Abstract
Isocitrate dehydrogenase 1 (IDH1) is the most frequently mutated gene in World Health Organization grade II–III and secondary glioma. The majority of IDH1 mutation cases involve the substitution from arginine to histidine at codon 132 (IDH1-R132H). Although the oncogenic role of IDH1-R132H has been confirmed, patients with IDH1-R132H brain tumors exhibit a better response to radiotherapy compared with those with wild-type (WT) IDH1. In the present study, the potential mechanism of radiosensitization mediated by IDH1-R132H was investigated by overexpressing IDH1-R132H in U87MG glioma cells. The results demonstrated decreased clonogenic capacity of IDH1-R132H-expressing cells, as well as delayed repair of DNA double-strand breaks compared with IDH1-WT. Data from The Cancer Genome Atlas were analyzed, which demonstrated that the expression of TP53-induced glycolysis and apoptosis regulator (TIGAR) was lower in patients with glioma harboring IDH1 mutations compared with that in patients with IDH1-WT. TIGAR-knockdown increases the radiosensitivity of glioma cells; in U87MG cells, IDH1-R132H suppressed TIGAR expression. Chromatin immunoprecipitation assays revealed increased levels of repressive H3K9me3 markers at the TIGAR promoter in IDH1-R132H compared with IDH1-WT. These data indicated that IDH1-R132H may overcome radioresistance in glioma cells through epigenetic suppression of TIGAR expression. However, these favorable effects were not observed in U87MG glioma stem-like cells. The results of the present study provide an improved understanding of the functionality of IDH1 mutations in glioma cells, which may improve the therapeutic efficacy of radiotherapy.
Keywords: isocitrate dehydrogenase 1, mutation, glioma cells, glioma stem cells, TP53-induced glycolysis and apoptosis regulator, radiosensitivity, histone H3 lysine 9 trimethylation
Introduction
Isocitrate dehydrogenase 1 (IDH1) catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) and generates NADPH from NADP+. Genome-wide analysis has demonstrated that ~12% of patients with glioblastoma multiforme exhibit mutations in the IDH1 gene (1). In addition, >70% of World Health Organization grade II–III and secondary gliomas carry heterozygous missense mutations at the IDH1 codon 132, and the majority of the mutations are arginine to histidine substitutions (IDH1-R132H) (2–4). The residue encoded by codon 132 of the IDH1 gene is crucial for the formation of the isocitrate binding site; thus, the R132H mutation enables neomorphic catalytic activity, by which α-KG is converted to the putative oncometabolite R-2-hydroxyglutarate (2-HG). The reduced bioavailability of α-KG and supraphysiological levels of 2-HG inhibit DNA and histone demethylation and promote malignant transformation through reshaping the epigenetic and transcriptional landscape (5,6). In addition, as IDH1 is one of the rate-limiting enzymes in the tricarboxylic acid cycle, IDH1 mutations elicit global metabolic reprogramming (7,8). Despite the oncogenic role of IDH1 mutations, prospective studies have demonstrated that patients with brain tumor harboring IDH1 mutations exhibit more favorable prognosis and are more responsive to clinical treatment compared with those with wild-type (WT) IDH1 (9–11). The detailed mechanisms of the roles of the IDH1-R132H mutation remain to be fully determined.
The prevalence and prominence of IDH1 mutations makes them a highly attractive focus of research to clarify the molecular mechanisms underlying the biological behavior of glioma cells and to develop novel targeted therapeutics (4,12). The IDH1-R132H mutation is an early event in gliomagenesis (13); however, once the malignant transformation is complete, glioma cell proliferation does not rely on IDH1-R132H, which implicates that the role of IDH1-R132H diminishes during this process (14). Since radiotherapy represents an indispensable therapeutic approach for patients with glioma, one feasible method to enhance its therapeutic benefits is to overcome the radioresistance of glioma cells. IDH1-R132H is considered to be a radiosensitizing gene based on its ability to catalyze the oxidation of NADPH to NADP+, whereas IDH1-WT reduces NADP+ to NADPH. The decreased generation of NADPH may inhibit the production of glutathione, resulting in increased levels of reactive oxygen species (ROS) induced by ionizing radiation (IR) exposure and more DNA double-strand breaks (DSBs) (15–17).
Our previous studies demonstrated the radiosensitization of glioma cells by knockdown of TP53-induced glycolysis and apoptosis regulator (TIGAR) (18,19). TIGAR is a p53 target gene that functions as a fructose 2,6-bisphosphatase to decrease the levels of fructose 2,6-bisphophate, which leads to allosteric inhibition of phosphofructokinase 1 activity (20). By directing the metabolic flux from glycolysis to the pentose phosphate pathway (PPP), TIGAR serves an important role in the regulation NADPH production. Gene set enrichment analysis and metabolic profiling have revealed elevated levels of glycolysis and reduced oxidative phosphorylation in IDH1-R132H-mutated U87MG cells compared with IDH1-WT (21). Therefore, it was hypothesized that IDH1-R132H may be associated with the regulation of TIGAR functions and consequently mediate the cytotoxic effects of IR on glioma cells. The present study aimed to investigate the radiosensitizing effects of IDH1-R132H on U87MG cells and U87MG glioma stem-like cells (GSCs).
Materials and methods
Cell culture and irradiation
Human malignant glioma U87MG cells (ATCC; Cat. no. HTB-14; glioblastoma of unknown origin) and T98G cells (ATCC; Cat. no. CRL-1690; glioblastoma multiforme) were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone; GE Healthcare Life Sciences) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences), 100 µg/ml streptomycin and 100 U/ml penicillin (Hyclone; GE Healthcare Life Sciences) at 37°C in a humidified atmosphere containing 5% CO2. The cell lines were authenticated by short tandem repeat profiling. For enrichment of U87MG-GSCs, U87MG cells were cultured in DMEM/F12 supplemented with B27 Plus Supplement (Invitrogen; Thermo Fisher Scientific, Inc.), 20 ng/ml epidermal growth factor (EGF; PeproTech, Inc.) and basic fibroblast growth factor (bFGF; PeproTech, Inc.) as previously described (22). The culture flasks were coated with 10 µg/ml laminin (Sigma-Aldrich; Merck KGaA) and incubated at 37°C for 3 h prior to use. At 90% confluence, the cells were dissociated by Accutase (Merck KGaA) and subcultured at 1:3-5 in fresh laminin-coated flasks. Cells were cultured for 10 passages, then 1×106 cells were washed and suspended in 100 µl PBS, and incubated for 30 min at 4°C with phycoerythrin-conjugated CD133 antibody (1:100, Thermo Fisher Scientific, Inc.; cat. no. 12-1338-42). U87MG-GSCs were sorted using a FACScalibur flow cytometer (BD Biosciences). Cells were irradiated with 160 kV X-ray at a dose rate of 1.18 Gy/min by a biological research irradiator (Rad Source Technologies).
Construction of the IDH1-R132H plasmid and transfection
The wild-type (WT) IDH1 fragment was obtained by using Phusion High-Fidelity DNA Polymerase (New England BioLabs, Inc.). The forward primer was 5′-TACCGGACTCAGATCTCGAGCGCCACCATGTCCAAAAAAATCAGTGGC-3′ and the reverse primer was 5′-GATCCCGGGCCCGCGGTACCGTAAGTTTGGCCTGAGCTAGTTTG-3′. The thermocycling conditions were as follows: 1 cycle at 98°C for 30 sec; 35 cycles at 98°C for 10 sec, 60°C for 30 sec, 72°C for 40 sec; and 1 cycle at 72°C for 10 min. The fragment was then subcloned into the pCMV-GV230 vector (Shanghai GeneChem Co., Ltd.) between the XhoI and KpnI sites, and IDH1 was tagged with green fluorescent protein. IDH1-R132H was generated from IDH1-WT using a Quick Change Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) according to the manufacturer's instructions and confirmed by DNA sequencing. The pCMV–IDH1-R132H plasmid and the corresponding control empty vector were transfected by Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature. Subsequent experiments were performed 48 h after transfection unless otherwise stated.
Western blot analysis
U87MG cells or U87MG-GSCs were transfected with pCMV–IDH1-R132H plasmid and/or treated with the selective IDH1-R132H inhibitor AGI-5198 (Selleck Chemicals) for 72 h at 37°C. Cells were lysed by RIPA buffer (Beyotime Institute of Biotechnology) on ice for 20 min and the protein concentration was measured by the BCA method. Cell lysate containing 20 µg protein were separated by 10% SDS-PAGE and electrically transferred to PVDF membranes. Non-specific binding was blocked by incubation with Tris-buffered saline containing 5% (w/v) bovine serum albumin (Beyotime Institute of Biotechnology) at room temperature for 1 h. The membranes were then incubated with primary antibodies against IDH1 and IDH1-R132H (1:1,000, Cell Signaling Technology, Inc.; cat. no. 3997), TIGAR (1:500, Abcam; cat. no. ab37910), histone H3 lysine 9 trimethylation (H3K9me3, 1:1,000, Abcam; cat. no. ab8898) and tubulin (1:2,000, Santa Cruz Biotechnology, Inc.; cat. no. sc-5286) at 4°C overnight. Membranes were then incubated with a horseradish peroxidase-conjugated goat anti-mouse or rat antibody (1:1,000, Beyotime Institute of Biotechnology; cat. no. A0216, A0192) at room temperature for 1 h. Tubulin was used as a loading control. Immunoblotting signals were detected using an enhanced chemiluminescence kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocols. Densitometric analysis was performed by using ImageJ software 1.52 (National Institutes of Health).
Clonogenic survival assay
U87MG cells or U87MG-GSCs were plated in triplicate into six-well plates and irradiated with different doses of X-ray (0.3×103 cells for 0 Gy, 0.8×103 cells for 2 Gy, 3×103 cells for 4 Gy, 9×103 cells for 6 Gy, 15×103 cells for 8 Gy). Cells were subsequently cultured for 10 days, fixed with methanol at room temperature for 15 min and stained with 1% crystal violet at room temperature for 30 min. Colonies consisting of >50 cells were counted. Plating efficiency (PE) was calculated as the ration of the number of colonies to the seeded cell number. Surviving fraction was calculated as PE(treated)/PE(control).
Immunofluorescence analysis
U87MG-GSCs were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. Immunostaining was performed using antibodies against glial fibrillary acidic protein (GFAP, 1:500 Abcam; cat. no. ab7260) or nestin (1:100, Beyotime; AN203) at 4°C overnight. For visualization of IR-induced foci, cells were irradiated with 4 Gy X-ray and cultured for 12 h prior to immunostaining with an antibody against phosphorylated histone H2A variant (γ-H2AX, 1:500, Abcam; cat. no. ab2893). Cell nuclei were stained with 4′6-diamidino-2-phenylindole (DAPI). Images were captured on Olympus FV1200 confocal microscopy at a magnification of ×200 in three random fields.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed by using the ChIP Assay kit (Beyotime) and the aforementioned antibody against H3K9me3. U87MG cells were transfected with pCMV–IDH1-R132H for 72 h; subsequently, 37% formaldehyde was added directly to the homogenate of cultured cells at a final concentration of 1% followed by incubation for 10 min at 37°C to crosslink DNA and histones. Following extensive washing with ice-cold phosphate-buffered saline supplemented with 1 mM phenylmethysulphonyl fluoride, the cells were lysed using the ChIP Assay kit and sonicated for 4 cycles (10 sec on, 10 sec off) on ice to generate DNA fragments (200–1,000 bp). Following centrifugation at 12,000 × g for 5 min at 4°C, the supernatant was diluted 10-fold with ChIP dilution buffer and blocked with salmon sperm DNA/Protein A/G-agarose beads. Protein-DNA complexes were immunoprecipitated with 3 µg H3K9me3 antibody overnight at 4°C. A total of 5% of the extract was used for PCR amplification as the input control. Following recovery, the DNA fragments were purified and subjected to PCR amplification using Taq DNA polymerase (Beyotime Institute of Biotechnology). The primers for TIGAR promoter: Forward, 5′-CCTCATAGAACACAGTCTGTTGGTCG-3′ and reverse, 5′-AGTCCGCGGTAGGATTCCTT-3′. The thermocycling conditions were as follows: 1 cycle at 94°C for 3 min; 35 cycles at 94°C for 30 sec, 55°C for 30 sec, 72°C for 60 sec; and 1 cycle at 72°C for 10 min. The PCR products were separated by 2% agarose gel electrophoresis.
The Cancer Genome Atlas (TCGA) data analysis
IDH1 mutation and TIGAR expression data were obtained from TCGA PanCancer Atlas and TCGA Provisional datasets from the cBioportal for Cancer Genomics (http://www.cbioportal.org) (23,24). TIGAR mRNA expression was quantified by RNA-Seq using the Expectation Maximization (RSEM) method and converted to z-scores (25). The data were visualized as violin plots using the Python 3.6 (Python Software Foundation) plotting library Matplotlib 3.0.2 (26).
Statistical analysis
Data are expressed as mean ± standard deviation. Analysis of differences between two groups was performed by Student's t-test. Comparisons of z-score values for TIGAR expression was performed by Wilcoxon rank-sum test. P<0.05 was considered to indicate a statistically significant difference.
Results
IDH1-R132H increases the radiosensitivity of U87MG cells, but not U87MG-GSCs
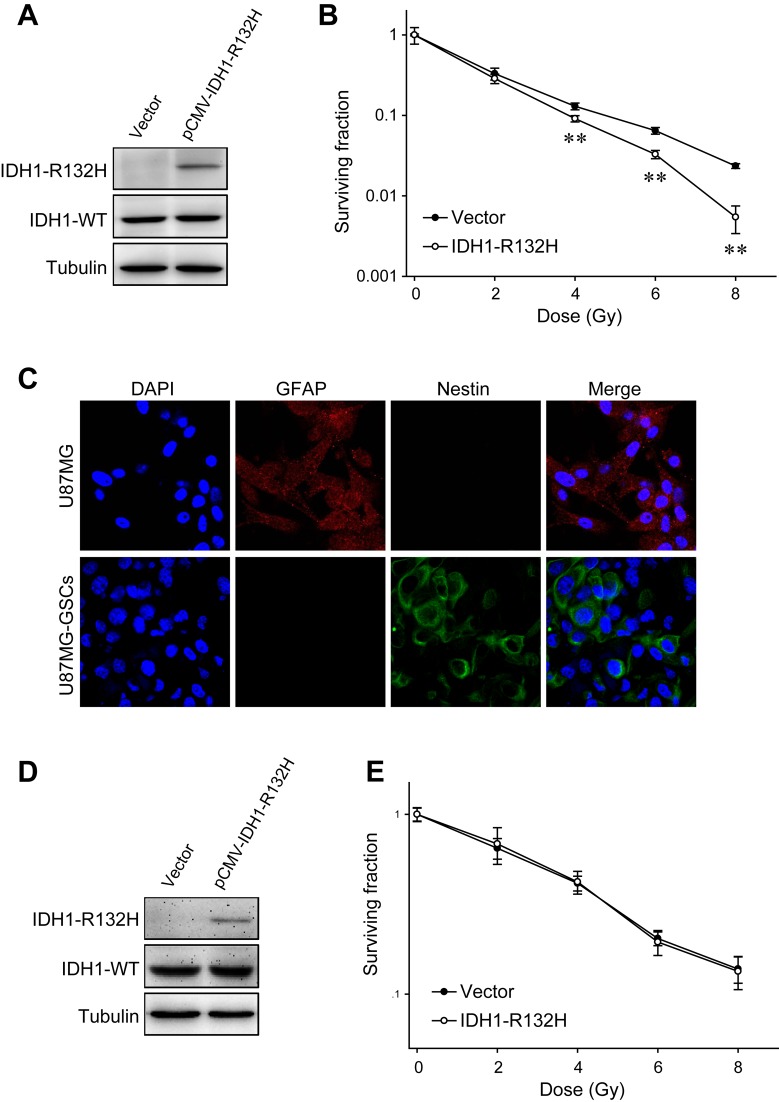
The IDH1-R132H plasmid was transfected into U87MG cells, and expression of the mutated protein was confirmed by western blotting (Fig. 1A). Clonogenic assays were performed to evaluate the survival fraction following exposure to different doses of X-ray; U87MG cells transfected with IDH1-R132H formed significantly fewer clones compared with the control groups, suggesting the radiosensitizing effects of IDH1-R132H on U87MG cells (Fig. 1B). In addition, the radioresistance of another glioma cell line, T98G, was also suppressed by IDH1-R132H (Fig. S1).
Figure 1.
Effects of IDH1-R132H on U87MG cell and U87MG-GSC radiosensitivity. (A) Expression of IDH1-R132H in U87MG cells transfected with the pCMV–IDH1-R132H plasmid was examined by western blot analysis. (B) The radiosensitivity of U87MG cells expressing IDH1-R132H was evaluated by clonogenic assay. (C) The differentiation marker GFAP and stem cell marker nestin were visualized in U87MG cells and U87MG-GSCs by immunofluorescence. (D) IDH1-R132H expression in U87MG-GSCs following transfection with the pCMV–IDH1-R132H plasmid. (E) Clonogenic assay of U87MG-GSCs transfected with an empty vector or the pCMV–IDH1-R132H plasmid. **P<0.01 vs. empty vector-transfected cells. IDH1, isocitrate dehydrogenase 1; GSC, glioma stem-like cell; WT, wild-type; GFAP, glial fibrillary acidic protein.
GSCs are highly radioresistant and may be responsible for tumor recurrence following radiotherapy (27,28); therefore, the potential impact of IDH1-R132H expression on U87MG-GSCs was investigated. U87MG cells were cultured in serum-free medium containing EGF, bFGF and B27 supplement on an adherent substrate, and CD133-positive U87MG-GSCs were isolated by fluorescence-activated cell sorting (29). Immunostaining demonstrated that U87MG-GSCs expressed lower levels of the differentiation marker GFAP, and higher levels of the stem cell marker nestin compared with untreated U87MG cells (Fig. 1C). In contrast to U87MG cells, the radiosensitivity of U87MG-GSCs was unaltered when IDH1-R132H was expressed ectopically (Fig. 1D and E). Additionally, in accordance with previous reports, the radioresistance of U87MG-GSCs was higher compared with that of U87MG cells (Fig. 1B and E).
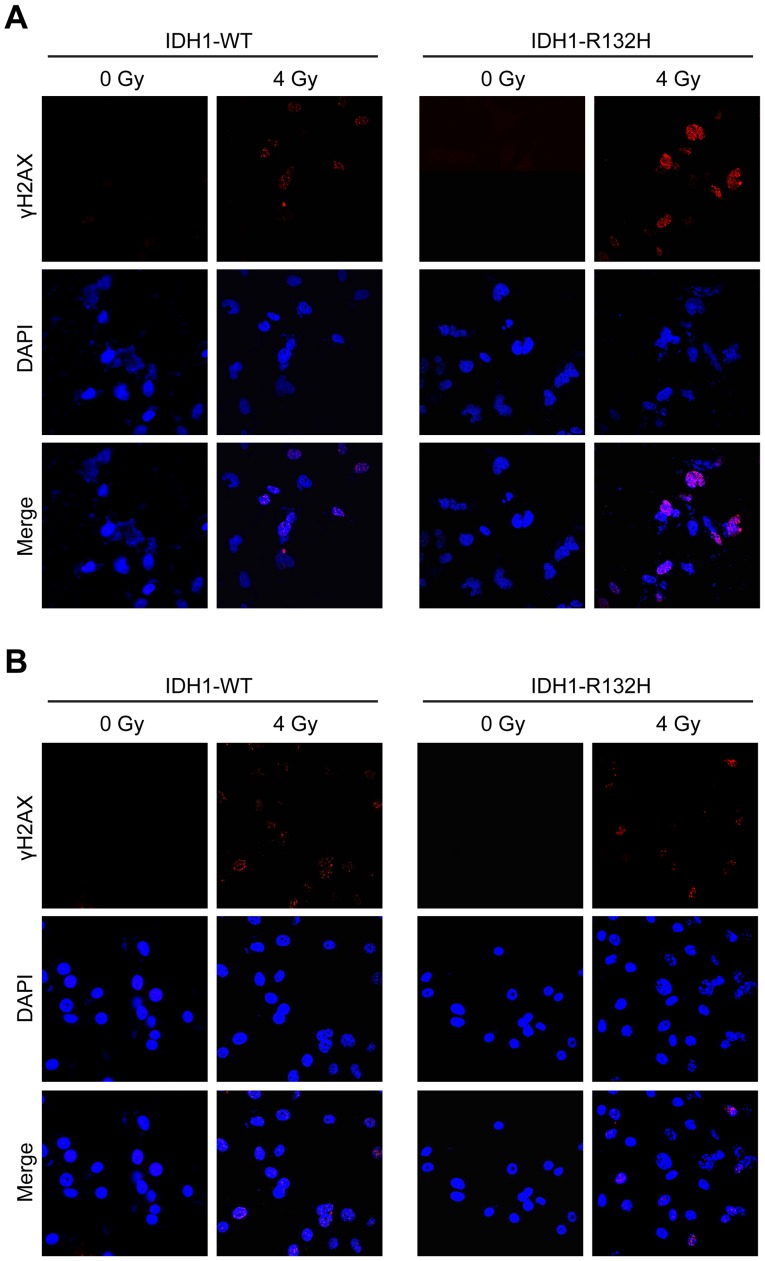
IDH1-R132H suppresses repair of IR-induced DSBs in U87MG cells
DSBs are among the most dangerous DNA lesions induced by IR; thus, their formation and repair determine the fate of irradiated cells (30). DSBs were visualized by fluorescent staining of nuclear γ-H2AX, which forms microscopically discernible foci at DSB sites. As presented in Fig. 2A, the majority of γ-H2AX foci in the control cells were resolved 12 h post-4 Gy X-ray irradiation, whereas a substantial fraction of IDH1-R132H-expressing U87MG cells retained γ-H2AX foci, suggesting inhibition of the DSB repair process. By contrast, no differences were observed in the IR-induced γ-H2AX foci in IDH1-WT- and IDH1-R132H-expressing U87MG-GSCs (Fig. 2B).
Figure 2.
IDH1-R132H delays IR-induced DSB repair in U87MG cells. Immunostaining of γ-H2AX foci of (A) U87MG cells and (B) U87MG-GSCs. IDH1, isocitrate dehydrogenase 1; GSC, glioma stem-like cell; WT, wild-type; GFAP, glial fibrillary acidic protein.
IDH1-R132H is associated with low TIGAR expression
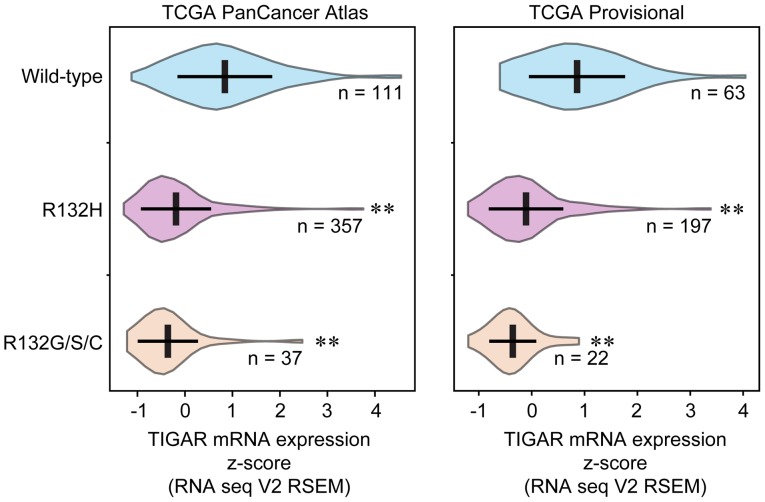
To investigate the relationship between IDH1-R132H and TIGAR expression, IDH1 mutation status and TIGAR expression values were collected from TCGA. Analysis of two glioma studies (TCGA PanCancer Atlas and TCGA Provisional) revealed that 70.69% (357/505) and 69.86% (197/282) of patients with glioma, respectively, were IDH1-R132H-positive. Other minor types of IDH1 mutation at codon 132, such as histidine to glycine, serine or cysteine (R132G/S/C), were included as a consolidated group. Glioma samples with IDH1-R132H, as well as IDH2-R132G/S/C, exhibited significantly lower levels of TIGAR expression compared with IDH1-WT (Fig. 3), which suggested that TIGAR may be one of the IDH1-R132H target genes.
Figure 3.
TCGA data analysis. TIGAR expression in two datasets of IDH1 mutations and TIGAR expression in patients with glioma with wild-type IDH1, IDH1-R132H and IDH2-R132G/S/C were compared. **P<0.01 vs. wild-type. IDH1, isocitrate dehydrogenase 1; R132H, patients with the IDH1-R132H mutation; TCGA, The Cancer Genome Atlas; TIGAR, TP53-induced glycolysis and apoptosis regulator; RSEM, RNA-Seq by Expectation Maximization.
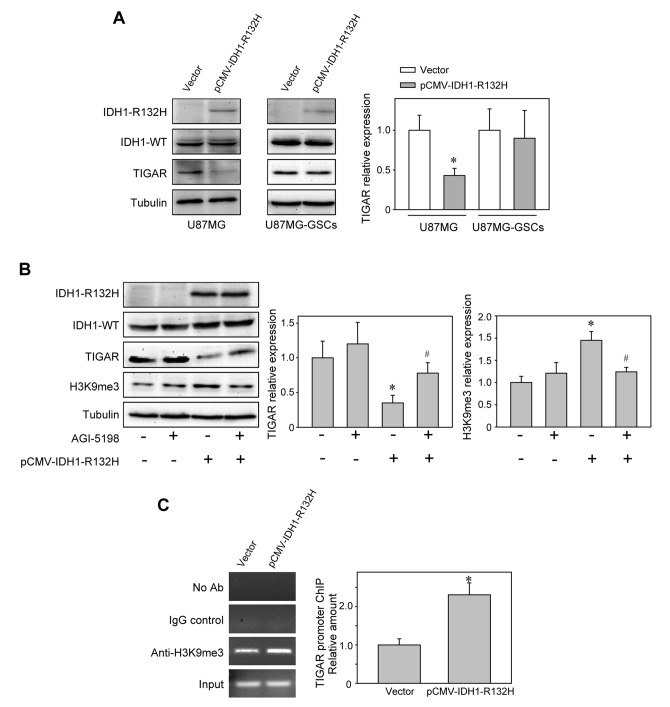
Epigenetic regulation of TIGAR expression by IDH1-R132H in U87MG cells
The downregulation of TIGAR expression by IDH1-R132H was confirmed by western blotting in U87MG and T98G cells (Figs. 4A and S1A). When IDH1-R132H activity was repressed by the selective inhibitor AGI-5198, TIGAR expression was upregulated (Fig. 4B), which suggested that the suppression of TIGAR expression may depend on IDH1-R132H. By contrast, IDH1-R132H (Fig. 4A) or AGI-5198 (Fig. S2) treatment did not alter TIGAR expression in U87MG-GSCs. The involvement of IDH1-R132H in H3K9me3 modification has been reported to serve an important role in the malignant transformation of glioma (14,31). Western blot analysis revealed increased levels of H3K9me3 in U87MG cells expressing IDH1-R132H compared with those expressing IDH1-WT (Fig. 4B), and ChIP assays further demonstrated the augmentation of the inhibitory H3K9me3 markers at the TIGAR promoter (Fig. 4C). These results indicated that IDH1-R132H may inhibit TIGAR expression by epigenetic silencing.
Figure 4.
IDH1-R132H epigenetically decreases TIGAR expression. (A) TIGAR expression was reduced in IDH1-R132H-expressing U87MG cells, but not in U87MG-GSCs. *P<0.05 vs. empty vector-transfected group. (B) The effects of IDH1-R132H inhibitor AGI-5198 on the expression of TIGAR and H3K9me3. *P<0.05 vs. control group, #P<0.05 vs. pCMV–IDH1-R132H-transfected group. (C) Chromatin immunoprecipitation assay demonstrated the interaction between H3K9me3 and the TIGAR promoter. *P<0.05 vs. empty vector-transfected group. IDH1, isocitrate dehydrogenase 1; WT, wild-type; TCGA, The Cancer Genome Atlas; TIGAR, TP53-induced glycolysis and apoptosis regulator; GSC, glioma stem-like cells; IgG, immunoglobulin G; H3K9me3, histone H3 lysine 9 trimethylation.
Discussion
Fractionated radiotherapy is one of the major treatments for patients with glioma; however, the therapeutic effects are often undermined by the radioresistance of glioma cells (32). Of note, several studies have confirmed that tumor cells with IDH1-R132H are more susceptible to oxidative damage compared with IDH1 WT cells, thus exhibiting increased sensitivity to IR (15,16,33). Treatment with the IDH1-R132H inhibitor AGI-5198 exhibits protective effects against IR-induced damage in IDH1-R132H-mutated cells (34). These findings indicated that a better understanding of the functional consequences of IDH1-R132H may help determine the mechanism underlying the radiosensitization of glioma cells. In the present study, the radiosensitizing effects of IDH1-R132H, as well as the inhibitory effects on DSB repair were confirmed using IDH1-R132H-expressing U87MG cells. The reduced generation of NADPH may account for the altered radiosensitivity (15,16). The capacity for DSB repair determines the cellular response to IR exposure and participates in malignant transformation of tumors (35). Contradictory results have been reported on the influence of IDH1 mutation on homologous recombination (36,37). The effects of IDH1 mutation on DSB repair appear to be dependent on genetic context, such as loss of function mutations of TP53 and X-linked helicase II (38).
TCGA data analysis in the present study demonstrated that TIGAR expression was significantly decreased in samples with IDH1-R132H, which is the dominant type of IDH1 codon 132 mutation, compared with those with IDH1-WT. Other mutations (R132G/S/C) also exhibited lower TIGAR expression compared with that in IDH1-WT. In addition, the suppressive effects of IDH1-R132H on TIGAR expression were confirmed in U87MG cells. These results suggested that IDH1-R132H may counteract glioma cell radioresistance by reducing TIGAR expression. TIGAR increases the entry of glucose 6-phosphate into the PPP to generate NADPH, which is necessary to maintain normal levels of reduced glutathione, the major cellular ROS scavenger; hence, TIGAR knockdown may result in radiosensitization of glioma cells (18,19,39). A previous study has demonstrated that IDH1-R132H leads to a metabolic shift toward glycolysis in U87MG cells (21). Therefore, it may be speculated that IDH1-R132H-reduced TIGAR expression may inhibit the PPP and promote glycolysis, serving as a potential initiator of metabolic reprogramming. The irreversible oxidative steps catalyzed by glucose 6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in the PPP, together with the oxidation of isocitrate catalyzed by IDH1, are the major sources of NADPH (40,41). Therefore, the IDH1-R132H mutation and TIGAR downregulation may act synergistically to reduce NADPH production and attenuate cellular defense against oxidative stress. Following exposure to IR, the inability of NADPH-deficient cells to eliminate ROS formed during water radiolysis makes them vulnerable to IR-induced cytotoxicity.
Numerous reports have confirmed the association between 2-HG, the product of IDH1-R132H, and epigenetic dysfunction. Increased levels of 2-HG competitively suppress α-KG-dependent dioxygenases, including lysine demethylases and the members of the ten-eleven translocation family, which results in transcriptional silencing of tumor-suppressor genes via histone and DNA hypermethylation, therefore suggesting one mechanism by which elevated 2-HG levels exert their oncogenic effects (4,5,42–44). In hematopoietic stem cells, increased H3K9me3 markers have been identified in the promoter of the ataxia telangiectasia mutated gene, which encodes an essential sensor kinase for DSB repair (45). Similarly, the present study demonstrated higher levels of H3K9me3 markers at the TIGAR promoter in U87MG cells with IDH1-R132H compared with IDH1-WT, which indicated a condensed chromatin state and transcriptional inactivation of TIGAR.
Glioma stem cells are radioresistant, and glioma recurrence after radiotherapy originates from these cells. To monitor the role of IDH1-R132H in glioma stem cells, an enriched population of U87MG-GSCs was successfully obtained, which was confirmed by the expression of nestin and reduced expression of GFAP. U87MG-GSCs expressing IDH1-R132H exhibited similar clonogenic ability following X-ray irradiation to that of IDH1-WT-expressing U87MG-GSCs. In addition, IDH1-R132H did not inhibit TIGAR expression in U87MG-GSCs. Little is currently known about the functions of IDH1 mutations in glioma stem cells; these preliminary data indicate that the radiosensitivity of U87MG-GSCs may be unaffected by IDH1-R132H, although further studies are required to define the exact functional relationship between IDH1-R132H and glioma stem cells.
In conclusion, the results of the present study provide a novel mechanism by which IDH1-R132H exerts radiosensitizing effects on glioma cells. Specifically, IDH1-R132H epigenetically lowers the expression of TIGAR, which is a crucial controller of intracellular redox homeostasis; therefore, glioma cells with IDH1-R132H fail to eliminate the IR-induced ROS. Although radiotherapy is the major form of therapy administered post-surgery to patients with glioma, glioma cells remain highly refractory to IR. The results of the present study may provide new insights into the radiosensitizing effects of IDH1-R132H in glioma cells.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of China (grant nos. 31770912, 31270897 and 81271682), the Project of State Key Laboratory of Radiation Medicine and Protection, Soochow University (grant no. GZK1201808), Jiangsu Provincial Key Laboratory of Radiation Medicine and Protection, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
NY, TX, HZ and JC performed the experiments. JY analyzed The Cancer Genome Atlas data. JC provided technical assistance. FL designed the research, analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 3.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waitkus MS, Diplas BH, Yan H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell. 2018;34:186–195. doi: 10.1016/j.ccell.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philip B, Yu DX, Silvis MR, Shin CH, Robinson JP, Robinson GL, Welker AE, Angel SN, Tripp SR, Sonnen JA, et al. Mutant IDH1 promotes glioma formation in vivo. Cell Rep. 2018;23:1553–1564. doi: 10.1016/j.celrep.2018.03.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, Slocum KL, Pu M, Lin F, Vickers C, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74:3317–3331. doi: 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izquierdo-Garcia JL, Viswanath P, Eriksson P, Chaumeil MM, Pieper RO, Phillips JJ, Ronen SM. Metabolic reprogramming in mutant IDH1 glioma cells. PLoS One. 2015;10:e0118781. doi: 10.1371/journal.pone.0118781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 10.Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, Buckner JC, Fink KL, Souhami L, Laperriere NJ, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32:783–790. doi: 10.1200/JCO.2013.49.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran AN, Lai A, Li S, Pope WB, Teixeira S, Harris RJ, Woodworth DC, Nghiemphu PL, Cloughesy TF, Ellingson BM. Increased sensitivity to radiochemotherapy in IDH1 mutant glioblastoma as demonstrated by serial quantitative MR volumetry. Neuro Oncol. 2014;16:414–420. doi: 10.1093/neuonc/not198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waitkus MS, Diplas BH, Yan H. Isocitrate dehydrogenase mutations in gliomas. Neuro Oncol. 2016;18:16–26. doi: 10.1093/neuonc/nov136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannessen TA, Mukherjee J, Viswanath P, Ohba S, Ronen SM, Bjerkvig R, Pieper RO. Rapid conversion of mutant IDH1 from driver to passenger in a model of human gliomagenesis. Mol Cancer Res. 2016;14:976–983. doi: 10.1158/1541-7786.MCR-16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Chou AP, Chen W, Chen R, Deng Y, Phillips HS, Selfridge J, Zurayk M, Lou JJ, Everson RG, et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro Oncol. 2013;15:57–68. doi: 10.1093/neuonc/nos261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Sun B, Shi W, Zuo H, Cui D, Ni L, Chen J. Decreasing GSH and increasing ROS in chemosensitivity gliomas with IDH1 mutation. Tumour Biol. 2015;36:655–662. doi: 10.1007/s13277-014-2644-z. [DOI] [PubMed] [Google Scholar]
- 17.Kessler J, Güttler A, Wichmann H, Rot S, Kappler M, Bache M, Vordermark D. IDH1(R132H) mutation causes a less aggressive phenotype and radiosensitizes human malignant glioma cells independent of the oxygenation status. Radiother Oncol. 2015;116:381–387. doi: 10.1016/j.radonc.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Chen F, Tai G, Wang J, Shang J, Zhang B, Wang P, Huang B, Du J, Yu J, et al. TIGAR knockdown radiosensitizes TrxR1-overexpressing glioma in vitro and in vivo via inhibiting Trx1 nuclear transport. Sci Rep. 2017;7:42928. doi: 10.1038/srep42928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Gu C, Yu J, Wang Z, Yuan X, Yang L, Wang J, Jia Y, Liu J, Liu F. Radiosensitization of glioma cells by TP53-induced glycolysis and apoptosis regulator knockdown is dependent on thioredoxin-1 nuclear translocation. Free Radic Biol Med. 2014;69:239–248. doi: 10.1016/j.freeradbiomed.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 20.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 21.Karpel-Massler G, Ishida CT, Bianchetti E, Zhang Y, Shu C, Tsujiuchi T, Banu MA, Garcia F, Roth KA, Bruce JN, et al. Induction of synthetic lethality in IDH1-mutated gliomas through inhibition of Bcl-xL. Nat Commun. 2017;8:1067. doi: 10.1038/s41467-017-00984-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 27.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 28.Rizzo AE, Yu JS. Radiation therapy for glioma stem cells. Adv Exp Med Biol. 2015;853:85–110. doi: 10.1007/978-3-319-16537-0_6. [DOI] [PubMed] [Google Scholar]
- 29.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 30.Santivasi WL, Xia F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid Redox Signal. 2014;21:251–259. doi: 10.1089/ars.2013.5668. [DOI] [PubMed] [Google Scholar]
- 31.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han X, Xue X, Zhou H, Zhang G. A molecular view of the radioresistance of gliomas. Oncotarget. 2017;8:100931–100941. doi: 10.18632/oncotarget.21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohrenz IV, Antonietti P, Pusch S, Capper D, Balss J, Voigt S, Weissert S, Mukrowsky A, Frank J, Senft C, et al. Isocitrate dehydrogenase 1 mutant R132H sensitizes glioma cells to BCNU-induced oxidative stress and cell death. Apoptosis. 2013;18:1416–1425. doi: 10.1007/s10495-013-0877-8. [DOI] [PubMed] [Google Scholar]
- 34.Molenaar RJ, Botman D, Smits MA, Hira VV, van Lith SA, Stap J, Henneman P, Khurshed M, Lenting K, Mul AN, et al. Radioprotection of IDH1-mutated cancer cells by the IDH1-mutant inhibitor AGI-5198. Cancer Res. 2015;75:4790–4802. doi: 10.1158/0008-5472.CAN-14-3603. [DOI] [PubMed] [Google Scholar]
- 35.Khanna KK, Jackson SP. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 36.Ohba S, Mukherjee J, See WL, Pieper RO. Mutant IDH1-driven cellular transformation increases RAD51-mediated homologous recombination and temozolomide resistance. Cancer Res. 2014;74:4836–4844. doi: 10.1158/0008-5472.CAN-14-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, Liu Y, Sundaram RK, Hegan DC, Fons NR, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Núñez FJ, Mendez FM, Kadiyala P, Alghamri MS, Savelieff MG, Garcia-Fabiani MB, Haase S, Koschmann C, Calinescu AA, Kamran N, et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aaq1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peña-Rico MA, Calvo-Vidal MN, Villalonga-Planells R, Martínez-Soler F, Giménez-Bonafé P, Navarro-Sabaté À, Tortosa A, Bartrons R, Manzano A. TP53 induced glycolysis and apoptosis regulator (TIGAR) knockdown results in radiosensitization of glioma cells. Radiother Oncol. 2011;101:132–139. doi: 10.1016/j.radonc.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Bleeker FE, Atai NA, Lamba S, Jonker A, Rijkeboer D, Bosch KS, Tigchelaar W, Troost D, Vandertop WP, Bardelli A, Van Noorden CJ. The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010;119:487–494. doi: 10.1007/s00401-010-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atai NA, Renkema-Mills NA, Bosman J, Schmidt N, Rijkeboer D, Tigchelaar W, Bosch KS, Troost D, Jonker A, Bleeker FE, et al. Differential activity of NADPH-producing dehydrogenases renders rodents unsuitable models to study IDH1R132 mutation effects in human glioblastoma. J Histochem Cytochem. 2011;59:489–503. doi: 10.1369/0022155411400606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M Gagné L, Boulay K, Topisirovic I, Huot MÉ, Mallette FA. Oncogenic activities of IDH1/2 mutations: From epigenetics to cellular signaling. Trends Cell Biol. 2017;27:738–752. doi: 10.1016/j.tcb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, Lemonnier F, Kron KJ, Cescon DW, Hao Z, et al. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30:337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.