Abstract
Stress is associated with increased Crohn’s Disease (CD) activity. This pilot study tested whether pediatric patients with CD reporting higher levels of perceived stress exhibited differences in the fecal microbiome and metabolome. The perceived stress scale (PSS) questionnaire was administered within 2 days of collecting a stool sample for microbiome (using 16S rRNA gene sequencing) and metabolome (using NMR metabolomics) analyses. Higher levels of perceived stress were correlated with increased disease activity on the short Pediatric Crohn’s Disease Activity Index (sPCDAI). Patients with High PSS scores vs. Low PSS scores based on a median split had significantly lower relative abundances of Firmicutes and Anaerostipes, as well as higher relative abundances of Parabacteroides. Fecal alanine and nicotinate were also significantly different in patients with High vs. Low PSS Scores. This pilot study suggests that the fecal microbiome and metabolome differs in pediatric patients with CD and high perceived stress.
Keywords: Microbiome, metabolome, stress, pediatric Crohn’s disease
1. Introduction
The inflammatory bowel diseases (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are thought to be due to disrupted homeostasis between the intestinal immune system and the gut microbiota in genetically susceptible individuals (Kostic, Xavier, & Gevers, 2014). Alterations in the microbiota can be both a cause and a consequence of mucosal inflammation, illustrating a complex cycle of host-microbe interactions during IBD, and multiple studies demonstrate abnormal microbial communities in IBD patients (Gevers et al., 2014). The onset and severity of the disease can be heavily influenced by environmental factors, with one environmental factor being exposure to stress (Bernstein et al., 2010; Camara et al., 2009; Traue & Kosarz, 1999). Multiple studies in laboratory animals demonstrate that stressor exposure can change the composition of the gut microbiota (Mackos, Maltz, & Bailey, 2016). However, it is not yet known whether stress is associated with altered gut microbiota composition, and/or function, in pediatric IBD patients.
In adults with IBD, levels of stress, depression, or anxiety are predictors of future disease activity (Bernstein et al., 2010; Bitton et al., 2008; Langhorst, Hofstetter, Wolfe, & Hauser, 2013; Mardini, Kip, & Wilson, 2004; Mikocka-Walus, Pittet, Rossel, von Kanel, & Swiss, 2016). In studies investigating both psychological and disease predictors, psychological factors are better predictors of future disease activity than disease factors, including baseline inflammatory markers, baseline mucosal healing, baseline disease activity, disease duration, previous relapses, medications, and history of surgery (Bernstein et al., 2010; Bitton et al., 2008; Langhorst et al., 2013). Moreover, children and adolescents with IBD are at increased risk for depression, anxiety and poor quality of life (QOL), with up to 31% reporting clinically significant symptoms in our previous work (Mackner & Crandall, 2006). Worse QOL is associated with higher disease severity (Herzer, Denson, Baldassano, & Hommel, 2011; Kunz, Hommel, & Greenley, 2010). It is not yet known how stress and low QOL contribute to worsened disease activity, but indigenous gut microbes may be involved. Studies in mice show that the microbiota are necessary for stressor-induced dysregulation of the immune response, and that stressor-induced changes to the composition of the gut microbiota contribute to dysregulated colonic inflammatory responses (Mackos et al., 2016). Thus, one purpose of this study was to test the hypothesis that CD patients reporting higher levels of perceived stress would have worsened disease activity and gut microbiota compositional differences compared to those with lower levels of stress.
Microbial-produced metabolites in the intestines can impact immune system functioning (Skelly, Sato, Kearney, & Honda, 2019), and IBD is associated with significant differences in the fecal metabolome. In both adult and pediatric IBD patients, amino acid levels are increased in the stool and serum in comparison to healthy controls (Bjerrum et al., 2015; Santoru, Piras, Murgia, Palmas, Camboni, Liggi, Ibba, Lai, Orru, Loizedda, et al., 2017). Stress exposure can also modify the gut metabolome, including robust shifts in amino acid and vitamin metabolites, which are together associated with an aberrant immune response to an ex vivo challenge in mice (Allen et al., Under Review). However, the relationship between stress and the gut metabolome has not been explored in a human clinical population. Therefore, an additional purpose of this study was to test the hypothesis that CD patients with high levels of perceived stress would exhibit differences in the fecal metabolome compared to those with lower stress.
2. Methods
2.1. Study Participants
Participants were patients ages 11 – 21 years who were participating in a larger, 4 year longitudinal study investigating psychosocial outcomes in adolescents with IBD. Eligibility criteria for the longitudinal study included ages 10 – 17 years and recent diagnosis of IBD. All participants in the current study had been enrolled in the longitudinal study for at least one year. For the current pilot study, 38 participants ages 11 – 21 years with Crohn’s disease were asked to participate in additional study procedures investigating “how stress affects your intestines” while being scheduled for a regular longitudinal study visit. Of the 38 patients asked to participate, 27 agreed, and after informed consent, 23 submitted a stool sample. Stool and data from one participant was not included due to failing to meet study criteria for substance use (demographics are in Table 1).
Table 1.
Patient demographics, disease severity, and medication usage.
| n=22 | |
|---|---|
| Mean age (yrs; s.d.) | 17.00 (2.75) |
| Min-max | 11.58–21.00 |
| % Male | 36% |
| % Caucasian | 67% |
| % Married (parents) | 73% |
| Median family income | $65,000 |
| Disease duration (yrs; s.d.) | 3.68 (1.60) |
| Min-max | 1.17–5.92 |
| Disease severitya | |
| Remission | 68% |
| Mild | 21% |
| Moderate/severe | 11% |
| Current corticosteroid | 0% |
| Current biologic | 57% |
| Current antidepressant | 18% |
| Clinic visits past 6 mo. (median) (Min-max) | 2.00 (0–4) |
| Hospitalized past 6 mo. | 5% |
| Surgery past 6 mo. | 0% |
Short Pediatric Crohn’s Disease Activity Index
2.2. Procedures
Participants in this pilot study were sent stool collection supplies and instructions in the mail to obtain a stool sample within 24 hours of the study visit. Participants were asked to store the samples in their freezers before they were transferred to a −80°C freezer at the hospital. During the longitudinal study visit, participants in this pilot study completed a Block Food Frequency Questionnaire (FFQ) and Perceived Stress Scale (PSS) in addition to the psychosocial measures for the longitudinal study. Chart reviews were conducted to obtain information about disease severity, medications and healthcare use.
2.3. Measures
2.3.1. Block Food Frequency Questionnaire
The Block Questionnaire —2005 FFQ (Block, Woods, Potosky, & Clifford, 1990) and the Block Questionnaire for Ages 8–17 — 2004 FFQ (Cullen, Watson, & Zakeri, 2008) estimate intake of a wide array of nutrients and food groups. Individual portion size is asked, and pictures are provided to enhance accuracy of quantification. Median correlations with 2-day food records are 0.8 for males and 0.7 for females (range = 0.5 – 0.9) (Maresperlman et al., 1993). FFQ data were used to calculate a Healthy Eating Index (HEI-2010) score, which is a measure of diet quality in terms of conformance with federal dietary guidance (Guenther et al., 2013).
2.3.2. Perceived Stress Scale
The PSS is a widely used measure for assessing the perception of stress in one’s life (Cohen, Kamarck, & Mermelstein, 1983). Items tap how unpredictable, uncontrollable, and overloaded respondents find their lives. Internal consistency ranges from 0.84 – 0.86, and the total score is significantly correlated with number and impact of life events, depression symptoms, and physical symptoms (Cohen et al., 1983). It has been used in children as young as 11 years (Cartwright et al., 2003). For analyses of the microbial community structure, the sample was divided into high and low stress groups via median split; otherwise, the PSS total score was used as a continuous variable.
2.3.3. Short Pediatric Crohn’s Disease Activity Index (sPCDAI)
The sPCDAI is a standardized, validated measure of disease activity that is derived from the longer Pediatric Crohn’s Disease Activity Index (PCDAI) (Kappelman et al., 2011). It correlates well with the PCDAI, r = 0.84, and its correlation with Physician Global Assessment (PGA; r = 0.60) is similar to the correlation of the PCDAI and PGA (r = 0.61) (Kappelman et al., 2011).
2.4. Stool Sample Collection
A stool sample was collected at home using a stool collection kit that we created consisting of a commode specimen collector, stool collection tube without preservative, biohazard bag, gloves and alcohol prep pad to clean any contacted surfaces. Stool samples were collected within 24 hours of completing the PSS and FFQ. Stool samples were kept frozen until transported to the laboratory where they were stored at −80°C.
2.5. DNA isolation and preparation
Genomic DNA was isolated from stool samples using the PowerSoil® DNA Isolation Kit (MoBio) following the manufacturer’s instructions. As an alternative to the recommended 250 mg of soil, approximately 250 mg of stool was added to the PowerBeads tube to undergo cell lysis. The purified DNA was eluted from the spin filter using 50 μL of solution C6 and stored at −20°C until PCR amplification.
2.6. 16S rRNA gene sequencing and analysis
The 16S rRNA gene V4-V5 region was amplified using 515f and 806r primers. The 515f primer contained a DNA barcode for sample identification and samples were amplified using a 28 cycle PCR using the HotStarTaq Plus Master Mix kit (Qiagen, USA) with cycling conditions of 94°C for 3 min, followed by 28 cycles of 94°C for 30 sec, 53°C for 40 sec, and 72°C for 1 min. A final elongation step of 72°C for 5 min was also performed. Success of PCR amplification was verified using 2% agarose gel electrophoresis. Samples were purified using Ampure XP beads and pooled together in equimolar concentrations. Samples were sequenced on an Illumina MiSeq at MRDNA (Shallowater, TX, USA) following manufacturer’s guidelines. Sequence data were processed using custom pipeline (MRDNA, Shallowater, TX, USA), including removal of barcodes and low quality reads (<150 bp and sequences with ambiguous base calls), denoising and chimera removal. Forward and reverse reads were joined and operational taxonomic units (OTU) were binned by clustering sequences at 3% divergence (i.e., 97% similarity). Final OTU’s were taxonomically classified using BLASTn against a curated database derived from GreenGenes/RDPII/NCBI (DeSantis et al., 2006). The QIIME pipeline was used for measures of α and β diversity.
2.7. Stool sample preparation for NMR analysis
50 mg of thawed feces were mixed with 1.0 ml of PBS (0.1M Na+/K+, NaN3 30 mM, pH 7.4, 50% v/v D2O (99.9% in D, Cambridge Isotope Laboratories, Tewksbury, MA)) containing sodium 3-trimethylsilyl [2,2,3,3-d4] propionate (TSP-d4) (Merck, Darmstadt, Germany) 0.0005 w/v as chemical shift internal standard. 300 mg of 0.1 mm zirconia beads were added and the mixture was vortexed for 30 s. Samples were then homogenized using a Biospec mini-beadbeater (Bartlesville, OK) for 90 s followed by two freeze thaw cycles in liquid nitrogen.
The fecal slurry was centrifuged at 11180 g for 10 mins at 4 °C and supernatants were transferred into 2 ml new EP tubes. 0.6 ml of PBS solution was added to the pellets followed by 30s of vortexing and then centrifuged at 11180 g for 10 mins at 4 °C. The supernatants were combined and centrifuged at 4°C and 16099g for 10 mins. 0.6 ml of the supernatants was transferred into 5mm NMR tubes.
2.8. NMR Spectroscopic Analysis
1H NMR spectra of aqueous feces extracts were acquired at 298K on a Bruker Avance III 600 MHz spectrometer (operating at 600.06 MHz for 1H and 150.90 MHz for 13C) equipped with a Bruker TCI probe (Bruker Biospin, Germany). For aqueous feces extracts, a typical one-dimensional 1H NMR spectrum was acquired for each sample, employing the first increment of the NOESY pulse sequence (NOESYPR1D; Relaxation delay-90-t1-90-mixing time-90-FID) with presaturation for watert suppression. The acquisition parameters were as follows: 64 scans and 4 dummy scans, 32K data points, 90° pulse angle (11.4 us), relaxation delay 3 s, 150 ms mixing time and a spectral width of 12 ppm. The spectra were acquired without spinning the NMR tube in order to avoid artifacts, such as spinning side bands of the first or higher order. All free induction decays (FIDs) were multiplied by a decaying exponential function with a 1 Hz line broadening factor prior to Fourier transformation. The 1H NMR spectra were corrected manually for phase and a polynomial fourth-order function was applied for base-line correction in order to achieve accurate and reproducible measurements upon integration of the signals of interest. Chemical shifts are reported in ppm as referenced to TSP (δ = 0). To confirm NMR signal assignments, a range of 2D NMR spectra were conducted and processed for certain samples, including 1H−1H correlation spectroscopy (gCOSY), 1H−1H total correlation spectroscopy (gTOCSY), 1H−13C edited heteronuclear single quantum correlation (gHSQC-DEPT), and 1H−13C heteronuclear multiple bond correlation spectra (gHMBC). 2D NMR experiments were acquired using the 600 MHz instrument mentioned above and an 800 MHz instrument (operating at 800.13 MHz for 1H and 201.21 MHz for 13C) equipped with a Bruker TCI probe (Bruker Biospin, Germany) as described previously. 1D and 2D NMR spectra were processed using TopSpin 3.2 (Bruker Biospin, Germany).
2.9. Spectral Data Processing and Multivariate Data Analysis
The spectral region δ 0.50−10.0 was integrated into regions with equal width of 0.004 ppm (2.4 Hz) using the AMIX software package (V3.8, Bruker-Biospin). The region δ 4.70−4.95 was discarded due to imperfect water saturation. Prior to statistical data analysis, each bucketed region was normalized to the total sum of the spectral intensities to compensate for the overall concentration differences.
3. Data Analysis
3.1. Bacterial taxa and metabolites
A median split of PSS scores was used to create “High PSS” and “Low PSS” groups. A Student’s t-test was used to compare bacterial taxa and to compare fecal metabolites between High PSS and Low PSS groups using SPSS (version 25). The Benjamini-Hochberg method (implemented in R) was used to correct for multiple comparisons.
4. Results
4.1. Perceived stress is associated with CD disease activity
Perceived stress and diet were not significantly associated, including diet measures such as HEI-2010 score, total calories, protein, carbohydrates, fat, or sugar (controlling for age), or percent of calories from protein, carbohydrates, fat, or sweets. Disease activity and perceived stress were significantly correlated, r=0.477, p=.039.
4.2. Composition differences in the gut microbiome are evident in patients with higher perceived stress
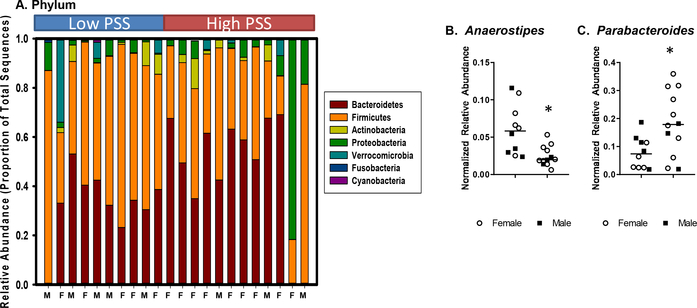
A median split of PSS scores was used to create a High PSS group (n=12) and a Low PSS group (n=10). Overall measures of microbial community composition (α- and β-diversity) were not significantly different in CD patients reporting higher levels of perceived stress vs. those reporting lower levels of perceived stress (Supplemental Figure 1). However, the relative abundances of bacteria in the phylum Firmicutes was significantly lower in the High PSS CD patients when compared to the Low PSS patients (t(20) = 2.45, p < .05) (Fig. 1A). Relative abundances of bacterial genera are shown in Table 2. These relative abundances were normalized by finding the arcsin of the square root of the proportion of the total sequences assigned to each genus (Galley et al., 2017). A total of 70 comparisons were made between the bacteria genera in the High vs. Low PSS groups, and after correcting for multiple comparisons, the relative abundance of two bacterial genera were found to be different in individuals reporting high levels of PSS compared to those reporting lower levels of PSS. In particular, Anaerostipes (t(20) = 4.68, p<.001, FDR p = 0.028) was significantly lower in the High PSS group, whereas Parabacteroides (t(20) = 4.27, p = .001, FDR p = 0.035) was significantly higher in the High PSS group (Fig. 1B). Firmicutes, Firmicutes/Bacteroidetes ratio, Anaerostipes, and Parabacteroides abundances were not related to medication use, disease severity scores, length of disease, or diet (HEI-2010 score, total calories, protein, carbohydrates, fat, or sugar). Although there was a trend for individuals in the High PSS group to have a higher body mass index than individuals in the Low PSS group (BMI not available for 2 individuals, t(18) = 1.93, p = .07; data not shown), BMI was also not correlated with Firmicutes, Firmicutes/Bacteroidetes ratio, Anaerostipes, or Parabacteroides abundances. Four individuals (2 in the High PSS group and 2 in the Low PSS group) were taking medication for depression. However, with this small sample size, there was no evidence that antidepressant medication affected the composition of the gut microbiome. None of the patients were taking medication for any other psychiatric illness.
Figure 1: Crohn’s disease patients with higher perceived stress have differences in specific bacterial taxa compared to patients with lower perceived stress.
A) Crohn’s disease patients in the High PSS group had significantly lower relative abundances of Firmicutes (as determined by percentage of total sequences) (p<.05). B) Crohn’s disease patients in the High PSS group had a significant decrease in the relative abundance of Anaerostipes and a significant increase in Parabacteroides (FDR p<.05). Data show the arcsin of the square root of the proportion for each bacterial genus.
Table 2:
Fecal microbes in Crohn’s disease patients with Low or High PSS Scores
| Low PSS | High PSS | |||||||
|---|---|---|---|---|---|---|---|---|
| Anaerostipes ** | 0.060 | +/− | 0.011 | 0.025 | +/− | 0.004 | ||
| Parabacteroides ** | 0.079 | +/− | 0.018 | 0.184 | +/− | 0.033 | ||
| Anaerotruncus | 0.019 | +/− | 0.004 | 0.009 | +/− | 0.002 | ||
| Blautia | 0.331 | +/− | 0.037 | 0.263 | +/− | 0.042 | ||
| Eggerthella | 0.015 | +/− | 0.004 | 0.008 | +/− | 0.001 | ||
| Eisenbergiella | 0.017 | +/− | 0.005 | 0.013 | +/− | 0.003 | ||
| Tyzzerella | 0.052 | +/− | 0.018 | 0.016 | +/− | 0.002 | ||
| Ruminoclostridium | 0.090 | +/− | 0.025 | 0.041 | +/− | 0.010 | ||
| Shigella | 0.088 | +/− | 0.020 | 0.049 | +/− | 0.010 | ||
| Parasutterella | 0.047 | +/− | 0.017 | 0.069 | +/− | 0.025 | ||
| Clostridium | 0.294 | +/− | 0.034 | 0.218 | +/− | 0.032 | ||
| Subdoligranulum | 0.101 | +/− | 0.020 | 0.067 | +/− | 0.016 | ||
| Fusobacterium | 0.024 | +/− | 0.013 | 0.005 | +/− | 0.001 | ||
| Erysipelatoclostridium | 0.019 | +/− | 0.005 | 0.011 | +/− | 0.002 | ||
| Barnesiella | 0.019 | +/− | 0.010 | 0.019 | +/− | 0.012 | ||
| Faecalitalea | 0.015 | +/− | 0.008 | 0.008 | +/− | 0.002 | ||
| Oscillospira | 0.055 | +/− | 0.015 | 0.040 | +/− | 0.014 | ||
| Turicibacter | 0.019 | +/− | 0.005 | 0.036 | +/− | 0.009 | ||
| Eubacterium | 0.180 | +/− | 0.034 | 0.213 | +/− | 0.038 | ||
| Intestimonas | 0.014 | +/− | 0.005 | 0.007 | +/− | 0.002 | ||
| Butyricimonas | 0.005 | +/− | 0.000 | 0.028 | +/− | 0.018 | ||
| Lactococcus | 0.005 | +/− | 0.001 | 0.007 | +/− | 0.002 | ||
| Akkermansia | 0.134 | +/− | 0.063 | 0.064 | +/− | 0.025 | ||
| Lachnoclostridium | 0.115 | +/− | 0.025 | 0.120 | +/− | 0.027 | ||
| Desulfovibrio | 0.003 | +/− | 0.001 | 0.015 | +/− | 0.007 | ||
| Lachnobacterium | 0.021 | +/− | 0.015 | 0.009 | +/− | 0.002 | ||
| Sporobacter | 0.016 | +/− | 0.007 | 0.008 | +/− | 0.003 | ||
| Megasphaera | 0.093 | +/− | 0.070 | 0.048 | +/− | 0.023 | ||
| Roseburia | 0.058 | +/− | 0.014 | 0.080 | +/− | 0.024 | ||
| Fusicatenibacter | 0.091 | +/− | 0.019 | 0.056 | +/− | 0.011 | ||
| Lactobacillus | 0.037 | +/− | 0.011 | 0.019 | +/− | 0.004 | ||
| Anaerovorax | 0.008 | +/− | 0.002 | 0.009 | +/− | 0.002 | ||
| Parasporobacterium | 0.021 | +/− | 0.010 | 0.009 | +/− | 0.003 | ||
| Cellulosilyticum | 0.005 | +/− | 0.002 | 0.018 | +/− | 0.013 | ||
| Sutterella | 0.020 | +/− | 0.004 | 0.096 | +/− | 0.039 | ||
| Collinsella | 0.027 | +/− | 0.010 | 0.011 | +/− | 0.004 | ||
| Entercococcus | 0.012 | +/− | 0.001 | 0.046 | +/− | 0.031 | ||
| Intestinibacter | 0.079 | +/− | 0.029 | 0.056 | +/− | 0.015 | ||
| Alistipes | 0.120 | +/− | 0.041 | 0.155 | +/− | 0.034 | ||
| Dialister | 0.085 | +/− | 0.037 | 0.052 | +/− | 0.014 | ||
| Klebsiella | 0.006 | +/− | 0.001 | 0.009 | +/− | 0.003 | ||
| Bacteroides | 0.520 | +/− | 0.057 | 0.616 | +/− | 0.085 | ||
| Lachnospira | 0.019 | +/− | 0.007 | 0.013 | +/− | 0.005 | ||
| Sporobacterium | 0.003 | +/− | 0.001 | 0.003 | +/− | 0.001 | ||
| Prevotella | 0.049 | +/− | 0.032 | 0.052 | +/− | 0.029 | ||
| Pantoea | 0.031 | +/− | 0.006 | 0.124 | +/− | 0.092 | ||
| Acidominococcus | 0.019 | +/− | 0.010 | 0.038 | +/− | 0.029 | ||
| Haemophilus | 0.067 | +/− | 0.032 | 0.041 | +/− | 0.014 | ||
| Coprococcus | 0.036 | +/− | 0.014 | 0.026 | +/− | 0.007 | ||
| Flavonifactor | 0.031 | +/− | 0.006 | 0.039 | +/− | 0.010 | ||
| Faecilbacterium | 0.269 | +/− | 0.070 | 0.136 | +/− | 0.035 | ||
| Actinomyces | 0.011 | +/− | 0.001 | 0.007 | +/− | 0.001 | ||
| Phascolarctoba | 0.036 | +/− | 0.012 | 0.054 | +/− | 0.021 | ||
| Allisonella | 0.003 | +/− | 0.001 | 0.008 | +/− | 0.006 | ||
| Porphyromonas | 0.005 | +/− | 0.001 | 0.004 | +/− | 0.001 | ||
| Odoribacter | 0.033 | +/− | 0.010 | 0.037 | +/− | 0.011 | ||
| Lactonifactor | 0.007 | +/− | 0.002 | 0.007 | +/− | 0.002 | ||
| Veillonella | 0.032 | +/− | 0.012 | 0.044 | +/− | 0.012 | ||
| Ruminococcus | 0.119 | +/− | 0.023 | 0.091 | +/− | 0.024 | ||
| Hespellia | 0.004 | +/− | 0.002 | 0.004 | +/− | 0.001 | ||
| Rothia | 0.003 | +/− | 0.001 | 0.003 | +/− | 0.001 | ||
| Bilophila | 0.024 | +/− | 0.008 | 0.032 | +/− | 0.008 | ||
| Gemella | 0.003 | +/− | 0.001 | 0.003 | +/− | 0.001 | ||
| Dorea | 0.072 | +/− | 0.011 | 0.063 | +/− | 0.012 | ||
| Streptococcus | 0.038 | +/− | 0.005 | 0.031 | +/− | 0.005 | ||
| Bifidobacterium | 0.118 | +/− | 0.038 | 0.112 | +/− | 0.033 | ||
| Sporoanaerobacterium | 0.013 | +/− | 0.003 | 0.009 | +/− | 0.003 | ||
| Anaerospora | 0.010 | +/− | 0.004 | 0.007 | +/− | 0.002 | ||
| Paludibacter | 0.053 | +/− | 0.021 | 0.030 | +/− | 0.015 | ||
| Adlercreutzia | 0.006 | +/− | 0.002 | 0.005 | +/− | 0.002 | ||
| 4.074 | 3.833 | |||||||
Data are the normalized relative abundances calculated by the arcsin of the square root of the sequence proportions assigned to each genus.
FDR corrected p < .05
4.3. Higher perceived stress is associated with an altered gut metabolome
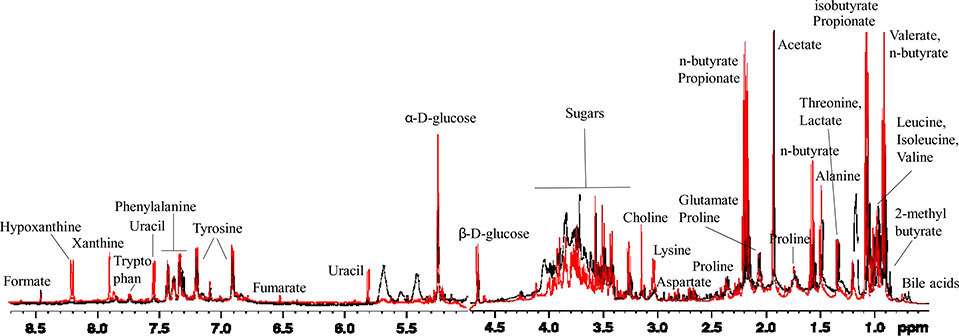
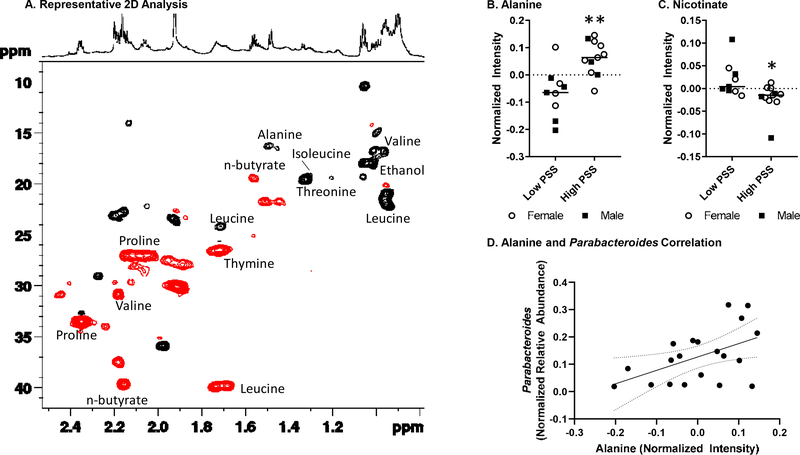
Patients in the High PSS group also exhibited a distinct fecal metabolome as assessed with NMR. Representative superimposed 1H NMR spectra of the aqueous extracts of stool samples from pediatric CD patients in the Low PSS and High PSS groups (black and red spectrum respectively), are shown in Fig. 2. As indicated by their spectra, the water extracts of feces samples contain multiple metabolites, including short chain fatty acids, amino acids, sugars, organic acids, as well as purine/pyrimidine derivatives (Kostidis et al., 2017; Lin et al., 2016). The chemical shift assignment of various metabolites was conducted on the basis of literature, the Human Metabolome Database (HMDB) and a range of 2D experiments. The use of 2D NMR was incorporated to ensure the correct chemical shift assignment of the compounds of interest. As an example, Fig. 3 is a representative HSQC-DEPT spectrum, a multiplicity edited HSQC, of a fecal aqueous extract sample, showing one bond proton–carbon correlations; positive signals (black) for the CH3/CH carbons and negative signals (red) for the CH2 carbons.
Figure 2: NMR spectra differ in CD patients with higher vs. lower levels of perceived stress.
Superimposed 1D 1H NMR spectra of the aqueous extracts of stool samples from pediatric CD patients in the Low PSS and High PSS groups (black and red spectrum respectively).
Figure 3: Fecal metabolites differ in CD patients with higher vs lower perceived stress.
A) Multiplicity edited HSQC of a stool aqueous extract sample, displaying one bond 1H-13C correlations; positive signals (black) for the CH3/CH carbons and negative signals (red) for the CH2 carbons. B) Relative intensities of alanine and C) nicotinate are significant different in CD patients in the High PSS group. D) Normalized relative abundance of Parabacteroides was significantly correlated with alanine intensity (p < .05).
** p and FDR<.05. *p<.05 prior to FDR correction; FDR p=.13.
When the metabolome of patients in the High PSS group was compared to the metabolome of patients in the Low PSS group, four metabolites were significantly different: alanine (t(20)=3.82, p<.001), nicotinate (t(20)=2.60, p<.05), lysine (t(20)=2.20, p<.05), and phenylalanine (t(20)=2.18, p<.05) (see Table 3). However, a total of 37 statistical tests were conducted on the metabolites, and only alanine remained significantly different after correcting for multiple tests (FDR p=.03) (Fig. 3B). Correlation analyses were then conducted to determine whether the increased alanine was related to the observed changes Parabacteroides or Anaerostipes. Alanine was positively correlated with Parabacteroides (r(20) = .501, p < .02) (Fig. 3D), but was not related to Anaerostipes (data not shown).
Table 3:
Fecal metabolites in CD patients with Low or High PSS scores.
| Low PSS | High PSS | |||||||
|---|---|---|---|---|---|---|---|---|
| Alanine ** | −0.067 | +/− | 0.030 | 0.063 | +/− | 0.019 | ||
| Nicotinate * | 0.020 | +/− | 0.013 | −0.020 | +/− | 0.010 | ||
| Lysine * | −0.047 | +/− | 0.036 | 0.043 | +/− | 0.017 | ||
| Phenylalanine * | −0.093 | +/− | 0.039 | 0.030 | +/− | 0.039 | ||
| Tyrosine | −0.056 | +/− | 0.026 | 0.031 | +/− | 0.033 | ||
| Uracil | −0.019 | +/− | 0.009 | 0.018 | +/− | 0.015 | ||
| Fumerate | 0.004 | +/− | 0.004 | −0.007 | +/− | 0.004 | ||
| Adenosine | 0.026 | +/− | 0.020 | −0.015 | +/− | 0.017 | ||
| Bile Acids | 0.059 | +/− | 0.030 | −0.004 | +/− | 0.026 | ||
| Uroconate | −0.014 | +/− | 0.007 | 0.020 | +/− | 0.020 | ||
| Xanthine | −0.007 | +/− | 0.008 | 0.010 | +/− | 0.011 | ||
| UDP glucoronate | 0.039 | +/− | 0.051 | −0.032 | +/− | 0.040 | ||
| Glutamine | 0.032 | +/− | 0.042 | −0.027 | +/− | 0.035 | ||
| Hypoxanthine | −0.010 | +/− | 0.015 | 0.013 | +/− | 0.015 | ||
| Adenosine | 0.010 | +/− | 0.010 | −0.013 | +/− | 0.017 | ||
| 2-methylbutyrate | 0.012 | +/− | 0.018 | −0.010 | +/− | 0.017 | ||
| n-butyrate | 0.054 | +/− | 0.064 | −0.025 | +/− | 0.066 | ||
| Iso-caproate | 0.000 | +/− | 0.018 | 0.022 | +/− | 0.017 | ||
| Asparagine | 0.058 | +/− | 0.056 | −0.010 | +/− | 0.057 | ||
| Methionine | 0.006 | +/− | 0.006 | −0.001 | +/− | 0.006 | ||
| Galactose | 0.026 | +/− | 0.030 | −0.002 | +/− | 0.019 | ||
| Glycine | −0.027 | +/− | 0.017 | −0.006 | +/− | 0.023 | ||
| 3-hydroxyphenylacetate | −0.042 | +/− | 0.004 | −0.034 | +/− | 0.011 | ||
| Tryptophan | −0.025 | +/− | 0.005 | −0.014 | +/− | 0.015 | ||
| Methylamine | 0.009 | +/− | 0.012 | −0.001 | +/− | 0.011 | ||
| Ribose | −0.026 | +/− | 0.032 | −0.009 | +/− | 0.017 | ||
| p-Cresol | −0.004 | +/− | 0.017 | −0.018 | +/− | 0.029 | ||
| Histidine | −0.003 | +/− | 0.008 | −0.006 | +/− | 0.006 | ||
| 5-aminopentanoate | 0.006 | +/− | 0.004 | 0.003 | +/− | 0.007 | ||
| Succinate | 0.020 | +/− | 0.081 | −0.009 | +/− | 0.057 | ||
| Beta-D-glucose | 0.033 | +/− | 0.061 | 0.016 | +/− | 0.028 | ||
| Formate | 0.002 | +/− | 0.005 | −0.001 | +/− | 0.011 | ||
| Alpha-D-glucose | 0.001 | +/− | 0.030 | −0.005 | +/− | 0.018 | ||
| Fructose, lactate | 0.015 | +/− | 0.048 | 0.022 | +/− | 0.055 | ||
| Aspartate | 0.027 | +/− | 0.120 | 0.012 | +/− | 0.125 | ||
p < .05 prior to FDR correction.
FDR corrected p < .05.
5. Discussion
Consistent with our hypotheses, pediatric patients with CD who reported higher levels of stress had greater disease activity and significant differences in the composition of their intestinal microbiota when compared to patients with lower reported stress. Anaerostipes spp. abundance was lower, whereas Parabacteroides spp. abundance was higher in patients with high perceived stress. There are a number of factors that have the potential to contribute to differences in the gut microbiome. For example, diet is well known to affect microbiome composition, including Firmicutes relative abundances (David et al, 2014), which were reduced in our study. Food sensitivities, and thus, restricted diets, are common in CD patients (Prince et al., 2011; Vidarsdottir et al., 2016), but the differences in the microbiome in CD patients with higher perceived stress were not related to differences in diet. Current therapies for CD, including anti-inflammatory and immunomodulatory medications (such as corticosteroids, thiopurines, methotrexate and anti-TNF) also have the potential to affect the microbiome. For example, the anti-TNF treatment, Infliximab, has been shown to increase α diversity and shift overall microbial community composition (Wang et al., 2018). Despite these potential effects, differences in the microbiome between CD patients with high vs. low perceived stress were independent of medication use, as well as disease severity and duration. Similarly, the gut metabolome was different in CD patients with higher perceived stress compared to patients with lower stress. The metabolites alanine, nicotinate, lysine, and phyenylalanine all differed between high- and low-perceived stress CD patients. Although only alanine remained significantly different after correcting for multiple comparisons, similar changes in nicotinate, lysine, and phenylalanine were evident in an animal model of stress (Allen et al., 2019).
There are now multiple studies comparing the microbiome in CD patients to non-CD controls, with the most consistent findings being reduced α-diversity, lower levels of bacteria in the phylum Firmicutes, and higher levels of bacteria in the phylum Gammaproteobacteria. Although we did not observe statistically significant differences in α diversity or in Gammaproteobacteria, we did observe that patients with higher perceived stress had significantly lower Firmicutes than patients with lower stress. At the genus level, Anaerostipes was significantly decreased in samples from the higher stress group. Anaerostipes is a member of the Firmicutes phylum, and the reduction in Anaerostipes contributed to the observed reduction in Firmicutes in high stress individuals. Interestingly, pediatric CD patients also have lower levels of Anaerostipes in fecal samples than do healthy controls (Wang et al., 2018).
While it is not known if more severe intestinal inflammation leads to a loss of Anaerostipes or if the loss of Anaerostipes directly contributes to disease severity, it is known that Anaerostipes produces the short chain fatty acid (SCFA) butyrate (Duncan, Holtrop, et al., 2004; Duncan, Louis, & Flint, 2004; Schwiertz et al., 2002). Low levels of colonic butyrate, and butyrate-producing taxa, have been identified in CD and UC (Laserna-Mendieta et al., 2018; Machiels et al., 2014). Although our NMR method was able to detect butyrate, GC-MS is a preferred method for assessing volatile compounds like SCFAs. Thus, we did not observe reductions in butyrate (or other SCFAs) in the high stress group. However, we have previously used GC-MS to show that exposing healthy mice to an experimental stressor reduces colonic SCFAs, including butyrate (Maltz et al., 2018; Maltz et al., 2019). Thus, future studies will utilize GC-MS to assess fecal butyrate as a possible mechanism linking the gut microbiome and disease severity in CD patients with higher levels of perceived stress.
In contrast to reductions in Anaerostipes, there were significant increases in Parabacteroides in CD patients with higher PSS scores. Some research has suggested that Parabacteroides is lower in IBD patients compared to controls and is associated with less inflammation (Zitomersky et al., 2013). This is seemingly inconsistent with our observations, but some species of Parabacteroides may increase inflammation. Administering P. distasonis to mice with existing inflammation (induced by dextran sulfate sodium (DSS)) had a significant increase in colonic inflammation (Dziarski, Park, Kashyap, Dowd, & Gupta, 2016), indicating that Parabacteroides spp. have the potential to exacerbate existing colonic inflammation. Moreover, mice exposed to a social stressor that enhances colonic inflammation (Mackos et al., 2016), have increased abundances of Parabacteroides (Allen et al., 2019), suggesting that increases in Parabacteroides is a consequence of the stress response that is conserved across animal species. One limitation of this study is that the cross-sectional design does not allow for a prediction of whether these microbiome changes are a cause or a consequence of more severe intestinal inflammation in CD patients with higher perceived stress. Follow-up, longitudinal studies with multiple sampling time points are needed to determine whether changes in the abundance of Parabacteroides (or Anaerostipes) are a cause or a consequence of increased intestinal inflammation during periods of high perceived stress.
It is becoming increasingly evident that the gut metabolome (which includes small molecule metabolites produced by both the host and the resident microbiota) has far reaching effects on host health. In fact, metabolomic analyses have demonstrated that gut microbes contribute to metabolites found in blood (Wikoff et al., 2009), urine (Holmes et al., 2008; Li et al., 2008), as well as stool (Franzosa et al., 2019; Saric et al., 2008). Metabolomic profiles in stool clearly distinguish active IBD from IBD in remission and healthy controls, in part due to malabsorption that results from intestinal inflammation. This malabsorption results in higher levels of amino acids in the intestinal lumen, or stool, compared to healthy controls (Dawiskiba et al., 2014; Santoru, Piras, Murgia, Palmas, Camboni, Liggi, Ibba, Lai, Orru, Blois, et al., 2017).
In our study, fecal levels of alanine were significantly higher in individuals with higher stress. Alanine is a nonessential amino acid that can be produced by the host and/or derived from the diet and the commensal microbiota. The majority of alanine is dependent upon pyruvate, which is produced by the breakdown of glucose during glycolysis. Multiple bacteria contain enzymes that can be used to convert pyruvate into alanine (Oikawa, 2006). Thus, it is possible that the increase in fecal alanine is the result of altered bacterial metabolism, such as metabolism in Parabacteroides whose abundance was increased in high stress individuals and correlated with alanine levels in the stool. This hypothesis warrants further testing. However, it is equally possible that the alanine is host-derived. Increased glycolysis in skeletal muscle is a hallmark of sympathetic nervous system activation and leads to an accumulation of pyruvate. As in bacteria, mammals convert pyruvate to alanine via alanine aminotransferase. Thus, it is also a possibility that increased alanine in the stool of CD patients with higher stress is a result of increased sympathetic nervous system activity. Alternatively, the higher levels of alanine may reflect malabsorption in the small intestine; multiple studies have reported amino acid malabsorption and higher fecal amino acids (including alanine) in CD patients (Bjerrum et al., 2015; Santoru, Piras, Murgia, Palmas, Camboni, Liggi, Ibba, Lai, Orru, Blois, et al., 2017). While increased disease activity in patients with higher stress would be consistent with reduced amino acid absorption in the intestine (and thus, higher amino acids in the lumen of the intestine/feces), further studies are needed to identify mechanisms by which high stress is associated with increased fecal alanine levels.
Mice exposed to a social stressor have reductions in colonic B1, B3, and B6 vitamins (Allen et al., 2019), which led us to test whether B vitamins would also be reduced in CD patients with higher stress. Nicotinate, a B3 vitamer, was reduced in individuals with high stress. And, when our a priori hypothesis that B vitamins would be reduced was tested, we found a reduction in nicotinate in individuals with higher stress. Reductions in nicotinate may have important biological functions. This B3 vitamer is the anion form of niacin, which can significantly reduce experimental colitis in mice (Salem & Wadie, 2017). Colonic bacteria are important sources of essential vitamins, including the B vitamins, and our studies in stressor-exposed mice demonstrate that stressor exposure reduces both the abundance and the diversity of bacteria capable of producing B vitamins (Allen et al., 2019). The current pilot/proof of concept study suggests that stress in human patients may also lead to reductions in intestinal B vitamins and provides the rationale for mechanism-based studies to determine whether bacterial-produced B vitamins link stressor-induced changes in the gut microbiome and disease severity in CD patients.
An important limitation of this study is that the microbiome and metabolome in patients with IBD were not compared to the microbiome and metabolome in healthy controls. Moreover, multiple comparisons were conducted to compare the microbiomes and metabolomes of CD patients with high vs. low levels of perceived stress. Because of the multiple comparisons, we used the Benjamini Hochberg method of correcting p values and maintained a conservative corrected p value of < .05 to indicate statistical significance. The benefit of this approach is that it reduces the likelihood of a Type I error (due to the multiple comparisons), but it may lead to increased Type II error. Nonetheless, we detected changes in both the microbiome and metabolome, and our results are consistent with other studies that have found that stressful periods contribute to increased severity of inflammatory diseases and conditions like IBD. Mechanisms by which the stress response enhances inflammation are not yet well understood, but murine studies indicate that gut microbes are crucial links between stress exposure and increased intestinal inflammation. This led us to assess whether there is any evidence of disrupted microbiota in IBD patients with higher levels of perceived stress compared to patients with lower levels of perceived stress. This pilot study suggests that the gut microbiome and metabolome differ in CD patients with higher perceived stress. However, additional longitudinal studies are needed to determine whether the changes in the microbiome and metabolome contribute to intestinal inflammation, or are the result of increased disease activity during stressful periods.
Supplementary Material
Supplementary Figure 1: Overall microbial community composition is not significantly different in Crohn’s disease patients with higher perceived stress. A. Principal coordinate plot of unweighted UniFrac distances did not show significant differences in beta diversity between High PSS and Low PSS CD patients (Adonis, p=0.15). Similar findings were evident for weighted UniFrac distances. B. Shannon’s diversity index, a measure of community alpha diversity, was not significantly different between High PSS and Low PSS CD patients. Similar findings were evident with other measures of alpha diversity (including Ace, Chao1, PD Whole Tree, Observed Features).
Highlights.
Higher levels of perceived stress were associated with worse disease activity
Lower relative abundances of bacteria in phylum Firmicutes in higher stress group
Genus Anaerostipes was lower and Parabacteroides was higher in the high stress group
Differences were independent of medications, disease severity, disease duration, diet
Levels of the metabolite alanine were higher in youth with higher stress
ACKNOWLEDGMENTS
The authors gratefully acknowledge the expert technical assistance of Mr. Robert (Max) Jaggers. R21HD073609 and start-up funds and bridge funds from the Research Institute at Nationwide Children’s Hospital were used for this study. Jacob Allen was supported by National Institute of Dental and Craniofacial Research Training Grant T32-DE014320-19.
Footnotes
Conflict of Interests
The authors do not have any conflicts of interests that affect the conduct or interpretation of this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen JM, Jaggers RM, Solden LM, Loman BR, Davies RH, Mackos AR, Ladaika CA, Berg BM, Chichlowski M, & Bailey MT (2019). Dietary oligosaccharides attenuate stress-induced disruptions in immune reactivity and microbial B-vitamin metabolism. Front Immunol, 10:1774, doi: 10.3389/fimmu.2019.01774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein CN, Singh S, Graff LA, Walker JR, Miller N, & Cheang M (2010). A prospective population-based study of triggers of symptomatic flares in IBD. Am J Gastroenterol, 105(9), 1994–2002. doi: 10.1038/ajg.2010.140 [DOI] [PubMed] [Google Scholar]
- Bitton A, Dobkin PL, Edwardes MD, Sewitch MJ, Meddings JB, Rawal S,… Wild GE (2008). Predicting relapse in Crohn’s disease: a biopsychosocial model. Gut, 57(10), 1386–1392. doi: 10.1136/gut.2007.134817 [DOI] [PubMed] [Google Scholar]
- Bjerrum JT, Wang Y, Hao F, Coskun M, Ludwig C, Gunther U, & Nielsen OH (2015). Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics, 11, 122–133. doi: 10.1007/s11306-014-0677-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block G, Woods M, Potosky A, & Clifford C (1990). VALIDATION OF A SELFADMINISTERED DIET HISTORY QUESTIONNAIRE USING MULTIPLE DIET RECORDS. Journal of Clinical Epidemiology, 43(12), 1327–1335. doi: 10.1016/0895-4356(90)90099-b [DOI] [PubMed] [Google Scholar]
- Camara RJ., Ziegler R., Begre S., Schoepfer AM., von Kanel R., & Swiss Inflammatory Bowel Disease Cohort Study, group. (2009). The role of psychological stress in inflammatory bowel disease: quality assessment of methods of 18 prospective studies and suggestions for future research. Digestion, 80(2), 129–139. doi: 10.1159/000226087 [DOI] [PubMed] [Google Scholar]
- Cartwright M, Wardle J, Steggles N, Simon AE, Croker H, & Jarvis MJ (2003). Stress and dietary practices in adolescents. Health Psychol, 22(4), 362–369. [DOI] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, & Mermelstein R (1983). A global measure of perceived stress. J Health Soc Behav, 24(4), 385–396. [PubMed] [Google Scholar]
- Cullen KW, Watson K, & Zakeri I (2008). Relative reliability and validity of the Block Kids Questionnaire among youth aged 10 to 17 years. Journal of the American Dietetic Association, 108(5), 862–866. doi: 10.1016/j.jada.2008.02.015 [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505(7484), 559–563. doi: 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawiskiba T, Deja S, Mulak A, Zabek A, Jawien E, Pawelka D,… Mlynarz P (2014). Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol, 20(1), 163–174. doi: 10.3748/wjg.v20.i1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K,… Andersen GL (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol, 72(7), 5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, & Flint HJ (2004). Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr, 91(6), 915–923. doi: 10.1079/BJN20041150 [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, & Flint HJ (2004). Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol, 70(10), 5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Park SY, Kashyap DR, Dowd SE, & Gupta D (2016). Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS One, 11(1), e0146162. doi: 10.1371/journal.pone.0146162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S,… Xavier RJ (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol, 4(2), 293–305. doi: 10.1038/s41564-018-0306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B,… Xavier RJ (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe, 15(3), 382–392. doi: 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther PM, Casavale KO, Reedy J, Kirkpatrick SI, Hiza HAB, Kuczynski KJ,… Krebs-Smith SM (2013). Update of the Healthy Eating Index: HEI-2010. Journal of the Academy of Nutrition and Dietetics, 113(4), 569–580. doi: 10.1016/j.jand.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzer M, Denson LA, Baldassano RN, & Hommel KA (2011). Patient and parent psychosocial factors associated with health-related quality of life in pediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr, 52(3), 295–299. doi: 10.1097/MPG.0b013e3181f5714e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q,… Elliott P (2008). Human metabolic phenotype diversity and its association with diet and blood pressure. Nature, 453(7193), 396–400. doi: 10.1038/nature06882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappelman MD, Crandall WV, Colletti RB, Goudie A, Leibowitz IH, Duffy L,… Margolis P (2011). Short pediatric Crohn’s disease activity index for quality improvement and observational research. Inflamm Bowel Dis, 17(1), 112–117. doi: 10.1002/ibd.21452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Xavier RJ, & Gevers D (2014). The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology, 146(6), 1489–1499. doi: 10.1053/j.gastro.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostidis S, Kokova D, Dementeva N, Saltykova IV, Kim HK, Choi YH, & Mayboroda OA (2017). (1)H-NMR analysis of feces: new possibilities in the helminthes infections research. BMC Infect Dis, 17(1), 275. doi: 10.1186/s12879-017-2351-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz JH, Hommel KA, & Greenley RN (2010). Health-related quality of life of youth with inflammatory bowel disease: a comparison with published data using the PedsQL 4.0 generic core scales. Inflamm Bowel Dis, 16(6), 939–946. doi: 10.1002/ibd.21128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhorst J., Hofstetter A., Wolfe F., & Hauser W. (2013). Short-term stress, but not mucosal healing nor depression was predictive for the risk of relapse in patients with ulcerative colitis: a prospective 12-month follow-up study. Inflamm Bowel Dis, 19(11), 2380–2386. doi: 10.1097/MIB.0b013e3182a192ba [DOI] [PubMed] [Google Scholar]
- Laserna-Mendieta EJ, Clooney AG, Carretero-Gomez JF, Moran C, Sheehan D, Nolan JA,… Claesson MJ (2018). Determinants of Reduced Genetic Capacity for Butyrate Synthesis by the Gut Microbiome in Crohn’s Disease and Ulcerative Colitis. J Crohns Colitis, 12(2), 204–216. doi: 10.1093/ecco-jcc/jjx137 [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H,… Zhao L (2008). Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A, 105(6), 2117–2122. doi: 10.1073/pnas.0712038105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Ma C, Liu C, Wang Z, Yang J, Liu X,… Wu R (2016). NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget, 7(20), 29454–29464. doi: 10.18632/oncotarget.8762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V,… Vermeire S (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut, 63(8), 1275–1283. doi: 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- Mackner LM, & Crandall WV (2006). Brief report: psychosocial adjustment in adolescents with inflammatory bowel disease. J Pediatr Psychol, 31(3), 281–285. doi: 10.1093/jpepsy/jsj023 [DOI] [PubMed] [Google Scholar]
- Mackos AR, Maltz R, & Bailey MT (2016). The role of the commensal microbiota in adaptive and maladaptive stressor-induced immunomodulation. Horm Behav. doi: 10.1016/j.yhbeh.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltz RM, Keirsey J, Kim SC, Mackos AR, Gharaibeh RZ, Moore CC,… Bailey MT (2018). Prolonged restraint stressor exposure in outbred CD-1 mice impacts microbiota, colonic inflammation, and short chain fatty acids. PLoS One, 13(5), e0196961. doi: 10.1371/journal.pone.0196961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltz RM, Keirsey J, Kim SC, Mackos AR, Gharaibeh RZ, Moore CC,… Bailey MT (2019). Social Stress Affects Colonic Inflammation, the Gut Microbiome, and Short-chain Fatty Acid Levels and Receptors. J Pediatr Gastroenterol Nutr, 68(4), 533–540. doi: 10.1097/MPG.0000000000002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardini HE, Kip KE, & Wilson JW (2004). Crohn’s disease: a two-year prospective study of the association between psychological distress and disease activity. Dig Dis Sci, 49(3), 492–497. [DOI] [PubMed] [Google Scholar]
- Maresperlman JA, Klein BEK, Klein R, Ritter LL, Fisher MR, & Freudenheim JL (1993). A DIET HISTORY QUESTIONNAIRE RANKS NUTRIENT INTAKES IN MIDDLE-AGED AND OLDER MEN AND WOMEN SIMILARLY TO MULTIPLE FOOD RECORDS. Journal of Nutrition, 123(3), 489–501. [DOI] [PubMed] [Google Scholar]
- Mikocka-Walus A, Pittet V, Rossel JB, von Kanel R, & Swiss IBD Cohort Study Group. (2016). Symptoms of Depression and Anxiety Are Independently Associated With Clinical Recurrence of Inflammatory Bowel Disease. Clin Gastroenterol Hepatol, 14(6), 829–835 e821. doi: 10.1016/j.cgh.2015.12.045 [DOI] [PubMed] [Google Scholar]
- Oikawa T (2006). Alanine, Aspartate, and Asparagine Metabolism in Microorganisms In Wendisch WF (Ed.), Amino Acids Biosynthesis - Pathways, Regulation, and Metabolic Engineering (pp. 273–288). Berlin, Heidelberg: Springer. [Google Scholar]
- Prince A, Whelan K, Moosa A, Lomer MC, and Reidlinger DP (2016). Nutritional problems in inflammatory bowel disease: the patient perspective. J Crohns Colitis, 5(5), 443–450. [DOI] [PubMed] [Google Scholar]
- Salem HA, & Wadie W (2017). Effect of Niacin on Inflammation and Angiogenesis in a Murine Model of Ulcerative Colitis. Sci Rep, 7(1), 7139. doi: 10.1038/s41598-017-07280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S,… Manzin A (2017). Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep, 7(1), 9523. doi: 10.1038/s41598-017-10034-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S,… Manzin A (2017). Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep, 7(1), 9523. doi: 10.1038/s41598-017-10034-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric J, Wang Y, Li J, Coen M, Utzinger J, Marchesi JR,… Holmes E (2008). Species variation in the fecal metabolome gives insight into differential gastrointestinal function. J Proteome Res, 7(1), 352–360. doi: 10.1021/pr070340k [DOI] [PubMed] [Google Scholar]
- Schwiertz A., Hold GL., Duncan SH., Gruhl B., Collins MD., Lawson PA.,… Blaut M (2002). Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst Appl Microbiol, 25(1), 46–51. doi: 10.1078/0723-2020-00096 [DOI] [PubMed] [Google Scholar]
- Skelly AN, Sato Y, Kearney S, & Honda K (2019). Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol, 19(5), 305–323. doi: 10.1038/s41577-019-0144-5 [DOI] [PubMed] [Google Scholar]
- Traue HC, & Kosarz P (1999). Everyday stress and Crohn’s disease activity: a time series analysis of 20 single cases. Int J Behav Med, 6(2), 101–119. doi: 10.1207/s15327558ijbm0602_1 [DOI] [PubMed] [Google Scholar]
- Vidarsdottir JB, Johannsdottir SE, Thorsdottir I, Bjornsson E, Ramel A (2011). A cross-sectional study on nutrient intake and status in inflammatory bowel disease patients. Nutr J, 15 (1), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao X, Ghozlane A, Hu H, Li X, Xiao Y,… Zhang T (2018). Characteristics of Faecal Microbiota in Paediatric Crohn’s Disease and Their Dynamic Changes During Infliximab Therapy. J Crohns Colitis, 12(3), 337–346. doi: 10.1093/ecco-jcc/jjx153 [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, & Siuzdak G (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A, 106(10), 3698–3703. doi: 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomersky NL., Atkinson BJ., Franklin SW., Mitchell PD., Snapper SB., Comstock LE., & Bousvaros A. (2013). Characterization of adherent bacteroidales from intestinal biopsies of children and young adults with inflammatory bowel disease. PLoS One, 8(6), e63686. doi: 10.1371/journal.pone.0063686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Overall microbial community composition is not significantly different in Crohn’s disease patients with higher perceived stress. A. Principal coordinate plot of unweighted UniFrac distances did not show significant differences in beta diversity between High PSS and Low PSS CD patients (Adonis, p=0.15). Similar findings were evident for weighted UniFrac distances. B. Shannon’s diversity index, a measure of community alpha diversity, was not significantly different between High PSS and Low PSS CD patients. Similar findings were evident with other measures of alpha diversity (including Ace, Chao1, PD Whole Tree, Observed Features).