Abstract
Hepatocellular carcinoma (HCC) is the most commonly diagnosed liver cancer, accounting for ~90% of all primary malignancy of the liver. Although various medical treatments have been used as systemic therapies, patient survival time may be extended by only a few months. Moreover, the underlying mechanisms of HCC development and progression remain poorly understood. In the present study, the single-cell transcriptome of one in vivo HCC tumor sample, two in vitro HCC cell lines and normal peripheral blood mononuclear cells were analysed in order to identify the potential mechanism underlying the development and progression of HCC. Interestingly, JunB proto-oncogene was identified to serve a role in the immune response and in development and progression of HCC, potentially contributing to the development of novel therapeutics for HCC patients.
Keywords: JunB proto-oncogene, hepatocellular carcinoma, single-cell RNA-sequencing, immune environment
Introduction
Hepatocellular carcinoma (HCC) is a malignant tumor with high incidence and the number of novel cases is expected to increase by 1 million every year in the next decade (1). The main causative factors include alcoholic liver disease, hepatitis B and hepatitis C viral infections, and non-alcoholic fatty liver disease (2). However, the pathogenesis and exact molecular mechanism of HCC are not fully understood. Although international guidelines suggest HCC screening for patients with cirrhosis, regular monitoring presents various limitations in clinical practice (3). The large number of undiagnosed patients leads to a low HCC monitoring rate and high late diagnosis rate, causing the patients with HCC to be diagnosed only when the tumor exhibits a large size, leading to poor prognosis (4).
During tumor development and progression, multiple cell types interact with tumor cells, including astrocytes, B cells, lymphocytes, macrophages, monocytes, natural killer cells and T cells, constituting the tumor immune microenvironment (5,6). These cells, together with the fibroblasts, vascular endothelial cells and other factors, which are collectively called the tumor stroma, as well as the extracellular matrix, oxygen levels and pH values, constitute the tumor microenvironment (7,8). Notably, the interactions between immune cells and tumor cells affect the growth and remodeling of the tumor microenvironment (9). Immune cells can stimulate tumor cells to secrete cytokines, which mediate the tumor growth by promoting the growth of new blood and lymphatic vessels (6). Different cell types may serve pro- and anti-tumorigenic roles in the tumor microenvironment. For example, targeting T cell activation is considered as an important novel strategy to repress tumor growth (10,11). S100A4 has been shown to be an oncogene able to promote inflammation (12) and affect angiogenesis (13). Accumulating evidence showed that expression of S100A4 in tumor cells is related to the tumor-associated T cell deficiency (14). The rich blood supply and unique sinusoid structure of the liver provide a plastic environment for the formation and function of the tumor immune microenvironment (15). Therefore, it is of great significance to study the molecular characteristics and intercellular interactions in the HCC immune microenvironment.
Massively parallel sequencing data have provided novel insights in the field of cancer research. In particular, RNA-sequencing (RNA-seq) has been used to detect genomic mutations and rearrangement signatures in the human genome and transcriptome. However, conventional bulk RNA-seq can only provide the average expression signal of transcripts in the whole tumor tissue, without considering the tumor heterogeneity. By contrast, single-cell RNA-seq may facilitate the identification of complex and rare cells populations, thus allowing investigation of the tumor immune microenvironment (16), especially in HCC. For example, using single-cell RNA-seq, a previous study identified 11 HCC-related T cell subpopulations, which provided valuable insights for the understanding of the cancer immune microenvironment (17). In addition, a previous study has described the molecular characteristics of immune cells that infiltrate HCC to determine whether certain types of drugs may be effective against liver cancer (18).
Moreover, chromatin immunoprecipitation (ChIP)-seq results and protein-protein interaction (PPI) network may facilitate the detection of gene regulatory networks and interaction events, such as the bindings between transcription factors (TFs) and promoters. The present study compared the single-cell RNA-seq data of normal peripheral blood mononuclear cells (PBMCs) with that of in vivo tumor cells and in vitro cell lines using single cell classification and identification. By integrating differential expression analysis, ChIP-seq data and PPI networks, the present results suggested that the JunB proto-oncogene (JUNB) may serve an important role in the development and progression of HCC and the immune response. In addition, apolipoprotein A2 (APOA2), which encodes a genetically susceptible protein in HCC (19), was found to exhibit the same expression pattern as JUNB. The present results may contribute to the identification of novel therapeutic targets for the treatment of HCC patients.
Materials and methods
Data collection
In vivo tumor cells were isolated from a patient who had undergone resection at the National Institutes of Health (NIH) Clinical Center. The tissue acquisition procedures were approved by the Institutional Review Board of The NIH (20). In total, two in vitro cell lines (HuH1 and HuH7) from The Health Science Research Resources Bank (cat. nos. JCRB0199 and JCRB0403) were pooled and used for 10× Genomics single-cell RNA-seq. These data were collected and downloaded from the GEO database (database no. GSE103867) (20). Single-cell data of PBMCs were downloaded from the GEO database (database no. GSE111360) (21). Gene expression profile and clinical information of a cohort of 360 patients with HCC were collected from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/). The PPI network was obtained using STRING (v11.0) with only highly strong interactions (score, 0.4) being used (22).
Preprocessing for 10× Genomics single-cell RNA-seq data
Seurat v2.1 (http://satijalab.org/seurat/) was used to analyze the 10× Genomics data (23). Genes whose expression was detected in ≥3 cells and cells with ≥10 genes were used in this study. Variable genes were identified using cutoffs (x.low.cutoff = 0.05; y.cutoff = 0.1). The top 20 principal components were used in the clustering analysis (resolution = 0.6). Gene expression levels were quantified using the unique molecular identifier counts. Dimensionality reduction was based on the t-SNE algorithm. Subsequently, cell populations were clustered by principal component analysis.
Differential expression gene and pathway enrichment analysis
Log2Fold-Change represented the ratio of gene expression between one cluster of cells and all the other cells. P-values were calculated using the negative binomial test and adjusted by the Benjamini-Hochberg method. Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using DAVID (version 6.8) (24).
Correlation analysis
Gene expression of 360 patients with HCC, obtained from the TCGA database, was used as an independent external test set to validate the putative genes of interest. Co-expression analysis based on Spearman's Correlation was performed using cBioPortal (http://www.cbioportal.org/) (25).
Survival analysis
Data from patients with HCC derived from TCGA were divided into two groups according to the expression of APOA2 (high or low, with an mRNA Z-score >0 or <0, respectively) and the expression of JUNB (high or low, with an mRNA Z-score >0 or <0, respectively). Survival curves were estimated by the Kaplan-Meier method and compared with the log-rank test.
Transcriptional regulation analysis
For each gene, interactions between proteins and their promoters and enhancers were obtained from r GeneHancer (26), a database of ChIP-seq data classify by inferred target genes. The interactions between TFs and their binding sites in the promoter and enhancer regions were supported by ChIP-sequencing. Subsequently, the TFs predicted to regulate both JUNB and APOA2 were used.
Results
Identification of significant differences in gene expression between in vivo and in vitro cells
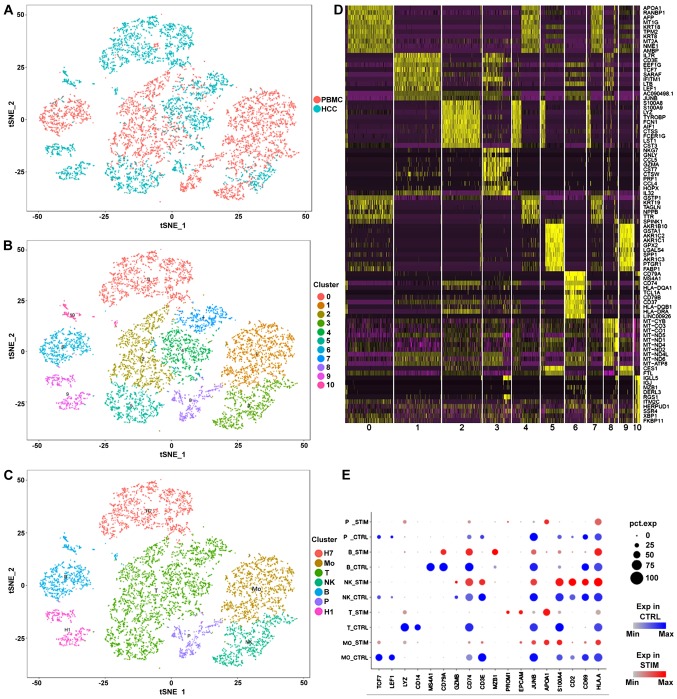
Differential expression analysis was performed between the transcriptomes of in vivo tumor cells and two in vitro cell lines (Fig. S1). Top 2 cell population-specific marker gene expression is presented in Fig. S2A. Top 9 cell type-specific expressed genes are presented in Fig. S2B (HuH7 cells, Fig. S2Ba; P1T and P1C cells, Fig. S2Bb; P1B cells, Fig. S2Bc; and HuH1 cells; Fig. S2Bd). The specific expression was potentially due to the invasion of immune cells in the tumor and the immune response elicited by tumor cells (27). These significant differences suggested that the validation of the RNA-seq data should be a crucially important component for the in vitro analysis of the immune response in tumors. Cell population identifications of PBMCs are presented in Fig. S3A-D. Subsequently, an integrated comparison among tumor cells, cell lines and PBMCs was performed (Fig. 1). As expected, HuH1 and HuH7 cells clustered together in the analysis (Fig. 1A). Interestingly, the clustering analysis showed that a small number of epithelial cells were observed among the PBMCs (Fig. 1A-C). According to previous studies (28–30), these epithelial cells may be vascular endothelial cells, which have the potential to regulate the tumor cells. In addition to B cells and T cells, numerous types of mononuclear cells were detected among the PBMCs (Fig. 1C). A heat map was used to observe the expression of the differentially expressed genes (marker genes) in the comparisons (Fig. 1D). The expression of the marker genes in each group of single cells is presented in Fig. 1E.
Figure 1.
Single-cell transcriptomic data of PBMCs, HCC tumor cells and liver cancer cell lines integration. (A) Integration of PMBC and tumor samples. (B) Identification of cell populations by PCA. (C) Definition of cell types according to the marker genes. (D) Heatmap of the markers identified by PCA. (E) Comparison between expression of marker genes in PBMC and tumor samples. The color blue indicates the expression levels in PBMC samples, the color red in tumor samples. The size of each dot represents the percentage of the cells that expressed the corresponding gene. H1, HuH1; H7, HuH7; Mo, monocytes; NK, natural killer cells; P, epithelial cells; T, T cells; B, B cells; PBMC, peripheral blood mononuclear cells; PCA, principal component analysis; STIM, tumor samples; CTRL, PBMC samples; HCC, hepatocellular carcinoma.
Tumor-infiltrating immune cells exhibit a higher transcription level
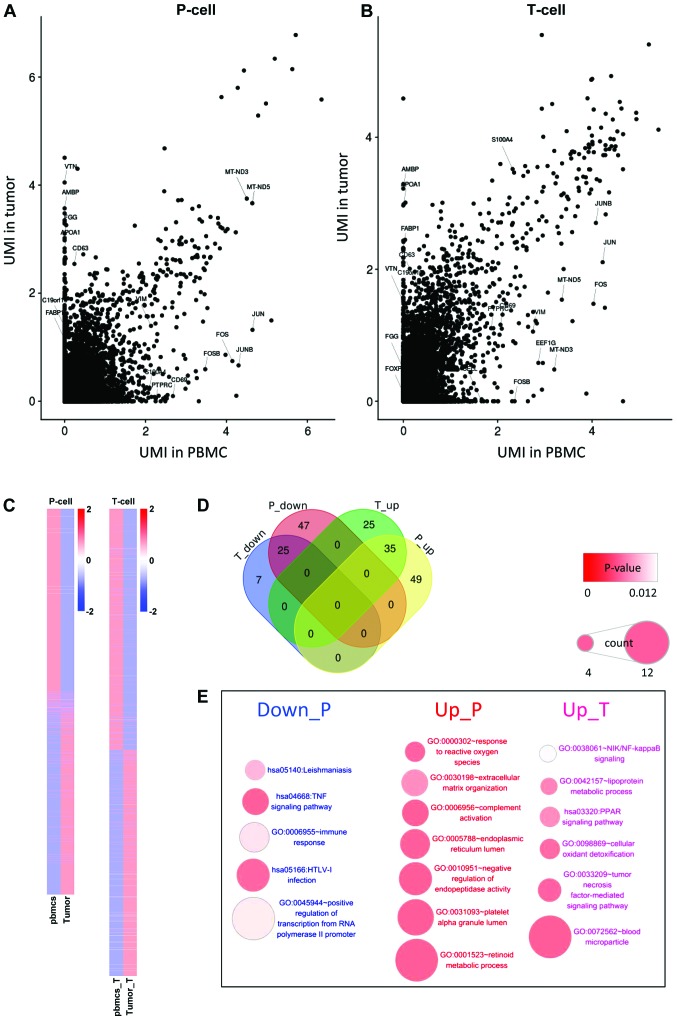
In the analysis of differentially expressed genes, the gene expression levels between tumor cells and vascular epithelial cells in peripheral blood (Fig. 2A) were compared. In addition, the differentially expressed genes between T cells infiltrated in tumors and T cells in peripheral blood were examined (Fig. 2B). In the two comparisons, the number of the differentially expressed genes in T cells was significantly decreased compared with those in epithelial cells, while most of the differentially expressed genes in T cells were due to differences among cell types (Fig. 2C and D). Enrichment analysis of these differentially expressed genes showed that the differentially expressed genes in epithelial cells were significantly enriched in ‘inflammation’ and ‘immune response’ (Fig. 2E). The inhibition of some positive regulators of T cells, such as tumor necrosis factor (TNF), NF-κB inhibitor α (NFKBIA), Fos proto-oncogene (FOS), JUN and DEAD-box helicase 3 X-linked (DDX3X) may lead to the downregulation of the T cell receptor signaling pathway, and the inhibition of the hepatitis B and cellular oxidant detoxification pathways, which may be caused by the abnormal growth of tumor cells and the accumulation of stress-associated factors. In this condition, tumor cells may require more energy to sustain their proliferation and to adapt to the hypoxic micro-environment.
Figure 2.
Differentially expressed analysis in tumor cells and T cells. (A) A scatter plot of the gene expression correlation between tumor cells and vascular epithelial cells in the peripheral blood. (B) A scatter plot showing the gene expression correlation between T cells infiltrated in the tumor and T cells in the peripheral blood. (C) A heatmap of differentially expressed genes in the comparisons. (D) A Venn diagram of the number of the differential genes with |log2Fold-Change|>2 in the comparisons. (E) Enriched Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways of these differential genes. UMI, unique molecular identifier; PBMC, peripheral blood mononuclear cells.
JUNB may serve a crucial role in HCC tumor immune microenvironment
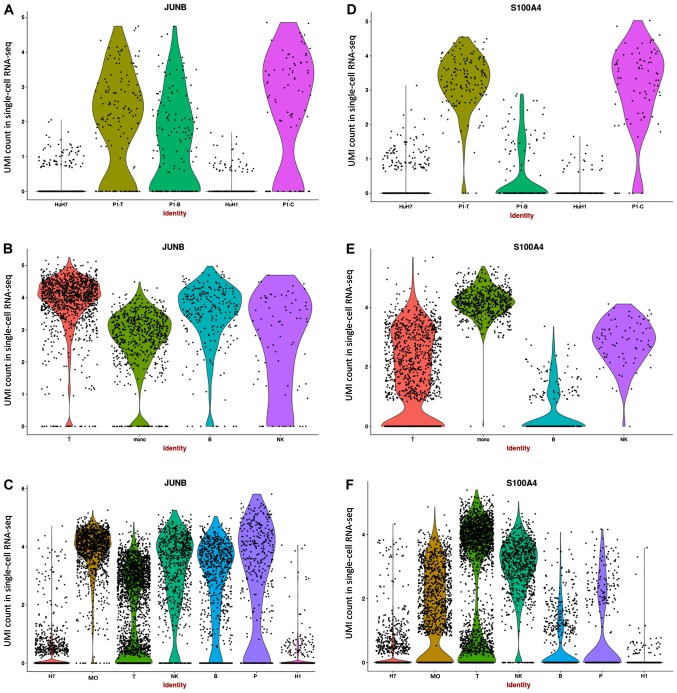
JUNB, which was reported as highly expressed in cancer cells in previous studies (22,31–33), was observed to be downregulated in numerous tumor cells, although it was observed to be highly expressed in certain tumor cell lines (Fig. 3A). Additionally, all PBMCs presented high expression levels of JUNB (Fig. 3B). Since JUNB is associated with lipid metabolism, high expression of JUNB was expected in activated cells. S100A4, which is associated with the development and progression of the tumor and immune infiltration (34,35), exhibited high expression in in vivo epithelial cells and T cells, but low expression in in vitro cell lines (Fig. 3D and E). Although the indirect interaction between JUNB and S100A4 was found to function through annexin A2 (ANXA2; Fig. S3E), a positive association between ANXA2, JUNB and S100A4 was detected in patients with HCC from the TCGA dataset (Fig. S3F). In addition, the functional roles of ANXA2, JUNB and S100A4 in tumor cells were found to be associated with T cells (14,36,37), suggesting that the function of JUNB in HCC tumor cells may be associated with the interaction between these genes in the tumor immune microenvironment. By combining the data from PBMCs and tumor samples, JUNB and S1004A were found to be decreased in epithelial cells (Fig. 3C and F). However, the expression level of S100A4 was found to be increased in tumor-associated T cells, while the expression of JUNB was decreased. JUNB was identified to be downregulated in various tumors in the previous studies (31).
Figure 3.
Violin plots of the gene expression levels of JUNB and S100A4 in different cell populations. (A) Gene expression of JUNB in tumor samples. (B) Gene expression of JUNB in PBMC samples. (C) Gene expression of JUNB in integrated samples. (D) Gene expression of S1004A in tumor samples. (E) Gene expression of S1004A in PBMC samples. (F) Gene expression of S1004A in integrated samples. PBMC, peripheral blood mononuclear cells; UMI, unique molecular identifier.
Validation of the key role of JUNB in an independent dataset
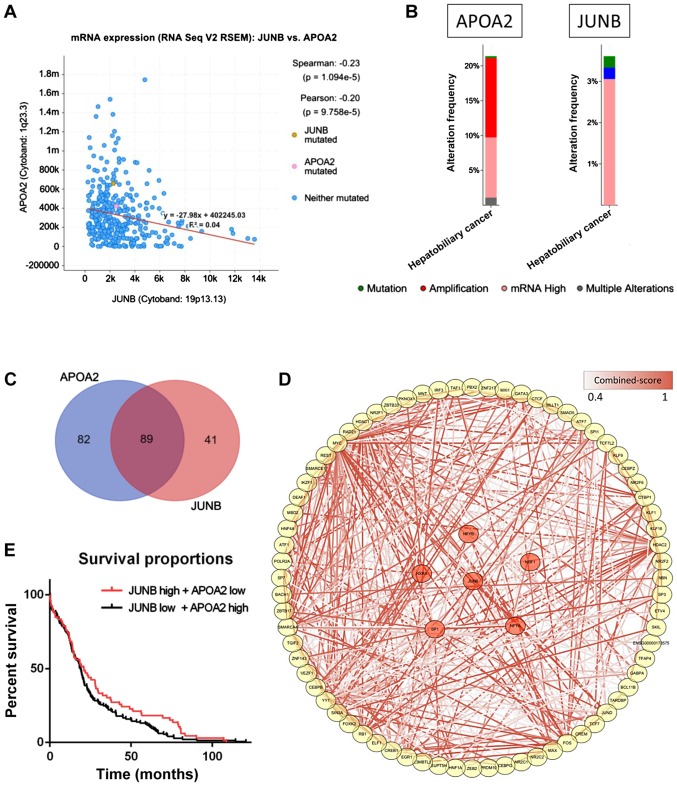
The potential role of JUNB was further investigated by integrating a cohort of 360 HCC patients from the TCGA database. APOA2 was found to be significantly negatively associated with JUNB in the dataset (P=1.094×10−5; Fig. 4A). Among the 360 HCC samples, >20% of the samples showed APOA2 mRNA upregulation, which may be explained by the low expression of JUNB in HCC samples (Fig. 4B). The high expression of JUNB was previously predicted as a poor survival indicator in patients with tumors (32,33), while APOA2 was found to serve a role in the development and progression of the tumor through the peroxisome proliferator activated receptor α (PPARα) pathway (38). Moreover, >50% of the TFs that interact with the promoters and enhancers of these two genes were found to regulate both APOA2 and JUNB (Fig. 4C). Further data enrichment analysis showed that the pathways associated with the TFs regulating APOA2 and JUNB were involved in the development and progression of HCC and immune response, which may represent a potential novel mechanism underlying HCC (Fig. 4D), although further experimental evidence is required to test this hypothesis. Although survival analysis showed a slightly different result when analyzing the overall survival time in two groups of patients (P=0.11), a significant difference was observed when analyzing only longer survival time (>15 months; Fig. 4E), indicating that JUNB and APOA2 may play a key role in improving the survival time in patients with HCC.
Figure 4.
Validation of JUNB expression in the TCGA dataset. (A) A scatter plot of the gene expression correlation of APOA2 and JUNB. (B) Mutation and expression level of APOA2 and JUNB in the HCC dataset from TCGA. (C) A Venn diagram of the number of the regulatory elements of APOA2 and JUNB detected in GeneHancer. (D) TFs that coregulated APOA2 and JUNB. Color of the edge indicates the confidence level of the interaction between two proteins. The nodes, in red, indicate the PPAR signaling pathway associated proteins. (E) A survival curve for patients from TCGA with high/low expression of APOA2 and low/high expression of JUNB. TF, transcription factor; TCGA, The Cancer Genome Atlas; APOA2, apolipoprotein A2; PPAR, peroxisome proliferator activated receptor; HCC, hepatocellular carcinoma; JUNB, JunB proto-oncogene.
Discussion
Increased understanding of tumor-host interactions has accelerated the development of novel cancer immunotherapies. In addition, drug resistance in biomarker therapy and immunotherapy have recently been investigated (21). Despite their success, immune-checkpoint inhibitors present certain limitations. Therefore, in addition to improving the treatment of patients presenting with tumors at an advanced stage, the identification of driver biomarkers may contribute to immunotherapy in an early clinical stage (39). Since immune-checkpoint inhibitors could be used in HCC treatment, combining molecular targeted therapy with immunotherapy has become a therapeutic method to stimulate the immune response. The previously described mechanisms underlying tumor development based on cell lines studies have been found to be unreliable, particularly for immune response and immunotherapy-related studies. In order to investigate the role of the immune response in tumor progression, the tumor microenvironment and the balance between tumor cells and immune cells must be considered.
The present results suggested that the inhibition of JUNB may be a key indicator of the regulation of the APOA2-associated PPARα pathway in HCC (31). APOA2 is a well-known member of the apolipoprotein family (40), which is functionally involved in triglyceride, fatty acid and glucose metabolism. This gene family has been previously reported to be overexpressed in HCC and regulated by the PPARα pathway (41). In addition, together with the co-expression of APOA2 and JUNB observed in the TCGA dataset and the transcriptional regulation analysis, a potential regulation of APOA2 and JUNB by the PPARα pathway was identified. Finally, the present survival analysis in HCC patients suggested that the investigation of JUNB may facilitate the identification of novel therapeutic targets for HCC patients. The inhibition of some positive regulators of T cells, such as TNF, NFKBIA, FOS, JUN and DDX3X may lead to the downregulation of the T cell receptor signaling pathway (42–44), affecting the hepatitis B and cellular oxidant detoxification response, which may be caused by the abnormal growth of tumor cells and the accumulation of stress-associated factors. In this condition, tumor cells may require more energy to sustain their proliferation and to adapt to the hypoxic micro-environment.
Although the present results could potentially contribute to the development of novel therapeutics to treat patients with HCC, the lack of experimental validations on mRNA levels are still the main limitations for the study. In addition, investigating the expression levels of the products of candidate mRNAs, the proteins, is equally critical to validate the results, as they play a central role in biological processes. Furthermore, validations using both in vivo and in vitro models should be further investigated, such as ChIP to detect the potential regulatory regions on JUNB, Junb defective mice to study the important functions of Junb.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HCC
hepatocellular carcinoma
- PBMC
peripheral blood mononuclear cells
- TCGA
The Cancer Genome Atlas
- TF
transcription factors
- UMI
unique molecular identifier
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81501561); the Natural Science Foundation of Guangdong Province (grant nos. 2014A030310043 and 2017A030313873) and the Science and Technology Planning Project of Zhuhai (grant no. 20171009E030008).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
PY, BZ, PP and JM conceived and designed the experiments. PY, YM, AW, XH, YL, YY and YW analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable. No patients were enrolled in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Harris PS, Hansen RM, Gray ME, Massoud OI, McGuire BM, Shoreibah MG. Hepatocellular carcinoma surveillance: An evidence-based approach. World J Gastroenterol. 2019;25:1550–1559. doi: 10.3748/wjg.v25.i13.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bou-Nader M, Caruso S, Donne R, Celton-Morizur S, Calderaro J, Gentric G, Cadoux M, L'Hermitte A, Klein C, Guilbert T, et al. Polyploidy spectrum: A new marker in HCC classification. Gut. 2019;(pii) doi: 10.1136/gutjnl-2018-318021. gutjnl-2018-318021. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HL, An J, Park JA, Park SH, Lim YS, Lee EK. Magnetic resonance imaging is cost-effective for hepatocellular carcinoma surveillance in high-risk patients with cirrhosis. Hepatology. 2019;69:1599–1613. doi: 10.1002/hep.30330. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Zou R, Zheng Y, Qiu J, Shen J, Liao Y, Zhang Y, Wang C, Wang Y, Yuan Y, et al. Lipiodol deposition in portal vein tumour thrombus predicts treatment outcome in HCC patients after transarterial chemoembolisation. Eur Radiol. 2019;29:5752–5762. doi: 10.1007/s00330-019-06157-0. [DOI] [PubMed] [Google Scholar]
- 5.Al-Zoughbi W, Huang J, Paramasivan GS, Till H, Pichler M, Guertl-Lackner B, Hoefler G. Tumor macroenvironment and metabolism. Semin Oncol. 2014;41:281–195. doi: 10.1053/j.seminoncol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 7.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 8.Hirata E, Sahai E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb Perspect Med. 2017;7:a026781. doi: 10.1101/cshperspect.a026781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Ma L, Shen S, Guo Y, Cao Q, Cai X, Feng J, Yan Y, Hu T, Luo S, et al. Intestinal dysbacteriosis-induced IL-25 promotes development of HCC via alternative activation of macrophages in tumor microenvironment. J Exp Clin Cancer Res. 2019;38:303. doi: 10.1186/s13046-019-1456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis JM, Kiezun A, Ramos AH, Serra S, Pedamallu CS, Qian ZR, Banck MS, Kanwar R, Kulkarni AA, Karpathakis A, et al. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat Genet. 2013;45:1483–1486. doi: 10.1038/ng.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang HJ, Oh JH, Chun SM, Kim D, Ryu YM, Hwang HS, Kim SY, An J, Cho EJ, Lee H, et al. Immunogenomic landscape of hepatocellular carcinoma with immune cell stroma and EBV-positive tumor-infiltrating lymphocytes. J Hepatol. 2019;71:91–103. doi: 10.1016/j.jhep.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Q, Hou S, Zhai J, Tian T, Wu Y, Wu Z, He J, Chen Z, Zhang J. S100A4 promotes inflammation but suppresses lipid accumulation via the STAT3 pathway in chronic ethanol-induced fatty liver. J Mol Med (Berl) 2019;97:1399–1412. doi: 10.1007/s00109-019-01808-7. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt-Hansen B, Ornås D, Grigorian M, Klingelhöfer J, Tulchinsky E, Lukanidin E, Ambartsumian N. Extracellular S100A4(mts1) stimulates invasive growth of mouse endothelial cells and modulates MMP-13 matrix metalloproteinase activity. Oncogene. 2004;23:5487–5495. doi: 10.1038/sj.onc.1207720. [DOI] [PubMed] [Google Scholar]
- 14.Grum-Schwensen B, Klingelhöfer J, Grigorian M, Almholt K, Nielsen BS, Lukanidin E, Ambartsumian N. Lung metastasis fails in MMTV-PyMT oncomice lacking S100A4 due to a T-cell deficiency in primary tumors. Cancer Res. 2010;70:936–947. doi: 10.1158/0008-5472.CAN-09-3220. [DOI] [PubMed] [Google Scholar]
- 15.Ringelhan M, Pfister D, O'Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222–232. doi: 10.1038/s41590-018-0044-z. [DOI] [PubMed] [Google Scholar]
- 16.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:96. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. Landscape of infiltrating t cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356 e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 18.Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, Putra J, Camprecios G, Bassaganyas L, Akers N, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology. 2017;153:812–826. doi: 10.1053/j.gastro.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Zhong DN, Ning QY, Wu JZ, Zang N, Wu JL, Hu DF, Luo SY, Huang AC, Li LL, Li GJ. Comparative proteomic profiles indicating genetic factors may involve in hepatocellular carcinoma familial aggregation. Cancer Sci. 2012;103:1833–1838. doi: 10.1111/j.1349-7006.2012.02368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng H, Pomyen Y, Hernandez MO, Li C, Livak F, Tang W, Dang H, Greten TF, Davis JL, Zhao Y, et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127–140. doi: 10.1002/hep.29778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988 e16. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KH, Kim JR. Regulation of HGF-mediated cell proliferation and invasion through NF-κB, JunB, and MMP-9 cascades in stomach cancer cells. Clin Exp Metastasis. 2012;29:263–272. doi: 10.1007/s10585-011-9449-x. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Fan J, Francis JM, Georghiou G, Hergert S, Li S, Gambe R, Zhou CW, Yang C, Xiao S, et al. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res. 2017;27:1300–1311. doi: 10.1101/gr.217331.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:ppl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M, et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. 2017 doi: 10.1093/database/bax028. Database (Oxford) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fennemann FL, de Vries IJM, Figdor CG, Verdoes M. Attacking tumors from all sides: Personalized multiplex vaccines to tackle intratumor heterogeneity. Front Immunol. 2019;10:824. doi: 10.3389/fimmu.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali M, Khan SY, Vasanth S, Ahmed MR, Chen R, Na CH, Thomson JJ, Qiu C, Gottsch JD, Riazuddin SA. Generation and proteome profiling of pbmc-originated, ipsc-derived corneal endothelial cells. Invest Ophthalmol Vis Sci. 2018;59:2437–2444. doi: 10.1167/iovs.17-22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Wijst MGP, Brugge H, de Vries DH, Deelen P, Swertz MA, LifeLines Cohort Study BIOS Consortium. Franke L. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet. 2018;50:493–497. doi: 10.1038/s41588-018-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Q, Hawkins GA, Wudel L, Chou PC, Forbes E, Pullikuth AK, Liu L, Jin G, Craddock L, Topaloglu U, et al. Dissecting intratumoral myeloid cell plasticity by single cell RNA-seq. Cancer Med. 2019;8:3072–3085. doi: 10.1002/cam4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo C, Liu QG, Zhang L, Song T, Yang X. Expression and clinical significance of p53, JunB and KAI1/CD82 in human hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2009;8:389–396. [PubMed] [Google Scholar]
- 32.Guo C, Liu Q, Zhang L, Yang X, Song T, Yao Y. Double lethal effects of fusion gene of wild-type p53 and JunB on hepatocellular carcinoma cells. J Huazhong Univ Sci Technolog Med Sci. 2012;32:663–668. doi: 10.1007/s11596-012-1014-6. [DOI] [PubMed] [Google Scholar]
- 33.Chang YS, Yeh KT, Yang MY, Liu TC, Lin SF, Chan WL, Chang JG. Abnormal expression of JUNB gene in hepatocellular carcinoma. Oncol Rep. 2005;13:433–438. [PubMed] [Google Scholar]
- 34.Zhai X, Zhu H, Wang W, Zhang S, Zhang Y, Mao G. Abnormal expression of EMT-related proteins, S100A4, vimentin and E-cadherin, is correlated with clinicopathological features and prognosis in HCC. Med Oncol. 2014;31:970. doi: 10.1007/s12032-014-0970-z. [DOI] [PubMed] [Google Scholar]
- 35.Dukhanina EA, Lukyanova TI, Romanova EA, Guerriero V, Gnuchev NV, Georgiev GP, Yashin DV, Sashchenko LP. A new role for PGRP-S (Tag7) in immune defense: Lymphocyte migration is induced by a chemoattractant complex of Tag7 with Mts1. Cell Cycle. 2015;14:3635–3643. doi: 10.1080/15384101.2015.1104440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Ma S, Hotz-Wagenblatt A, Angel P, Mohr K, Schlimbach T, Schmitt M, Cui G. Regulatory T cells sense effector T-cell activation through synchronized JunB expression. FEBS Lett. 2019;593:1020–1029. doi: 10.1002/1873-3468.13393. [DOI] [PubMed] [Google Scholar]
- 37.Kim VM, Blair AB, Lauer P, Foley K, Che X, Soares K, Xia T, Muth ST, Kleponis J, Armstrong TD, et al. Anti-pancreatic tumor efficacy of a listeria-based, annexin A2-targeting immunotherapy in combination with anti-PD-1 antibodies. J Immunother Cancer. 2019;7:132. doi: 10.1186/s40425-019-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagasawa M, Akasaka Y, Ide T, Hara T, Kobayashi N, Utsumi M, Murakami K. Highly sensitive upregulation of apolipoprotein A-IV by peroxisome proliferator-activated receptor alpha (PPARalpha) agonist in human hepatoma cells. Biochem Pharmacol. 2007;74:1738–1746. doi: 10.1016/j.bcp.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 39.Harris WP, Wong KM, Saha S, Dika IE, Abou-Alfa GK. Biomarker-driven and molecular targeted therapies for hepatobiliary cancers. Semin Oncol. 2018;45:116–123. doi: 10.1053/j.seminoncol.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Ballester M, Revilla M, Puig-Oliveras A, Marchesi JA, Castelló A, Corominas J, Fernández AI, Folch JM. Analysis of the porcine APOA2 gene expression in liver, polymorphism identification and association with fatty acid composition traits. Anim Genet. 2016;47:552–559. doi: 10.1111/age.12462. [DOI] [PubMed] [Google Scholar]
- 41.Thulin P, Glinghammar B, Skogsberg J, Lundell K, Ehrenborg E. PPARdelta increases expression of the human apolipoprotein A-II gene in human liver cells. Int J Mol Med. 2008;21:819–824. [PubMed] [Google Scholar]
- 42.Lu G, Zhang G, Zheng X, Zeng Y, Xu Z, Zeng W, Wang K. c9, t11- conjugated linoleic acid induces HCC cell apoptosis and correlation with PPAR-γ signaling pathway. Am J Transl Res. 2015;7:2752–2763. [PMC free article] [PubMed] [Google Scholar]
- 43.Kahraman DC, Kahraman T, Cetin-Atalay R. Targeting PI3K/Akt/mTOR pathway identifies differential expression and functional role of IL-8 in liver cancer stem cell enrichment. Mol Cancer Ther. 2019;18:2146–2157. doi: 10.1158/1535-7163.MCT-19-0004. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, Wilhelm S, Lynch M, Carter C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.