Abstract
Bovine tuberculosis (TB) is a zoonotic disease, mainly caused by Mycobacterium bovis. France was declared officially TB free in 2001, however, the disease persists in livestock and wildlife. Among wild animals, deer are particularly susceptible to bovine TB. Here, a whole genome sequence (WGS) analysis was performed on strains with the same genetic profile—spoligotype SB0121, Multiple Loci VNTR Analysis (MLVA) 6 4 5 3 11 2 5 7—isolated from different types of outbreaks, including from deer or cattle herds, or zoological or hunting parks where the presence of infected deer was a common trait in most of them. The results of the phylogeny based on the SNP calling shows that two sub-clusters co-exist in France, one related to deer bred to be raised as livestock, and the other to hunting parks and zoos. The persistence over almost 30 years of sporadic cases due to strains belonging to these clusters highlights the deficiency in the surveillance of captive wildlife and the need for better monitoring of animals, especially before movement between parks or herds.
Keywords: bovine tuberculosis, Mycobacterium bovis, deer herd, animal park, WGS, SNP calling
1. Introduction
Bovine tuberculosis (bTB), mainly caused by Mycobacterium bovis, is an important re-emergent zoonotic disease in Europe [1]. Although France has been officially bTB free (OTF) since 2001, the persistence of the disease in livestock and its occurrence in some areas of wildlife are matters of great concern [2]. This ancient disease is no longer considered just a cattle-borne problem, but a concern for multihost communities that include wildlife species such as wild boar (Sus scrofa), red deer (Cervus elaphus), and badgers (Meles meles) [3].
In Europe, red and fallow deer (Dama dama) are frequently reported to be infected by M. bovis or M. caprae [4]. In the wild population, red deer are considered to play an important role in the dissemination of the disease as an important number of infected animals disclose generalized patterns of clinical tuberculosis [5]. Farmed deer are also very susceptible to M. bovis, as a high incidence of animals with generalized infection is commonly observed in deer herds [6]. Gross lesions are generally observed in lymph nodes of the head, thorax and abdomen [4]; most frequently in the retropharyngeal lymph nodes for red deer, and the thoracic region for fallow deer [7].
Interaction among different wildlife species, and between wildlife and human beings, may now be more frequent due to the increasing amount of captive wildlife in zoos or other types of animal parks [8]. The zoonotic transmission of Mycobacterium tuberculosis complex (MTBC) pathogens has been reported worldwide: e.g., an M. bovis infection in zookeepers exposed to a white rhinoceros [9], an M. pinnipedii infection in animal handlers linked to a sea lion colony [10], and an M. tuberculosis infection of staff members exposed to elephants [11,12]. Infrastructure where the public has potential contact with captive wildlife is a serious emerging concern for zoonotic transmission, especially for immunocompromised individuals [8]. The same concern arose after the discovery in an Iranian zoo of an M. bovis infection in free-roaming fallow deer that had been in close contact with the public [13].
MTBC members are excellent microorganisms for conducting molecular epidemiology studies on, by virtue of their low mutation rate nature. In the case of bTB, two genotyping methods are widely used—spoligotyping [14] and Multiple Loci VNTR Analysis (MLVA) typing [15]—both of which establish differentiation between M. bovis strains, taking into account relative genetic stability. The combination of these two techniques allows for a very fine differentiation of strains. In France, 497 different genotypes could be identified between 1978 and 2013, in a collection of M. bovis strains isolated from cattle and wild animals [2,16]. The vast diversity of the molecular profiles obtained by combining these two techniques makes it possible, by comparing the profiles of isolated strains of cattle and wild animals, to determine the origin of the infection of a very large number of outbreaks and to highlight possible inter-species transmission [17]. However, in areas where the disease is still highly prevalent, unique dominant genetic profiles-per-zone are shared by almost all isolates, which makes the reconstitution of a transmission chain impossible [17,18]. Molecular techniques with finer resolutions are necessary in such contexts. Analyses of mycobacterial whole genome sequences (WGS), which detect genomic changes at a very small scale, hold promise as a strain discrimination alternative [19]. This technique has particularly been used to better understand the human-to-human transmission of different mycobacteria, such as M. leprae [20] or M. tuberculosis [21]. WGS data have also been used to reconstruct transmission scenarios of M. bovis infection [22,23].
The aim of this study was to use WGS analysis in order to investigate the transmission network of M. bovis with a particular genotype, mainly identified in infected red deer belonging to French farms, zoos, or hunting or other type of parks.
2. Material and Methods
2.1. Ethical Statement
In France, bTB is a notifiable disease with an eradication program in cattle and surveillance in free range wildlife. The official methods for the diagnosis of this disease are culture, PCR and histopathology. Therefore, all the samples included in this study are issued from animals analyzed within an official context. No purposeful killing of animals was performed for this study. All samplings were in complete agreement with national and European regulations. No ethical approval was necessary.
2.2. Bacterial Strains
This study included 24 field M. bovis isolates belonging to the French bTB National Reference Laboratory (NRL, Anses) collection (Table 1). The source population was animals whose samples were analyzed because (i) they presented non-negative skin tests and/or γ-interferon tests results and were culled for diagnostic purposes; (ii) they were culled as a result of the total or partial slaughter of their herd of origin after confirmation of the herd’s infection, or (iii) they presented macroscopic bTB-like lesions at routine abattoir inspection. Bacterial culture was performed following the protocol established by the French NRL (NF U 47–104) for the isolation of M. bovis. The identity of the isolated Mycobacterium tuberculosis complex colonies was confirmed by DNA amplification as described by Hénault et al. [24], and M. bovis was confirmed by spoligotyping and VNTR typing as described below. For livestock breakdowns, a herd-based criterion was used for strain choice: at least 1 strain/outbreak was selected. Spoligotyping was performed as described by Zhang et al. [14], using TB-SPOL kits purchased from Beamedex® (Beamedex® SAS, Orsay, France) on BioPLex200/Luminex 200® as described by Hauer et al. [2]. Spoligotypes have been named according to an agreed international convention (www.mbovis.org). A Multi-Locus Variable numbers tandem repeats Analysis (MLVA) profile identification [25] was performed by Genoscreen (Lille, France), using PCR amplification targeting genetic loci including mycobacterial MIRU-VNTR. Analysis was based on 8 loci, ETR A (VNTR2165), ETR B (VNTR2461), ETR C (VNTR577), ETR D (MIRU4 or VNTR580), QUB 11a (VNTR2163a), QUB 11b (VNTR2163b), QUB 26 (VNTR4052) and QUB 3232 (VNTR3232), chosen in the framework of a European consortium based on their degree of polymorphism and their ability to discriminate local strains [26].
Table 1.
Description of the 24 sequenced M. bovis strains.
| ID | Species | Year of Isolation | Context | Spoligotype | VNTR Profile | Accession Number | ||
|---|---|---|---|---|---|---|---|---|
| 1 | 1979-02442 | S79-020 | Wild boar | 1979 | Park | GB54/SB0121 | 6 3 5 3 11 1 5 7 | ERS3860435 |
| 2 | 1990-08827 | F90-035 | Red deer | 1990 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860436 |
| 3 | 1990-09372 | S90-020 | Cattle | 1990 | French herd | F70/SB0295 | 6 4 5 3 11 2 5 7 | ERS3860437 |
| 4 | 1993-04379 | F93-039 | Cattle | 1993 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860438 |
| 5 | 1993-06259 | S93-007 | Bison | 1993 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860439 |
| 6 | 1993-11575 | F93-076 | Red deer | 1993 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860440 |
| 7 | 1993-11583 | S93-013 | Monkey | 1993 | Park | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860441 |
| 8 | 1995-00583 | S95-007 | Cattle | 1995 | Foreign-born animal | F070/SB0295 | 6 4 5 3 11 2 5 7 | ERS3860442 |
| 9 | 1996-07003 | S96-046 | Cattle | 1996 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860443 |
| 10 | 1996-08822 | S96-050 | Cattle | 1996 | Foreign-born animal | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860444 |
| 11 | 1997-00766 | S97-004 | Cattle | 1997 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860445 |
| 12 | 1998-12646 | S98-099 | Bison | 1998 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860446 |
| 13 | 00-06572 | S00-206 | Cattle | 2000 | Foreign-born animal | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860447 |
| 14 | 07-03151 | S07-524 | Wild boar | 2007 | Park | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860448 |
| 15 | 08-00202 | S08-044 | Red deer | 2008 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860449 |
| 16 | 08-02744 | S08-481 | Cattle | 2008 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860450 |
| 17 | D11-02906 | S11-496 | Cattle | 2011 | Foreign-born animal | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860451 |
| 18 | D12-01540 | S12-164 | Wild boar | 2012 | Park | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860452 |
| 19 | D12-02012 | S12-295 | Cattle | 2012 | Foreign-born animal | F070/SB0295 | 6 4 5 3 11 2 5 7 | ERS3860453 |
| 20 | D14-00543 | S14-130 | Cattle | 2014 | Foreign-born animal | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860454 |
| 21 | D14-02802 | S14-680 | Cattle | 2014 | French herd | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860455 |
| 22 | D17-02544 | S17-321 | Fallow deer | 2017 | Park | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860456 |
| 23 | D17-03607 | S17-426 | Wapiti | 2017 | Park | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860457 |
| 24 | D18-00301 | S18-037 | Bison | 2018 | Park | GB54/SB0121 | 6 4 5 3 11 2 5 7 | ERS3860458 |
2.3. Whole Genome Sequencing and Variant Calling
Whole genome paired end (2 × 250 bp) sequencing was performed on Rapid Run HiSeq 2500 (Illumina) using NextEra XT libraries (Illumina, San Diego, CA, United States), and indexed following the manufacturer’s recommendations (Illumina, San Diego, CA, United States) by Genoscreen (Lille, France). The theoretical coverage was expected to be higher than 50×. Raw sequences were aligned to the reference genome AF2122/97 (Genbank accession NC_002945.4) using BioNumerics v7.6 (Applied Maths). A wgSNP analysis was performed with the strict SNP filtering template, i.e., removing positions with at least one unreliable base, ambiguous base, or gap; removing nondiscriminatory positions; with a minimum of 10× coverage; with a minimum distance of 12 nucleotides between SNPs. The evolutionary history of this dataset was inferred on MEGA7 software [27], using the Maximum Likelihood method based on the General Time Reversible (GTR) model (model selected based on jmodeltest v2.1.10 [28]). A bootstrap analysis was performed with 500 replicates.
WGS data were submitted to the EBI/EMBL. The accession Numbers are listed in Table 1.
3. Results
Among the 86 strains available at the NRL with the genotype of interest, i.e., a spoligotype profile belonging to the European 2 complex (either GB54/SB0121 or F70/SB0295) and a unique VNTR profile (6 4 5 3 11 2 5 7), 23 strains were selected from different herds or other origins, to represent all potential outbreaks (Table 1). They were isolated between 1979 and 2018 from 12 cattle (Bos taurus), 3 bison (Bison bonasus), 3 wild boar (Sus scrofa), 3 red deer (Cervus elaphus), 1 fallow deer (Dama dama), 1 wapiti (Cervus canadensis) and 1 nonhuman primate (species not specified). The strain isolated in 1979 from a wild boar was selected as an outgroup, as it shared the same spoligotype but a slightly different VNTR profile (6 3 5 3 11 1 5 7).
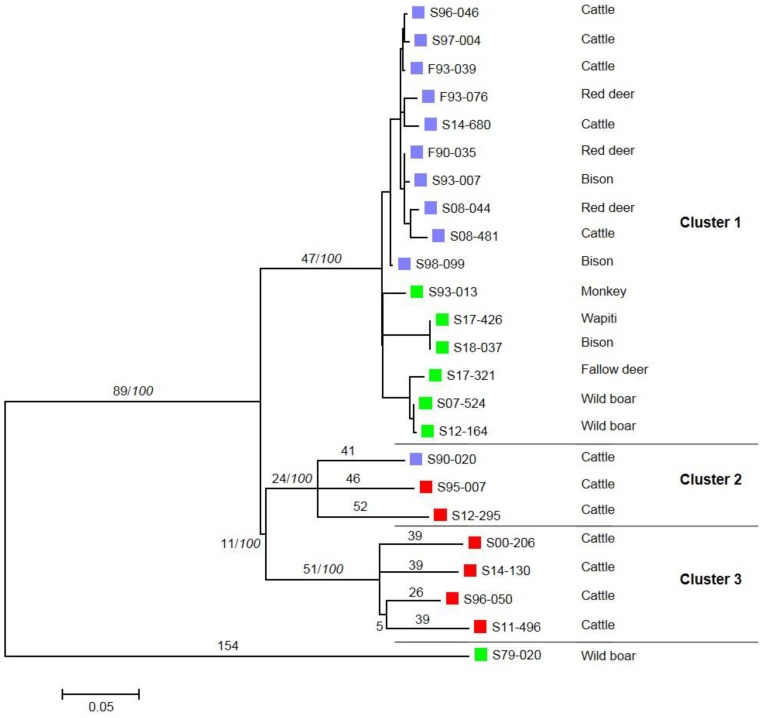
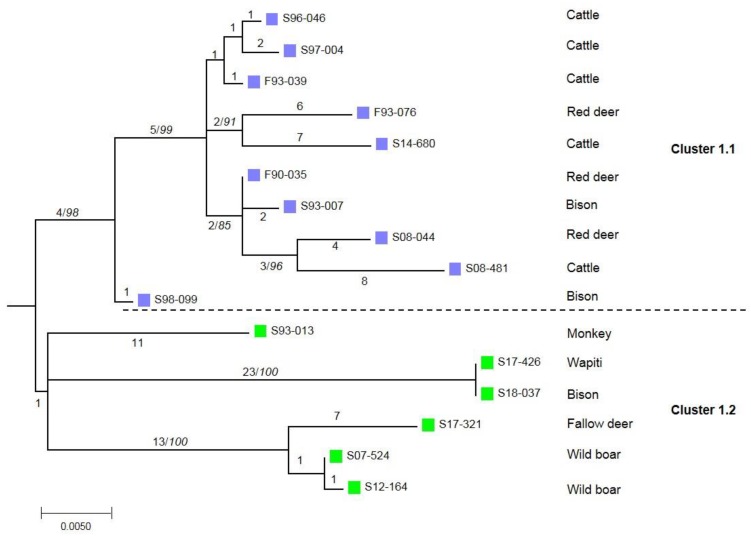
These isolates were analyzed by WGS. In total, we identified 770 SNPs compared to the M. bovis AF2122/97 reference genome (supplementary Table S1). The phylogenetic analysis highlighted different clusters, all enrooted by the wild boar strain isolated in 1979 (Figure 1 and Figure 2). A distance matrix of SNPs was generated using MEGA (Table 2) and the number of SNPs between each strain was determined. A first cluster supported by a bootstrap value of 100% is defined by 47 SNPs. This cluster is sub-divided into two sub-clusters: the first one, which is phylogenetically well supported (cluster 1.1, 98%), containing strains isolated from the cattle or deer of livestock herds; the second one, which is less robust (cluster 1.2, 67%), contains strains isolated from zoo animals or belonging to hunting parks (Figure 2). A low number of SNPs defined these sub-clusters—four and one respectively. Cluster 2 groups all the strains with spoligotype F70/SB0295, isolated from one French cow and from foreign-born animals, and is defined by 24 SNPs. Cluster 3, defined by 51 SNPs, contains strains isolated from foreign-born animals with spoligotype GB54/SB0121.
Figure 1.
Molecular Phylogenetic tree of the 24 strains using the Maximum Likelihood method. The evolutionary history was inferred on MEGA by using the Maximum Likelihood method based on the General Time Reversible model on a final dataset of 770 SNP positions for 24 strains [1]. The number above each branch corresponds to the number of SNPs. Bootstrap values (%) based on 500 replications are indicated in italics after a slash if greater than 75%. A violet square represents strains isolated from French herds. A red square represents strains isolated from foreign-born animals. A green square represents strains isolated from park animals.
Figure 2.
Zoomed-in view of the phylogenetic tree of cluster 1. The number above each branch corresponds to the number of SNPs. Bootstrap values (%) based on 500 replications are indicated in italics after a slash if greater than 75%. A violet square represents strains isolated from French herds. A green square represents strains isolated from park animals.
Table 2.
Estimates of Evolutionary Divergence between Sequences. The number of base differences between sequences are shown.
| F90-035 | F93-039 | S93-007 | F93-076 | S96-046 | S97-004 | S98-099 | S08-044 | S08-481 | S14-680 | S93-013 | S07-524 | S12-164 | S17-321 | S17-426 | S18-037 | S90-020 | S95-007 | S12-295 | S96-050 | S00-206 | S11-496 | S14-130 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1.1 | F90-035 | |||||||||||||||||||||||
| Cluster 1.1 | F93-039 | 4 | ||||||||||||||||||||||
| Cluster 1.1 | S93-007 | 2 | 6 | |||||||||||||||||||||
| Cluster 1.1 | F93-076 | 10 | 10 | 12 | ||||||||||||||||||||
| Cluster 1.1 | S96-046 | 5 | 3 | 7 | 11 | |||||||||||||||||||
| Cluster 1.1 | S97-004 | 6 | 4 | 8 | 12 | 3 | ||||||||||||||||||
| Cluster 1.1 | S98-099 | 8 | 8 | 10 | 14 | 9 | 10 | |||||||||||||||||
| Cluster 1.1 | S08-044 | 7 | 11 | 9 | 17 | 12 | 13 | 15 | ||||||||||||||||
| Cluster 1.1 | S08-481 | 11 | 15 | 13 | 21 | 16 | 17 | 19 | 12 | |||||||||||||||
| Cluster 1.1 | S14-680 | 11 | 11 | 13 | 13 | 12 | 13 | 15 | 18 | 22 | ||||||||||||||
| Cluster 1.2 | S93-013 | 23 | 23 | 25 | 29 | 24 | 25 | 17 | 30 | 34 | 30 | |||||||||||||
| Cluster 1.2 | S07-524 | 27 | 27 | 29 | 33 | 28 | 29 | 21 | 34 | 38 | 34 | 26 | ||||||||||||
| Cluster 1.2 | S12-164 | 28 | 28 | 30 | 34 | 29 | 30 | 22 | 35 | 39 | 35 | 27 | 1 | |||||||||||
| Cluster 1.2 | S17-321 | 32 | 32 | 34 | 38 | 33 | 34 | 26 | 39 | 43 | 39 | 31 | 9 | 10 | ||||||||||
| Cluster 1.2 | S17-426 | 35 | 35 | 37 | 41 | 36 | 37 | 29 | 42 | 46 | 42 | 34 | 38 | 39 | 43 | |||||||||
| Cluster 1.2 | S18-037 | 35 | 35 | 37 | 41 | 36 | 37 | 29 | 42 | 46 | 42 | 34 | 38 | 39 | 43 | 0 | ||||||||
| Cluster 2 | S90-020 | 134 | 134 | 136 | 140 | 135 | 136 | 128 | 141 | 145 | 141 | 135 | 139 | 140 | 144 | 147 | 147 | |||||||
| Cluster 2 | S95-007 | 139 | 139 | 141 | 145 | 140 | 141 | 133 | 146 | 150 | 146 | 140 | 144 | 145 | 149 | 152 | 152 | 87 | ||||||
| Cluster 2 | S12-295 | 145 | 145 | 147 | 151 | 146 | 147 | 139 | 152 | 156 | 152 | 146 | 150 | 151 | 155 | 158 | 158 | 93 | 98 | |||||
| Cluster 3 | S96-050 | 151 | 151 | 153 | 157 | 152 | 153 | 145 | 158 | 162 | 158 | 152 | 156 | 157 | 161 | 164 | 164 | 147 | 152 | 158 | ||||
| Cluster 3 | S00-206 | 159 | 159 | 161 | 165 | 160 | 161 | 153 | 166 | 170 | 166 | 160 | 164 | 165 | 169 | 172 | 172 | 155 | 160 | 166 | 70 | |||
| Cluster 3 | S11-496 | 164 | 164 | 166 | 170 | 165 | 166 | 158 | 171 | 175 | 171 | 165 | 169 | 170 | 174 | 177 | 177 | 160 | 165 | 171 | 65 | 83 | ||
| Cluster 3 | S14-130 | 159 | 159 | 161 | 165 | 160 | 161 | 153 | 166 | 170 | 166 | 160 | 164 | 165 | 169 | 172 | 172 | 155 | 160 | 166 | 68 | 78 | 81 | |
| Outgroup | S79-020 | 301 | 301 | 303 | 307 | 302 | 303 | 295 | 308 | 312 | 308 | 302 | 306 | 307 | 311 | 314 | 314 | 319 | 324 | 330 | 336 | 344 | 349 | 344 |
The 16 French strains gathered in cluster 1 seem to be closer (between 0 and 46 SNPs) than the strains of cluster 2 and 3. Indeed, some of them have very few genetic differences. Two strains isolated from a bison and a wapiti from the same animal park are identical. The WGS analysis allows us to confirm a link between two strains isolated from one wild boar in 2007, another in 2012 and a strain isolated from a fallow deer in 2017. The total number of SNPs for cluster 1 is 107, 50 for cluster 1.1, and 57 for cluster 1.2. On the other hand, the number of differences in the remaining clusters is higher with fewer strains, i.e., 163 SNPs for cluster 2 and 199 SNPs for cluster 3.
4. Discussion and Conclusions
This investigation, based on genomic data, allowed us to reveal the circulation of closely related M. bovis strains that seem to be confined to deer farms and animal parks. Although these types of strains have been regularly found since the 1990s, no geographical link seems to exist between most herd cases. WGS analysis showed a main cluster in France, divided into two very close sub-clusters (with just five SNP differences)—cluster 1.1 related to cattle and deer herds reared as livestock, and cluster 1.2 related to recreational or hunting animal parks. This analysis confirms the infection by the same strain of a bison and a wapiti from the same park. It also shows a link between the strain isolated recently in a fallow deer and two wild boar strains in 2007 and 2012 [29]. An epidemiological link exists between these three cases: the wild boars belonged to the same hunting park and the fallow deer, found infected but in another animal park, was originally issued from the same hunting park as that of the wild boar cases. Altogether, the common point in all these French cases was the presence of deer in the parks/herds or close to cattle herds. This genotype has been circulating in France for the last 30 years, however such strains have been isolated sporadically which clearly shows that there is a lack of appropriate surveillance in animal parks. Given that primates are not maintenance hosts of M. bovis, the case described in the monkey—acting as a sentinel of the infection—could be the result of the under-detection of another wild ungulate case in the park. The low number of screening tests, such as the skin test, conducted on deer is the result of several factors, such as the difficulty to perform the test on these wild animals, the potential cross-reaction with environmental mycobacteria, or the inherent technical variability of the test [30].
Our data highlight the limit of classical genotyping techniques that can be overcome by the use of whole genome SNP analysis. Strains that shared the same genotype (spoligotype-VNTR) from French and foreign-born animals were genetically different and very distant.
The use of WGS revealed an unsuspected transmission network between hunting parks and zoos that needs to be further explored. These results reinforce the suspicion of an infectious deer-breeding network that could be at the origin of the bacillus dissemination. Indeed, this transmission could have occurred through inter-breeding animal exchanges. Our results raise awareness about the infectious risk of wild animals in captivity and the consequences of trading non-appropriately-tested animals. P22 ELISA, a currently available, highly specific and sensitive new test, could contribute to a more accurate diagnosis of TB in this species [30].
This study raises the problem of the zoonotic transmission of the disease, especially for cases in animal parks where close contact between free-roaming animals and the public is allowed. The fact that deer often develop generalized TB, and thus that they probably excrete the bacillus at significant rates, increases the risk to humans. This study therefore shows the need for the strengthened monitoring of these animals with regular ante-mortem tests (as for livestock) before allowing animal movement between parks.
Acknowledgments
We thank Victoria Boschiroli for her useful comments on this article.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2076-2607/7/12/687/s1.
Author Contributions
Conceptualization, M.L.B., F.B. and L.M.; Software, C.C., M.B., T.C., L.M.; Formal Analysis, C.C., M.B., T.C., L.M.; Investigation, M.L.B. and L.M.; Writing—Original Draft Preparation, M.L.B. and L.M.; Writing—Review & Editing, C.C., M.B., T.C., F.B.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Muller B., Durr S., Alonso S., Hattendorf J., Laisse C.J., Parsons S.D., van Helden P.D., Zinsstag J. Zoonotic Mycobacterium bovis-induced tuberculosis in humans. Emerg. Infect. Dis. 2013;19:899–908. doi: 10.3201/eid1906.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauer A., De Cruz K., Cochard T., Godreuil S., Karoui C., Henault S., Bulach T., Banuls A.L., Biet F., Boschiroli M.L. Genetic evolution of Mycobacterium bovis causing tuberculosis in livestock and wildlife in France since 1978. PLoS ONE. 2015;10:e0117103. doi: 10.1371/journal.pone.0117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gortázar C., Delahay R.J., McDonald R.A., Boadella M., Wilson G.J., Gavier-Widen D., Acevedo P. The status of tuberculosis in European wild mammals. Mammal Rev. 2012;42:193–206. doi: 10.1111/j.1365-2907.2011.00191.x. [DOI] [Google Scholar]
- 4.Griffin J.F., Mackintosh C.G. Tuberculosis in deer: Perceptions, problems and progress. Vet. J. 2000;160:202–219. doi: 10.1053/tvjl.2000.0514. [DOI] [PubMed] [Google Scholar]
- 5.Zanella G., Duvauchelle A., Hars J., Moutou F., Boschiroli M.L., Durand B. Patterns of lesions of bovine tuberculosis in wild red deer and wild boar. Vet. Rec. 2008;163:43–47. doi: 10.1136/vr.163.2.43. [DOI] [PubMed] [Google Scholar]
- 6.Mackintosh C.G., de Lisle G.W., Collins D.M., Griffin J.F. Mycobacterial diseases of deer. N. Z. Vet. J. 2004;52:163–174. doi: 10.1080/00480169.2004.36424. [DOI] [PubMed] [Google Scholar]
- 7.Martin-Hernando M.P., Torres M.J., Aznar J., Negro J.J., Gandia A., Gortazar C. Distribution of lesions in red and fallow deer naturally infected with Mycobacterium bovis. J. Comp. Pathol. 2010;142:43–50. doi: 10.1016/j.jcpa.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Miller M., Olea-Popelka F. One Health in the shrinking world: Experiences with tuberculosis at the human-livestock-wildlife interface. Comp. Immunol. Microbiol. Infect. Dis. 2013;36:263–268. doi: 10.1016/j.cimid.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Dalovisio J.R., Stetter M., Mikota-Wells S. Rhinoceros’ rhinorrhea: Cause of an outbreak of infection due to airborne Mycobacterium bovis in zookeepers. Clin. Infect. Dis. 1992;15:598–600. doi: 10.1093/clind/15.4.598. [DOI] [PubMed] [Google Scholar]
- 10.Kiers A., Klarenbeek A., Mendelts B., Van Soolingen D., Koeter G. Transmission of Mycobacterium pinnipedii to humans in a zoo with marine mammals. Int. J. Tuberc. Lung Dis. 2008;12:1469–1473. [PubMed] [Google Scholar]
- 11.Montali R.J., Mikota S.K., Cheng L.I. Mycobacterium tuberculosis in zoo and wildlife species. Rev. Sci. Tech. 2001;20:291–303. doi: 10.20506/rst.20.1.1268. [DOI] [PubMed] [Google Scholar]
- 12.Murphree R., Warkentin J.V., Dunn J.R., Schaffner W., Jones T.F. Elephant-to-human transmission of tuberculosis, 2009. Emerg. Infect. Dis. 2011;17:366–371. doi: 10.3201/eid1703.101668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gharib Mombeni E., Mosavari N., Moradi Gravand M., Amir Rezai A., Keshavarz R., Tadayon K., Bakhshi R., Behmanesh R. First Report of Mycobacterium bovis Isolation from a European Fallow Deer (Dama Dama Dama) in Iran. Iran. J. Public Health. 2016;45:814–816. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., Abadia E., Refregier G., Tafaj S., Boschiroli M.L., Guillard B., Andremont A., Ruimy R., Sola C. Mycobacterium tuberculosis complex CRISPR genotyping: Improving efficiency, throughput and discriminative power of ‘spoligotyping’ with new spacers and a microbead-based hybridization assay. J. Med. Microbiol. 2010;59:285–294. doi: 10.1099/jmm.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 15.Skuce R.A., McDowell S.W., Mallon T.R., Luke B., Breadon E.L., Lagan P.L., McCormick C.M., McBride S.H., Pollock J.M. Discrimination of isolates of Mycobacterium bovis in Northern Ireland on the basis of variable numbers of tandem repeats (VNTRs) Vet. Rec. 2005;157:501–504. doi: 10.1136/vr.157.17.501. [DOI] [PubMed] [Google Scholar]
- 16.Hauer A., Michelet L., De Cruz K., Cochard T., Branger M., Karoui C., Henault S., Biet F., Boschiroli M.L. MIRU-VNTR allelic variability depends on Mycobacterium bovis clonal group identity. Infect. Genet. Evol. 2016;45:165–169. doi: 10.1016/j.meegid.2016.08.038. [DOI] [PubMed] [Google Scholar]
- 17.Boschiroli M.L., Michelet L., Hauer A., De Cruz K., Courcoul A., Hénault S., Palisson A., Karoui C., Biet F., Zanella G. Tuberculose bovine en France: Cartographie des souches de Mycobacterium bovis entre 2000–2013. Bull. Epid. St. Anim. Alim. 2015;70:2–8. [Google Scholar]
- 18.Cavalerie L., Courcoul A., Boschiroli M.L., Réveillaud E., Gay P. Bovine Tuberculosis in France in 2014: A stable situation. Bull. Epid. St. Anim. Alim. 2015;71:4–11. [Google Scholar]
- 19.Hauer A., Michelet L., Cochard T., Branger M., Nunez J., Boschiroli M.L., Biet F. Accurate Phylogenetic Relationships Among Mycobacterium bovis Strains Circulating in France Based on Whole Genome Sequencing and Single Nucleotide Polymorphism Analysis. Front. Microbiol. 2019;10:955. doi: 10.3389/fmicb.2019.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turankar R.P., Lavania M., Chaitanya V.S., Sengupta U., Darlong J., Darlong F., Siva Sai K.S., Jadhav R.S. Single nucleotide polymorphism-based molecular typing of M. leprae from multicase families of leprosy patients and their surroundings to understand the transmission of leprosy. Clin. Microbiol. Infect. 2014;20:O142–O149. doi: 10.1111/1469-0691.12365. [DOI] [PubMed] [Google Scholar]
- 21.Roetzer A., Diel R., Kohl T.A., Ruckert C., Nubel U., Blom J., Wirth T., Jaenicke S., Schuback S., Rusch-Gerdes S., et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: A longitudinal molecular epidemiological study. PLoS Med. 2013;10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biek R., O’Hare A., Wright D., Mallon T., McCormick C., Orton R.J., McDowell S., Trewby H., Skuce R.A., Kao R.R. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathog. 2012;8:e1003008. doi: 10.1371/journal.ppat.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trewby H., Wright D., Breadon E.L., Lycett S.J., Mallon T.R., McCormick C., Johnson P., Orton R.J., Allen A.R., Galbraith J., et al. Use of bacterial whole-genome sequencing to investigate local persistence and spread in bovine tuberculosis. Epidemics. 2016;14:26–35. doi: 10.1016/j.epidem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henault S., Karoui C., Boschiroli M.L. A PCR-based method for tuberculosis detection in wildlife. Dev. Biol. 2006;126:23–132. discussion 325–126. [PubMed] [Google Scholar]
- 25.Roring S., Scott A., Brittain D., Walker I., Hewinson G., Neill S., Skuce R. Development of variable-number tandem repeat typing of Mycobacterium bovis: Comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 2002;40:2126–2133. doi: 10.1128/JCM.40.6.2126-2133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rusch-Gerdes S., Willery E., Savine E., de Haas P., van Deutekom H., Roring S., et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richomme C., Rivière J., Hars J., Boschiroli M.L., Gueneau E., Fediaevsky A., Dufour H. Tuberculose bovine: Infection de sangliers dans un parc de chasse. Bull. Epid. St. Anim. Alim. 2013;56:14–16. [Google Scholar]
- 30.Thomas J., Infantes-Lorenzo J.A., Moreno I., Romero B., Garrido J.M., Juste R., Dominguez M., Dominguez L., Gortazar C., Risalde M.A. A new test to detect antibodies against Mycobacterium tuberculosis complex in red deer serum. Vet. J. 2019;244:98–103. doi: 10.1016/j.tvjl.2018.12.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.