Abstract
Antibiotic resistance is a global issue which is magnified by interspecies horizontal gene transfer. Understanding antibiotic resistance in bacteria in a natural setting is crucial to check whether they are multidrug resistant (MDR) and possibly avoid outbreaks. In this study, we have isolated several antibiotic-resistant bacteria (ARB) (n = 128) from the mangroves in Kerala, India. ARBs were distributed based on antibiotics (p = 1.6 × 10−5). The 16S rRNA gene characterization revealed dominance by Bacillaceae (45%), Planococcaceae (22.5%), and Enterobacteriaceae (17.5%). A high proportion of the isolates were MDR (75%) with maximum resistance to methicillin (70%). Four isolates affiliated to plant-growth promoters, probiotics, food, and human pathogens were resistant to all antibiotics indicating the seriousness and prevalence of MDR. A significant correlation (R = 0.66; p = 2.5 × 10−6) was observed between MDR and biofilm formation. Antagonist activity was observed in 62.5% isolates. Gram-positive isolates were more susceptible to antagonism (75.86%) than gram-negative (36.36%) isolates. Antagonism interactions against gram-negative isolates were lower (9.42%) when compared to gram-positive isolates (89.85%). Such strong antagonist activity can be harnessed for inspection of novel antimicrobial mechanisms and drugs. Our study shows that MDR with strong biofilm formation is prevalent in natural habitat and if acquired by deadly pathogens may create havoc in public health.
Keywords: multidrug resistance, mangrove, biofilm, antibiotic-resistant genes, antagonism
1. Introduction
The discovery of antibiotics has been a breakthrough in the medical field, which has saved millions of lives. However, the emergence of antibiotic resistance over time has rendered pathogenic microbes resistant to single and multidrugs. Such evolution has made the treatment of infectious diseases extremely difficult. Given the importance of pathogenic microbes in public welfare, most of the research has focused mainly on the human, agricultural, and veterinary pathogens. However, there is an increasing evidence of several non-pathogenic drug-resistant microbes in the environmental microbial community. This is of serious concern since those non-pathogenic microbes could serve as a reservoir of antibiotic-resistant genes, which could ultimately spread to pathogens through horizontal gene transfer and give rise to numerous super bugs. Despite such threats, the natural environment microbial community resistome has received relatively less focus [1]. Antibiotic resistance of a bacterium in clinical terms is classified into breakpoints such as susceptible, intermediate, or resistant [2]. However, such breakpoints do not exist in environmental bacteria because clinical breakpoints are defined based on several factors such as pharmacokinetics and pharmacodynamics [1]. However, attempts have also been made by considering the growth of a soil bacterium at 20 µg/mL antibiotic as a resistant isolate [3,4]. In addition, studies on the multidrug resistance of environmental isolates are hampered by the fact that ~99% of the microbes are unculturable and to tap the maximum members of the microbial community would require non-cultivable techniques such as metagenomics and pan-metagenomics. However, the downside of such techniques is that they are limited to known isolates registered in the database with the exception of expression-based functional metagenomics. Most of the clinically used antibiotics are derived from actinomycetes and other soil dwelling microbes. Hence, resistance has to develop to avoid self-killing. In addition, microbes around producers would eventually develop resistance to antibiotics in order to coexist or compete for food and space. Such origination of resistant genes, which has been found in the soil, is clinically relevant and causes difficulties in treating patients. Treatment of antibiotic-resistant infections is further complicated by biofilm activity that serves as an extra protective measure from antibiotics as well as other biotic and abiotic stress. Interestingly, it has been found that the concentration of antibiotics such as sulfamerazine, sulfamethazine, ofloxacin, norfloxacin, ciprofloxacin, oxytetracycline, and tetracycline are lower in sediments with mangrove vegetation compared to bare mudflats [5]. In addition, ARGs (antibiotic-resistant genes) were also found to be lower in mangrove areas compared to non-mangrove ones [6]. Mangroves are saline tolerant forests that are distributed in the tropical and subtropical coastal regions. Mangroves have also served as a reservoir of novel species and bioactive secondary metabolites against pathogens such as MRSA (methicillin-resistant Staphylococcus aureus) [7,8]. Mangrove extracts have also been shown to have antimicrobial activities [9]. However, with the increasing rate of pollution from anthropogenic sources, mangroves are continuously exposed to pollution. In addition, the rate of mangrove deforestation has increased alarmingly in the recent decade. Hence, in this paper, we have elucidated the multidrug resistant nature, biofilm, and the community antagonism from several antibiotic-resistant bacteria of mangrove sediment origin.
2. Materials and Methods
2.1. Sampling, Enrichment, and Isolation of Antibiotic-Resistant Bacteria
Mangrove sediment samples were collected from eight different locations along the coastal region of Kerala according to our previous study on the Kerala mangrove resistome (Table 1) [10,11]. These mangrove ecosystems have indirect mild exposure to sewage from anthropogenic activities. All the sediments were collected using sterile gloves and polythene bags from the upper 5–20 cm. The sediment samples were transported at 4 °C and stored in −80 °C for further study. Antibiotic-resistant microbes in the sediment samples were enriched by inoculating 1 g sediment in 10 mL LB (Luria-Bertani) broth with seven different antibiotics separately (Table 2) for each location. The broth was incubated overnight at 37 °C in shaker set to 120 rpm. Overnight grown cultures were serially diluted in different concentrations (10−1, 10−2, 10−3) and 20 μL of the diluted cultures was spread onto antibiotic-specific LB agar plates and further incubated overnight at 37 °C. Bacterial colonies obtained the following day were selected and sub-cultured based upon their morphology and pigmentation patterns. After several sub-cultures, pure colonies were affirmed. Pure cultures were stored in 40% glycerol at −80 °C for further studies.
Table 1.
Sampling locations spanning the north and central Kerala (India) were surveyed according to our previous studies [10,11] and mangrove sediments were collected in triplicate from the upper 5–10 cm in sterile sampling bags. The sampling locations and their codes are provided in first and second column, respectively.
| Location | Co-Ordinates (Latitude, Longitude) | Sample ID |
|---|---|---|
| Payannur | 12.1050687, 75.2058 | PYN |
| Bangramanjeshwar | 12.708333, 74.900754 | BNH |
| Kumbla | 12.594195, 74.946623 | KMA |
| Kavvayi | 12.088286, 75.176029 | KVY |
| Valapattanam | 9.996566, 76.247189 | VPM |
| Panangod | 9.8959941, 76.326094 | PGD |
| Madakal | 9.9091896, 76.30629 | MAL |
| Vallarpadam | 9.9994138, 76.253705 | VPDM |
Table 2.
The antibiotics used for screening are provided in the first column and their respective concentrations in the second column were used as the final concentration for screening of antibiotic-resistant microbes.
| Antibiotics | Working Concentration (µg/mL) |
|---|---|
| Ampicillin | 100 |
| Gentamicin | 10 |
| Chloramphenicol | 25 |
| Ciprofloxacin | 10 |
| Tetracycline | 10 |
| Vancomycin | 50 |
| Methicillin | 1 |
2.2. Multidrug-Resistance Profiling
Isolates were screened for multidrug resistance against all seven antibiotics. In order to differentiate the resistance level, the growth (optical density at 600 nm) of each isolate was measured based on a positive control (LB broth without antibiotic selection). A profile was generated based on the level of resistance which was categorized into three parts i.e., >80% growth was considered resistant, >30% growth was considered as partially resistant, and >0% growth was considered as partially susceptible. Profile was screened for unique resistance profile and morphology by removing the identical profiles. MDR (multidrug resistance) heat map of unique isolates was plotted with R using the “pheatmap” library [12].
2.3. Molecular Identification and Phylogenetic Analysis
Genomic DNA was isolated using the phenol chloroform method and confirmed in 1% agarose gel electrophoresis. Polymerase chain reaction (PCR) amplification of full-length 16S rRNA gene with 8F and 1492R was carried out at 55 °C annealing temperature. The amplicons were sequenced at Eurofins Scientific (Bangalore, India). Raw Sanger sequences were checked for their quality using Sequence Scanner Software v2.0 (Applied Biosystems, Foster City, CA, USA). Low quality reads at the 5’ and 3’ ends were trimmed and sequences below 500 base pairs after trimming were excluded. The 16s rRNA gene sequences generated in this study are available in the National Center for Biotechnology Information (NCBI) gene bank under the accession number MN629977–MN630016. The resultant sequences were further processed for top hit taxonomy similarity by considering “valid name only” criteria in EzTaxon. Sequences in FASTA format of the top hit against each isolate were retrieved from EzTaxon and multiple sequence alignment (MSA) was performed using ClustalW [13] in Molecular Evolutionary Genetics Analysis X (MEGA) [14]. The phylogenetic tree was constructed using maximum-likelihood algorithm with 1000 bootstraps.
2.4. Biofilm Assay
Biofilm formation by the isolates was quantified based on Microtiter Dish Biofilm Formation Assay as described by [15]. We followed this protocol because of its rapid procedure and its high-throughput nature. In brief, 125 µL of 0.1 optical density (OD) overnight culture was diluted 1:150 in LB broth and incubated at 37 °C for 24 h. The culture was decanted and shaken gently in Milli-Q water (Millipore, Tokyo, Japan) and air dried on a tissue paper. Then, 125 μL of 1% (w/v) crystal violet solution was added and incubated for 15 min at room temperature followed by gently shaking in sub-merge Milli-Q water that was further discarded and air dried on a tissue paper. Moreover, 125 μL of 30% (v/v) acetic acid was added and incubated for 15 min at room temperature. The contents were transferred to a clean and dry 96-well plate and optical density was determined by measuring at 550 nm. Blanks in triplicates were prepared by adding LB broth instead of bacterial culture.
2.5. Antagonism
A pairwise antagonistic activity was evaluated for each isolate. In total, we evaluated 1560 (40 × 39) interactions using the agar well method. Wells were punched into LB agar plates, and 100 µL of overnight culture of test organism was spread and air dried in the aseptic condition under laminar airflow. Five-millimeter wells were punched on to the plate surface. Twenty microliters of overnight indicator culture was added into the well. The plates were incubated for 1–3 days until a zone of inhibition was observed around the well.
3. Results
3.1. Distribution of Antibiotic-Resistant Bacteria, MDR Profiles, and Phylogenetic Analysis
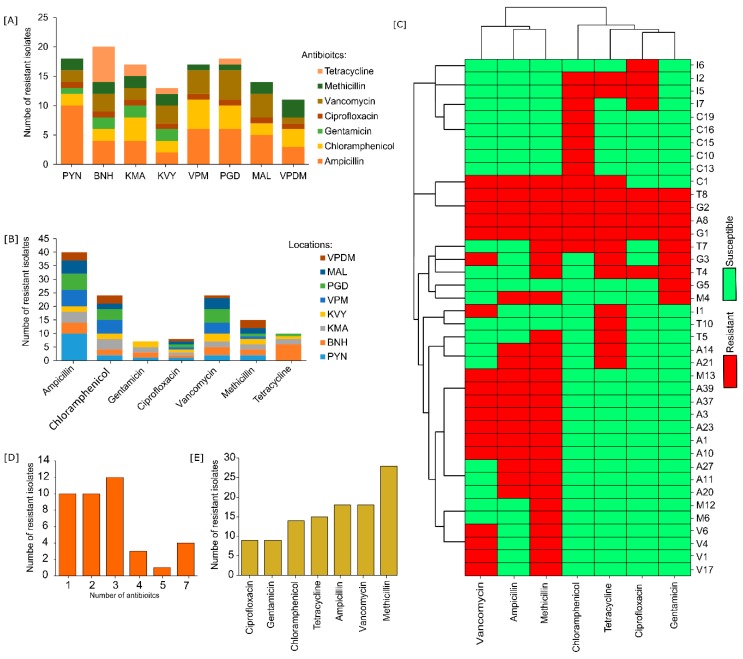
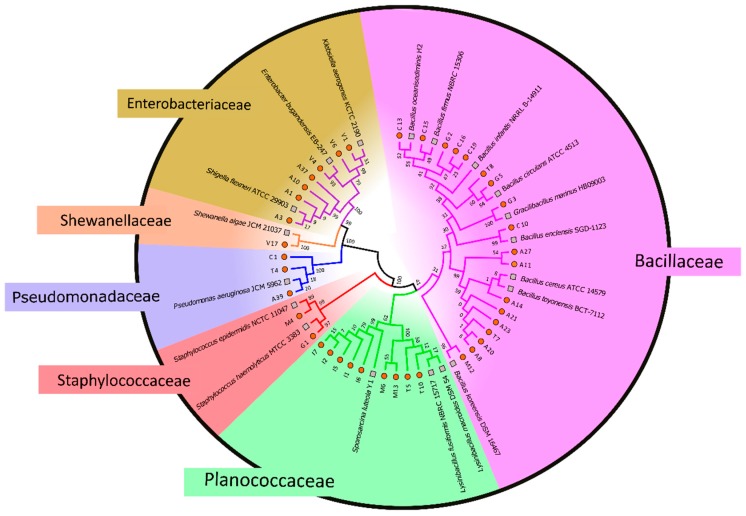
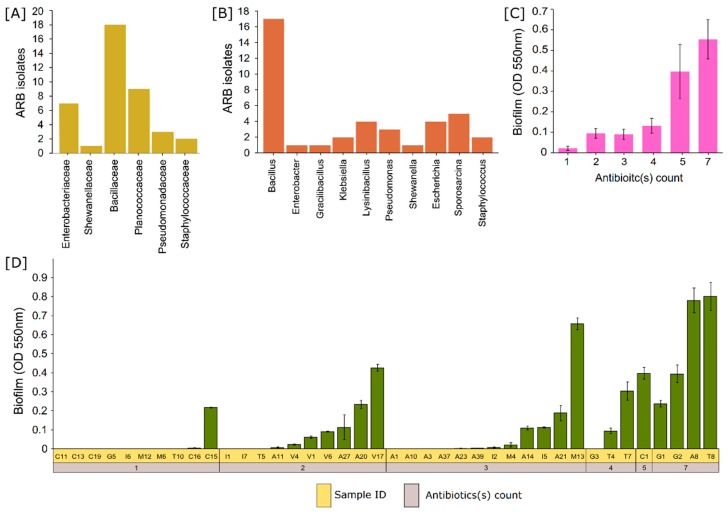
Screening of antibiotic-resistant microbes from mangrove sediments in LB plates fortified separately with various antibiotics obtained 128 pure cultures from multiple subcultures based on morphology and pigmentation. More than 10 antibiotic-resistant bacteria were isolated from each location (Figure 1A). Tetracycline resistance was highest in BNH (Bangramanjeshwar) (n = 6) as compared to other locations (n = 0.57 ± 0.27). Number of antibiotic-resistant isolates based on location had no significant (p = 0.8) difference. However, significant (p = 1.6 × 10−5) difference was observed based on antibiotics (Figure 1B). Resistance to ampicillin, methicillin, and vancomycin was among the highest and common to all locations indicating that the resistance to beta-lactam antibiotic is highly prevalent. Multidrug-resistance profiling was carried out to determine the prevalence of multidrug resistance among the isolates and to screen out repeating isolates. Several isolates exhibited similar resistance profiles/patterns. We selected 40 unique isolates for further study based on MDR profile and morphology. The isolates exhibited a high number (75%; 30 out of 40) of multidrug resistance (resistance to two or more antibiotics) (Figure 1C). Among the 40 isolates, four isolates (T8, G2, A8, and G1) exhibited resistance to all antibiotics, indicating the seriousness and prevalence of multidrug resistance. The phylogenetic relationship of the four isolates indicates that G1 has the closest phylogeny to Staphylococcus haemolyticus under the family Staphylococcaceae while A8, T8, and G2 have the closest hit to Bacillus cereus, Bacillus circulans, and Bacillus infantis, respectively, under family Bacillaceae (Figure 2). Multidrug resistance was significantly prevalent for up to three antibiotics but reduced significantly for higher number of antibiotics (Figure 1D). In addition, resistance to methicillin was highest (70%) among all antibiotics tested, followed by vancomycin and ampicillin (45% each) (Figure 1E). The lowest number of resistance (22.5%; 9 out of 40 isolates) was observed against ciprofloxacin and gentamicin. On the other hand, a moderate level of resistance was observed against tetracycline (37.5%) and chloramphenicol (35%). The 16S rRNA gene identification revealed that the antibiotic-resistant isolates belonged to five families dominated by the Bacillaceae family (45%; 18 out of 40) followed by Planococcaceae (22.5%), Enterobacteriaceae (17.5%), Pseudomonadaceae (7.5%), Staphylococcaceae (5%), and Shewanellaceae (2.5%) (Figure 3A,B). Several groups of isolates such as C16, C19, and G2 had the same top hit taxonomy as B. infantis although with different levels of similarity, 99.74%, 99.75%, and 99.73%, respectively, indicating that they could belong to different sub-strains (Figure 2; Table 3).
Figure 1.
Number of antibiotic-resistant isolates according to (A) location and (B) antibiotics. (C) Clustered heat map of unique isolates based on Euclidean distance. (D) Bar chart of antibiotic-resistant isolates against the number of antibiotics shows high prevalence of resistance to up to three antibiotics followed by drastic reduction on more antibiotics. (E) Prevalence of antibiotic resistance based on antibiotics.
Figure 2.
Phylogenetic tree was constructed using Molecular Evolutionary Genetics Analysis (MEGA) and maximum-likelihood method. The red circles represent the antibiotic-resistant isolates and the grey square represents the nearest representative strain. The colored pie and enlarged text against each pie represent the respective family.
Figure 3.
Bar chart representation of the number of isolated antibiotic-resistant bacteria (ARB) on the y-axis and the x-axis representing the (A) families and (B) genera of ARB. (C) Biofilm forming (OD (optical density) 550 nm) potential of the isolates grouped according to the number of multidrug resistance/antibiotic(s) count shows a direct correlation between biofilm and multidrug resistance (MDR). (D) Biofilm formation (OD 550 nm) of each isolate sorted according to the number of multidrug resistance/antibiotic(s) count.
Table 3.
Identification of the isolates based on EzBioCloud’s 16S identify service works on similarity-based searches in quality-controlled 16S rRNA databases sequences. The top-hit of each isolate with valid prokaryotic names having the nearest match is listed in the table.
| Isolate Sample ID | Species | Strain | Similarity (%) | Phylum | Family | Genus |
|---|---|---|---|---|---|---|
| V6 | Klebsiella aerogenes | KCTC 2190 | 100 | Proteobacteria | Enterobacteriaceae | Klebsiella |
| V4 | Enterobacter bugandensis | EB–247(T) | 100 | Proteobacteria | Enterobacteriaceae | Enterobacter |
| V1 | K. aerogenes | KCTC 2190 | 100 | Proteobacteria | Enterobacteriaceae | Klebsiella |
| V17 | Shewanella algae | JCM 21037 | 100 | Proteobacteria | Shewanellaceae | Shewanella |
| T8 | Bacillus circulans | ATCC 4513(T) | 99.15 | Firmicutes | Bacillaceae | Bacillus |
| T7 | Bacillus cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| T5 | Lysinibacillus macroides | DSM 54(T) | 100 | Firmicutes | Planococcaceae | Lysinibacillus |
| T4 | Pseudomonas aeruginosa | JCM 5962 | 100 | Proteobacteria | Pseudomonadaceae | Pseudomonas |
| T10 | L. macroides | DSM 54(T) | 100 | Firmicutes | Planococcaceae | Lysinibacillus |
| M6 | Lysinibacillus fusiformis | NBRC 15717(T) | 99.88 | Firmicutes | Planococcaceae | Lysinibacillus |
| M4 | Staphylococcus epidermidis | NCTC 11047(T) | 99.88 | Firmicutes | Staphylococcaceae | Staphylococcus |
| M13 | L. fusiformis | NBRC 15717(T) | 99.84 | Firmicutes | Planococcaceae | Lysinibacillus |
| M12 | Bacillus koreensis | DSM 16467 | 100 | Firmicutes | Bacillaceae | Bacillus |
| I7 | Sporosarcina luteola | Y1 | 99.75 | Firmicutes | Planococcaceae | Sporosarcina |
| I6 | S. luteola | Y1 | 99.72 | Firmicutes | Planococcaceae | Sporosarcina |
| I5 | S. luteola | Y1 | 99.75 | Firmicutes | Planococcaceae | Sporosarcina |
| I2 | S. luteola | Y1 | 99.75 | Firmicutes | Planococcaceae | Sporosarcina |
| I1 | S. luteola | Y1 | 99.74 | Firmicutes | Planococcaceae | Sporosarcina |
| G5 | B. circulans | ATCC 4513(T) | 99.66 | Firmicutes | Bacillaceae | Bacillus |
| G3 | Gracilibacillus marinus | HB09003 | 100 | Firmicutes | Bacillaceae | Gracilibacillus |
| G2 | Bacillus infantis | NRRL B–14911 | 99.73 | Firmicutes | Bacillaceae | Bacillus |
| G1 | Staphylococcus haemolyticus | MTCC 3383 | 100 | Firmicutes | Staphylococcaceae | Staphylococcus |
| C1 | P. aeruginosa | JCM 5962 | 100 | Proteobacteria | Pseudomonadaceae | Pseudomonas |
| C19 | B. infantis | NRRL B–14911 | 99.75 | Firmicutes | Bacillaceae | Bacillus |
| C16 | B. infantis | NRRL B–14911 | 99.74 | Firmicutes | Bacillaceae | Bacillus |
| C15 | Bacillus firmus | NBRC 15306 | 100 | Firmicutes | Bacillaceae | Bacillus |
| C13 | Bacillus oceanisediminis | H2(T) | 99.3 | Firmicutes | Bacillaceae | Bacillus |
| C10 | Bacillus enclensis | SGD–1123(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A8 | B. cereus | ATCC 14579 | 100 | Firmicutes | Bacillaceae | Bacillus |
| A3 | Shigella flexneri | ATCC 29903 | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
| A39 | P. aeruginosa | JCM 5962 | 100 | Proteobacteria | Pseudomonadaceae | Pseudomonas |
| A37 | S. flexneri | ATCC 29903(T) | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
| A27 | Bacillus toyonensis | BCT–7112 | 99.85 | Firmicutes | Bacillaceae | Bacillus |
| A23 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A21 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A20 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A1 | S. flexneri | ATCC 29903(T) | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
| A14 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A11 | B. cereus | ATCC 14579(T) | 99.88 | Firmicutes | Bacillaceae | Bacillus |
| A10 | S. flexneri | ATCC 29903(T) | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
3.2. Biofilm Formation in Environmental Isolates
We observed biofilm formation in 60% of the isolates (24 out of 40) (Figure 3D). Biofilm formation was not observed in several MDR isolates. For instance, multidrug-resistant isolates such as A1, A10, A3, A37, and G3 had resistance to three or more different drugs but did not exhibit biofilm formation. Isolate V17, resistant to vancomycin and methicillin, had a higher biofilm formation (OD5500.42 ± 0.017) as compared to the highly resistant isolate G1 (OD5500.017 ± 0.23) (resistant to all seven drugs). However, we also observed a relationship between biofilm activity and multidrug resistance. All isolates having resistance to five or more drugs exhibited biofilm formation. Average biofilm activity from all the isolates vs. the number of drug resistance indicated a clear trend that biofilm formation is indeed a strong weapon for MDR (Figure 3C). A strong statistically significant correlation (R = 0.66; p = 2.5 × 10−6) was also observed between the biofilm formation and MDR.
3.3. Antagonist Interactions
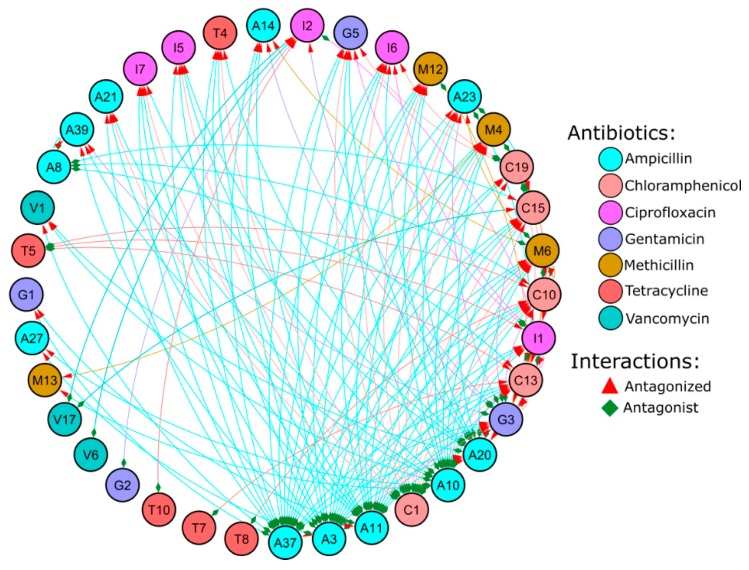
Antagonist activity of 1560 interactions was evaluated. There were 137 (8.78%) antagonist interactions (Figure 4). Out of 40 isolates, 23 (57.5%) isolates had antagonist activity against at least one of the other isolates. The highest antagonist activity was exhibited by A37 (Shigella flexneri) against 19 isolates and inhibited by only one isolate (I1 i.e., Sporosarcina luteola). Second highest antagonist activity was exhibited by A3 (Bacillus cereus) against 18 isolates which was not antagonized by any isolates. Among the susceptible isolates, Bacillus oceanisediminis, Bacillus enclensis, and Bacillus firmus (antagonized by 12, 11, and 9 isolates, respectively) were among the highest which exhibited antagonist activity against only one isolate. Such observation shows a pattern between the antagonisms and antagonized isolates, where an isolate with strong antagonist activity was generally antagonized by a lower number of isolates and vice versa. A correlation test between the two, by excluding the isolates that neither showed antagonism nor susceptibility, revealed a mild yet statistically significant negative correlation (R = −0.36; p = 0.03). Classification of the isolates based on their Gram nature revealed an interesting pattern where both gram positive and negative had similar number of antagonist isolates (54.5% and 58.6% for gram negative and positive, respectively) but gram-positive isolates were antagonized (75.86%) to a much larger extent compared to gram-negative isolates (36.36%) (Table 4 and Table 5). With respect to biofilm forming attribute, the non-biofilm formers were antagonized slightly more (68.75%) compared to bio-film formers (62.5%) but exhibited a stronger antagonist activity (68.75%) compared to biofilm formers (50%).
Figure 4.
Network plot of the antagonist interaction between antibiotic-resistant isolates was constructed using Cytoscape. Nodes are represented by circles. Edges are represented by the lines connecting the nodes. The color of the nodes and edges represents the respective antibiotics (as shown in graphical legend) that were used for screening the respective isolate. The interaction is undirected, the source of the interaction (antagonist) is denoted by green diamond shapes and the target nodes (antagonized) are denoted by red arrows.
Table 4.
Antagonism on the basis of biofilm and cell wall (Gram nature) was evaluated. Antagonism was further categorized based on isolates and interactions. In this study, antagonist interactions take all interaction from all isolates into consideration; however, calculation of antagonist isolates considered whether the insolate exhibited antagonism to at least one other isolate irrespective of the number of interactions. Isolates and interactions are expressed in percentage based on biofilm and gram nature. Higher number of biofilm forming isolates and interactions exhibited higher resistance but exhibited lower antagonism activity. Similarly, gram-negative isolates had higher resistance.
| Antagonized (%) | Antagonist (%) | ||
|---|---|---|---|
| Isolates | Non-Biofilm | 68.75 | 68.75 |
| Biofilm | 62.5 | 50 | |
| Interactions | Non-Biofilm | 53.28 | 57.66 |
| Biofilm | 46.71 | 42.33 | |
| Isolates | Gram negative | 36.36 | 54.54 |
| Gram positive | 75.86 | 58.62 | |
| Interactions | Gram negative | 9.42 | 52.17 |
| Gram positive | 89.85 | 47.10 |
Table 5.
Antagonist interactions were sorted based on all possible combinations of gram nature. Antagonism against gram-positive isolates was 90.51% while antagonism against gram-negative ones was only 9.49%.
| Interactions (Antagonist vs. Antagonized) |
Antagonist (%) |
|---|---|
| Gram negative vs. gram positive | 47.45 |
| Gram negative vs. gram negative | 5.11 |
| Gram positive vs. gram negative | 4.38 |
| Gram positive vs. gram positive | 43.07 |
4. Discussion
Mangroves in coastal forest are tolerant to saline environments. Owing to their location, they face pollutions from both land and sea. Mangrove environments are inhabited by various animals and birds, which could serve as a source as well as the dissemination factor of antibiotic-resistance genes (ARGs). This could lead to the enrichment of harmful genes such as ARGs. In this study, we have isolated antibiotic-resistant bacteria from Kerala mangrove sediments, which have indirect exposure to moderate anthropogenic activities. We isolated 128 antibiotic-resistant bacteria from eight different mangrove sediments. There was a significant (p < 0.05) pattern in the distribution of ARB (antibiotic-resistant bacteria) based on antibiotics rather than on the sampling location (p < 0.05). In line with this, a previous study on Chinese mangroves [16] noted that the abundance of antibiotic resistance had no significant difference based on location. Number of isolated bacteria based on various antibiotics showed dominance of ampicillin, chloramphenicol, and vancomycin (p < 0.05). According to the MDR profile, a high proportion (66.6%) of the isolates were MDR. This is in corroboration to the MDR observation by [16] from Chinese mangroves. The majority of the bacteria isolated using ampicillin, gentamicin, and vancomycin were resistant to methicillin. Hence, after the MDR profiling, resistance to methicillin was highest followed by resistance to ampicillin and vancomycin. Similarly, in a previous mangroves study from the Calicut zone of Kerala (India), resistance to penicillin (68%) and vancomycin (32%) was the highest followed by resistance to erythromycin (28%), gentamicin (20%), tetracycline (16%), and chloramphenicol (12%) [17]. It is noteworthy that all three antibiotics (methicillin, vancomycin, and ampicillin), having the least antibacterial activity within the study, target cell wall synthesis although they belong to different classes. In other words, resistance seems to have a relationship with the target of the antibiotic irrespective of the antibiotic class. The similarity of the resistance based on the mode of action was evident from the clustering of beta lactam antibiotics (methicillin and ampicillin) within a clade and vancomycin as the nearest clade. Such resistance to antibiotics of similar structure has also been reported in the past [18,19]. A previous report from Gaoqiao Mangrove (China) observed a similar abundance of resistance to beta lactam antibiotics [16]. Beta lactamases have been found in several studies both in pristine and human-intervened locations [20,21]. This could be explained by the cross-resistance to antibiotics of the same class having similar targets and mechanisms. However, resistance to multiple classes of antibiotics can be explained more plausibly by the selective enrichment of ARGs through horizontal gene transfer (HGT) [22]. Moderate level of inhibition was observed against semisynthetic antibiotics tetracycline (37.5%) and chloramphenicol (35%), belonging to different antibiotic classes, derived from different species but the same genus Streptomyces. Gentamicin (aminoglycoside) and ciprofloxacin (fluoroquinolone) had the highest inhibition. In contrast to our result, Matang mangrove in Malaysia had high occurrence of aminoglycosides (83%) resistance compared to beta-lactams [23] resistance. The comparable level of strong antimicrobial activity exhibited by synthetic antibiotic ciprofloxacin and Micromonospora spp.-derived antibiotic gentamicin could be a hint that the development of synthetic antibiotic, as well as bio-prospection of antibiotic from natural source is inexhaustive. Although antibiotic-resistant bacteria are found in the mangrove, it should be noted that mangrove environment is known to harbor lower antibiotic-resistant types, mechanism, and abundance [6].
The 16S rRNA gene sequencing of the resistant isolates identified maximum entries from Bacillus genus followed by Sporosarcina. In another study, Bacillus species were also the dominant members of antibiotic-resistant bacteria in shrimp aquacultures of Vietnamese mangrove [24] and Northern China aquaculture environment [25]. The high abundance of Bacillus among the resistant isolates could be because they are spore formers ubiquitously present in water and soil [26]. Bacillus has over 200 species including pathogenic ones as well as probiotics. Among the bacilli, we identified human pathogens such as B. cereus, B. circulans, and B. infantis. Detection of B. cereus is of major concern for public health since they produce a heat-stable toxin that causes food poisoning with two distinct types: diarrheal and emetic syndrome [27]. Another detected pathogenic Bacillus, B. circulans, a gram-positive spore forming opportunistic bacteria, causes nosocomial and food infection [28]. B. infantis is known to cause sepsis [29] and bacterial myocarditis in mouse models [30]. Out of several isolated strains of B. cereus (A8, T7, A21, A14, A23, A20, A11), B. circulans (T8, G5, C10), and B. infantis (G2, C16, C19), at least one isolate from each species (A8, T8, and G2, respectively) was resistant to all seven antibiotics under study. In addition to Bacillus species, S. haemolyticus (G1) exhibited resistance to all the antibiotics. S. haemolyticus is a known opportunistic pathogen that often carries enterotoxin genes such as hla [31] and causes nosocomial infections [32]. Mangroves are a major source of food for coastal communities and migratory birds. The presence of such multidrug-resistant opportunistic pathogens raises an alarming concern for the health of the coastal communities and livestock and as well as the food products derived from mangroves.
Plant-growth-promoting bacteria (PGPB) and probiotics such as B. firmus, Bacillus koreensis, and Bacillus toyonensis were also detected in the mangrove sediment. B. firmus promotes plant growth by protecting roots against plant pathogens [33] and serves as a bionematicide [34] by production of serine protease, which degrades vitals proteins associated with intestinal tissues and physical barriers [35]. B. koreensis has also been shown to have multiple desirable plant-growth-promotion properties such has IAA (indole-3-acetic acid) production, nitrogenase activity, and antifungal activity [36]. In addition, B. toyonensis is used as a probiotic in post-weaning piglets to protect from enteric pathogens [37]. Probiotics have been shown to have strong benefits for human health as well as agriculture, aquaculture, and animal farming [38]. However, antibiotic-resistance properties of such plant-growth-promoting bacteria and probiotics can cause more harm than good since they could serve as an excellent source of ARGs. Hence, selection of strains for probiotic consumption would be an essential criterion to minimize ARG dissemination. However, previous antibiotic-resistance studies on human dietary probiotics [39] have shown resistance to antibiotics in a batch-dependent manner indicating that the maintenance of suitable probiotic strains should be taken with utmost care, and quality control has to be taken strictly. Dissemination of ARGs from such commercial probiotics can be rapid and deadly since they are consumed at a high rate in animal and agricultural settings [38].
Resistance to antibiotics can be strongly complemented by the formation of biofilms. A statistically significant correlation (R = 0.66; p = 2.5 × 10−6) was observed between biofilm formation and the number of resistant drugs indicating that biofilm formation is a crucial tool for a majority of the MDR isolates. However, biofilm formation was not obligatory for the MDR since several MDR isolates did not produce biofilm. Biofilm formation and its correlation to antibiotic resistance have received a mixed observation in the literature. This could be due to the variations in biofilm-forming potential even at strain level. For instance, significant variations within same species in biofilm-forming potential were observed in several genera such as environmental Escherichia coli isolates [40], group B Streptococcus [41], S. aureus [42], Vibrio fischeri [43], Candida albicans [44], etc. Such variation within species would make it unreliable to establish a concentrate relationship on the biofilm formation and genus. Moreover, significant correlation was found between antibiotic resistance and biofilm formation in Pseudomonas aeruginosa [45,46] while other studies have found otherwise [47]. A study between P. aeruginosa from environmental and human sources observed significantly weaker biofilm formation and susceptibility to most antibiotics from environmental isolates as compared to human sources [48]. Similarly, MDR Acinetobacter baumannii [49], E. coli [50], and isolates from urinary tract infections [51] that produced biofilms had significantly higher resistance to antibiotics. On the contrary, a previous study [52] could not find any relation to the MDR and biofilm producers. Interestingly, Klebsiella pneumoniae susceptible to ciprofloxacin had a strong biofilm formation [53]. Biofilms are microbes’ self-produced extracellular matrix that attaches the bacterial cells to a surface or encompasses themselves with a layer of extracellular matrix, which serves as a blanket of defense that prevents the entry of antibiotics or toxic compounds into the biofilm [54]. Such blockage of antibiotics renders the bacterial cells up to 1000-fold more resistant than the planktonic cells [55]. This further creates a heterogeneous population within the biofilm due to the gradient of nutrient and waste products [56,57]. The densely packed structure can be composed of single or multiple species and increases the changes of HGT owing to the physical proximity. This ultimately serves not only as a protective layer but also as a hot spot for ARGs [54]. Although a biofilm is a dreadful bacterial attribute that takes thousands of lives annually, it is a normal characteristic of the environment. They play a crucial role in nutrient as well as biogeochemical cycling [54] and could be used as an environmental bio indicator [58]. Since they have medical importance, biofilms are also considered as an indicator of ARG pollution [59]. Nonetheless, the high prevalence of biofilm producers in the mangrove sediment with a strong correlation to MDR warrants further study on their role in HGT of ARGs and its biogeochemical functions.
In order to understand the interaction among the isolates, we evaluated a pair-wise antagonist activity among all the isolates. Compared to the pair-wise antagonist activity of isolates from sponge (18%) [60], mangrove sediment in our study exhibited relatively lower antagonist activity (8.78%) similar to previous study (6.6%) [61] on isolates from various sources such as maize leaf, algae, forest topsoil, small stones, streambed, roots of Aegopodium podagraria, etc. Similarly, the percentage population of isolates with antagonist activity (57.5%) was on the lower side as compared to the marine sponge (98.2%) [60], Antarctic sponges (62.2% and 90%) [62], and coastal waters (66.7%) [63]. However, our finding was significantly higher than clinical origin (22%) [64], free-living isolates from unfiltered seawater (17%) [65] and coastal waters (40.9%) [64]. This could be due to the strong competition in the environmental settings compared to clinical setup [60,66]. Particle-attached bacteria [63] have been known to have higher frequency of activity and a wider range of targets indicating the expression of broad range bacterial inhibitors. Inhibitory compounds from particle-attached bacteria have been hypothesized as a weapon to colonies, defend the dominance in the particle by attached bacteria, and play a crucial role in the degradation of the particle, which in turn plays a major role in biogeochemical recycling [63]. Hence, according to the literature and current study, it can be established that the particle-attached bacteria have a significantly stronger arsenal of defense, which can be harnessed for medical applications. In addition to the attachment nature, cell wall and membrane permeability have been postulated as a strong factor for microbial interactions and resistance. In previous studies, antagonism was observed most frequently between same Gram nature except for alpha-proteobacterium isolates which inhibited both gram +ve and −ve [67]. However, such patterns were not observed in subsequent studies [68,69]. Similarly, in our study, antagonism based on the Gram nature revealed an interesting pattern where both gram positive and negative had similar antagonist activity (58.6% and 54.5%, respectively) and antagonist interactions (47.1% and 52.17%, respectively). However, there was a stark difference between gram positive and negative when it comes to being antagonized (susceptibility). A high (75.86%) percentage of the gram-positive isolates were antagonized by at least one of the isolates while it was only 36.36% for gram-negative isolates. Similarly, antagonism by gram positive and negative against gram negative was observed in 4.35% and 5.07%, respectively. However, antagonism by gram positive and negative against gram positive was several folds higher (42.75% and 47.1%, respectively) (Table 4 and Table 5).
Another aspect of antagonism was the assessment of inhibition based on phylogenetic similarity. Previous reports have shown that isolates belonging to related open taxonomic units (OTUs) rarely had antagonist activity [62,69,70,71]. In contrast, other reports have shown inhibition of closely related species [61,65]. Similarly, strain level difference was also observed in such studies [62,65,72]. In corroboration with such findings, we also observed several isolates from the same species to have inhibitory action against other isolates of the same species. For instance, A23 was inhibited by A11 although both belong to the same OTU B. cereus. Similarly, T4 was inhibited by C1 although both belongs to P. aeruginosa. Prolific antagonist taxa vary based on literature such as Arthrobacter (Actinobacteria) [65], Pseudomonas (Proteobacteria) [63,71], and Bacillus (Firmicutes) [68,71,72]. Actinobacteria [71] and Flavobacterium [72] were most susceptible to antagonism. Similarly, in our study, the most prolific antagonist was A37 and A3 (S. flexneri; Proteobacteria) and A11 (B. cereus; Firmicutes) which inhibited 19, 18, and 17 isolates, respectively. However, the most susceptible isolates were also from Firmicutes phyla such as C13 (B. oceanisediminis), C10 (B. enclensis), and C15 (B. firmus) which were inhibited by 12, 11, and 9 isolates, respectively. In other words, there was a wide variation in the antagonistic activity within similar open taxonomic units (OTUs).
We also considered antagonism interaction based on biofilm and non-biofilm producers. In a previous study [72], a correlation was observed in selected strains between biofilm and antagonistic activity. The authors hypothesize that the biofilm formation could be an advantage to protect from the microbial community. Biofilm formation can also be induced by the presence of other bacteria or antibiotics [73]. Similarly, in our study, the percentage of susceptible isolates was slightly higher for non-biofilm (68.75%) compared to biofilm (62.5%) producers. However, isolates with antagonist activity from biofilm (50%) was lower than non-biofilm (68.75%) producers. The higher resistance to antagonism by biofilm producers is in agreement with our finding that biofilms are directly correlated to MDR. Hence, understanding the antagonism mechanism from insolates could provide a new window for the treatment of biofilm-based pathogens.
It should be taken into consideration during the interpretation of this work that antagonism interaction in this study as well as literature are based on pure culture in vitro growth on artificial agar media which could differ from the natural environment. In addition, the antagonism observation was solely based on the inhibition. There could be several ways of competition in nature in addition to direct inhibition. Production of inhibitory compounds can vary from direct killing to chelation of essential nutrients and quorum sensing [60]. It can also vary based on the availability and abundance of nutrients such as carbon source [71]. Inhibition by secondary metabolites could also be observed only if the expression is induced in the media and are diffusible in the agar media [56]. In addition, expression of several metabolites could be based on quorum sensing which are activated in the presence of several other microbes.
5. Conclusions
In this study, we have identified 128 antibiotic-resistant bacteria from mangrove sediments in Kerala (India) with a high incidence of clinically relevant multidrug resistance (66%). The action of such multidrug resistance was significantly (R = 0.66; p = 2.5 × 10−6) complemented by biofilm formation. Upon molecular 16S rRNA gene characterization, the isolates were found to be associated with 10 genera dominated by Bacillus. Alarming incidences of MDR resistance to ampicillin, gentamicin, chloramphenicol, ciprofloxacin, tetracycline, vancomycin, and methicillin were observed in several isolates that are potentially used as probiotics, plant-growth regulators, and also known food and human pathogens. This is of serious threat to the natural livestock in mangroves and a major concern to public welfare posed by the derived food products. Antibiotic resistance is a global issue, and its incidence has increased exponentially in the past decades. Sustainable development and strict regulatory measures should be implemented globally to minimize the occurrence and dissemination of such antibiotic resistance.
Author Contributions
Conceptualization, R.K. and M.I.; methodology, R.K., M.I., and R.K.V.; software, R.K. and M.I.; validation, R.K., M.I., and R.K.V.; formal analysis, R.K. and M.I.; investigation, R.K. and M.I.; resources, R.K.; data curation, R.K., M.I., R.K.V., and P.G.; writing—original draft preparation, R.K. and M.I.; writing—review and editing, R.K., M.I., R.K.V., and P.G.; visualization, R.K., M.I., and R.K.V.; supervision, R.K.; project administration, R.K.; funding acquisition, R.K.
Funding
Authors thank Science and Engineering Research Board (SERB) - Empowerment and Equity Opportunities for Excellence in Science (EMEQ) /051/2014 and Empowerment and Equity (EEQ)/2018/001085 for partial financial assistance, M.I. thanks University Grants Commission - National Fellowship for Higher Education of ST Students, Government of India and the research facilities supported by Central University of Kerala.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Walsh F., Duffy B. The culturable soil antibiotic resistome: A community of multi-drug resistant bacteria. PLoS ONE. 2013;8:e65567. doi: 10.1371/journal.pone.0065567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wikler M.A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved standard. CLSI (NCCLS) 2006;26:M7-A7. [Google Scholar]
- 3.D’Costa V.M., McGrann K.M., Hughes D.W., Wright G.D. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 4.Bhullar K., Waglechner N., Pawlowski A., Koteva K., Banks E.D., Johnston M.D., Barton H.A., Wright G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE. 2012;7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Li Q., Zhou K., Sun X.L., Zhao L.R., Zhang Y.B. Occurrence and distribution of the environmental pollutant antibiotics in Gaoqiao mangrove area, China. Chemosphere. 2016;147:25–35. doi: 10.1016/j.chemosphere.2015.12.107. [DOI] [PubMed] [Google Scholar]
- 6.Zhao H., Yan B., Mo X., Li P., Li B., Li Q., Li N., Mo S., Ou Q., Shen P., et al. Prevalence and proliferation of antibiotic resistance genes in the subtropical mangrove wetland ecosystem of South China Sea. MicrobiologyOpen. 2019;8:e871. doi: 10.1002/mbo3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee L.H., Zainal N., Azman A.S., Eng S.K., Goh B.H., Yin W.F., Ab Mutalib N.S., Chan K.G. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014;2014 doi: 10.1155/2014/698178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu D.B., Ye W.W., Han Y., Deng Z.X., Hong K. Natural products from mangrove actinomycetes. Mar. Drugs. 2014;12:2590–2613. doi: 10.3390/md12052590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhayanithi N.B., Kumar T.A., Murthy R.G., Kathiresan K. Isolation of antibacterials from the mangrove, Avicennia marina and their activity against multi drug resistant Staphylococcus aureus. Asian Pac. J. Trop. Biomed. 2012;2:S1892–S1895. doi: 10.1016/S2221-1691(12)60516-4. [DOI] [Google Scholar]
- 10.Imchen M., Kumavath R., Barh D., Azevedo V., Ghosh P., Viana M., Wattam A.R. Searching for signatures across microbial communities: Metagenomic analysis of soil samples from mangrove and other ecosystems. Sci. Rep. 2017;7:8859. doi: 10.1038/s41598-017-09254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imchen M., Kumavath R., Barh D., Vaz A., Góes-Neto A., Tiwari S., Ghosh P., Wattam A.R., Azevedo V. Comparative mangrove metagenome reveals global prevalence of heavy metals and antibiotic resistome across different ecosystems. Sci. Rep. 2018;8:11187. doi: 10.1038/s41598-018-29521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 13.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Toole G.A. Microtiter dish biofilm formation assay. JoVE (J. Vis. Exp.) 2011:e2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Li Q., Sun X.L., Zhao L.R., Fan H.Q. Occurrence and Distribution of Antibiotic-Resistant Bacteria Isolated in Gaoqiao Mangrove Wetland. China J. Antibiot. Res. 2017;1:202. [Google Scholar]
- 17.Sayd M., Dev A., Sahr Z., Sajisha S., Safna M., Anu Ahmmad B. Prevalence of high multiple antibiotic resistance index (mar) bacterial isolates from mangroves with reference to Calicut zones, kerala. Int. J. Adv. Sci. Eng. Technol. 2017;5:1–6. [Google Scholar]
- 18.Chopra I., Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaman S.B., Hussain M.A., Nye R., Mehta V., Mamun K.T., Hossain N. A review on antibiotic resistance: Alarm bells are ringing. Cureus. 2017;9:e1403. doi: 10.7759/cureus.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen H.K., Moe L.A., Rodbumrer J., Gaarder A., Handelsman J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009;3:243. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 21.Udikovic-Kolic N., Wichmann F., Broderick N.A., Handelsman J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. USA. 2014;111:15202–15207. doi: 10.1073/pnas.1409836111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laroche E., Pawlak B., Berthe T., Skurnik D., Petit F. Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France) FEMS Microbiol. Ecol. 2009;68:118–130. doi: 10.1111/j.1574-6941.2009.00655.x. [DOI] [PubMed] [Google Scholar]
- 23.Ghaderpour A., Ho W.S., Chew L.L., Bong C.W., Chong V.C., Thong K.L., Chai L.C. Diverse and abundant multi-drug resistant E. coli in Matang mangrove estuaries, Malaysia. Front. Microbiol. 2015;6:977. doi: 10.3389/fmicb.2015.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le T.X., Munekage Y., Kato S.I. Antibiotic resistance in bacteria from shrimp farming in mangrove areas. Sci. Total Environ. 2005;349:95–105. doi: 10.1016/j.scitotenv.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Gao P., Mao D., Luo Y., Wang L., Xu B., Xu L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012;46:2355–2364. doi: 10.1016/j.watres.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 26.De WBlackburn C., McClure P.J., editors. Foodborne Pathogens: Hazards, Risk Analysis and Control. Elsevier; Amsterdam, The Netherlands: 2009. [Google Scholar]
- 27.Taylor J.M., Sutherland A.D., Aidoo K.E., Logan N.A. Heat-stable toxin production by strains of Bacillus cereus, Bacillus firmus, Bacillus megaterium, Bacillus simplex and Bacillus licheniformis. FEMS Microbiol. Lett. 2005;242:313–317. doi: 10.1016/j.femsle.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Sanyal S., Karmaker M., Sultana M., Hossain M. Association of Bacillus circulans with non-diabetic foot infection in Bangladeshi patient. Indian J. Med. Microbiol. 2015;33 doi: 10.4103/0255-0857.167346. [DOI] [PubMed] [Google Scholar]
- 29.Ko K.S., Oh W.S., Lee M.Y., Lee J.H., Lee H., Peck K.R., Lee N.Y., Song J.H. Bacillus infantis sp. nov. and Bacillus idriensis sp. nov., isolated from a patient with neonatal sepsis. Int. J. Syst. Evol. Microbiol. 2006;56:2541–2544. doi: 10.1099/ijs.0.64213-0. [DOI] [PubMed] [Google Scholar]
- 30.Krishnan B., Massilamany C., Smith T., Loy J., Barletta R., Reddy J. Bacillus infantis NRRL B−14911-induced myocarditis: A novel disease model to study the autoimmune mechanisms of myocardial injuries in the myocarditis-susceptible, A/J mice (BA15P. 221) J. Immunol. 2014;192 [Google Scholar]
- 31.Pinheiro L., Brito C., de Oliveira A., Martins P., Pereira V., da Cunha M. Staphylococcus epidermidis and Staphylococcus haemolyticus: Molecular detection of cytotoxin and enterotoxin genes. Toxins. 2015;7:3688–3699. doi: 10.3390/toxins7093688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czekaj T., Ciszewski M., Szewczyk E.M. Staphylococcus haemolyticus—An emerging threat in the twilight of the antibiotics age. Microbiology. 2015;161:2061–2068. doi: 10.1099/mic.0.000178. [DOI] [PubMed] [Google Scholar]
- 33.Mendis H.C., Thomas V.P., Schwientek P., Salamzade R., Chien J.T., Waidyarathne P., Kloepper J., De La Fuente L. Strain-specific quantification of root colonization by plant growth promoting rhizobacteria Bacillus firmus I–1582 and Bacillus amyloliquefaciens QST713 in non-sterile soil and field conditions. PLoS ONE. 2018;13:e0193119. doi: 10.1371/journal.pone.0193119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendoza A.R., Kiewnick S., Sikora R.A. In vitro activity of Bacillus firmus against the burrowing nematode Radopholus similis, the root-knot nematode Meloidogyne incognita and the stem nematode Ditylenchus dipsaci. Biocontrol Sci. Technol. 2008;18:377–389. doi: 10.1080/09583150801952143. [DOI] [Google Scholar]
- 35.Geng C., Nie X., Tang Z., Zhang Y., Lin J., Sun M., Peng D. A novel serine protease, Sep1, from Bacillus firmus DS–1 has nematicidal activity and degrades multiple intestinal-associated nematode proteins. Sci. Rep. 2016;6:25012. doi: 10.1038/srep25012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rafikova G.F., Korshunova T.Y., Minnebaev L.F., Chetverikov S.P., Loginov O.N. A new bacterial strain, Pseudomonas koreensis IB−4, as a promising agent for plant pathogen biological control. Microbiology. 2016;85:333–341. doi: 10.1134/S0026261716030115. [DOI] [Google Scholar]
- 37.Kantas D., Papatsiros V.G., Tassis P.D., Giavasis I., Bouki P., Tzika E.D. A feed additive containing Bacillus toyonensis (Toyocerin®) protects against enteric pathogens in postweaning piglets. J. Appl. Microbiol. 2015;118:727–738. doi: 10.1111/jam.12729. [DOI] [PubMed] [Google Scholar]
- 38.Imperial I.C., Ibana J.A. Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 2016;7:1983. doi: 10.3389/fmicb.2016.01983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong A., Saint Ngu D.Y., Dan L.A., Ooi A., Lim R.L. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 2015;14:95. doi: 10.1186/s12937-015-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reisner A., Krogfelt K.A., Klein B.M., Zechner E.L., Molin S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: Impact of environmental and genetic factors. J. Bacteriol. 2006;188:3572–3581. doi: 10.1128/JB.188.10.3572-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker R.E., Laut C., Gaddy J.A., Zadoks R.N., Davies H.D., Manning S.D. Association between genotypic diversity and biofilm production in group B Streptococcus. BMC Microbiol. 2016;16:86. doi: 10.1186/s12866-016-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avila-Novoa M.G., Iñíguez-Moreno M., Solís-Velázquez O.A., González-Gómez J.P., Guerrero-Medina P.J., Gutiérrez-Lomelí M. Biofilm Formation by Staphylococcus aureus Isolated from Food Contact Surfaces in the Dairy Industry of Jalisco, Mexico. J. Food Qual. 2018;2018 doi: 10.1155/2018/1746139. [DOI] [Google Scholar]
- 43.Chavez-Dozal A., Nishiguchi M.K. Variation in biofilm formation among symbiotic and free-living strains of Vibrio fischeri. J. Basic Microbiol. 2011;51:452–458. doi: 10.1002/jobm.201000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X., Yan Z., Xu J. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology. 2003;149:353–362. doi: 10.1099/mic.0.25932-0. [DOI] [PubMed] [Google Scholar]
- 45.Saha S., Devi K.M., Damrolien S., Devi K.S., Krossnunpuii K.T. Biofilm production and its correlation with antibiotic resistance pattern among clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital in north-east India. Int. J. Adv. Med. 2018;5:964. doi: 10.18203/2349-3933.ijam20183129. [DOI] [Google Scholar]
- 46.Namuq A.O., Ali K.O., Al-Ani A.H. Correlation between Biofilm Formation, Multi-Drug Resistance and AlgD Gene among Pseudomonas aeruginosa Clinical Isolates. J. Univ. Babylon Pure Appl. Sci. 2019;27:143–150. [Google Scholar]
- 47.Roshani-Asl P., Rashidi N., Shokoohizadeh L., Zarei J. Relationship Among Antibiotic Resistance, Biofilm Formation and lasB Gene in Pseudomonas Aeruginosa Isolated from Burn Patients. Clin. Lab. 2018;64:1477–1484. doi: 10.7754/Clin.Lab.2018.180331. [DOI] [PubMed] [Google Scholar]
- 48.Tabatabaei M., Sohrabi N. Comparison of biofilm formation and antibiotic resistance pattern of Pseudomonas aeruginosa in human and environmental isolates. Microb. Pathog. 2017;109:94–98. doi: 10.1016/j.micpath.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Rao R.S., Karthika R.U., Singh S.P., Shashikala P., Kanungo R., Jayachandran S., Prashanth K. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J. Med. Microbiol. 2008;26:333. doi: 10.4103/0255-0857.43566. [DOI] [PubMed] [Google Scholar]
- 50.Karigoudar R.M., Karigoudar M.H., Wavare S.M., Mangalgi S.S. Detection of biofilm among uropathogenic Escherichia coli and its correlation with antibiotic resistance pattern. J. Lab. Physicians. 2019;11:17. doi: 10.4103/JLP.JLP_98_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shahidul K.M., Farahnaaz F., Sunjukta A. Determination of antibiotic resistance pattern of biofilm producing pathogenic bacteria associated with UTI. Int. J. Drug Dev. Res. 2013;5:312–319. [Google Scholar]
- 52.Cepas V., López Y., Munoz E., Rolo D., Ardanuy C., Martí S., Xercavins M., Horcajada J.P., Bosch J., Soto S.M. Relationship between Biofilm Formation and Antimicrobial Resistance in Gram-Negative Bacteria. Microb. Drug Resist. 2019;25:72–79. doi: 10.1089/mdr.2018.0027. [DOI] [PubMed] [Google Scholar]
- 53.Türkel İ., Yıldırım T., Yazgan B., Bilgin M., Başbulut E. Relationship between antibiotic resistance, efflux pumps, and biofilm formation in extended-spectrum β-lactamase producing Klebsiella pneumoniae. J. Chemother. 2018;30:354–363. doi: 10.1080/1120009X.2018.1521773. [DOI] [PubMed] [Google Scholar]
- 54.Balcázar J.L., Subirats J., Borrego C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015;6:1216. doi: 10.3389/fmicb.2015.01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert P., Maira-Litran T., McBain A.J., Rickard A.H., Whyte F. The Physiology and Collective Recalcitrance of Microbial Biofilm Communities–2 Changes in susceptibility associated with cellular aggregates. Adv. Microb. Physiol. 2002;46:213–216. [PubMed] [Google Scholar]
- 56.Stewart P.S., Franklin M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008;6:199. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 57.Hall C.W., Mah T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 58.Burns A., Ryder D.S. Potential for biofilms as biological indicators in Australian riverine systems. Ecol. Manag. Restor. 2001;2:53–64. doi: 10.1046/j.1442-8903.2001.00069.x. [DOI] [Google Scholar]
- 59.Sabater S., Guasch H., Ricart M., Romaní A., Vidal G., Klünder C., Schmitt-Jansen M. Monitoring the effect of chemicals on biological communities. The biofilm as an interface. Anal. Bioanal. Chem. 2007;387:1425–1434. doi: 10.1007/s00216-006-1051-8. [DOI] [PubMed] [Google Scholar]
- 60.Laport M.S., Santos-Gandelman J.F., Muricy G., Giambiade-deMarval M., George I. Antagonistic interactions among bacteria isolated from either the same or from different sponges native to the Brazilian coast. J. Mar. Sci. Res. Dev. 2016;6:2. doi: 10.4172/2155-9910.1000185. [DOI] [Google Scholar]
- 61.Russel J., Røder H.L., Madsen J.S., Burmølle M., Sørensen S.J. Antagonism correlates with metabolic similarity in diverse bacteria. Proc. Natl. Acad. Sci. USA. 2017;114:10684–10688. doi: 10.1073/pnas.1706016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mangano S., Michaud L., Caruso C., Brilli M., Bruni V., Fani R., Giudice A.L. Antagonistic interactions between psychrotrophic cultivable bacteria isolated from Antarctic sponges: A preliminary analysis. Res. Microbiol. 2009;160:27–37. doi: 10.1016/j.resmic.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 63.Long R.A., Azam F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 2001;67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nair S.H., Simidu U.S. Distribution and significance of heterotrophic marine bacteria with antibacterial activity. Appl. Environ. Microbiol. 1987;53:2957–2962. doi: 10.1128/aem.53.12.2957-2962.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lo Giudice A., Brilli M., Bruni V., De Domenico M., Fani R., Michaud L. Bacterium–bacterium inhibitory interactions among psychrotrophic bacteria isolated from Antarctic seawater (Terra Nova Bay, Ross Sea) FEMS Microbiol. Ecol. 2007;60:383–396. doi: 10.1111/j.1574-6941.2007.00300.x. [DOI] [PubMed] [Google Scholar]
- 66.Kanagasabhapathy M., Nagata S. Cross-species induction of antibacterial activity produced by epibiotic bacteria isolated from Indian marine sponge Pseudoceratina purpurea. World J. Microbiol. Biotechnol. 2008;24:687–691. doi: 10.1007/s11274-007-9525-1. [DOI] [Google Scholar]
- 67.Hentschel U., Schmid M., Wagner M., Fieseler L., Gernert C., Hacker J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001;35:305–312. doi: 10.1111/j.1574-6941.2001.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 68.Grossart H.P., Schlingloff A., Bernhard M., Simon M., Brinkhoff T. Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol. Ecol. 2004;47:387–396. doi: 10.1016/S0168-6496(03)00305-2. [DOI] [PubMed] [Google Scholar]
- 69.Vetsigian K., Jajoo R., Kishony R. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 2011;9:e1001184. doi: 10.1371/annotation/1d584443-c6b8-423b-8027-5f9034d4599f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cordero O.X., Wildschutte H., Kirkup B., Proehl S., Ngo L., Hussain F., Le Roux F., Mincer T., Polz M.F. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 71.Deng Y.J., Wang S.Y. Complex carbohydrates reduce the frequency of antagonistic interactions among bacteria degrading cellulose and xylan. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujimoto M., Lovett B., Angoshtari R., Nirenberg P., Loch T.P., Scribner K.T., Marsh T.L. Antagonistic Interactions and Biofilm Forming Capabilities Among Bacterial Strains Isolated from the Egg Surfaces of Lake Sturgeon (Acipenser fulvescens) Microb. Ecol. 2018;75:22–37. doi: 10.1007/s00248-017-1013-z. [DOI] [PubMed] [Google Scholar]
- 73.Hoffman L.R., D’Argenio D.A., MacCoss M.J., Zhang Z., Jones R.A., Miller S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]