Abstract
Thyroid cancer (TC) is one of the most common types of malignancy of the endocrine-system. At present, there is a lack of effective methods to predict neck lymph node metastasis (LNM) in TC. The present study compared the expression profiles from The Cancer Genome Atlas between N1M0 and N0M0 subgroups in each T1-4 stages TC in order to identify the four groups of TC LNM-associated differentially expressed genes (DEGs). Subsequently, DEGs were combined to obtain a total of 493 integrated DEGs by using the method of Robust Rank Aggregation. Furthermore, the underlying mechanisms of LNM were investigated. The results from Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses demonstrated that the identified DEGs may promote LNM via numerous pathways, including extracellular matrix-receptor interaction, PI3K-AKT signaling pathway and focal adhesion. Following construction of a protein-protein interaction network, the significance score for each gene was calculated and seven hub genes were screened, including interleukin 6, actinin α2, collagen type I α 1 chain, actin α1, calbindin 2, thrombospondin 1 and parathyroid hormone. These genes were predicted to serve crucial roles in TC with LNM. The results from the present study could therefore improve the understanding of LNM in TC. In addition, the seven DEGs identified may be considered as potential novel targets for the development of biomarkers that could be used in the diagnosis and therapy of TC.
Keywords: thyroid cancer, bioinformatics analysis, Robust Rank Aggregation, lymph node metastasis, hub gene
Introduction
Thyroid cancer (TC) is a common endocrine malignancy for all age groups worldwide and affects three times as many women as men (1). In the past decade, the incidence of TC has rapidly increased in numerous countries, particularly the USA, Croatia, Korea and China (2). In 2018, an estimated 562,000 patients with TC were newly diagnosed in 185 countries (3). The most common pathological type of TC is papillary thyroid carcinoma (PTC), which accounts for >90% of all cases (4,5). Although the 10-year survival rate of TC is 90% (6,7), lymph node metastasis (LNM) occurs in 24–63% of patients with TC and is considered as an independent risk factor for recurrence and distant metastases (6–11). However, to the best of our knowledge, the fundamental pathophysiology of LNM in TC has not been completely elucidated, which is a major obstacle for clinical diagnosis and treatment. It is therefore crucial to identify the key genes associated with LNM in TC.
In the last decade, microarray technology has been widely applied to identify TC-associated genes, including those involved in LNM. For example, high expression levels of Cbp/P300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1 and γ-aminobutyric acid type A receptor β 2 have been identified as promoters of PTC migration (12,13). However, to obtain a more accurate result, the present study performed a subgroup analysis according to T stage, including T1N0M0 vs. T1N1M0, T2N0M0 vs. T2N1M0, T3N0M0 vs. T3N1M0 and T4N0M0 vs. T4N1M0 comparisons, by using the novel stable analysis method Robust Rank Aggregation.
The present study used the expression profiling data from The Cancer Genome Atlas (TCGA) database (14) to identify the DEGs between N1M0 and N0M0 in each individual T1-4 stage of TC. Combined with the results of Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction (PPI) network analyses, the hub genes and pathways involved in TC with LNM were identified.
Materials and methods
Identification of DEGs
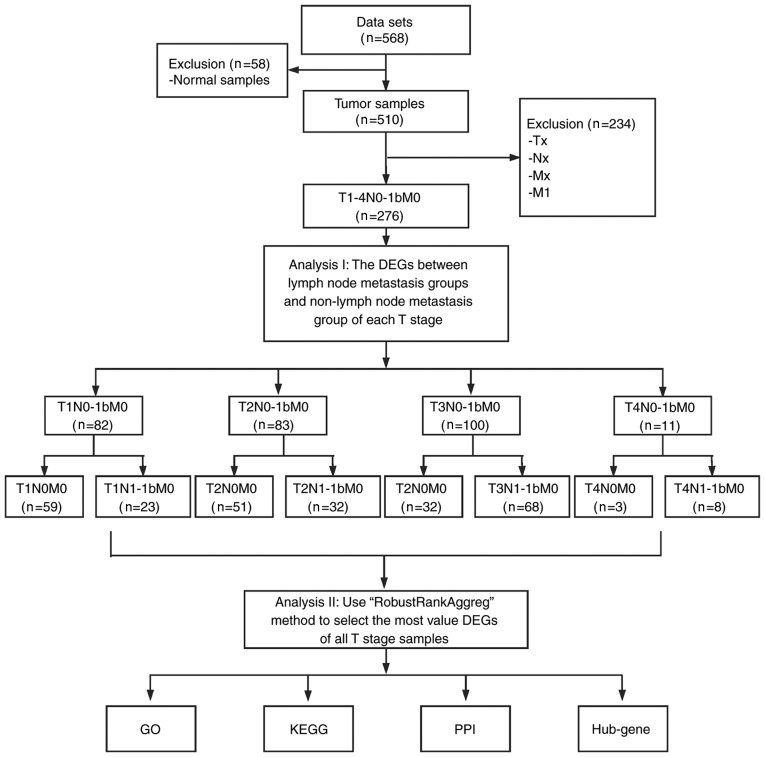
The present study analyzed the RNA-sequencing (RNA-seq) data from TC and normal tissues that were downloaded from the publicly available TCGA database (https://cancergenome.nih.gov). The RNA-seq data were from 568 samples, including 510 TC samples and 58 normal samples. A total of 276 TC samples with T1-4N0-1M0 information were selected. N0 and M0 represent patients with TC without regional nodes and distant metastasis, respectively. Similarly, N1 and M1 represent patients with TC with regional nodes and distant metastasis, respectively (14). According to different T and N status, the samples were divided into 8 subgroups, including T1N0M0 vs. T1N1M0, T2N0M0 vs. T2N1M0, T3N0M0 vs. T3N1M0 and T4N0M0 vs. T4N1M0 (Fig. 1), and made differentially expressed analyses using ‘edgeR’ package (15). Genes that met the conditions of log2 fold change [log fold change (FC)] ≥0.4 and P<0.05 were considered as DEGs. Subsequently, the variable DEGs lists of the different groups were integrated using probabilistic models and a significance score for each gene was calculated using RobustRankAggreg (RRA) package (16) in order to identify the integrated DEGs (iDEGs). Due to the limited size of gene symbol and figure, only the top 40 genes were presented. In addition to the RRA method, Venn diagram was used to determine an intersection among genes in Bioinformatics and Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Figure 1.
Study flowchart. DEG, differentially expressed gene; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction.
GO and KEGG enrichment analysis of the DEGs
GO (http://geneontology.org; release 2019-10-07) terms were used to describe the biological process (BP) of genes. The iDEGs list was uploaded to the Database for Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov/) (17) for GO enrichment analysis. The results were visualized in the GOplot package (18). KEGG (https://www.genome.jp/kegg; release 92.0), which is a comprehensive knowledge database, can be used for functional interpretation and application of genomic information. In the present study, KEGG pathway analysis was performed using the clusterProfiler package (19). P<0.05 was considered to indicate a statistically significant difference.
Construction and assessment of a PPI network
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) is an online server used for evaluating the interactions among proteins, including 9,643,763 proteins from 2,031 organisms (20). To assess the interactions among DEGs, DEGs were mapped into STRING in order to construct a PPI network. Only the interactions with a combined score >0.4 were considered as significant. The PPI network was visualized using Cytoscape software (version 3.5.1) (21). The weight of each gene was ranked using the cytoHubba plug-in (22). The top 30 genes identified in the 12 topological algorithms were used for interaction analysis. Only genes that overlapped in at least nine algorithms were identified as hub genes. In addition, overall survival analysis for all the seven hub genes (IL6, ACTN2, COL1A1, ACTA1, CALB2, THBS1 and PTH) was performed in 276 patients with TC using Kaplan Meier method and the statistical significance was evaluated using the log-rank test. Median expression level was defined as the cut-off value, above this value was defined as high expression, and below this value was defined as low expression.
Analysis of hub genes
To verify the value of the seven aforementioned hub genes, multivariable logistic regression analysis was used to construct a predictive model of LNM in TC and the different T stages. The response variable of this model was the status of lymph node metastasis, with 1 representing LNM and 0 representing LNM, whereas predictive variables were the expression levels of seven hub genes. The prediction efficiency of this model was assessed by determining the area under curve (AUC) of receiver operating characteristic (ROC) curves using the pROC package (23).
Results
Overview of the expression levels of the DEGs
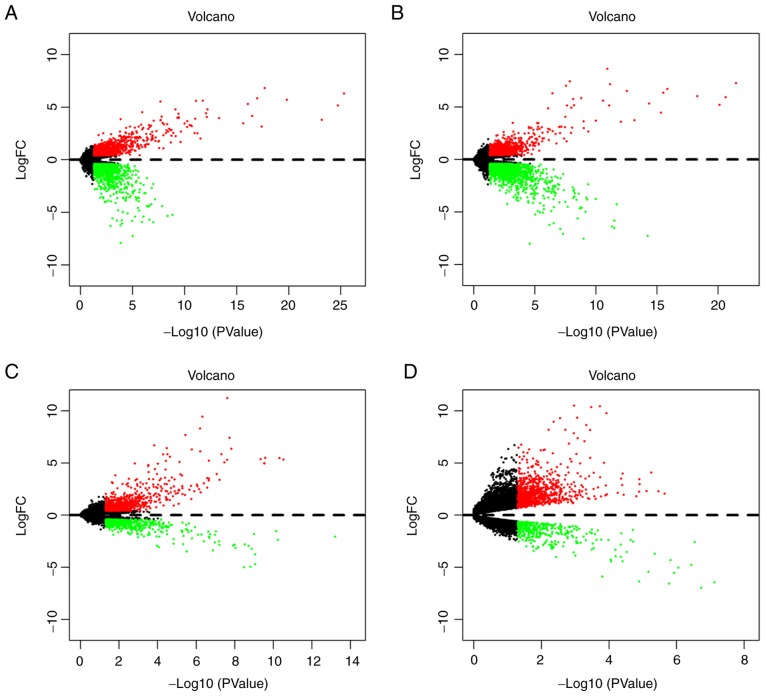
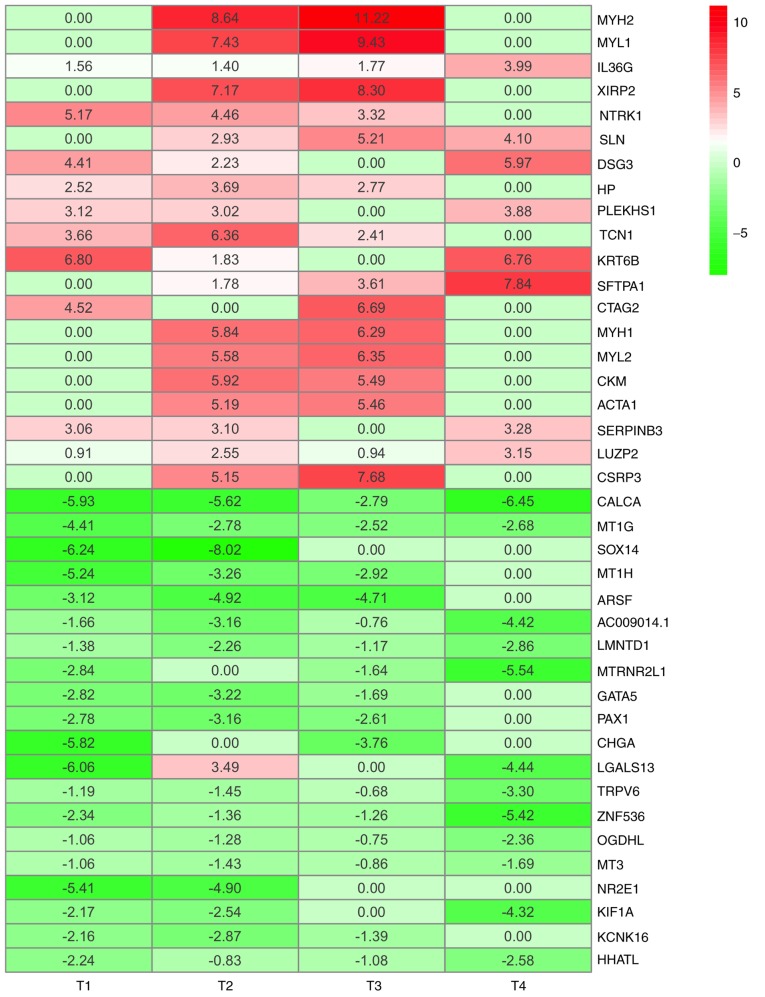
In order o identify gene signatures present during the development of TC, the present study compared the expression profiles between TC, LNM and non-metastasis samples in T1-4 disease stages. Based on the criteria of P<0.05 and logFC ≥0.4, the present study identified 2,369 DEGs (1,057 upregulated and 1,312 downregulated) in T1N0M0 vs. T1N1M0, 2,581 DEGs (968 upregulated and 1,613 downregulated) in T2N0M0 vs. T2N1M0, 1,394 DEGs (872 upregulated and 522 downregulated) in T3N0M0 vs. T3N1M0 and 1,281 DEGs (889 upregulated and 392 downregulated) in T4N0M0 vs. T4N1M0 (Fig. 2). According to the significance score for each gene, a total of 493 iDEGs were eventually identified, including 246 upregulated genes and 247 downregulated genes. Among these iDEGs, details of the top 20 upregulated (Table I) and downregulated genes (Table II) are presented (Fig. 3). In addition, by using the Venn diagram method, six upregulated genes and 24 downregulated genes were identified (Fig. S1A and B). These genes have all been contained in RRA result (Fig. S1C and D). These results indicated that LNM process in TC may involve the interaction of multiple genes.
Figure 2.
Volcano plots of DEGs obtained from the T1-T4 subgroups. (A) DEGs between T1N0M0 and T1N1M0. (B) DEGs between T2N0M0 and T2N1M0. (C) DEGs between T3N0M0 and T3N1M0. (D) DEGs between T4N0M0 and T4N1M0. Red indicates upregulation in LNM and green indicates downregulation in LNM. DEGs, differentially expressed genes; FC, fold change; LNM, lymph node metastasis.
Table I.
Top 20 upregulated integrated differentially expressed genes identified using the robust rank aggregation method.
| Gene symbol | LogFC | P-value |
|---|---|---|
| MYH2 | 4.9649980 | 8.87×107 |
| MYL1 | 4.2157600 | 3.55×106 |
| IL36G | 2.1786312 | 4.56×106 |
| XIRP2 | 3.8684034 | 1.42×105 |
| NTRK1 | 3.2382082 | 1.50×105 |
| SLN | 3.0614500 | 2.43×105 |
| DSG3 | 3.1533312 | 3.24×105 |
| HP | 2.2461089 | 4.03×105 |
| PLEKHS1 | 2.5055146 | 4.38×105 |
| TCN1 | 3.1083532 | 8.73×105 |
| KTR6B | 3.8472305 | 9.31×105 |
| SFTPA1 | 3.3075095 | 0.000105 |
| CTAG2 | 2.8039948 | 0.000127 |
| MYH1 | 3.0319229 | 0.000127 |
| MYL2 | 2.9843667 | 0.000173 |
| CKM | 2.8526071 | 0.000199 |
| ACTA1 | 2.6647653 | 0.000226 |
| SERPINB3 | 2.3597234 | 0.000229 |
| LUZP2 | 1.8885731 | 0.000236 |
| CSRP3 | 3.2079334 | 0.000255 |
FC, fold change.
Table II.
Top 20 downregulated integrated differentially expressed genes identified using the robust rank aggregation method.
| Gene symbol | LogFC | P-value |
|---|---|---|
| CALCA | −5.1963956 | 3.58×10−10 |
| MT1G | −3.0975483 | 4.64×10−7 |
| SOX14 | −3.5652297 | 1.42×10−5 |
| MT1H | −2.8551229 | 2.43×10−5 |
| ARSF | −3.1873280 | 2.43×10−5 |
| AC009014.1 | −2.5009319 | 2.53×10−5 |
| LMNTD1 | −1.9169332 | 4.03×10−5 |
| MTRNR2L1 | −2.5038958 | 4.03×10−5 |
| GATA5 | −1.9320512 | 4.38×10−5 |
| PAX1 | −2.1391495 | 5.13×10−5 |
| CHGA | −2.3957941 | 5.66×10−5 |
| LGALS13 | −1.7503245 | 8.85×10−5 |
| TRPV6 | −1.6544989 | 0.000106 |
| ZNF536 | −2.5953115 | 0.000137 |
| OGDHL | −1.3620672 | 0.000214 |
| MT3 | 1.2591934 | 0.000220 |
| NR2E1 | −2.5790365 | 0.000255 |
| KIF1A | −2.2569354 | 0.000286 |
| KCNK16 | −1.6063756 | 0.000293 |
| HHATL | −1.6816666 | 0.000299 |
FC, fold change.
Figure 3.
Top 20 upregulated iDEGs and top 20 downregulated iDEGs. Red indicates upregulation and green indicates downregulation in LNM. Dimensions and extrathyroidal extension of tumor determine the classification of T status. iDEGs, integrated differentially expressed genes; T, tumor (14).
Enrichment analysis of iDEGs
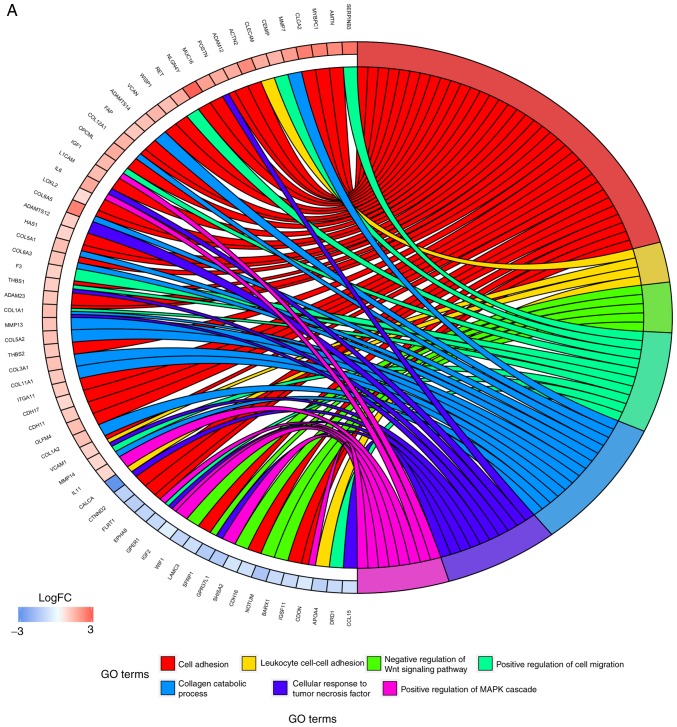
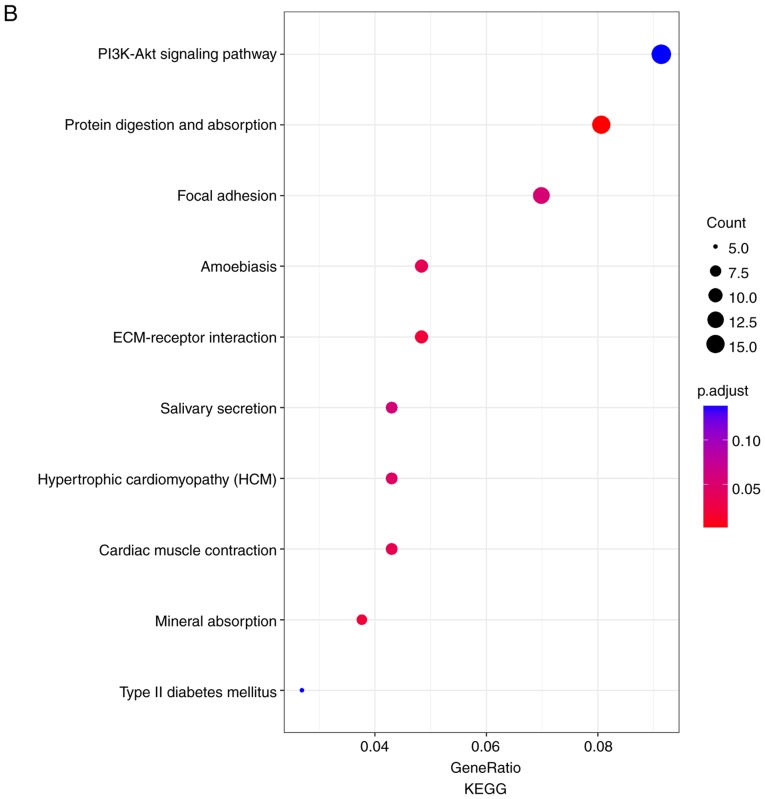
In order to further investigate the potential mechanisms of LNM in TC, GO functional annotation and KEGG pathway enrichment analyses were performed. As presented in Fig. 4A, numerous biological processes were identified, including ‘cell adhesion’, ‘positive regulation of cell migration’, ‘cellular response to tumor necrosis factor’ and ‘positive regulation of MAPK cascade’. The results from KEGG pathway enrichment analyses demonstrated that the DEGs were predominantly involved in the pathways ‘PI3K-Akt signaling pathway’, ‘protein digestion and absorption’, ‘focal adhesion’ and ‘ECM-receptor interaction’ (Fig. 4B). These results indicated that the iDEGs were associated with LNM and require further investigation.
Figure 4.
Results of GO enrichment analysis and KEGG pathway analysis. (A) GO enrichment analysis. Different colors on the right side of the image represent different GO terms, whereas the left side of the image represents the expression levels of the genes. Lines represent the associations between genes and corresponding biological processes. (B) KEGG pathway analysis. Size of the points represents the count of genes enriched in each pathway. Colors of the point represent the P-value. FC, fold change; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
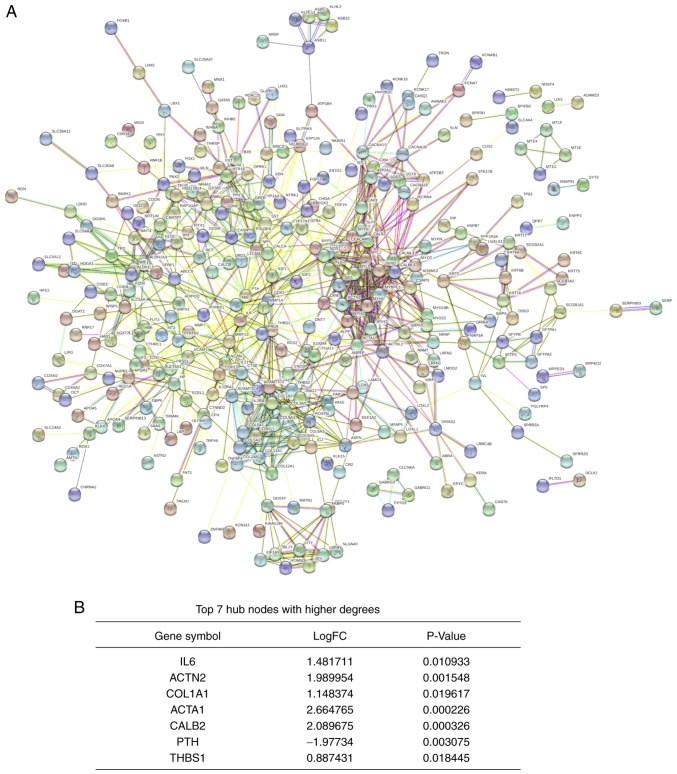
PPI network and hub genes identification
A PPI network was constructed using the STRING database. The network consisted of 988 edges and 483 nodes (Fig. 5A). According to the significance score of each node, seven hub genes were identified using the cytoHubba plug-in. These genes included the upregulated genes interleukin 6 (IL6), actinin α2 (ACTN2), collagen type I α 1 chain (COL1A1), actin α1 (ACTA1), calbindin 2 (CALB2) and thrombospondin 1 (THBS1), and the downregulated gene parathyroid hormone (PTH; Fig. 5B).
Figure 5.
PPI network for integrated differentially expressed genes and hub genes analyses. (A) PPI network. Circles represent proteins and lines represent interactions between proteins. The thickness of the line represents the significance of the interaction. (B) List of the hub genes. ACTA1, actin α1; ACTN2, actinin α2; CALB2, calbindin 2; COL1A1, collagen type I α 1 chain; FC, fold change; IL-6, interleukin 6; PPI, protein-protein interaction; PTH, parathyroid hormone; THBS1, thrombospondin 1.
Analysis of hub genes
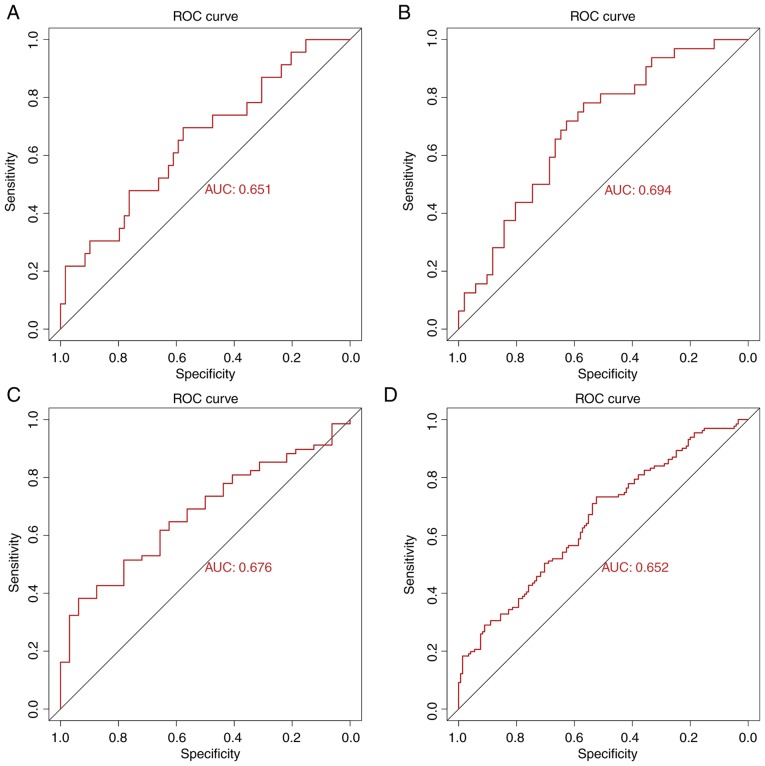
To validate the predictive power of the seven identified hub genes, ROC curve analysis for each T-stage subgroup and total ROC curve analysis for all samples were performed (due to the small sample size of the T4 subgroup, the ROC curve could not be presented). The AUC values for each subgroup were 0.651 (T1), 0.694 (T2), 0.676 (T3) and 0.652 (all samples; Fig. 6). These results indicated that the seven hub genes may serve crucial role in the prediction of LNM in TC. In addition, among these hub genes, high expression of ACTN2 indicated a worse prognosis for patients with TC (Fig. S2). To the best of our knowledge, the present study was the first to investigate the prognosis value of hub genes in TC.
Figure 6.
ROC curves for the seven hub genes to predict LNM in TC obtained from the T1-T3 subgroups and all-samples group. (A) ROC curve for predicting LNM of TC had an AUC of 0.651 in the T1 group. (B) ROC curve for predicting LNM of TC had an AUC of 0.694 in the T2 group. (C) ROC curve for predicting LNM of TC had an AUC of 0.676 in the T3 group. (D) ROC curve for predicting LNM of TC had an AUC of 0.652 in the all-samples group. AUC, area under the curve; LNM, lymph node metastasis; ROC, receiver operating characteristic; TC, thyroid cancer.
Discussion
In the present study, 493 iDEGs were identified from four subgroups of TC using the RRA method. Compared with the results from the Venn diagram, all genes were included in RRA result. These iDEGs were predominantly enriched in the pathways ‘PI3K-Akt signaling pathway’, ‘focal adhesion’, ‘ECM-receptor interaction’ and the ‘calcium signaling pathway’ (24–26). A total of 7 hub genes were identified from the PPI network, which was composed of 493 iDEGs. ROC curve analysis revealed that the combination of these 7 hub genes was a good predictive model for LNM in TC.
Among the identified hub genes, IL6, COL1A1 and THBS1 have been reported to participate in the process of LNM. For example, CXCR7 may induce COL1A1 upregulation and subsequently accelerate the LNM of PTC via the PI3K/AKT pathway (27). Furthermore, COL1A1 expression level is higher in poorly differentiated thyroid cancer (PDTC) and anaplastic thyroid cancer (ATC) compared with PTC (10). Both PDTC and ATC are known as invasive pathological types of TC (10). Similarly, the present study demonstrated that COL1A1 expression was increased in TC tissues from the LNM group. In addition, the results from enrichment analysis suggested that COL1A1 may promote LNM in TC through ‘ECM-receptor interaction’, ‘focal adhesion’, ‘PI3K-Akt signaling pathway’ and ‘positive regulation of cell migration’.
The role of THBS1 in tumor metastasis remains unclear. THBS1 is a multifunctional molecule that can affect biological activities both positively and negatively. Duquette et al (28) reported that THBS1 high expression could promote ATC cell metastasis; however, THBS1 knockdown can decrease the levels of thrombospondin 1, integrin (ITG)α3, ITGα6 and ITGβ1 in the tumor microenvironment, thus inhibiting ATC cell migration. In other types of cancer, including breast cancer, gastric cancer and melanoma, it was demonstrated that the occurrence of LNM is significantly higher in THBS1-positive samples compared with THBS1-negative samples, which was meditated by the PI3K/Akt/mTOR signaling pathway and epithelial-to-mesenchymal transition (EMT) (29–32). In the present study, THBS1 was also highly expressed and involved in cell adhesion and positive regulation of cell migration, according to the GO analysis. However, it has been reported that importin-11 overexpression could promote the invasion, migration and progression of bladder cancer via the upregulation of THBS1 (33–35). Heterogeneity among the types of cancer may explain these differences.
In addition, IL-6 overexpression can accelerate LNM in cervical cancer, colorectal cancer and lung cancer by stimulating cell proliferation, migration or EMT (36–38). This is consistent with the results from the present study, which demonstrated that IL-6 was upregulated and enriched in the PI3K-Akt signaling pathway, indicating that IL-6 may promote LNM progression in TC. In addition, ACTN2 downregulation was demonstrated to cause pancreatic endocrine neoplasms metastasis in IGF-signaling cascade, leading to cell separation and migration (39,40). The other hub genes identified in the present study, including ACTA1, CALB2 and PTH, may serve important role in TC with LNM; however, this has not yet been reported. The results from the present bioinformatics analyses require further experimental confirmation.
In summary, the hub genes identified in this study may exhibit crucial functions in LNM of TC via numerous mechanisms, including ECM-receptor interaction, focal adhesion and PI3K-Akt signaling pathway. The present study provided novel insights on the prediction of LNM in TC, and may serve at determining the appropriate extent of surgical resection needed to decrease recurrence rate.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by The Zhejiang Xinmiao Innovative Program (grant no. 2018R410053), The Key Project of Scientific and Technological Innovation in Hangzhou (grant no. 20131813A08) and The Science Research Program of Hangzhou (grant no. 20160533B05).
Availability of data and materials
The datasets analyzed during the present study are available in The Cancer Genome Atlas (cancergenome.nih.gov/).
Authors' contributions
DL and WH conceived and conducted the study. SL and RZ partially responsible for the design of study and drafted the manuscript. JS and YZ made substantial contributions to the acquisition and interpretation of data. JJS and CX programmed R codes and analyzed the data. JC and RL contributed to the design of the study and revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA. 2017;317:1338–1348. doi: 10.1001/jama.2017.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014;140:317–322. doi: 10.1001/jamaoto.2014.1. [DOI] [PubMed] [Google Scholar]
- 5.Morris LG, Sikora AG, Tosteson TD, Davies L. The increasing incidence of thyroid cancer: The influence of access to care. Thyroid. 2013;23:885–891. doi: 10.1089/thy.2013.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Wei WJ, Ji QH, Zhu YX, Wang ZY, Wang Y, Huang CP, Shen Q, Li DS, Wu Y. Risk factors for neck nodal metastasis in papillary thyroid microcarcinoma: A study of 1066 patients. J Clin Endocrinol Metab. 2012;97:1250–1257. doi: 10.1210/jc.2011-1546. [DOI] [PubMed] [Google Scholar]
- 7.Adam MA, Pura J, Goffredo P, Dinan MA, Reed SD, Scheri RP, Hyslop T, Roman SA, Sosa JA. Presence and number of lymph node metastases are associated with compromised survival for patients younger than age 45 years with papillary thyroid cancer. J Clin Oncol. 2015;33:2370–2375. doi: 10.1200/JCO.2014.59.8391. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Wang Z, Meng X, Duh QY, Chen G. Predictors for central lymph node metastases in CN0 papillary thyroid microcarcinoma (mPTC): A retrospect analysis of 1304 cases. Asian J Surg. 2019;42:571–576. doi: 10.1016/j.asjsur.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Pisanu A, Saba A, Podda M, Reccia I, Uccheddu A. Nodal metastasis and recurrence in papillary thyroid microcarcinoma. Endocrine. 2014;48:575–581. doi: 10.1007/s12020-014-0350-7. [DOI] [PubMed] [Google Scholar]
- 10.Jin WX, Ye DR, Sun YH, Zhou XF, Wang OC, Zhang XH, Cai YF. Prediction of central lymph node metastasis in papillary thyroid microcarcinoma according to clinicopathologic factors and thyroid nodule sonographic features: A case-control study. Cancer Manag Res. 2018;10:3237–3243. doi: 10.2147/CMAR.S169741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia E, Wang Y, Bhandari A, Niu J, Yang F, Yao Z, Wang O. CITED1 gene promotes proliferation, migration and invasion in papillary thyroid cancer. Oncol Lett. 2018;16:105–112. doi: 10.3892/ol.2018.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Y, Jin W, Zheng Z, Chen E, Wang Q, Wang Y, Wang O, Zhang X. GABRB2 plays an important role in the lymph node metastasis of papillary thyroid cancer. Biochem Biophys Res Commun. 2017;492:323–330. doi: 10.1016/j.bbrc.2017.08.114. [DOI] [PubMed] [Google Scholar]
- 13.Collins A. The cancer genome atlas (TCGA) pilot project. Cancer Res. 2007;67 [Google Scholar]
- 14.Perrier ND, Brierley JD, Tuttle RM. Differentiated and anaplastic thyroid carcinoma: Major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2018;68:55–63. doi: 10.3322/caac.21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Walter W, Sánchez-Cabo F, Ricote M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang N, Li Y, Wei J, Pu J, Liu R, Yang Q, Guan H, Shi B, Hou P, Ji M. TBX1 functions as a tumor suppressor in thyroid cancer through inhibiting the activities of the PI3K/AKT and MAPK/ERK pathways. Thyroid. 2019;29:378–394. doi: 10.1089/thy.2018.0312. [DOI] [PubMed] [Google Scholar]
- 25.Choi C, Thi Thao Tran N, Van Ngu T, Park SW, Song MS, Kim SH, Bae YU, Ayudthaya PDN, Munir J, Kim E, et al. Promotion of tumor progression and cancer stemness by MUC15 in thyroid cancer via the GPCR/ERK and integrin-FAK signaling pathways. Oncogenesis. 2018;7:85. doi: 10.1038/s41389-018-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vitale G, Dicitore A, Pepe D, Gentilini D, Grassi ES, Borghi MO, Gelmini G, Cantone MC, Gaudenzi G, Misso G, et al. Synergistic activity of everolimus and 5-aza-2′-deoxycytidine in medullary thyroid carcinoma cell lines. Mol Oncol. 2017;11:1007–1022. doi: 10.1002/1878-0261.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Teng X, Liu Z, Zhang L, Liu Z. Gene expression profile analyze the molecular mechanism of CXCR7 regulating papillary thyroid carcinoma growth and metastasis. J Exp Clin Cancer Res. 2015;34:16. doi: 10.1186/s13046-015-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duquette M, Sadow PM, Lawler J, Nucera C. Thrombospondin-1 silencing down-regulates integrin expression levels in human anaplastic thyroid cancer cells with BRAF(V600E): New insights in the host tissue adaptation and homeostasis of tumor microenvironment. Front Endocrinol (Lausanne) 2013;4:189. doi: 10.3389/fendo.2013.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yee KO, Connolly CM, Duquette M, Kazerounian S, Washington R, Lawler J. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Res Treat. 2009;114:85–96. doi: 10.1007/s10549-008-9992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang T, Wang L, Liu D, Li P, Xiong H, Zhuang L, Sun L, Yuan X, Qiu H. FGF7/FGFR2 signal promotes invasion and migration in human gastric cancer through upregulation of thrombospondin-1. Int J Oncol. 2017;50:1501–1512. doi: 10.3892/ijo.2017.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borsotti P, Ghilardi C, Ostano P, Silini A, Dossi R, Pinessi D, Foglieni C, Scatolini M, Lacal PM, Ferrari R, et al. Thrombospondin-1 is part of a Slug-independent motility and metastatic program in cutaneous melanoma, in association with VEGFR-1 and FGF-2. Pigment Cell Melanoma Res. 2015;28:73–81. doi: 10.1111/pcmr.12319. [DOI] [PubMed] [Google Scholar]
- 32.Jayachandran A, Anaka M, Prithviraj P, Hudson C, McKeown SJ, Lo PH, Vella LJ, Goding CR, Cebon J, Behren A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget. 2014;5:5782–5797. doi: 10.18632/oncotarget.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Shi L, Zeng S, Ma C, Xu W, Zhang Z, Liu Q, Zhang P, Sun Y, Xu C. Importin-11 overexpression promotes the migration, invasion, and progression of bladder cancer associated with the deregulation of CDKN1A and THBS1. Urol Oncol. 2018;36:311.e1–311.e13. doi: 10.1016/j.urolonc.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Teraoku H, Morine Y, Ikemoto T, Saito Y, Yamada S, Yoshikawa M, Takasu C, Higashijima J, Imura S, Shimada M. Role of thrombospondin-1 expression in colorectal liver metastasis and its molecular mechanism. J Hepatobiliary Pancreat Sci. 2016;23:565–573. doi: 10.1002/jhbp.376. [DOI] [PubMed] [Google Scholar]
- 35.Tzeng HT, Tsai CH, Yen YT, Cheng HC, Chen YC, Pu SW, Wang YS, Shan YS, Tseng YL, Su WC, et al. Dysregulation of Rab37-mediated cross-talk between cancer cells and endothelial cells via thrombospondin-1 promotes tumor neovasculature and metastasis. Clin Cancer Res. 2017;23:2335–2345. doi: 10.1158/1078-0432.CCR-16-1520. [DOI] [PubMed] [Google Scholar]
- 36.Han J, Meng Q, Xi Q, Zhang Y, Zhuang Q, Han Y, Jiang Y, Ding Q, Wu G. Interleukin-6 stimulates aerobic glycolysis by regulating PFKFB3 at early stage of colorectal cancer. Int J Oncol. 2016;48:215–224. doi: 10.3892/ijo.2015.3225. [DOI] [PubMed] [Google Scholar]
- 37.Luan S, An Z, Bi S, Chen L, Fan J. Interleukin 6 receptor (IL-6R) was an independent prognostic factor in cervical cancer. Histol Histopathol. 2018;33:269–276. doi: 10.14670/HH-11-920. [DOI] [PubMed] [Google Scholar]
- 38.Shang GS, Liu L, Qin YW. IL-6 and TNF-α promote metastasis of lung cancer by inducing epithelial-mesenchymal transition. Oncol Lett. 2017;13:4657–4660. doi: 10.3892/ol.2017.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansel DE, Rahman A, House M, Ashfaq R, Berg K, Yeo CJ, Maitra A. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res. 2004;10:6152–6158. doi: 10.1158/1078-0432.CCR-04-0285. [DOI] [PubMed] [Google Scholar]
- 40.Guvakova MA, Adams JC, Boettiger D. Functional role of alpha-actinin, PI 3-kinase and MEK1/2 in insulin-like growth factor I receptor kinase regulated motility of human breast carcinoma cells. J Cell Sci. 2002;115:4149–4165. doi: 10.1242/jcs.00104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the present study are available in The Cancer Genome Atlas (cancergenome.nih.gov/).