Abstract
Lung adenocarcinoma (LUAD) is a common malignancy; however, the majority of its underlying molecular mechanisms remain unknown. In the present study, weighted gene co-expression network analysis was applied to construct gene co-expression networks for the GSE19804 dataset, in order to screen hub genes associated with the pathogenesis of LUAD. In addition, with the aid of the Database for Annotation, Visualization and Integrated Discovery, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes, pathway enrichment analyses were performed on the genes in the selected module. Using the GSE40791 dataset and The Cancer Genome Atlas database, the hub genes were identified. It was discovered that the turquoise module was the most significant module associated with the tumor stage of LUAD. After performing functional enrichment analyses, it was indicated that the turquoise module was mainly enriched in signal transduction. Additionally, at the transcriptional and translational level, nine hub genes were identified and validated: Carbonic anhydrase 4 (CA4), platelet and endothelial cell adhesion molecule 1 (PECAM1), DnaJ member B4 (DNAJB4), advanced glycosylation end-product specific receptor (AGER), GTPase, IMAP family member 6 (GIMAP6), chromosome 10 open reading frame 54 (C10orf54), dedicator of cytokinesis 4 (DOCK4), Golgi membrane protein 1 (GOLM1) and platelet activating factor acetylhydrolase 1b catalytic subunit 3 (PAFAH1B3). CA4, PECAM1, DNAJB4, AGER, GIMAP6, C10orf54 and DOCK4 were expressed at lower levels in the tumor samples, whereas GOLM1 and PAFAH1B3 were highly expressed in tumor samples. In addition, all hub genes were associated with prognosis. In conclusion, one module and nine genes were recognized to be associated with the tumor stage of LUAD. These findings may enhance the understanding of the progression and prognosis of LUAD.
Keywords: lung adenocarcinoma, weighted gene co-expression network analysis, hub genes, clinical prognosis, Gene Expression Omnibus, The Cancer Genome Atlas
Introduction
The incidence and mortality of lung cancer rank the highest among all types of cancer worldwide. In 2018, lung cancer was the most commonly diagnosed cancer (11.6% of all cancer cases) and the leading cause of cancer-associated mortality (18.4% of all cancer-associated mortality cases) across 20 world regions (1). Malignant epithelial tumors are the most frequently observed in lung cancer, and can be grouped into non-small cell lung carcinoma (NSCLC) and small cell lung carcinoma (2). NSCLC accounts for 85–90% of lung cancer cases, and lung adenocarcinoma (LUAD) is a common type of NSCLC (3). Although positive outcomes have been achieved following early diagnosis, the recurrence rate remains unacceptably high, and the 5-year overall survival rate of patients with LUAD remains low (4). Without sufficient early detection methods and effective therapeutic strategies during the early tumor stages, the mortality rate of patients with LUAD has not markedly decreased in recent years (5). Therefore, further insight into the mechanisms responsible for the development and progression of LUAD is urgently required (6).
Due to the development of high-throughput microarray technology, an increasing number of genes have been identified to serve an important role in tumor occurrence and in the progression of LUAD (7). Gene expression profiles were used to identify important genes associated with tumor progression (8). However, the majority of studies have focused on differentially expressed genes (DEGs) and not on the interconnection between genes (9–11). In order to obtain further information on the association between gene expression levels and important clinical features, scale-free gene co-expression networks were constructed using co-expression analysis. Previous studies have applied weighted gene co-expression network analysis (WGCNA) to analyze gene expression datasets and screen hub genes (12,13). Tumor stage is crucial to the clinical prognosis of patients with LUAD, and the survival status of patients at different tumor stages differs significantly (14). Therefore, tumor stage was selected as a main clinical feature. Subsequently, co-expression networks of the association between genes were constructed, and network-centric genes associated with the clinical features were identified. Finally, GSE40791 and UALCAN were applied to investigate the value of the candidate hub genes.
Materials and methods
Data sources and processing
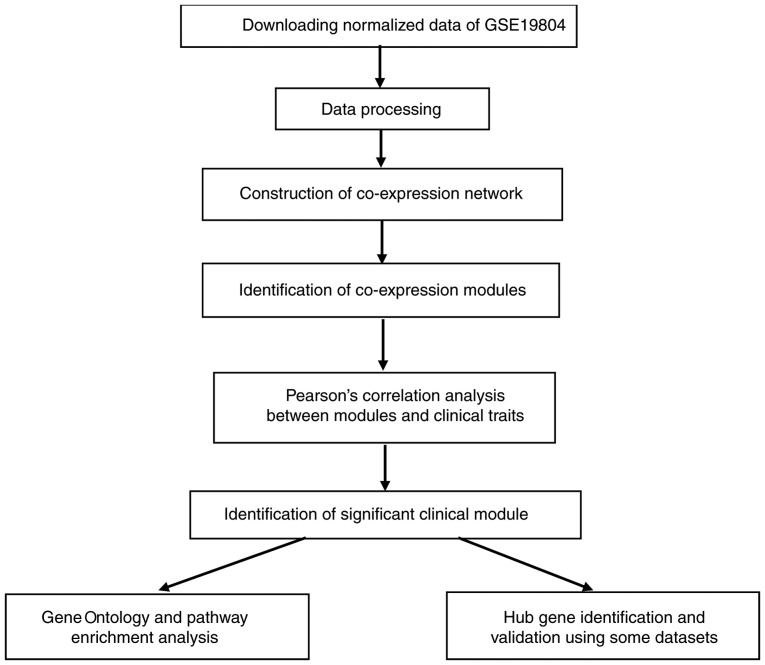
The brief study flow is presented in Fig. 1. The gene expression profile GSE19804 dataset associated with LUAD was downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). GSE19804, which was based on the GPL96 platform (Affymetrix Human Genome U133A Array), contains 120 samples (60 normal and 60 LUAD samples) and 54,675 genes (15). The dataset was normalized with quantile normalization by the R package ‘affy’ (16). The top 25% most variant genes (13,669 genes) were then selected by analysis of variance for further study in R 3.5.1.
Figure 1.
Flow chart of data preparation, processing, analysis and validation.
Co-expression network construction
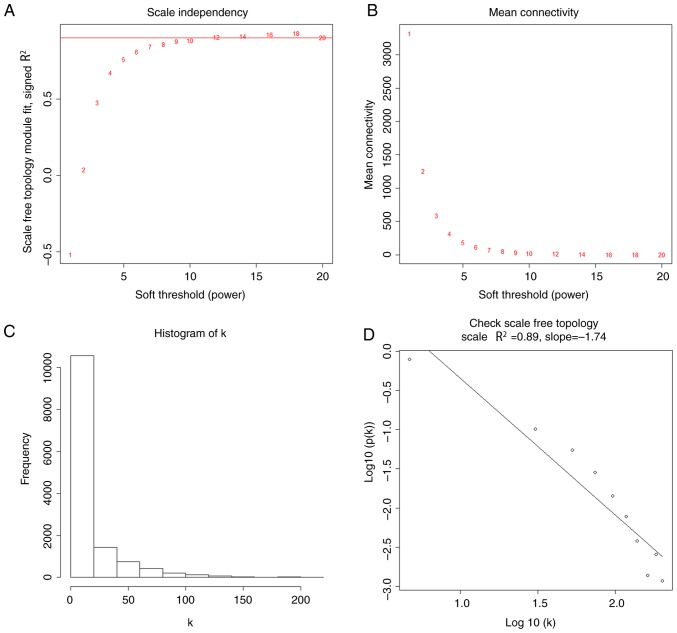
The R (R 3.5.1; http://www.r-project.org/) package ‘WGCNA’ (7) was used to construct gene co-expression networks for the filtered gene expression matrix. To construct a scale-free network, the power of β=12 (scale-free R2=0.89) was selected as the soft-thresholding parameter. After transforming the adjacency into a topological overlap measurement (TOM), the corresponding dissimilarity (1-TOM) was calculated and the dissimilarity of module eigengenes (MEs) was estimated. Using the DynamicTreeCut algorithm (17), the genes, which had similar expression profiles, were categorized into the same module.
Identification of clinically significant modules
The clinical trait of focus was the T stage of LUAD. The association between the clinical phenotype and MEs was determined to identify clinically significant modules. MEs were deemed to represent the expression levels of all genes in the associated module. In addition, the mediated P-value of each gene was calculated and the gene significance (GS=lg P) was identified. Finally, the most clinically significant module was selected according to module significance, which was the average GS of genes involved in the associated module.
Functional and pathway enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery 5 (https://david-d.ncifcrf.gov/) (DAVID) is a database for several types of functional annotation. With the aid of DAVID, the biological relevance of the genes in a given module was identified according to false discovery rate (FDR) <0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of the genes in the hub module were performed by DAVID.
Identification and validation of hub genes
The connectivity of the module can be measured by the absolute value of the Pearson's correlation. Additionally, the association between clinical traits and genes can be measured by the absolute value of the Pearson's correlation. The genes that had a high connectivity with the module and selected phenotype were regarded to be hub genes in hub module (cor.geneModuleMembership >0.8 and cor.geneTraitSignificance >0.2) (18). Survival analysis was performed to explore the association between the expression level of hub genes and overall survival rate in lung adenocarcinoma (based on The Cancer Genome Atlas data in Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/). Furthermore, other data of LUAD from GSE40791 (19) and UALCAN (http://ualcan.path.uab.edu/) were used for validation. GSE40791 was used to identify DEGs between normal tissues and LUAD tissues by using the ‘limma’ package in R (20). There are 100 normal samples and 94 tumor samples in the GSE40791 dataset. To overlap the genes in the turquoise module and DEGs, a Venn diagram was constructed using the online tool jvenn (http://jvenn.toulouse.inra.fr/app/example.html). UALCAN is a useful online tool for analyzing cancer transcriptome data, which is based on public cancer transcriptome data from The Cancer Genome Atlas (https://portal.gdc.cancer.gov) (TCGA) and MET500 (21) transcriptome sequencing (22). Independent-sample t-test was used to validate the hub genes in UALCAN. Validation of the genes that were selected from protein levels using The Human Protein Atlas (http://www.proteinatlas.org) was also performed.
Results
Weighted co-expression network construction and key module identification
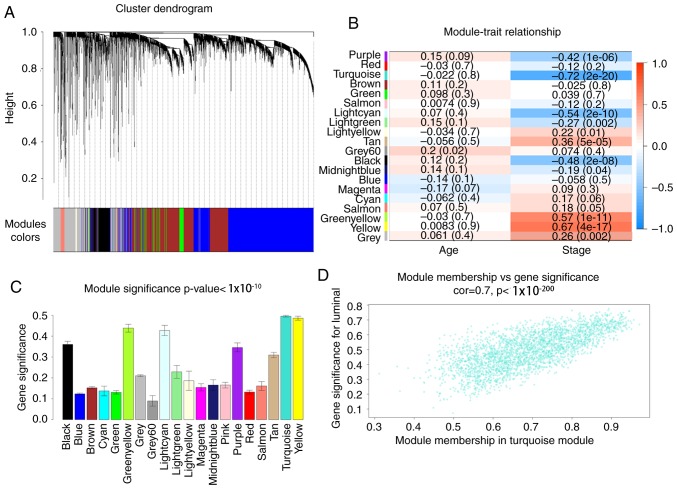
Using the method of average linkage hierarchical clustering, 120 samples from the GSE19804 dataset were clustered (Fig. 2). Using the ‘WGCNA’ package, genes with similar expression levels were divided into modules to construct co-expression networks. The power of β=12 (scale free R2=0.89) was selected as the soft-thresholding parameter (Fig. 3). In total, 20 modules were identified and the turquoise module exhibited the highest association with the T stage of LUAD (Fig. 4). Therefore, the turquoise module was selected for further analysis as the clinically significant module.
Figure 2.
Clustering dendrogram of 120 samples with the method of average linkage hierarchical clustering, and no samples were removed from subsequent analysis in GSE19804.
Figure 3.
Determination of soft-thresholding power in the weighted gene co-expression network analysis. (A) Analysis of the scale-free fit index for various soft-thresholding powers. (B) Analysis of the mean connectivity for various soft-thresholding powers. (C) Histogram of connectivity distribution when β=12. (D) Checking the scale-free topology when β=12.
Figure 4.
Identification of modules associated with the clinical traits of LUAD. (A) Dendrogram of all differentially expressed genes clustered based on a dissimilarity measure (1-TOM). (B) Heatmap of the association between MEs and clinical traits of lung adenocarcinoma. (C) Distribution of average gene significance and errors in the modules associated with tumor grades of LUAD. (D) Scatter plot of MEs in the turquoise module. LUAD, lung adenocarcinoma; MEs, module eigengenes; TOM, topological overlap measurement.
GO and pathway enrichment analysis
To obtain further information on the function of candidate genes, the genes from the turquoise module were categorized into biological process (BP), cellular component (CC) and molecular function (MF) terms. The outcome of GO enrichment analysis is presented in Table I. The BP terms were generally enriched in the ‘positive regulation of transcription from RNA polymerase II promoter’, ‘signal transduction’, ‘negative regulation of transcription from RNA polymerase II promoter’, ‘cell adhesion’ and ‘positive regulation of GTPase activity’; the CC terms were mainly focused on ‘cytoplasm’, ‘plasma membrane’, ‘extracellular exosome’, ‘extracellular region’ and ‘integral component of plasma membrane’; and the MF terms were focused on ‘protein binding’, ‘calcium ion binding’, ‘actin binding’, ‘transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding’ and ‘heparin binding’. In addition, KEGG analysis was performed to obtain the pathways in the turquoise module. The results presented in Table II indicated that these genes were included in ‘focal adhesion’, ‘cGMP-PKG signaling pathway’ and ‘tight junction’. Overall, the genes in the turquoise module were primarily associated with signal transduction.
Table I.
GO enrichment analysis of the turquoise module.
| Category | Term | Count, n | % | FDR |
|---|---|---|---|---|
| BP | GO:0045944-positive regulation of transcription from RNA polymerase II promoter | 157 | 8.26751 | 3.54×10−7 |
| BP | GO:0007165-signal transduction | 157 | 8.26751 | 2.14×10−2 |
| BP | GO:0000122-negative regulation of transcription from RNA polymerase II promoter | 114 | 6.00316 | 2.66×10−4 |
| BP | GO:0007155-cell adhesion | 98 | 5.16061 | 1.54×10−10 |
| BP | GO:0043547-positive regulation of GTPase activity | 96 | 5.05529 | 1.08×10−4 |
| CC | GO:0005737-cytoplasm | 577 | 30.38441 | 1.68×10−2 |
| CC | GO:0005886-plasma membrane | 531 | 27.96209 | 1.33×10−12 |
| CC | GO:0070062-extracellular exosome | 367 | 19.32596 | 5.00×10−8 |
| CC | GO:0005576-extracellular region | 213 | 11.21643 | 6.62×10−4 |
| CC | GO:0005887-ntegral component of plasma membrane | 206 | 10.84781 | 5.04×10−7 |
| MF | GO:0005515-protein binding | 953 | 50.18431 | 8.86×10−7 |
| MF | GO:0005509-calcium ion binding | 112 | 5.89784 | 2.45×10−4 |
| MF | GO:0003779-actin binding | 54 | 2.84360 | 1.10×10−3 |
| MF | GO:0001077-transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 45 | 2.36967 | 1.72×10−2 |
| MF | GO:0008201-heparin binding | 36 | 1.895735 | 3.57×10−3 |
GO, Gene Ontology; FDR, false discovery rate; BP, biological process; CC, cellular component; MF, molecular function.
Table II.
KEGG pathway enrichment analysis of the turquoise module.
| Category | Term | Count, n | Percentage | FDR |
|---|---|---|---|---|
| KEGG | hsa04510: focal adhesion | 47 | 2.474987 | 0.001208 |
| KEGG | hsa04022: cGMP-PKG signaling pathway | 37 | 1.948394 | 0.036805 |
| KEGG | hsa04530: tight junction | 34 | 1.790416 | 0.007826 |
FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; cGMP-PKG, cyclic guanosine monophosphate-protein kinase G.
Identification and validation of hub genes
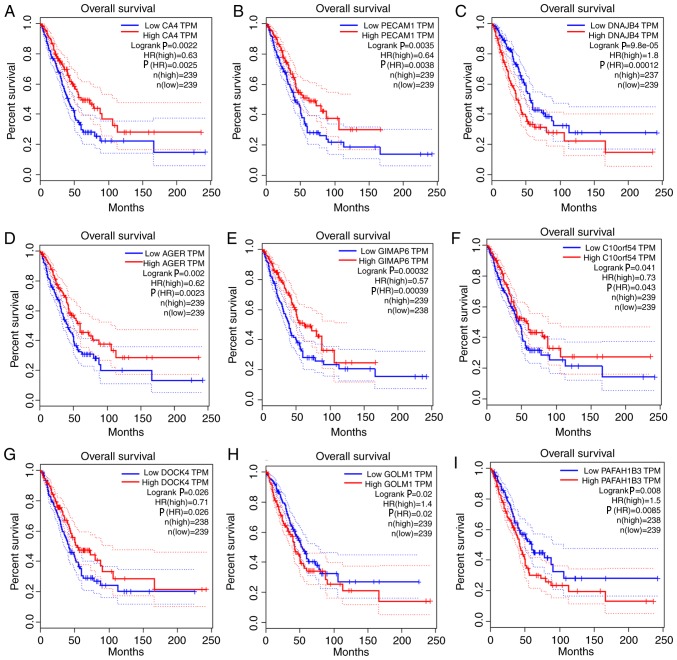
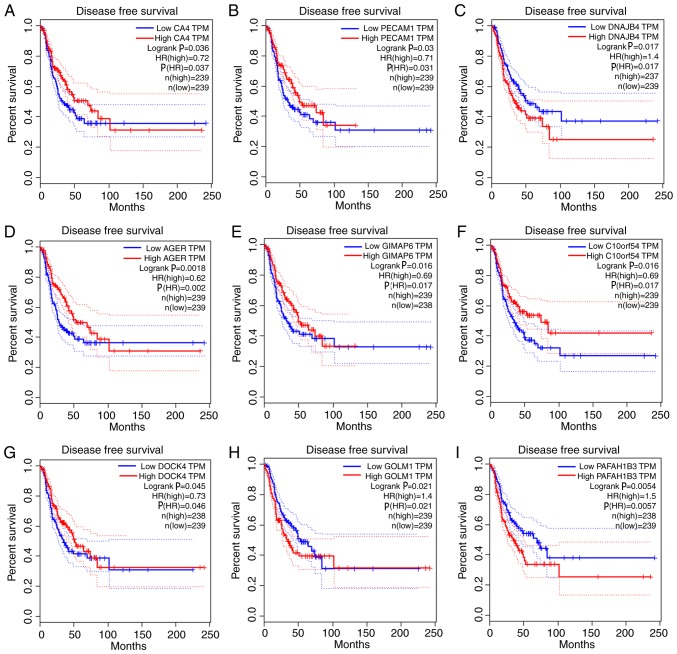
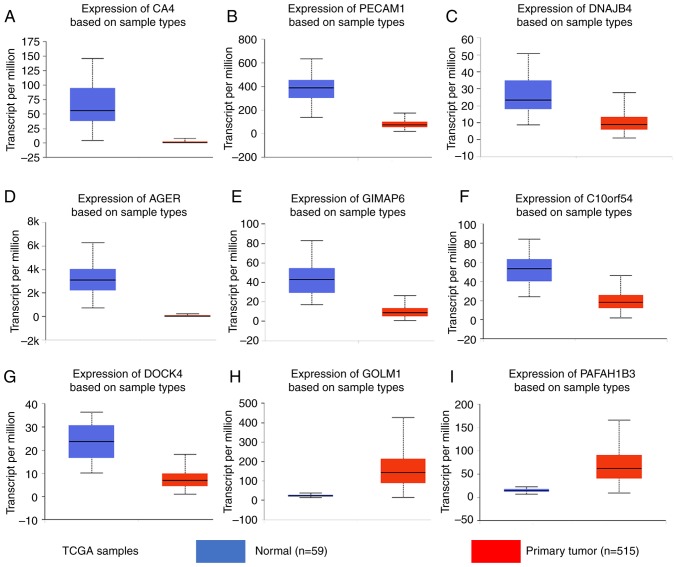
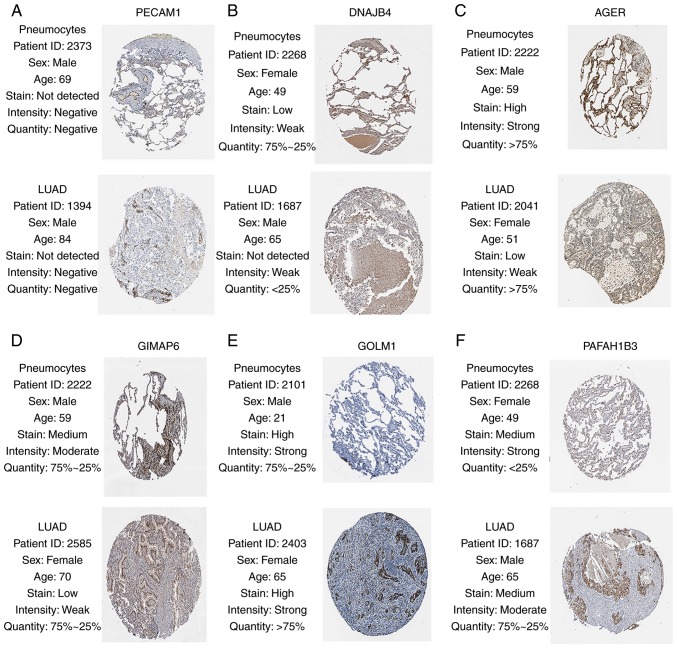
Given the threshold of |module membership (MM)|>0.8 and |GS|>0.2, a total of 415 genes in the turquoise module were recognized as hub genes. Additionally, carbonic anhydrase 4 (CA4), platelet and endothelial cell adhesion molecule 1 (PECAM1), DnaJ heat shock protein family (Hsp40) member B4 (DNAJB4), advanced glycosylation end-product specific receptor (AGER), GTPase, IMAP family member 6 (GIMAP6), chromosome 10 open reading frame 54 (C10orf54), dedicator of cytokinesis 4 (DOCK4), Golgi membrane protein 1 (GOLM1) and platelet activating factor acetylhydrolase 1b catalytic subunit 3 (PAFAH1B3) were associated with overall survival and relapse-free survival (Figs. 5 and 6). After clarifying the hub genes, some methods were used for validation. Firstly, the ‘limma’ package in R was used to identify DEGs between normal tissues and LUAD tissues in the GSE40791 dataset. Defined by the threshold of |log2 fold change|≥2 and FDR ≤0.05, 3,295 DEGs were obtained. To overlap the genes in the turquoise module and DEGs, a Venn diagram was constructed using the online tool jvenn (Fig. 7). Secondly, the expression levels of these nine genes differed between the normal and LUAD samples in UALCAN (Fig. 8). CA4, PECAM1, DNAJB4, AGER, GIMAP6, C10orf54 and DOCK4 were expressed at lower levels in the tumor samples, whereas GOLM1 and PAFAH1B3 were highly expressed in the tumor samples. Finally, in the Human Protein Atlas database, the protein expression levels of six genes (PECAM1, DNAJB4, AGER, GIMAP6, GOLM1 and PAFAH1B3) in the LUAD samples were distinct from the normal samples (Fig. 9). There were no associated IHC samples of CA4, C10orf54 and DOCK4 in the database.
Figure 5.
Survival analysis of the association between the expression levels of hub genes and overall survival rate in lung adenocarcinoma (based on The Cancer Genome Atlas data in Gene Expression Profiling Interactive Analysis). (A) CA4. (B) PECAM1. (C) DNAJB4. (D) AGER. (E) GIMAP6. (F) C10orf54. (G) DOCK4. (H) GOLM1. (I) PAFAH1B3. The red line indicates the samples with a high gene expression, and the blue line indicates the samples with low gene expression. P≤0.05 was regarded as statistically significant. CA4, carbonic anhydrase 4; PECAM1, platelet and endothelial cell adhesion molecule 1; DNAJB4, DnaJ member B4; AGER, advanced glycosylation end-product specific receptor; GIMAP6, GTPase, IMAP family member 6; C10orf54, chromosome 10 open reading frame 54; DOCK4, dedicator of cytokinesis 4; GOLM1, Golgi membrane protein 1; PAFAH1B3, platelet activating factor acetylhydrolase 1b catalytic subunit 3; HR, hazard ratio; TPM, transcripts per million.
Figure 6.
Survival analysis of the association between the expression levels of hub genes and disease-free survival time in lung adenocarcinoma (based on The Cancer Genome Atlas data in Gene Expression Profiling Interactive Analysis). (A) CA4. (B) PECAM1. (C) DNAJB4. (D) AGER. (E) GIMAP6. (F) C10orf54. (G) DOCK4. (H) GOLM1. (I) PAFAH1B3. Red line indicates the samples with highly expressed genes, and blue line shows the samples with lowly expressed gene. P≤0.05 was regarded as statistically significant. HR, hazard ratio; CA4, carbonic anhydrase 4; PECAM1, platelet and endothelial cell adhesion molecule 1; DNAJB4, DnaJ member B4; AGER, advanced glycosylation end-product specific receptor; GIMAP6, GTPase, IMAP family member 6; C10orf54, chromosome 10 open reading frame 54; DOCK4, dedicator of cytokinesis 4; GOLM1, Golgi membrane protein 1; PAFAH1B3, platelet activating factor acetylhydrolase 1b catalytic subunit 3; TPM, transcripts per million.
Figure 7.
Validation of hub genes in GSE40791. (A) Volcano plot visualizing DEGs in GSE40791 (100 normal samples and 94 lung adenocarcinoma samples). The vertical lines demarcate the fold change values. The right vertical line corresponds to ≥2-fold change (upregulation) and the left vertical line to ≥2-fold change (downregulation), whereas the horizontal line marks a -log10 adjusted P-value of 0.01. (B) Identification of common genes between DEGs and the turquoise module by overlapping them. The nine hub genes in the turquoise module were also DEGs in the GSE40791 dataset. DEGs, differentially expressed genes; CA4, carbonic anhydrase 4; PECAM1, platelet and endothelial cell adhesion molecule 1; DNAJB4, DnaJ member B4; AGER, advanced glycosylation end-product specific receptor; GIMAP6, GTPase, IMAP family member 6; C10orf54, chromosome 10 open reading frame 54; DOCK4, dedicator of cytokinesis 4; GOLM1, Golgi membrane protein 1; PAFAH1B3, platelet activating factor acetylhydrolase 1b catalytic subunit 3.
Figure 8.
Gene expression levels in normal lung and tumor samples (based on The Cancer Genome Atlas data in UALCAN). mRNA levels of (A) CA4, (B) PECAM1, (C) DNAJB4, (D) AGER, (E) GIMAP6, (F) C10orf54, (G) DOCK4, (H) GOLM1 and (I) PAFAH1B3. (A-I) P<0.0001. TCGA, The Cancer Genome Atlas; CA4, carbonic anhydrase 4; PECAM1, platelet and endothelial cell adhesion molecule 1; DNAJB4, DnaJ member B4; AGER, advanced glycosylation end-product specific receptor; GIMAP6, GTPase, IMAP family member 6; C10orf54, chromosome 10 open reading frame 54; DOCK4, dedicator of cytokinesis 4; GOLM1, Golgi membrane protein 1; PAFAH1B3, platelet activating factor acetylhydrolase 1b catalytic subunit 3.
Figure 9.
Validation of six hub genes in the turquoise module using The Human Protein Atlas database. There were no associated immunohistochemistry samples of carbonic anhydrase 4, chromosome 10 open reading frame 54 and dedicator of cytokinesis 4 in the database. Expression of (A) PECAM1, (B) DNAJB4, (C) AGER, (D) GIMAP6, (E) GOLM1, (F) PAFAH1B3. (A-F) Translational expression levels of six hub genes were positively associated with the disease status, as they were upregulated in the LUAD samples. PECAM1, platelet and endothelial cell adhesion molecule 1; DNAJB4, DnaJ member B4; AGER, advanced glycosylation end-product specific receptor; GIMAP6, GTPase, IMAP family member 6; GOLM1, Golgi membrane protein 1; PAFAH1B3, platelet activating factor acetylhydrolase 1b catalytic subunit 3; LUAD, lung adenocarcinoma.
Discussion
The early diagnosis and recurrence prediction of LUAD are crucial for effective prevention and treatment. Therefore, a deeper understanding of the molecular mechanisms associated with the development of these tumors is of utmost importance. In the present study, free-scale gene co-expression networks were constructed to identify genes with a high connectivity with the T stage of LUAD. WGCNA is a method of analyzing the association between the expression levels of genes and important clinical features (12). Although co-expression does not mean causality, the module and the genes that are closely associated with a certain clinical phenotype in co-expression networks could be identified. Additionally, survival analysis was performed to screen hub genes, which were associated with overall survival and relapse-free survival rate. The hub genes (CA4, PECAM1, DNAJB4, AGER, GIMAP6, C10orf54, DOCK4, GOLM1 and PAFAH1B3) were differentially expressed between the normal and LUAD samples at the transcriptional and protein level, and were significantly associated with prognosis. Therefore, the hub genes serving an important role in tumor progression have the potential to be prognostic biomarkers for LUAD.
In the present study, the tumor stage of the patients with LUAD was of prime concern. The tumor samples of the GSE19804 dataset included different stages, and the findings of WGCNA for GSE19804 would be more convincing and more significant compared with other datasets in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo). Although the GSE19804 dataset refers to lung cancer in female non-smokers, the result of WGCNA, in which the stage of the tumor was selected as a main clinical feature, could not be significantly influenced. Furthermore, other data of LUAD from the GSE40791 dataset and TCGA (in UALCAN), which exhibited no significant differences in sex and smoking status, were used for the validation of the hub genes. Following adjustment for other predictors, the results demonstrated the important role of the hub genes in the progression and prognosis of LUAD. Validation of the hub genes based on protein levels in the Human Protein Atlas was also performed.
There were 100 normal samples and 94 tumor samples in the GSE40791 dataset. For validation, the GSE40791 dataset was used to identify DEGs between normal and LUAD tissues, and the dataset had a sufficient sample number. Additionally, the tumor tissues in the GSE40791 dataset included 69, 12 and 13 stage I, II and III LUAD frozen tissues, respectively. The tumor samples with various stages of LUAD provided more reliable results.
To obtain further information on the role of the hub module (turquoise module) in tumor progression, GO and KEGG pathway enrichment analyses were performed based on DAVID. The genes were generally enriched for ‘positive regulation of transcription from RNA polymerase II promoter’, ‘signal transduction’, ‘negative regulation of transcription from RNA polymerase II promoter’, ‘cell adhesion’ and ‘positive regulation of GTPase activity’. Currently, inhibitors of RNA polymerase II have the potential to be effective anticancer drugs (23). The hub genes, which are associated with regulating RNA polymerase II, may serve as therapeutic targets for drug design. Signaling by small GTPase, Ras-related protein 1/Rac1, is one of the major signaling pathways controlling cancer cell migration and tumor metastasis (24). The results revealed that the hub genes were involved in tumor progression by regulating GTPase activity. Additionally, a literature review of the hub genes was conducted, and identified that the hub genes led to the induction of apoptotic programs by regulating certain enzymes (25). For example, the molecular mechanisms by which CA4 carries out its anti-invasive functions have been identified to be mediated by the enhancement of E-cadherin expression and the inhibition of N-cadherin and vimentin expression (25). E-cadherin functions as a suppressor of invasion, whereas N-cadherin and vimentin promote cell motility and invasion in cancer (25–27).
The protein encoded by CA4 is one of 12 active human isozymes. It is also one of four existing on the extracellular surfaces of certain epithelial and endothelial cells (28). By interacting with Wilms' tumor 1-associating protein (WTAP), CA4 influences WTAP protein degradation through polyubiquitination (25). Furthermore, it has been demonstrated that a low expression of carbonic anhydrase IV can promote the proliferation of cancer cells (29). PECAM1 can code for CD31, which belongs to the adhesion molecule in the immunoglobulin superfamily (30). Via the wingless-related integration site signaling pathway, PECAM1 can maintain and restore vascular integrity (30). This indicates that PECAM1 is involved in the tumorigenesis of LUAD by regulating the expression of vascular endothelial growth factor. Additionally, CD31 is a member of I transmembrane glycoprotein that is enriched in platelets, monocytes, endothelial cells and discrete circulating lymphocytes (31). DNAJB4 belongs to the heat shock protein 40 family (Hsp40/DnaJ), and serves an important role in suppressing cancer metastasis (32). AGER is a member of the immunoglobulin superfamily of cell surface molecules. AGER engagement activates multiple intracellular signaling mechanisms to fuel chronic inflammatory conditions, which can lead to malignant transformation (33,34). Previous studies have demonstrated that long non-coding RNA AGER can influence the development of lung cancer by regulating the expression of AGER (34,35). GIMAP6 belongs to the GIMAP gene family, which includes one pseudogene and seven functional members. By regulating the activation of T cells and cell death, GIMAP6 can modulate immune function accurately (36). Although it has been indicated that the association between autophagy-related protein 8 and GIMAP6 is of importance to autophagy (37), a deeper understanding of the functions of GIMAP6 is required. C10orf54 encodes for V-set immunoregulatory receptor (VISTA), which belongs to I transmembrane proteins. There is an ~30-amino acid stalk, a single N-terminal immunoglobulin V domain, a 95 amino acid cytoplasmic tail and a transmembrane domain in VISTA (38,39). A recent study found that the expression levels of C10orf54 were closely associated with tumor immune evasion (40). The protein encoded by DOCK4 is a member of a large family of proteins (CED5/DOCK180/MYOBLAST CITY class), and a previous study suggested that it could suppress tumor growth in several types of cancer, including ovarian cancer, breast cancer and glioblastoma (41–43). GOLM1 is a resident cis-Golgi membrane protein. On the surface of the Golgi apparatus, there is an extensive C-terminal, coiled-coil domain and a single N-terminal transmembrane domain (44). It has been confirmed that the Golgi apparatus serves an active role in cell migration, which can be activated by post-translational modification and prominent alterations (45). The overexpression of microRNA-200a, which has been reported to be involved in blocking the increase in cell proliferation, can repress LUAD cell proliferation induced by the overexpression of GOLM1 (46). PAFAH1B3 serves an important role in tumorigenic features and aggressiveness. PAFAH1B3 encodes a catalytic subunit of platelet-activating factor (PAF) acetyl hydrolase 1b (Pafah1b) (47). By hydrolyzing PAF, Pafah1b can regulate the intracellular PAF levels and may lead to the evasion of apoptosis caused by high intracellular PAF concentrations (47,48). In addition, by regulating an optimal landscape of signaling lipids, PAFAH1B3 can weaken the aggressiveness of cancer and regulate cancer cell pathogenicity (49).
In conclusion, the present study identified the turquoise module and nine hub genes (CA4, PECAM1, DNAJB4, AGER, GIMAP6, C10orf54, DOCK4, GOLM1 and PAFAH1B3), which are of importance to the development of LUAD. Through the turquoise module, further information on the mechanisms of tumorigenesis in LUAD was obtained. In the future, these nine hub genes that serve a vital role in LUAD tumorigenesis may also contribute to early diagnosis and treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by Key Projects of Hubei Provincial Health and Family Planning Commission (grant no. WJ2017Z006), the Zhongnan Hospital of Wuhan University Science, Technology and Innovation Cultivating Fund (grant no. cxpy2017041), and the 351 Talent Project of Wuhan University (Luojia Young Scholars: SL).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
WH and SL conceived and designed the study. DY, JH and XL performed the analysis procedures. DY, JH, XR, CC and XL analyzed the results. XR, XL, WH and SL contributed to the analysis tools. DY and JH contributed to the writing of the manuscript. All authors reviewed the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Knight SB, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7(pii):170070. doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang JT, Lee YM, Huang RS. The impact of the cancer genome atlas on lung cancer. Transl Res. 2015;166:568–585. doi: 10.1016/j.trsl.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin P, Leighl NB. Review of the use of pretest probability for molecular testing in non-small cell lung cancer and overview of new mutations that may affect clinical practice. Ther Adv Med Oncol. 2017;9:405–413. doi: 10.1177/1758834017704329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R, Wu J, Feng J, Chen P. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci. 2018;109:3068–3079. doi: 10.1111/cas.13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan TQ, Chen WJ, Qin H, Huang SN, Yang LH, Fang YY, Pan LJ, Li ZY, Chen G. Clinical value and prospective pathway signaling of MicroRNA-375 in lung adenocarcinoma: A study based on the cancer genome atlas (TCGA), gene expression omnibus (GEO) and bioinformatics analysis. Med Sci Monit. 2017;23:2453–2464. doi: 10.12659/MSM.901460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi YX, Zhu T, Zou T, Zhuo W, Chen YX, Huang MS, Zheng W, Wang CJ, Li X, Mao XY, et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget. 2016;7:85235–85243. doi: 10.18632/oncotarget.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang J, Song Q, Yang Z, Li D, Chen W, Luo L, Wang Y, Yang J, Li S. Identification of lung adenocarcinoma specific dysregulated genes with diagnostic and prognostic value across 27 TCGA cancer types. Oncotarget. 2017;8:87292–87306. doi: 10.18632/oncotarget.19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kober KM, Olshen A, Conley YP, Schumacher M, Topp K, Smoot B, Mazor M, Chesney M, Hammer M, Paul SM, et al. Expression of mitochondrial dysfunction-related genes and pathways in paclitaxel-induced peripheral neuropathy in breast cancer survivors. Mol Pain. 2018;14:1744806918816462. doi: 10.1177/1744806918816462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Aarag SA, Mahmoud A, Hashem MH, Abd Elkader H, Hemeida AE, ElHefnawi M. In silico identification of potential key regulatory factors in smoking-induced lung cancer. BMC Med Genomics. 2017;10:40. doi: 10.1186/s12920-017-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan L, Yu X, Huang Z, Zheng S, Zhou Y, Lv H, Zeng Y, Xu JF, Zhu X, Yi X. Analysis of Microarray-identified genes and microRNAs associated with idiopathic pulmonary fibrosis. Mediators Inflamm. 2017;2017:1804240. doi: 10.1155/2017/1804240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Liu X, Liu T, Meng X, Yin X, Fang C, Huang D, Cao Y, Weng H, Zeng X, Wang X. Identification of biomarkers correlated with the TNM staging and overall survival of patients with bladder cancer. Front Physiol. 2017;8:947. doi: 10.3389/fphys.2017.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadara H, Choi M, Zhang J, Parra ER, Rodriguez-Canales J, Gaffney SG, Zhao Z, Behrens C, Fujimoto J, Chow C, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol. 2018;29:1072–1072. doi: 10.1093/annonc/mdx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu TP, Lee JM, Hsu CP, et al. Genome-wide screening of genomic alterations and transcriptional modulation in non-smoking female lung cancer in Taiwan. Cancer Res. 2009;69 [Google Scholar]
- 16.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy - analysis of affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 17.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 18.Tang J, Kong D, Cui Q, Wang K, Zhang D, Gong Y, Wu G. Prognostic genes of breast cancer identified by gene co-expression network analysis. Fron Oncol. 2018;8:474. doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Foreman O, Wigle DA, Kosari F, Vasmatzis G, Salisbury JL, van Deursen J, Galardy PJ. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest. 2012;122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oronsky B, Reid TR, Oronsky A, Carter CA. What's new in SCLC? A review. Neoplasia. 2017;19:842–847. doi: 10.1016/j.neo.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JW, Ryu YK, Ji YH, Kang JH, Moon EY. Hypoxia/reoxygenation-experienced cancer cell migration and metastasis are regulated by Rap1-and Rac1-GTPase activation via the expression of thymosin beta-4. Oncotarget. 2015;6:9820–9833. doi: 10.18632/oncotarget.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K, Go MY, Ng SC, Chan FK, Sung JJ, Yu J. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut. 2016;65:1482–1493. doi: 10.1136/gutjnl-2014-308614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci. 2013;126:393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
- 27.Pan TL, Wang PW, Huang CC, Yeh CT, Hu TH, Yu JS. Network analysis and proteomic identification of vimentin as a key regulator associated with invasion and metastasis in human hepatocellular carcinoma cells. J Proteomics. 2012;75:4676–4692. doi: 10.1016/j.jprot.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Waheed A, Sly WS. Membrane associated carbonic anhydrase IV (CA IV): A personal and historical perspective. Subcell Biochem. 2014;75:157–179. doi: 10.1007/978-94-007-7359-2_9. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Hu L, Zhang F, Wang J, Chen J, Wang Y. Downregulation of carbonic anhydrase IV contributes to promotion of cell proliferation and is associated with poor prognosis in non-small cell lung cancer. Oncol Lett. 2017;14:5046–5050. doi: 10.3892/ol.2017.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villar J, Zhang H, Slutsky AS. Lung repair and regeneration in acute respiratory distress syndrome: Role of PECAM1 and Wnt signaling. Chest. 2019;155:587–594. doi: 10.1016/j.chest.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren Q, Ren L, Ren C, Liu X, Dong C, Zhang X. Platelet endothelial cell adhesion molecule-1 (PECAM1) plays a critical role in the maintenance of human vascular endothelial barrier function. Cell Biochem Funct. 2015;33:560–565. doi: 10.1002/cbf.3155. [DOI] [PubMed] [Google Scholar]
- 32.Chen CH, Chang WH, Su KY, Ku WH, Chang GC, Hong QS, Hsiao YJ, Chen HC, Chen HY, Wu R, et al. HLJ1 is an endogenous Src inhibitor suppressing cancer progression through dual mechanisms. Oncogene. 2016;35:5674–5685. doi: 10.1038/onc.2016.106. [DOI] [PubMed] [Google Scholar]
- 33.Riehl A, Nemeth J, Angel P, Hess J. The receptor RAGE: Bridging inflammation and cancer. Cell Commun Signal. 2009;7:12. doi: 10.1186/1478-811X-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sparvero LJ, Asafu-Adjei D, Kang R, Tang D, Amin N, Im J, Rutledge R, Lin B, Amoscato AA, Zeh HJ, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE Ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Z, Liu L, Nie W, Miggin S, Qiu F, Cao Y, Chen J, Yang B, Zhou Y, Lu J, Yang L. Long non-coding RNA AGER-1 functionally upregulates the innate immunity gene AGER and approximates its anti-tumor effect in lung cancer. Mol Carcinog. 2018;57:305–318. doi: 10.1002/mc.22756. [DOI] [PubMed] [Google Scholar]
- 36.Ho CH, Tsai SF. Functional and biochemical characterization of a T cell-associated anti-apoptotic protein, GIMAP6. J Biol Chem. 2017;292:9305–9319. doi: 10.1074/jbc.M116.768689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascall JC, Rotondo S, Mukadam AS, Oxley D, Webster J, Walker SA, Piron J, Carter C, Ktistakis NT, Butcher GW. The immune system GTPase GIMAP6 interacts with the Atg8 homologue GABARAPL2 and is recruited to autophagosomes. PLoS One. 2013;8:e77782. doi: 10.1371/journal.pone.0077782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Rubinstein R, Lines JL, Wasiuk A, Ahonen C, Guo Y, Lu LF, Gondek D, Wang Y, Fava RA, et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208:577–592. doi: 10.1084/jem.20100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flies DB, Wang S, Xu H, Chen L. Cutting edge: A monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J Immunol. 2011;187:1537–1541. doi: 10.4049/jimmunol.1100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie S, Huang J, Qiao Q, Zang W, Hong S, Tan H, Dong C, Yang Z, Ni L. Expression of the inhibitory B7 family molecule VISTA in human colorectal carcinoma tumors. Cancer Immunol Immunother. 2018;67:1685–1694. doi: 10.1007/s00262-018-2227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J-R, Tai Y, Jin Y, Hammell MC, Wilkinson JE, Roe J-S, Vakoc CR, Van Aelst L. TGF-β/Smad signaling through DOCK4 facilitates lung adenocarcinoma metastasis. Genes Dev. 2015;29:250–261. doi: 10.1101/gad.248963.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westbrook JA, Wood SL, Cairns DA, McMahon K, Gahlaut R, Thygesen H, Shires M, Roberts S, Marshall H, Oliva MR, et al. Identification and validation of DOCK4 as a potential biomarker for risk of bone metastasis development in patients with early breast cancer. J Pathol. 2019;247:381–391. doi: 10.1002/path.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Debruyne DN, Turchi L, Burel-Vandenbos F, Fareh M, Almairac F, Virolle V, Figarella-Branger D, Baeza-Kallee N, Lagadec P, Kubiniek V, et al. DOCK4 promotes loss of proliferation in glioblastoma progenitor cells through nuclear beta-catenin accumulation and subsequent miR-302-367 cluster expression. Oncogene. 2018;37:241–254. doi: 10.1038/onc.2017.323. [DOI] [PubMed] [Google Scholar]
- 44.Donizy P, Kaczorowski M, Biecek P, Halon A, Szkudlarek T, Matkowski R. Golgi-related proteins GOLPH2 (GP73/GOLM1) and GOLPH3 (GOPP1/MIDAS) in cutaneous melanoma: Patterns of expression and prognostic significance. Int J Mol Sci. 2016;17(pii):E1619. doi: 10.3390/ijms17101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu L, Li L, Xie H, Gu Y, Peng T. The Golgi localization of GOLPH2 (GP73/GOLM1) is determined by the transmembrane and cytoplamic sequences. PLoS One. 2011;6:e28207. doi: 10.1371/journal.pone.0028207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arun a, Li LM. Overexpression of golgi membrane protein 1 promotes non-small-cell carcinoma aggressiveness by regulating the matrix metallopeptidase 13. Am J Cancer Res. 2018;8:551–565. [PMC free article] [PubMed] [Google Scholar]
- 47.McIntyre TM, Prescott SM, Stafforini DM. The emerging roles of PAF acetylhydrolase. J Lipid Res. 2009;50(Suppl):S255–S259. doi: 10.1194/jlr.R800024-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonin F, Ryan SD, Migahed L, Mo F, Lallier J, Franks DJ, Arai H, Bennett SAL. Anti-apoptotic actions of the platelet-activating factor acetylhydrolase I alpha2 catalytic subunit. J Biol Chem. 2004;279:52425–52436. doi: 10.1074/jbc.M410967200. [DOI] [PubMed] [Google Scholar]
- 49.Mulvihill MM, Benjamin DI, Ji X, Le Scolan E, Louie SM, Shieh A, Green M, Narasimhalu T, Morris PJ, Luo K, Nomura DK. Metabolic profiling reveals PAFAH1B3 as a critical driver of breast cancer pathogenicity. Chem Biol. 2014;21:831–840. doi: 10.1016/j.chembiol.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.