clades are colored to show the phylogenetic incongruity. Nodes are labeled with posterior probabilities with 95 per cent node height intervals shown. Time axis at the bottom is in years before 2017.
Figure 3.
(A) An RDP analysis (top) and a BootScan analysis (bottom) showing a putative recombination event between two R2 segment 1 genotypes. The red line compares the minor parent to the recombinant, blue line compares the major parent to the recombinant, and the green line compares the major parent to the minor parent. The Y-axis for RDP (top) is the pairwise identity, while the Y-axis for BootScan (bottom) is the bootstrap support. The X-axis is the sequence along segment 1. (B) The relevant sites shown above color coded to strain that the recombinant matches. The recombinant is the middle sequence, the minor parent is the bottom sequence, and the major parent is the top sequence. Mutations matching the major sequence are shown in blue, while mutations matching the minor parent are shown in purple. Yellow mutations show mutations not present in the recombinant sequence but which match the major and minor parents, possibly suggesting a second recombination event.
3.4 Segment 3 intragenic recombination
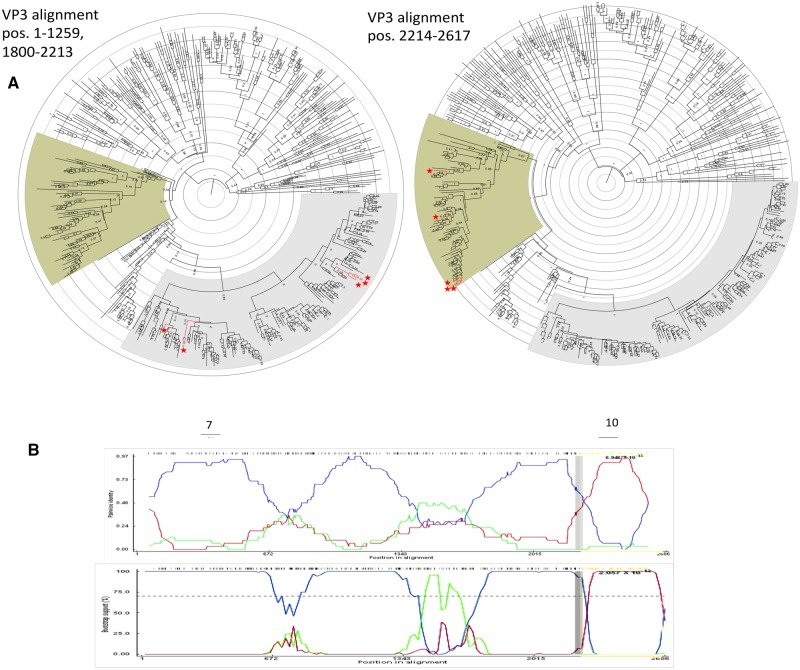
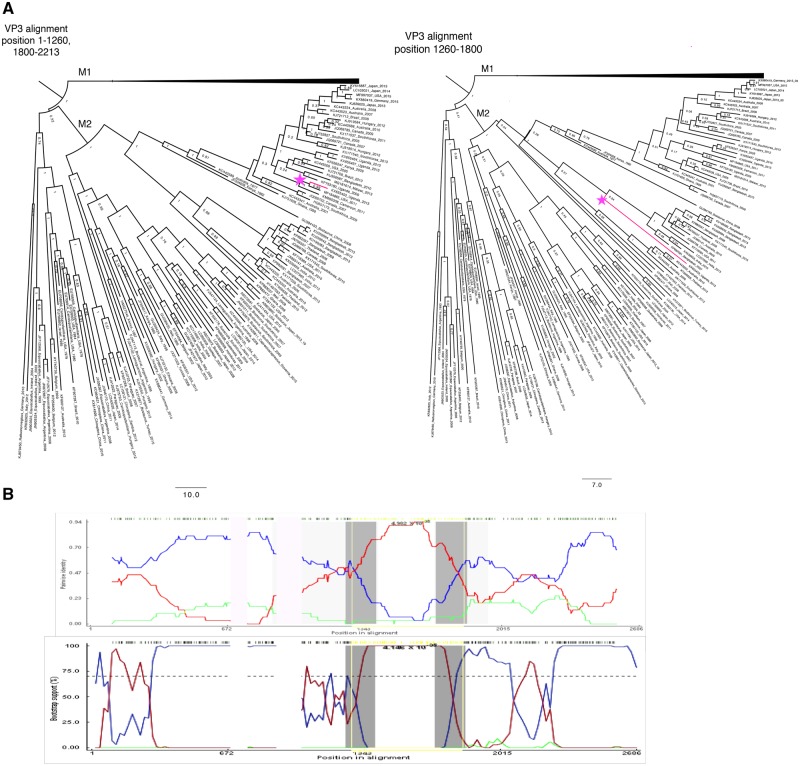
Two recombination events occurring in segment 3 (VP3) appear to have fixed in the population, and are seen in many descendant sequences (Table 2). The first strain identified as a putative recombinant is KJ753665, an isolate from South Africa in 2004. The recombinant region occurs between positions (2129–2174)—2531. This region is 98.7 per cent identical to a porcine strain isolated in Uganda in 2016 (KY055418) (Bwogi et al. 2017), while the rest of segment 3 is 95.8 per cent similar to a 2009 human isolate from Ethiopia (KJ752028). The Monte Carlo uncorrected and corrected probabilities were 2.491E-20 and 6.946E-13, respectively, with 107 isolates flagged as possibly derived from the recombination event. Phylogenetic analysis using 450 randomly selected VP3 sequences (excluding sequences lacking collection dates) within the putative recombinant region, along with an analysis of the genome excluding the two major recombination events, resulted in five sequences showing a significant phylogenetic incongruity (Fig. 4). The incongruity appeared between sublineages within the larger M1 lineage of VP3. Amino acid substitutions in the recombinant region included positions 748 (M → T) and 780 (T → M).
Figure 4.
Segment 3 recombination event supported by multiple isolates. (A) Phylogenetic trees made using alignments from (left) nucleotide positions 1-1259 and 1800-2213 representing the major parent region excluding second recombination event (Fig. 5), and (right) nucleotide position 2214-2617 representing the minor parent sequence. (B) BootScan (top) and RDP (bottom) analyses. The red line compares the minor parent to the recombinant, blue line compares the major parent to the recombinant, and the green line compares the major parent to the minor parent. The Y-axis for the BootScan analysis (top) is the bootstrap support, while the Y-axis for the RDP analysis (bottom) is the pairwise identity. The X-axis for both analyses is the sequence along segment 3.
A second potential recombination event was identified in segment 3 between sublineages within the M2 lineage (Table 2, Fig. 5). However, when we created split alignments and ran a phylogenetic analysis in BEAST v1.10.4, only one sequence showed a phylogenetic incongruity supporting this event (KX655453). Amino acid substitutions because of this event included positions 405 (I → V), 412 (V → M), 414 (N → D), 441 (N → D), 458 (I → V), 459 (I → T), 468 (L → F), 473 (N → D), 486 (M → I), 518 (N → S), and 519 (E → G).
Figure 5.
(A) Phylogenetic analysis of segment 3. Left tree (major parent) was made from an alignment of nucleotide positions 1-1260 and 1900-2213 and right tree (minor parent) was made from nucleotide positions 1260-1800. M2 strains have been collapsed, and recombinant is colored in pink. (B) BootScan (top) and RDP analyses (bottom of VP3 M2 putative recombinant). The red line compares the minor parent to the recombinant, blue line compares the major parent to the recombinant, and the green line compares the major parent to the minor parent. The Y-axis for BootScan is the bootstrap support, while the Y-axis for the RDP analysis is the pairwise identity. The X-axis in both analyses is the segment 3 sequence.
One possible complication with relying on phylogenetic incongruity as evidence of major recombination events is that a recombinant virus may evolve faster than the parental strains, and, despite possessing an initial fitness advantage such as immune avoidance, may eventually converge back toward the major parent’s sequence. Thus, phylogenetic analysis likely underestimates the frequency of transiently stable recombination events in a population. This phenomenon may account for the observation that the number of VP3 isolates flagged as deriving from a recombination event based on sequence analysis is greater than the prevalence of recombination predicted by the phylogenetic analysis.
3.5 Intergenotypic recombination
We found strong evidence for intergenotype recombination in all segments except segments 7 (NSP3) and 11 (NSP5) (Table 1, Supplementary Table S1). Instances of intergenotypic recombination in segment 1 (VP1) (KU714444, JQ988899) only occurred in regions where the amino acid sequence was highly conserved across genotypes, so these events resulted in few, if any, nonsynonymous mutations. Of the putative events observed in more than two environmental isolates with strong support from detection methods, only events in segment 9 (VP7) occurred between different genotypes (Table 2). The segment 9 recombinant region amino acid substitutions that match the minor parent and differ from the major parent are shown in Supplementary Table S2.
3.6 Structural prediction of recombinant proteins
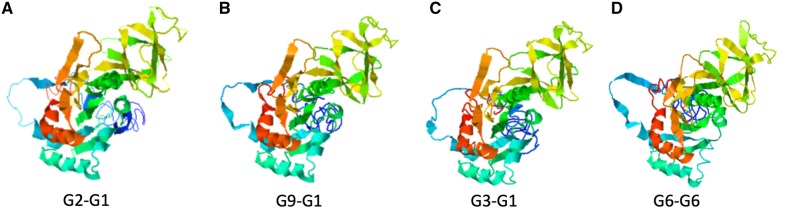
The protein models generated by I-TASSER for the intergenotypic recombinant G-proteins showed that although the amino acid changes for the G1-G2 recombinant were substantial (twenty-five changes) (Supplementary Table S2), the secondary and tertiary structures seemed largely intact (Fig. 6). The G3-G1 and G6-G6 recombinants each showed four amino acid changes. The G9-G1 recombinant showed the most secondary structure disruption, including a loss of beta sheets, a loss of antiparallel beta sheets, and slightly shorter beta sheets/helices, which could indicate lower stability. However, based on the structural modeling, the tertiary structure appeared to be maintained, suggesting that the putative recombinant glycoproteins were able to form properly folded G-proteins (Fig. 6).
Figure 6.
VP7 protein structures were predicted from amino acid alignments of the four strongly supported G recombinants using I-TASSER. C-scores are confidence scores estimating the quality of the predicted model, and range from [−5, 2], with higher scores indicate greater confidence (A) G2-G1, C-score = −1.38, KC443034; (B) G9-G1: AF281044, C-score = −1.29; (C) G3-G1, C-score = −1.27 KJ751729; (D) G6-G6: KF170899, C-score = −1.42.
Antigenic epitope predictions generated by IEDB (Vita et al. 2019) and SVMTriP (Yao et al. 2012), as well as a large study done on mammalian G-types (Ghosh et al. 2012), showed that VP7 recombination occasionally results in amino acid substitutions in conserved epitopes (Supplementary Table S2). For example, the amino acid sequence RVNWKKWWQV is usually flagged as, or part of, an epitope in most G-types including G2, G3, G4, G6, G9, but not in G1. In the G2-G1 recombination event flagged in multiple isolates (Table 2, Fig. 1), this region is altered (sometimes a KR substitution and sometimes multiple amino acid substitutions) (Supplementary Table S2). Moreover, despite containing highly variable regions, the region where the recombination occurred had low solvent accessibility. The G3-G1 recombinant had amino acid substitutions in two of four conserved epitope regions due to the recombination event. There was also a conserved epitope sequence around amino acids 297–316 in G9 proteins that was altered in the G9-G1 recombinant so it no longer appeared as an epitope.
Structural predictions generated by I-TASSER suggest that although the amino acid sequences may diverge, the protein folding and three-dimensional structures remain relatively conserved (Fig. 6). Thus, although the amino acid sequence substitutions do not result in significant changes to the protein structure, they may nevertheless reduce binding by antibodies or T-cell receptors, and may provide a selective advantage in allowing the virus to avoid immune surveillance.
3.7 Recombination junctions often correspond to RNA secondary structure elements
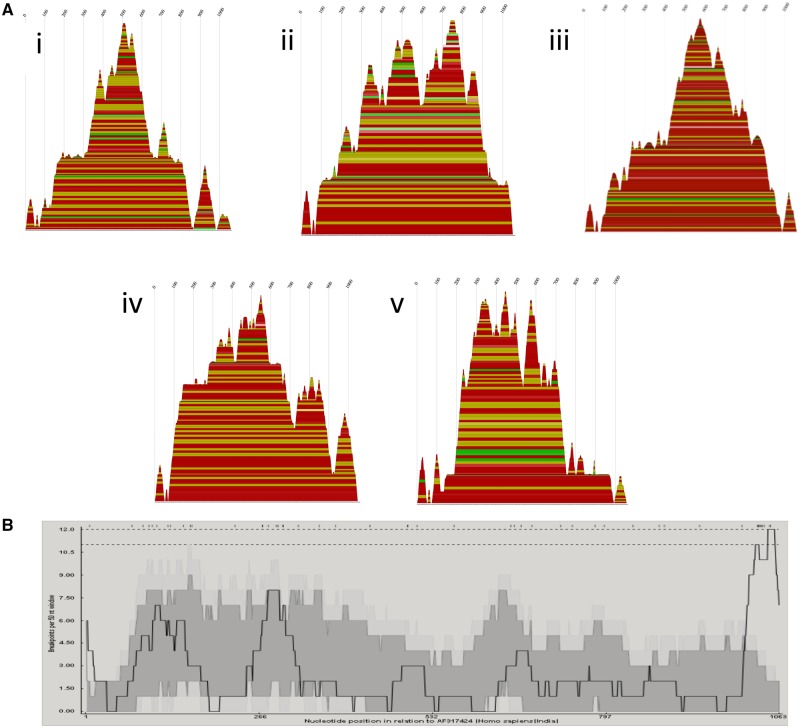
Breakpoint distribution plots showed the sequence regions with the most breakpoints. These breakpoints often corresponded to hairpins predicted by RNAalifold. Secondary RNA structure predictions for segment 9 (VP7) genotypes G1, G2, G6, and G9 are shown as mountain plots (Fig. 7). The breakpoints of the segment 9 recombination events correspond to areas leading to the peaks in the mountain plots (Fig. 7). The peaks indicate a conserved hairpin loop, with the sequences leading up to the peak being the double-stranded portion of the hairpin. Breakpoint distributions also appeared to correspond to secondary structure predictions made from alignments of segments 1 (VP1) and 6 (VP6) (Fig. 8). Segment 4 (VP4) RNA secondary structure predictions (Supplementary Fig. S1) showed greater variation across genotypes, so while breakpoints did coincide with secondary RNA structures as predicted by the models, there were not enough events in each genotype to provide strong support that the breakpoints were correlated with secondary structure. The breakpoint distributions for NSP1 and VP3 were also nonrandomly distributed across the sequence (Supplementary Fig. S1), however, secondary structure predictions from these alignments were not consistent, so are not shown (Supplementary Fig. S2).
Figure 7.
(A) Consensus mountain plots made in RNAalifold using the ViennaRNA package of the predicted RNA secondary structures based on alignments made using full sequences for each of the five segment 9G types involved in the intergenotypic recombination events: (1) G9, (2) G6, (3) G3, (4) G1, and (5) G2. Peaks represent hairpin loops, slopes correspond to helices, and plateaus correspond to loops. The X-axis corresponds to the sequence of the segment. Each base-pairing is represented by a horizontal box where the height of the box corresponds with the thermodynamic likelihood of the pairing. The colors correspond to the variation of base pairings at that position. Red indicates the base pairs are highly conserved across all the sequences, and black indicates the least conservation of those base pairings. (B) Breakpoint distribution plots made in RDP4 of putative recombinants in segment 9. The X-axis shows the position in the sequence, and Y-axis shows the number of breakpoints per 50-nucleotide window. The highest peaks are around X = 115, 285, 1,050.
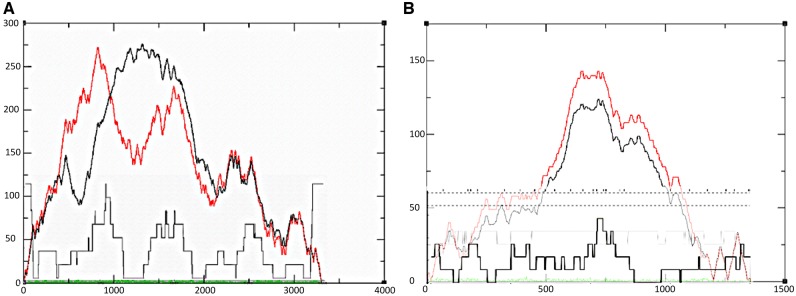
Figure 8.
Consensus mountain plots overlaid with breakpoint distribution plots for recombination events in (A) VP6 and (B) VP1. Position in the nucleotide sequence is on the X-axis. The entropy curve is represented in green. The black curve represents the pairing probabilities, and the red curve represents the minimum free energy structure with well-defined regions having low entropy.
4. Discussion
4.1 Detecting recombination: recognizing type I and type II error
Apparent instances of recombination may actually be the result of convergent evolution, lineage-specific rate variation, sequencing error, poor sequence alignment, laboratory contamination, or improper bioinformatics analysis (Worobey et al. 2002; Boni et al. 2010, 2012; Bertrand et al. 2012). Several steps can be taken to minimize incorrect attribution of viral recombination. Ideally, this process should begin at the time of sequencing. First, it should be confirmed that the originating sample did not come from a host infected with multiple genotypes of the same virus type. Prior to RNA extraction, single plaques should be repeatedly picked and plated (i.e., plaque purification) to ensure that multiple genotypes are not inadvertently sequenced. Similarly, care must be taken when sequencing multiple samples of the same virus to minimize the possibility of cross-contamination.
For sequences obtained from online repositories, such precautions are rarely possible. Instead, careful bioinformatics procedures can help minimize possible errors. As typical first step in identifying recombination events, virus genome sequences are analyzed with software such as RDP4 (Martin et al. 2015), but all software programs are prone to error. For example, programs may falsely identify a recombination event when none exists (type I error) or fail to detect a true recombination event when one exists (type II error). Several studies measured errors incurred by RDP4 in the analysis of the genomes of tick-borne encephalitis virus, a positive-sense RNA flavivirus that rarely recombines (Norberg et al. 2013; Bertrand, Johansson, and Norberg 2016). The results of the analyses indicated that recombination was overestimated in these viruses, and that certain detection methods were more prone to type I error (Norberg et al. 2013; Bertrand, Johansson, and Norberg 2016). MaxChi, Chimaera, and SiScan showed higher false positive rates than other RDP4 programs, but had greater power to detect true recombination events. In contrast, 3Seq and GENECONV displayed lower false positive rates, but had the lowest detection power of true events.
False positives using RDP4 are especially common among closely related strains (Bertrand, Johansson, and Norberg 2016). That said, recombination events are likely occur between closely related strains given their close spatial/temporal proximity and genetic compatibility, so caution should be used in inferring events between highly dissimilar genotypes. When a positive recombination signal has been detected, it is essential to assess its statistical significance. However, in RDP4, the P-value of 0.05 does not correspond to a 5 per cent rate of false positives (Bertrand, Johansson, and Norberg 2016), therefore we used a cut-off value of 10E-04 and focused only on events where at least six RDP4 programs detected the putative recombinant.
After identifying putative recombination events, additional strategies can be used to eliminate errors. For example, rates of type I and type II errors increase with shorter length recombination regions (Boni et al. 2008; Norberg et al. 2013; Bertrand, Johansson, and Norberg 2016). Therefore, we ignored any putative recombination events of <100 nt with the exception of one isolate in segment 4 (NSP4) and one isolate in segment 5 (NSP5) as the putative recombinant regions in these isolates were in conserved regions at the ends of the respective segments (Supplementary Table S1). In addition, we visually inspected sequence alignments to exclude misaligned sequences (Boni et al. 2010). Splitting alignments by major and minor parents followed by carefully parameterized BEAST runs may help distinguish genuine phylogenetic incongruity signals from spurious false positives. Furthermore, we checked for the presence of unique polymorphisms differing from the parent strains within the suspected recombination region as they may provide evidence that recombination events are not laboratory artifacts. Presumably, such substitutions would reflect subsequent adaptive evolution by the recombinant virus.
In addition, we noted how many times the same recombination event occurred across multiple samples since false positive recombinants are likely to be present as single isolates in phylogenetic trees (Boni et al. 2010). The more isolates showing the same event, the greater the probability that it represents a true recombination event, especially if the isolates were acquired and sequenced by different laboratories. Events that showed strong support, but were only isolated in one sequence are noted (Supplementary Table S1), but not discussed, as it is difficult to rule out the possibility of type I error due to PCR or mosaic contig assembly (Boni et al. 2010; Varsani et al. 2018).
Sequence metadata can also be used to identify unlikely recombination events. For an event to be plausible, the major and minor parents should have had opportunity to coinfect the same host, which is only possible if they are congruent in time and space (Boni et al. 2010). For example, one study identified influenza A virus strain A/Taiwan/4845/99 as a recombinant of A/Wellington/24/2000 and A/WSN/33 (He et al. 2008). Given that the two parents were isolated 77 years apart in different parts of the world, it is exceedingly unlikely that is a natural recombination event. Any putative recombination events should be carefully screened to determine if the parental strains could have plausibly interacted. In Supplementary Table S1, we include information on source species, year, and place of isolation, percent average nucleotide identity, and genogroup for all putative recombinants and their major and minor parents.
4.2 Naturally high coinfection in rotavirus A
Some features of rotavirus biology make recombination not only possible, but also relatively plausible. Rotaviruses are often released from cells as aggregates of approximately five to fifteen particles contained within extracellular vesicles (Santiana et al. 2018). While it is not yet clear whether these extracellular vesicles can contain different rotavirus genotypes, they do allow for rotavirus coinfection even at low multiplicities of infection. Thus, the physical barriers to recombination in dsRNA viruses (Lai 1992) may be offset by the high rates of coinfection resulting from vesicle transmission of rotaviruses.
Furthermore, infection of hosts by multiple rotavirus strains appears to be relatively common. In a study of 100 children in the Detroit area, G and P typing, which identifies the serotype of the VP7 and VP4 proteins, respectively, revealed that ∼10 per cent of patients were infected with multiple rotavirus A strains (Abdel-Haq et al. 2003). Similarly high frequencies of G and P mixed genotype infections were observed in children sampled in India (three studies showing multiple G types in 11.3%, 12% and 21% of samples) (Husain et al. 1996; Jain et al. 2001; Khetawat et al. 2002), Spain (>11.4% of samples) (Sánchez-Fauquier et al. 2006), Kenya (5.9%) (Kiulia et al. 2006), Africa (12%) (Mwenda et al. 2010), and Mexico (5.6% in 2010, 33.5% in 2012) (Anaya-Molina et al. 2018). Even higher frequencies of mixed genotype infections were observed in whole genome studies. For example, among thirty-nine Peruvian fecal samples genotyped using multiplexed PCR, thirty-three (84.6%) showed evidence of multiple rotavirus genotypes (Rojas et al. 2019). In another study, whole genome deep sequencing revealed that 15/61 (25%) samples obtained in Kenya contained multiple rotavirus genotypes (Mwanga et al. 2018). Given the high genetic diversity of rotavirus populations (Kirkwood 2010; Ghosh and Kobayashi 2011; Sadiq et al. 2018), and their proficiency in infecting a broad range of mammalian hosts including many domesticated animal species (Martella et al. 2010; Doro et al. 2015), the high frequencies of hosts infected with multiple genotypes are not entirely surprising. These coinfections present abundant opportunities for rotavirus recombination.
4.3 Rotavirus recombination generates genetic diversity
Homologous recombination previously has not been considered a significant driver in rotavirus genetic diversity and evolution (Ramig 1997; Woods 2015). Recombination is usually expected to be deleterious as the breakage of open reading frames may disrupt RNA secondary structure and alter protein functionality (Lai 1992; Simon-Loriere and Holmes 2011). However, recombination, as with reassortment (Ramig and Ward 1991; Iturriza-Gomara et al. 2001; Schumann et al. 2009; Ghosh and Kobayashi 2011; Jere et al. 2018), may further increase rotavirus genetic diversity due to epistatic interactions resulting in reassortant-specific or recombinant-specific mutations (Zeldovich et al. 2015). Formerly deleterious mutations may become beneficial when the genetic background changes, resulting in an increase in circulating pathogenically relevant viral strains.
In our study, most recombination events occurred between strains of the same genotype (Fig. 1, Table 2, Supplementary Table S1). This outcome is consistent with the expectation that intragenotypic is more common since it would less likely to disrupt protein or secondary RNA structure. Nonetheless, intragenotypic recombination can have long lasting effects on rotavirus genetic diversity. For example, we identified a recombinant sublineage within the R2 clade of segment 1, the polymerase-encoding segment (Figs 2 and 3, Table 2). As the same event is found in strains isolated years apart from geographically distant locations, we can infer that the resulting genotype was sufficiently fit enough to be maintained in the population and disperse widely (Supplementary Table S1). This finding suggests that this homologous recombination event has had a long-term effect on rotavirus diversity.
While comparatively less common, we observed instances of intergenotypic recombination in all segments with the exception of segment 7 (NSP3), the only segment where we observed no recombination events (Table 2; Supplementary Table S1). A previous study reported intergenotypic recombination events in segments 6 (VP6), 8 (NSP2), and 10 (NSP4) (Jere et al. 2011), so our study adds to the number of segments able to tolerate intergenotypic recombination. Interestingly, the serotype proteins, VP4 and VP7, have the most different genotypes, with fifty-one and thirty-six, respectively (Steger et al. 2019). Given this genetic diversity, the chances of two viruses with different G or P types coinfecting a cell is substantially higher than other segments. In addition, both VP4 and VP7 seem to be more prone to reassortment, and to tolerate more divergent genetic backgrounds or genome constellations (Martella et al. 2003; Gentsch et al. 2005; McDonald et al. 2009; Patton 2012). This diversity and tolerance of many different genetic backgrounds implies that VP4 and VP7 may be more tolerant of recombination between divergent strains than the other segments. Our data seem to support this claim.
Specifically, we observed numerous instances of intergenotypic recombination in segment 9, the VP7 coding segment (Table 2; Supplementary Table S1). These events appear to be beneficial because the recombinant genotypes persisted in populations long enough for multiple samples showing the same event to be sampled (Table 2). For example, the same mosaic VP7 G6 gene (Fig. 8) was sequenced in multiple bovine strains, one isolated in 2009 (HM591496) and another in 2012 (KF170899), by two separate research groups. While still differentiable, the parents are closely related. However, in some instances, events between highly divergent genotypes seem to have been able to persist in populations. Both a 2014 Malawi isolate (MG181727) and a 2006 isolate from the United States (KC443034) showed a similar G1-G2 VP7 mosaic gene (Fig. 8). The fact that these two G genotypes are highly divergent from one another and were identified in different years in different locations by different research groups supports the contention that it is a true recombination event. Altogether, 22/24 instances of recombination in segment 9 occurred between different G serotypes (Supplementary Table S1). These examples indicate that mosaic genes formed from two divergent genotypes are relatively common and increase the diversity of circulating VP7 genotypes.
In addition, 7/16 instances of recombination in segment 4 (VP4) were intergenotypic. Recombination was observed between P4-P8, P6-P8, and P8-P14 serotypes (Supplementary Table S1). P4, P6, and P8 are all relatively closely related being in the P[II] genogroup, while P14 is in the P[III] genogroup. These putative recombination events suggest that relevant serotype diversity in human hosts is expanding. However, as P4, P6, and P8 are also the dominant P types in human infections, there is a sampling bias toward this genogroup as the genotypes within this group are more likely to coinfect humans, so caution should be taken with this conclusion.
Segment 5 (NSP1) also showed recombination between highly divergent strains. Not only is NSP1 not strictly required for viral replication (Hua et al. 1994), it is also the least conserved of all rotavirus proteins, including even the serotype proteins VP4 and VP7 (Arnold and Patton 2011), suggesting that intergenotypic recombination disrupting the protein’s amino acid sequence may be less likely to be deleterious.
Segments 7 (NSP3), 8 (NSP2), 10 (NSP4), and 11 (NSP5) all had low rates of recombination. These segments are short thus recombination is expected to be less likely, however segment 9 (VP7), one of the smallest segments, defies this pattern, having a high number of events observed. The smaller segments may also be less able to tolerate recombination events due to the important roles they play during the formation and stabilization of the supramolecular RNA complex (Fajardo et al. 2015) during rotavirus packaging and assembly (Li et al. 2010; Suzuki 2015; Borodavka et al. 2017; Fajardo et al. 2017).
4.4 Generation of escape mutants
Recombination involving regions encoding conserved epitopes, especially in segments that encode proteins involved in host cell attachment and entry, may provide selective advantages to rotaviruses by allowing them to evade inactivation by host-produced antibodies. These escape mutants may be generally less fit than wild-type viruses, but competitively advantaged in hosts because of a lack of host recognition. Subsequent intrahost adaptation may then select for compensatory fitness-increasing mutations allowing these strains to be competitive with circulating rotavirus strains. Many of the recombination events observed in our study appeared to generate such escape mutants. For example, we found two instances of the same segment 9 (VP7) G1-G3 recombination event (Table 2; KJ751729 and KP752817) with amino acid changes in two of four conserved epitope regions due to the recombination event, which suggests that this strain prevailed in the population because it was better able to evade antibody neutralization.
Based on the I-TASSER structural predictions, the putative recombinant VP7 proteins detected in our survey appear able to fold properly and form functional proteins despite containing amino acid sequence from different ‘parental’ genotypes (Fig. 6). While the secondary structure appeared slightly altered (e.g., shorter beta sheets), the recombinant VP7 proteins generally maintained their three-dimensional shape. The selective advantage from swapping epitopes may outweigh any potential decrease in protein stability resulting from recombination.
We also identified many recombination events involving segment 4 (VP4). In order to infect cells, VP4 must be proteolytically cleaved to produce VP5* and VP8* (Arias et al. 1996). Most of the segment 4 recombination events involved the spike head of the VP8* protein or the spike body/stalk region of the VP5* protein (antigen domain). Escape mutant studies (Zhou et al. 1994; Ludert et al. 2002; Aoki et al. 2009; Nair et al. 2017) for VP4 show the VP8* spike head recognizes histo-blood group antigens, which is one of rotavirus’s main host range expansion barriers (Huang et al. 2012; Hu et al. 2018; Lee et al. 2018). VP5* mediates membrane penetration during cell entry (Yoder and Dormitzer 2006). VP4 recombinants therefore may help the virus expand host range and aid in immune evasion.
Segment 6 (VP6) also showed recombination events resulting in substantial amino acid changes. VP6 is a more conserved protein, but also an antigenic protein that interacts with naïve B cells (Parez et al. 2004). This feature suggests that there may be selection for VP6 escape mutants to evade host immune responses. Structure analyses of VP6 indicated that it is relatively conserved across genotypes (Jiang et al. 1992; Tang et al. 1997; Charpilienne et al. 2002), which may explain why VP6 seems to have more frequent intergenotypic recombination. Conserved epitopes in VP6 exist around amino acid positions 197–214 and 308–316 (Aiyegbo et al. 2014).
4.5 Recombination in other dsRNA viruses
Recombination has had a significant impact on the diversity of other dsRNA reoviruses (He et al. 2010). One study of 692 complete bluetongue virus segments found evidence for at least eleven unique recombinant genotypes (1.6%) (He et al. 2010). The case for recombination among bluetongue viruses is strengthened by the fact that viruses containing the same (or similar) recombinant segments were isolated by different research groups in different countries at different times, indicating that the recombinant viruses persisted and spread following the recombination event (Carpi et al. 2010; He et al. 2010). Another study found multiple possible instances of recombination in genome segment 8 (encoding NS2) of the epizootic hemorrhagic disease virus, a reovirus similar to bluetongue virus (Anthony et al. 2009). Several studies have reported recombination among the dsRNA rice black-streaked dwarf virus, which is also a member of the Reoviridae family, but infects plants (Li et al. 2013; Yin et al. 2013). Putative recombinants were identified in six of the ten Southern rice black-streaked dwarf virus segments (Li et al. 2013; Yin et al. 2013). Finally, intragenic recombination was observed in multiple isolates of the African horse sickness virus, an Oribivirus of the family Reoviridae (Ngoveni et al. 2019). At least one of these events appeared in multiple subsequent lineages (Ngoveni et al. 2019).
In study of the family Birnaviridae, 1,881 sequences were analyzed for evidence of recombination (Hon et al. 2008). While no interspecies recombination was observed, at least eight putative instances of intraspecies recombination were observed among the infectious bursal disease viruses and the aquabirnaviruses (Hon et al. 2008). Subsequent studies focusing on the infectious bursal disease viruses supported these results, and identified additional potential recombination events (He et al. 2009; Jackwood 2012; Vukea et al. 2014). We note that birnaviruses’ genetic material is in a complex with ribonucleoprotein, while the genetic material of Reoviridae members is free in the virion, which could be a factor in differing rates of recombination across these dsRNA viruses.
Recombination has also been observed in dsRNA mycoviruses, including in the Partitiviridae (Botella et al. 2015) and the Hypoviridae (Carbone et al. 2004; Linder-Basso et al. 2005; Feau et al. 2014) and the Totiviridae (Voth et al. 2006). Recombination in Gammapartititvirus, which infects the fungus Gremmeniella abietina, may have permitted the virus to cross species borders (Botella et al. 2015). In cryphonectria hypovirus 1, which infects chestnut blight, recombination was implicated in the spread of the virus in Europe (Feau et al. 2014). Collectively, these studies, and those of other dsRNA families, suggest that not only is recombination possible, but also significantly impacts virus evolution. There is little doubt that this conclusion will be strengthened as more dsRNA viruses are discovered and/or sequenced.
4.6 Possible mechanism of recombination in rotavirus
The precise mechanism for how rotavirus recombination occurs is unknown, but inferences can be made because many of the details regarding rotavirus replication and packaging have been resolved (McDonald and Patton 2011; Borodavka et al. 2018). The most-accepted hypothesis is that recombination takes place when the rotavirus +ssRNA is replicated after being packaged in the nucleocapsid (Esona et al. 2017; Jing et al. 2018). For packaging and replication to occur, the eleven +ssRNA segments must join a protein complex consisting of the VP1 polymerase and the VP3 capping enzyme. Secondary RNA structures in the nontranslated terminal regions (NTRs) aid in the formation of this supramolecular RNA complex (Fajardo et al. 2015; Borodavka et al. 2017) and determine whether the segments are packaged (Li et al. 2010; Suzuki 2015). Perhaps recombination occurs when multiple homologous RNA strands are joined in the same complex, allowing the homologous NTRs to partially hybridize. In this scenario, the VP1 polymerase replicates part of one strand before switching to the other, thus producing a recombined segment. This conjecture is supported by the fact that recombination tends to occur in segment regions where self-hybridization forms three-dimensional structures. Moreover, rotavirus do not seem constrained from packaging extra genetic material (Desselberger 1996). A similar form of template switching is seen in poliovirus, an ssRNA virus, which exhibits high rates of recombination, although the precise mechanism may be different in that case insofar as in poliovirus, the polymerase may be stalled due to a hairpin or other secondary structure, and switches to a different template (Tolskaya et al. 1987). Further study is needed to determine the precise mechanism of recombination in rotavirus.
Supplementary Material
Acknowledgements
We are grateful for helpful discussions with members of the Dennehy Lab. We are also grateful to Thomas Hoxie for editing drafts of this article. Comments from two anonymous reviewers significantly improved this manuscript. All genomic data used in this article are available from NCBI GenBank and accession numbers are provided in the Supplementary Material. The research presented in this article was not funded.
Conflict of interest: None declared.
References
- Abdel-Haq N. M. et al. (2003) ‘Increased Prevalence of G1P[4] Genotype among Children with Rotavirus-Associated Gastroenteritis in Metropolitan Detroit’, Journal of Clinical Microbiology, 41: 2680–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida S. et al. (2016) ‘Whole Genomic Analysis of G2P[4] Human Rotaviruses in Mymensingh, North-Central Bangladesh’, Heliyon, 2: e00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyegbo M. S. et al. (2014) ‘Differential Accessibility of a Rotavirus VP6 Epitope in Trimers Comprising Type I, II, or III Channels as Revealed by Binding of a Human Rotavirus VP6-Specific Antibody’, Journal of Virology, 88: 469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaya-Molina Y. et al. (2018) ‘A One-Step Real-Time RT-PCR Helps to Identify Mixed Rotavirus Infections in Mexico’, Diagnostic Microbiology and Infectious Disease, 92: 288–93. [DOI] [PubMed] [Google Scholar]
- Anthony S. J. et al. (2009) ‘Genetic and Phylogenetic Analysis of the Non-Structural Proteins NS1, NS2 and NS3 of Epizootic Haemorrhagic Disease Virus (EHDV)’, Virus Research, 145: 211–9. [DOI] [PubMed] [Google Scholar]
- Aoki S. T. et al. (2009) ‘Structure of Rotavirus Outer-Layer Protein VP7 Bound with a Neutralizing Fab’, Science, 324: 1444–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C. F. et al. (1996) ‘Trypsin Activation Pathway of Rotavirus Infectivity’, Journal of Virology, 70: 5832–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M. M., Patton J. T. (2011) ‘Diversity of Interferon Antagonist Activities Mediated by NSP1 Proteins of Different Rotavirus Strains’, Journal of Virology, 85: 1970–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai K. et al. (2017) ‘Candidate New Rotavirus Species in Schreiber's Bats, Serbia’, Infection, Genetics and Evolution, 48: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart S. H. et al. (2008) ‘RNAalifold: Improved Consensus Structure Prediction for RNA Alignments’, BMC Bioinformatics, 9: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand Y. et al. (2012) ‘First Dating of a Recombination Event in Mammalian Tick-Borne Flaviviruses’, PLoS One, 7: e31981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand Y. J. K., Johansson M., Norberg P. (2016) ‘Revisiting Recombination Signal in the Tick-Borne Encephalitis Virus: A Simulation Approach’, PLoS One, 11: e0164435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M. F. et al. (2008) ‘Homologous Recombination is Very Rare or Absent in Human Influenza A Virus’, Journal of Virology, 82: 4807–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M. F. et al. (2010) ‘Guidelines for Identifying Homologous Recombination Events in Influenza a Virus’, PLoS One, 5: e10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M. F. et al. (2012) ‘No Evidence for Intra-Segment Recombination of 2009 H1N1 Influenza Virus in Swine’, Gene, 494: 242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodavka A. et al. (2017) ‘Protein-Mediated RNA Folding Governs Sequence-Specific Interactions between Rotavirus Genome Segments’, eLife, 6: e27453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodavka A., Desselberger U., Patton J. T. (2018) ‘Genome Packaging in Multi-Segmented dsRNA Viruses: Distinct Mechanisms with Similar Outcomes’, Current Opinion in Virology, 33: 106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella L. et al. (2015) ‘The European Race of Gremmeniella abietina Hosts a Single Species of Gammapartitivirus Showing a Global Distribution and Possible Recombinant Events in Its History’, Fungal Biology, 119: 125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bwogi J. et al. (2017) ‘Whole Genome Analysis of Selected Human and Animal Rotaviruses Identified in Uganda from 2012 to 2014 Reveals Complex Genome Reassortment Events between Human, Bovine, Caprine and Porcine Strains’, PLoS One, 12: e0178855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Barro M., Hoshino Y. (2008) ‘Porcine Rotavirus Bearing an Aberrant Gene Stemming from an Intergenic Recombination of the NSP2 and NSP5 Genes is Defective and Interfering’, Journal of Virology, 82: 6073–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I. et al. (2004) ‘Recombination and Migration of Cryphonectria Hypovirus 1 as Inferred from Gene Genealogies and the Coalescent’, Genetics, 166: 1611–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpi G., Holmes E. C., Kitchen A. (2010) ‘The Evolutionary Dynamics of Bluetongue Virus’, Journal of Molecular Evolution, 70: 583–92. [DOI] [PubMed] [Google Scholar]
- Charpilienne A. et al. (2002) ‘Identification of Rotavirus VP6 Residues Located at the Interface with VP2 That Are Essential for Capsid Assembly and Transcriptase Activity’, Journal of Virology, 76: 7822–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U. (1996) ‘Genome Rearrangements of Rotaviruses’, Advances in Virus Research, 46: 69–95. [DOI] [PubMed] [Google Scholar]
- Desselberger U. (2014) ‘Rotaviruses’, Virus Research, 190: 75–96. [DOI] [PubMed] [Google Scholar]
- Donker N. C., Kirkwood C. D. (2012) ‘Selection and Evolutionary Analysis in the Nonstructural Protein NSP2 of Rotavirus A’, Infection, Genetics and Evolution, 12: 1355–61. [DOI] [PubMed] [Google Scholar]
- Donker N. C., Boniface K., Kirkwood C. D. (2011) ‘Phylogenetic Analysis of Rotavirus A NSP2 Gene Sequences and Evidence of Intragenic Recombination’, Infection, Genetics and Evolution, 11: 1602–7. [DOI] [PubMed] [Google Scholar]
- Doro R. et al. (2015) ‘Zoonotic Transmission of Rotavirus: Surveillance and Control’, Expert Review of Anti-Infective Therapy, 13: 1337–50. [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007) ‘BEAST: Bayesian Evolutionary Analysis by Sampling Trees’, BMC Evolutionary Biology, 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona M. D. et al. (2017) ‘Characterization of a Triple-Recombinant, Reassortant Rotavirus Strain from the Dominican Republic’, Journal of General Virology, 98: 134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo T. Jr., Sung P. Y., Roy P. (2015) ‘Disruption of Specific RNA–RNA Interactions in a Double-Stranded RNA Virus Inhibits Genome Packaging and Virus Infectivity’, PLoS Pathogens, 11: e1005321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo T. et al. (2017) ‘Rotavirus Genomic RNA Complex Forms via Specific RNA–RNA Interactions: Disruption of RNA Complex Inhibits Virus Infectivity’, Viruses, 9: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feau N. et al. (2014) ‘Multiple Introductions and Recombination in Cryphonectria Hypovirus 1: Perspective for a Sustainable Biological Control of Chestnut Blight’, Evolutionary Applications, 7: 580–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch J. R. et al. (2005) ‘Serotype Diversity and Reassortment between Human and Animal Rotavirus Strains: Implications for Rotavirus Vaccine Programs’, The Journal of Infectious Diseases, 192(Suppl. 1): S146–59. [DOI] [PubMed] [Google Scholar]
- Ghosh A. et al. (2012) ‘In Silico Study of Rotavirus VP7 Surface Accessible Conserved Regions for Antiviral Drug/Vaccine Design’, PLoS One, 7: e40749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Kobayashi N. (2011) ‘Whole-Genomic Analysis of Rotavirus Strains: Current Status and Future Prospects’, Future Microbiology, 6: 1049–65. [DOI] [PubMed] [Google Scholar]
- Hatcher E. L. et al. (2017) ‘Virus Variation Resource - Improved Response to Emergent Viral Outbreaks’, Nucleic Acids Research, 45: D482–d490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Q. et al. (2008) ‘Homologous Recombination Evidence in Human and Swine Influenza a Viruses’, Virology, 380: 12–20. [DOI] [PubMed] [Google Scholar]
- He C.-Q. et al. (2009) ‘Homologous Recombination is Apparent in Infectious Bursal Disease Virus’, Virology, 384: 51–8. [DOI] [PubMed] [Google Scholar]
- He C. Q. et al. (2010) ‘Intragenic Recombination as a Mechanism of Genetic Diversity in Bluetongue Virus’, Journal of Virology, 84: 11487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C. C. et al. (2008) ‘Phylogenetic Evidence for Homologous Recombination within the Family Birnaviridae’, Journal of General Virology, 89: 3156–64. [DOI] [PubMed] [Google Scholar]
- Hu L. et al. (2018) ‘Glycan Recognition in Globally Dominant Human Rotaviruses’, Nature Communications, 9: 2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Chen X., Patton J. T. (1994) ‘Deletion Mapping of the Rotavirus Metalloprotein NS53 (NSP1): The Conserved Cysteine-Rich Region Is Essential for Virus-Specific RNA Binding’, Journal of Virology, 68: 3990–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. et al. (2012) ‘Spike Protein VP8* of Human Rotavirus Recognizes Histo-Blood Group Antigens in a Type-Specific Manner’, Journal of Virology, 86: 4833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M. et al. (1996) ‘Classification of Rotavirus into G and P Types with Specimens from Children with Acute Diarrhea in New Delhi, India’, Journal of Clinical Microbiology, 34: 1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gomara M. et al. (2001) ‘Reassortment In Vivo: Driving Force for Diversity of Human Rotavirus Strains Isolated in the United Kingdom between 1995 and 1999’, Journal of Virology, 75: 3696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood D. J. (2012) ‘Molecular Epidemiologic Evidence of Homologous Recombination in Infectious Bursal Disease Viruses’, Avian Diseases, 56: 574–7. [DOI] [PubMed] [Google Scholar]
- Jain V. et al. (2001) ‘Great Diversity of Group a Rotavirus Strains and High Prevalence of Mixed Rotavirus Infections in India’, Journal of Clinical Microbiology, 39: 3524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jere K. C. et al. (2011) ‘Whole Genome Analysis of Multiple Rotavirus Strains from a Single Stool Specimen Using Sequence-Independent Amplification and 454(R) Pyrosequencing Reveals Evidence of Intergenotype Genome Segment Recombination’, Infection, Genetics and Evolution, 11: 2072–82. [DOI] [PubMed] [Google Scholar]
- Jere K. C. et al. (2018) ‘Emergence of Double- and Triple-Gene Reassortant G1P 8 Rotaviruses Possessing a DS-1-Like Backbone after Rotavirus Vaccine Introduction in Malawi’, Journal of Virology, 92: e01246‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B. et al. (1992) ‘Nucleotide Sequence of Gene 5 Encoding the Inner Capsid Protein (VP6) of Bovine Group C Rotavirus: Comparison with Corresponding Genes of Group C, A, and B Rotaviruses’, Virology, 190: 542–7. [DOI] [PubMed] [Google Scholar]
- Jing Z. et al. (2018) ‘A G3P 13 Porcine Group a Rotavirus Emerging in China is a Reassortant and a Natural Recombinant in the VP4 Gene’, Transboundary and Emerging Diseases, 65: e317–e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetawat D. et al. (2002) ‘Distribution of Rotavirus VP7 Genotypes among Children Suffering from Watery Diarrhea in Kolkata, India’, Virus Research, 87: 31–40. [DOI] [PubMed] [Google Scholar]
- Kirkwood C. D. (2010) ‘Genetic and Antigenic Diversity of Human Rotaviruses: Potential Impact on Vaccination Programs’, The Journal of Infectious Diseases, 202: S43–S48. [DOI] [PubMed] [Google Scholar]
- Kiulia N. M. et al. (2006) ‘Molecular Characterisation of the Rotavirus Strains Prevalent in Maua, Meru North, Kenya’, East African Medical Journal, 83: 360–5. [DOI] [PubMed] [Google Scholar]
- Lai M. M. (1992) ‘RNA Recombination in Animal and Plant Viruses’, Microbiological Reviews, 56: 61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. et al. (2018) ‘Histo-Blood Group Antigen Phenotype Determines Susceptibility to Genotype-Specific Rotavirus Infections and Impacts Measures of Rotavirus Vaccine Efficacy’, The Journal of Infectious Diseases, 217: 1399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. (2010) ‘Genomic Analysis of Codon, Sequence and Structural Conservation with Selective Biochemical-Structure Mapping Reveals Highly Conserved and Dynamic Structures in Rotavirus RNAs with Potential Cis-Acting Functions’, Nucleic Acids Research, 38: 7718–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. (2013) ‘Evidence of Recombination and Genetic Diversity in Southern Rice Black-Streaked Dwarf Virus’, Archives of Virology, 158: 2147–51. [DOI] [PubMed] [Google Scholar]
- Linder-Basso D., Dynek J. N., Hillman B. I. (2005) ‘Genome Analysis of Cryphonectria Hypovirus 4, the Most Common Hypovirus Species in North America’, Virology, 337: 192–203. [DOI] [PubMed] [Google Scholar]
- Lorenz R. et al. (2011) ‘ViennaRNA Package 2.0’, Algorithms for Molecular Biology, 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludert J. E. et al. (2002) ‘Antibodies to Rotavirus Outer Capsid Glycoprotein VP7 Neutralize Infectivity by Inhibiting Virion Decapsidation’, Journal of Virology, 76: 6643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev A. N. (2005) ‘Role of Recombination in Evolution of Enteroviruses’, Reviews in Medical Virology, 15: 157–67. [DOI] [PubMed] [Google Scholar]
- Martella V. et al. (2003) ‘Molecular Analysis of the VP7, VP4, VP6, NSP4, and NSP5/6 Genes of a Buffalo Rotavirus Strain: Identification of the Rare P[3] Rhesus Rotavirus-like VP4 Gene Allele’, Journal of Clinical Microbiology, 41: 5665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V. et al. (2010) ‘Zoonotic Aspects of Rotaviruses’, Veterinary Microbiology, 140: 246–55. [DOI] [PubMed] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Laso J. et al. (2009) ‘Diversity of the G3 Genes of Human Rotaviruses in Isolates from Spain from 2004 to 2006: Cross-Species Transmission and Inter-Genotype Recombination Generates Alleles’, Journal of General Virology, 90: 935–43. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J. et al. (2008a) ‘Full Genome-Based Classification of Rotaviruses Reveals a Common Origin between Human Wa-Like and Porcine Rotavirus Strains and Human DS-1-Like and Bovine Rotavirus Strains’, Journal of Virology, 82: 3204–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J. et al. (2008b) ‘Recommendations for the Classification of Group a Rotaviruses Using All 11 Genomic RNA Segments’, Archives of Virology, 153: 1621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J. et al. (2009) ‘Rotavirus Disease and Vaccination: Impact on Genotype Diversity’, Future Microbiology, 4: 1303–16. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J. et al. (2010) ‘Phylodynamic Analyses of Rotavirus Genotypes G9 and G12 Underscore Their Potential for Swift Global Spread’, Molecular Biology and Evolution, 27: 2431–6. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J. et al. (2011) ‘Uniformity of Rotavirus Strain Nomenclature Proposed by the Rotavirus Classification Working Group (RCWG)’, Archives of Virology, 156: 1397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J. et al. (2012) ‘VP6-Sequence-Based Cutoff Values as a Criterion for Rotavirus Species Demarcation’, Archives of Virology, 157: 1177–82. [DOI] [PubMed] [Google Scholar]
- McDonald S. M., Patton J. T. (2011) ‘Assortment and Packaging of the Segmented Rotavirus Genome’, Trends in Microbiology, 19: 136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald S. M. et al. (2009) ‘Evolutionary Dynamics of Human Rotaviruses: Balancing Reassortment with Preferred Genome Constellations’, PLoS Pathogens, 5: e1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald S. M. et al. (2016) ‘Reassortment in Segmented RNA Viruses: Mechanisms and Outcomes’, Nature Reviews Microbiology, 14: 448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalov-Kovacs E. et al. (2015) ‘Candidate New Rotavirus Species in Sheltered Dogs, Hungary’, Emerging Infectious Disease, 21: 660–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V. N., Bloomquist E. W., Suchard M. A. (2008) ‘Smooth Skyride through a Rough Skyline: Bayesian Coalescent-Based Inference of Population Dynamics’, Molecular Biology and Evolution, 25: 1459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwanga M. et al. (2018) ‘Unbiased Whole Genome Deep Sequencing of Rotavirus Group a Positive Samples from Rural Kenya, 2012–14 Reveals High Frequency of Coinfection and Genetic Reassortment’, International Journal of Infectious Diseases, 73: 203. [Google Scholar]
- Mwenda J. M. et al. (2010) ‘Burden and Epidemiology of Rotavirus Diarrhea in Selected African Countries: Preliminary Results from the African Rotavirus Surveillance Network’, The Journal of Infectious Diseases, 202: S5–S11. [DOI] [PubMed] [Google Scholar]
- Nair N. et al. (2017) ‘VP4- AND VP7-Specific Antibodies Mediate Heterotypic Immunity to Rotavirus in Humans’, Science Translational Medicine, 9: eaam5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoveni H. G., van Schalkwyk A., Koekemoer J. J. O. (2019) ‘Evidence of Intragenic Recombination in African Horse Sickness Virus’, Viruses, 11: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg P., Roth A., Bergström T. (2013) ‘Genetic Recombination of Tick-Borne Flaviviruses among Wild-Type Strains’, Virology, 440: 105–16. [DOI] [PubMed] [Google Scholar]
- Parez N. et al. (2004) ‘The VP6 Protein of Rotavirus Interacts with a Large Fraction of Human Naive B Cells via Surface Immunoglobulins’, Journal of Virology, 78: 12489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G. I. et al. (2004) ‘Evidence of Rotavirus Intragenic Recombination between Two Sublineages of the Same Genotype’, Journal of General Virology, 85: 1713–6. [DOI] [PubMed] [Google Scholar]
- Patton J. T. (2012) ‘Rotavirus Diversity and Evolution in the Post-Vaccine World’, Discovery Medicine, 13: 85–97. [PMC free article] [PubMed] [Google Scholar]
- Patton J. T. et al. (2007) ‘Coupling of Rotavirus Genome Replication and Capsid Assembly’, Advances in Virus Research, 69: 167–201. [DOI] [PubMed] [Google Scholar]
- Pérez-Losada M. et al. (2015) ‘Recombination in Viruses: Mechanisms, Methods of Study, and Evolutionary Consequences’, Infection, Genetics and Evolution, 30: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. G. et al. (2007a) ‘Evidence of Intragenic Recombination in G1 Rotavirus VP7 Genes’, Journal of Virology, 81: 10188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. G. et al. (2007b) ‘Genetic Heterogeneity, Evolution and Recombination in Emerging G9 Rotaviruses’, Infection, Genetics and Evolution, 7: 656–63. [DOI] [PubMed] [Google Scholar]
- Rahman M. et al. (2007) ‘Evolutionary History and Global Spread of the Emerging G12 Human Rotaviruses’, Journal of Virology, 81: 2382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2019) Figtree v1.4.4 <http://tree.bio.ed.ac.uk/software/figtree/> accessed 5 Dec 2019.
- Rambaut A. et al. (2018) ‘Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7’, Systematic Biology, 67: 901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig R. F. (1997) ‘Genetics of the Rotaviruses’, Annual Review of Microbiology, 51: 225–55. [DOI] [PubMed] [Google Scholar]
- Ramig R. F., Ward R. L. (1991) ‘Genomic Segment Reassortment in Rotaviruses and Other Reoviridae’, Advances in Virus Research, 39: 163–207. [DOI] [PubMed] [Google Scholar]
- Rojas M. et al. (2019) ‘Genetic Diversity and Zoonotic Potential of Rotavirus A Strains in the Southern Andean Highlands, Peru’, Transboundary and Emerging Diseases, 66: 1718–26. [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010) ‘I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction’, Nature Protocols, 5: 725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiq A. et al. (2018) ‘Rotavirus: Genetics, Pathogenesis and Vaccine Advances’, Reviews in Medical Virology, 28: e2003. [DOI] [PubMed] [Google Scholar]
- Sánchez-Fauquier A. et al. (2006) ‘Human Rotavirus G9 and G3 as Major Cause of Diarrhea in Hospitalized Children, Spain’, Emerging Infectious Diseases, 12: 1536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiana M. et al. (2018) ‘Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-Organismal Viral Transmission’, Cell Host & Microbe, 24: 208–20.e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann T. et al. (2009) ‘Evidence of Interspecies Transmission and Reassortment among Avian Group a Rotaviruses’, Virology, 386: 334–43. [DOI] [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E. C. (2011) ‘Why Do RNA Viruses Recombine?’, Nature Reviews Microbiology, 9: 617–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger C. L. et al. (2019) ‘Group a Rotavirus VP1 Polymerase and VP2 Core Shell Proteins: Intergenotypic Sequence Variation and In Vitro Functional Compatibility’, Journal of Virology, 93: e01642–01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M. A. et al. (2018) ‘Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10’, Virus Evolution, 4: vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. (2015) ‘A Candidate Packaging Signal of Human Rotavirus Differentiating Wa-Like and DS-1-Like Genomic Constellations’, Microbiology and Immunology, 59: 567–71. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Gojobori T., Nakagomi O. (1998) ‘Intragenic Recombinations in Rotaviruses’, FEBS Letters, 427: 183–7. [DOI] [PubMed] [Google Scholar]
- Tang B. et al. (1997) ‘Comparison of the Rotavirus Gene 6 from Different Species by Sequence Analysis and Localization of Subgroup-Specific Epitopes Using Site-Directed Mutagenesis’, Virology, 237: 89–96. [DOI] [PubMed] [Google Scholar]
- Tolskaya E. A. et al. (1987) ‘Studies on the Recombination between RNA Genomes of Poliovirus: The Primary Structure and Nonrandom Distribution of Crossover Regions in the Genomes of Intertypic Poliovirus Recombinants’, Virology, 161: 54–61. [DOI] [PubMed] [Google Scholar]
- Varsani A. et al. (2018) ‘Notes on Recombination and Reassortment in Multipartite/Segmented Viruses’, Current Opinion in Virology, 33: 156–66. [DOI] [PubMed] [Google Scholar]
- Vita R. et al. (2019) ‘The Immune Epitope Database (IEDB): 2018 Update’, Nucleic Acids Research, 47: D339–d343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth P. D. et al. (2006) ‘Phylogeography of Ustilago maydis Virus H1 in the USA and Mexico’, Journal of General Virology, 87: 3433–41. [DOI] [PubMed] [Google Scholar]
- Vukea P. R. et al. (2014) ‘Phylogenetic Analysis of the Polyprotein Coding Region of an Infectious South African Bursal Disease Virus (IBDV) Strain’, Infection, Genetics and Evolution, 21: 279–86. [DOI] [PubMed] [Google Scholar]
- Woods R. J. (2015) ‘Intrasegmental Recombination Does Not Contribute to the Long-Term Evolution of Group a Rotavirus’, Infection, Genetics and Evolution, 32: 354–60. [DOI] [PubMed] [Google Scholar]
- Worobey M. et al. (2002) ‘Questioning the Evidence for Genetic Recombination in the 1918 “Spanish Flu” Virus’, Science, 296: 211a–211, discussion 211. [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang Y. (2015) ‘Protein Structure and Function Prediction Using I-TASSER’, Current Protocols in Bioinformatics, 52: 5.8.1–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B. et al. (2012) ‘SVMTriP: A Method to Predict Antigenic Epitopes Using Support Vector Machine to Integrate Tri-Peptide Similarity and Propensity’, PLoS One, 7: e45152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X. et al. (2013) ‘Genetic Structure of Rice Black-Streaked Dwarf Virus Populations in China’, Archives of Virology, 158: 2505–15. [DOI] [PubMed] [Google Scholar]
- Yoder J. D., Dormitzer P. R. (2006) ‘Alternative Intermolecular Contacts Underlie the Rotavirus VP5* Two- to Three-Fold Rearrangement’, The EMBO Journal, 25: 1559–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich K. B. et al. (2015) ‘Positive Selection Drives Preferred Segment Combinations during Influenza Virus Reassortment’, Molecular Biology and Evolution, 32: 1519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008) ‘I-TASSER Server for Protein 3D Structure Prediction’, BMC Bioinformatics, 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. J. et al. (1994) ‘Localization of Rotavirus VP4 Neutralization Epitopes Involved in Antibody-Induced Conformational Changes of Virus Structure’, Journal of Virology, 68: 3955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.