Abstract
Background
Aptamers, single-stranded DNAs or RNAs, can be selected from a library containing random sequences using a method called Systematic Evolution of Ligands by EXponential Enrichment (SELEX). In SELEX, monitoring the enriching statuses of aptamer candidates during the process is a key step until today. Conformational change of an aptamer caused by target-binding in gel can be used to indicate its statuses of binding.
Results
In this study, an easy-to-implement gel-based diffusion method (GBDM) was developed to monitor the interaction between enriched aptamer candidates and their targets. In order to prove the concept, characterization of aptamers targeting their targets including protein (thrombin) and non-protein molecules (acetamiprid, ATP, atrazine, profenofos and roxithromycin), respectively, were performed using mini gels. Our method has advantages over the common methods including easy performed with labor- and time- saving in experimental operation. The concept has been proven by monitoring enrichment of dynamic aptamer candidate libraries targeting a small molecule 2,2-bis(4-chlorophenyl) acetic acid (DDA) during SELEX process. A mini gel cassette was designed and fabricated by our laboratory to make mini agarose gels for diffusion with different directions.
Conclusions
These results indicate that GBDM, in particular, chasing diffusion is suitable for monitoring the interaction between enriched aptamer candidates and their targets. These pioneering efforts are helpful for novel aptamer selection by breaking through the technical bottleneck of aptamer development and helpful for development of novel aptasensors.
Keywords: Aptasensor, Monitor, Aptamer, Interaction, Visualization, Diffusion
Background
Systematic Evolution of Ligands by EXponential Enrichment (SELEX) is a common method currently used for isolating high-affinity single-stranded (ss) DNAs or RNAs from a large library with random sequences [1–3]. These SELEX-derived DNAs and RNAs named aptamers, can be selected against a broad range of targets (e.g. proteins, cells, virus, microorganisms, toxins and chemical compounds) [4–6]. Aptamers have high affinity and specificity to their target molecules, similar to antigen-antibody interaction [7], therefore, they are also called “chemical antibodies”. Aptamers offer advantages over antibodies including smaller size, higher pH and thermal stability, lower immunogenicity and toxicity, better tissue penetration, lower synthesis costs and easier conjugation or modification [8]. In addition to their widespread use as stand-alone affinity binding reagents in analytical chemistry, aptamers have been engineered into a variety of ligand-specific biosensors, termed aptasensors [9]. Aptasensors can be designed to integrate with a variety of readout methods, such as electrochemical aptasensors [10–12] fluorescent aptasensors [13, 14], label-free aptasensors [15, 16], and aptasensors designed to interface with nucleic acid signaling cascades [17, 18] or specifically for detection of targets such as pathogens [19, 20] or small molecules [7, 21].
Compared with antibody-based immunosensors and enzyme-based general biosensors, aptasensors exhibited better specificity and sensitivity, higher stability, greater flexibility, easier artificial synthesis as well as longer shelf life [22–24]. Moreover, the production of conventional animal-based specific monoclonal antibodies utilized for immunosensors is tedious, very expensive and challenging, and there are batch-to-batch variations, which further limits their applications [25]. Therefore, benefits from the booming development of various aptasensors for fast, selective, sensitive and on-site targets detection have attracted much attention. Aptamers as the molecular recognitions are at the center of aptasensors, however, up to date, there are only several target-specific aptamers developed for development of aptasensors and some of those aptamers lack selectivity, which is not able to distinguish among analogues [26]. Thus, more highly target analytes-specific aptamers need to be selected.
Since SELEX emerged in 1990, many selection methods have been developed for isolating aptamers such as purified-protein-based SELEX, whole-cell-based SELEX, capillary electrophoresis (CE)-based SELEX, Capture-SELEX, and so on [27–30]. Due to the low successful rates for aptamer selection, several factors in selection process have still been studying [31–33]. Importantly, monitoring enriching progress of candidate aptamers has always been the key and restrictive step in SELEX process. To date, many methods such as dot blotting, enzyme-linked oligonucleotide assay (ELONA), surface plasmon resonance (SPR), electrophoretic mobility shift assay (EMSA) and real-time quantitative PCR (qPCR) have been evaluated for monitoring the enriching situation of library (Table 1). During the partition step of Capture-SELEX process [50], for example, candidate molecules that bind to their target can be eluted and theoretically separated from non-bound ssDNA molecules. In practice, however, eluted DNA sub-library almost always contain more or less non-candidate sequences. This is why iterative enrichment is needed in the SELEX process. Monitoring of overall signal of the eluted DNA sub-library results in confusion with the non-specific eluates. On the other hand, detection of candidate-target binding signal can better monitor the progress of the enrichment. We have investigated how to measure the interaction between enriched aptamer candidates and their target molecules. Herein, we report a simple gel-based approach, which provides an effective way for monitoring the enrichment of dynamic libraries in the process of SELEX.
Table 1.
Advantages and disadvantages of reported common methods for monitoring the aptamer-target binding in SELEX rounds
| Monitoring methods | Suitable targets | Advantages | Limitations | References |
|---|---|---|---|---|
| Dot blotting | Protein |
Focus on the candidate aptamers binding phase, which shows the real enrichment in SELEX; Relative ease of performance |
Sequence labeling required; Not suitable for the target molecules with the same electrostatic charge as NC membrane used |
[34] |
| qPCR | Protein & Small molecule | Relative ease of performance |
Focus on the elution phase, which could be confused by non-specific eluates; Error of nonspecific amplification; |
[35–38] |
| EMSA | Protein | Focus on the candidate aptamers binding phase, which shows the real enrichment in SELEX |
Sequence labeling required; Labor- and time-consuming; Not suitable for small molecule targets |
[39] |
| Gel-shifting | Protein |
Focus on the candidate aptamers binding phase, which shows the real enrichment in SELEX; Easy for performance |
Detection in none-binding conditions, I.E. electrophoretic buffer solution; Not suitable for small molecule targets |
[34, 36] |
| ELONA | Protein |
Focus on the candidate aptamers binding phase, which shows the real enrichment in SELEX; Relative ease of performance |
Sequence labeling required; Labor- and time-consuming; Non-specific binding of candidates to the plate, which confuse the enrichment; Not suitable for small molecule targets |
[40] |
| Agarose gel analysis | Protein & Small molecule | Relative ease of performance |
Focus on the elution phase, which could be confused by non-specific eluates; Error of nonspecific amplification |
[41] |
| HTS | Protein & Small molecule | Focus on the enrichment of candidate aptamers according to the SELEX rounds |
Expensive cost required; Focus on the elution phase, which could confuse by none-specific elution; Time-consuming for sample preparation |
[35, 42] |
| SPR | Protein & Small molecule | Focus on the candidate aptamers binding phase, which shows the real enrichment in SELEX |
Expensive sensor chip required; Labor- and time-consuming |
[42, 43] |
| UV quantification | Small molecule | Ease of performance | Focus on the elution phase, which could confuse by none-specific elution | [44] |
| Fluorescence quantification | Small molecule | Relative ease of performance |
Sequence labeled with fluorophore required; Focus on the elution phase, which could confuse by none-specific elution |
[45, 46] |
| Fluorescence binding assay | Small molecule |
Focus on the candidate aptamers binding phase, which shows the real enrichment in SELEX; Relative ease of performance |
The autofluorescence of target required | [47, 48] |
| Gel-elution assay | Small molecule | Focus on the candidate aptamers binding phase, which shows the real enrichment in SELEX |
Target-coupled column required; Labor- and time-consuming |
[49] |
| GBDM | Protein & Small molecule |
Focus on the candidate-target binding phase, which shows the real enrichment in SELEX; Easy for performance without expensive equipment; Suitable for monitoring every selection step during SELEX process |
Overnight diffusion required | Optimized in This work |
Notes: qPCR real-time quantitative PCR, EMSA Electrophoretic mobility shift assay, ELONA enzyme-linked oligonucleotide assay, HTS High throughput sequencing, SPR Surface plasmon resonance, GBDM gel-based diffusion method
Results & discussions
A variety of methods for monitoring of SELEX process have been reported. We have evaluated a range of approaches including the methods of EMSA [34], dot blotting, Eastern blotting and target-capture assay [51], quartz crystal microbalance (QCM) analysis [52], qPCR (data not shown), HTS technology (data not shown), and GBDM in our laboratory. We rank the methods. Chasing diffusion, one of GBDM, is the most practically useful to monitor the interaction between enriched aptamer candidates and their targets round by round during SELEX process.
A mini gel cassette was designed and fabricated for GBDM
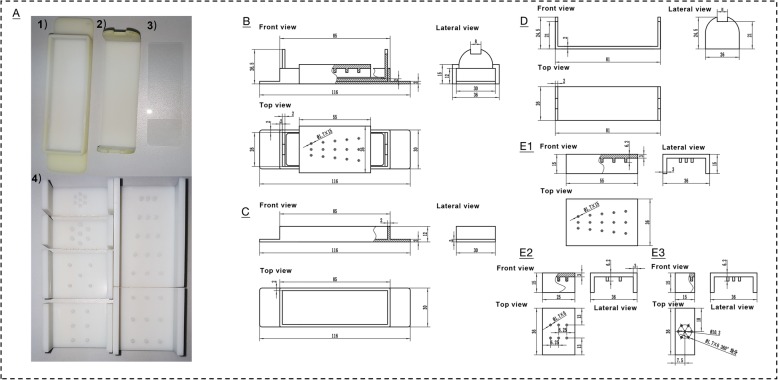
A mini gel cassette was designed and fabricated as a tool to cast mini gels used for the purpose of monitoring. The cassette consists of four parts: a base with a concave, a tray, a smooth glass plate (e.g. glass slide commonly used for microscopy), and one piece of a set of hole-making molds (Fig. 1a). In order to ensure a consistent distance between each bottom of the sample wells and the surface of the glass plate, the parts except glass plate were fabricated by 3D printing technology using Stereo Lithography Apparatus. Before making gel, these four parts can be assembled as shown in the diagram in Fig. 1b. The front, top and side views of the base and the tray are shown in Fig. 1c and d, respectively. Hole-making mold is a key aspect determining the diffusion profile of GBDM. The front, top and side views of three representative hole-making molds are shown in Fig. 1e (e1, e2 and e3).
Fig. 1.
The mini gel cassette and its diagrams. a Mini gel cassette with base (a1), tray (a2), smooth glass plate (a3) and a set of hole-making molds (a4); b Assembly drawing of the invented device. Diagrams of the base (c), the tray (d) and three representative hole-making molds (e). e1 hole-making mold with well spacing ranging from 3.5 to 7.0; e2 hole-making mold with six-well (parallel arrangement); e3 hole-making mold with seven-well (six-well around a centre). Units: mm
Excellent performance in double immunodiffusion (DID) assay by using mini gel cassette
DID was commonly used as a screening test for the monitoring of binding of antibody and antigen [53]. For conventional DID assay, the sample wells in gel are typically prepared by punching such as multiple-well patterns after the gel is solidified. These may create a damage on the bottom of wells, which may cause leakage. For our proposed device, a consistent distance between each bottom of the wells and the surface of the glass plate is ensured for preparation the well bottom-flat gels. We make mini gel using the cassette and perform DID assay routinely with mini gel. A representative gel is shown in Additional file 1: Figure S1. Thus, our device is a good alternative tool for immunodiffusion experiments.
Optimization of conditions for GBDM
Optimization of conditions for GBDM were conducted in four aspects, including well size, well positioning, gel concentration, and diffusion duration.
Giving the fact that 10 μL pipette tips are commonly used in aptamer laboratories. The well size shouldn’t be too large or too small. The optimization results showed that the well sizes in diameter of 0.85 mm (maximum volume of 5.0 μL) and of 1.0 mm (maximum volume of 6.0 μL) were ideal for sample loading (Additional file 2: Table S1). The well spacing is another important aspect for double diffusion. Hole-making molds with different distances between individual wells from 3.5 to 7.0 mm were investigated (Fig. 1e1). The results showed that there was a linear relationship between the intensity of the “precipitation line” and the spacing distance (Additional file 3: Figure S2). Distances of 5.0 and 6.0 mm were optimal for double diffusion.
There was no significant difference in the diffusion situation among different gel concentrations of 2.0% and 3.0% (data not shown) for the purpose of double diffusion. In general, gel concentration of 2.0% is suitable for larger target molecules in molecular weight and 3.0% for smaller target molecules. In addition, there was no significant difference between the diffusion profile of vertically placed gel (needs 5 min incubation in horizontal firstly) and that of horizontal gel (Additional file 4: Figure S3).
Aptamer-based double diffusion
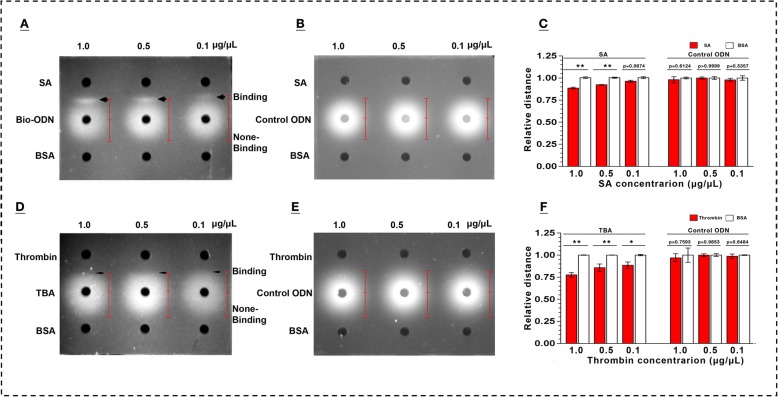
To test our established system, interactions of thrombin and its binding aptamer (TBA), streptavidin (SA) and biotinylated oligodeoxynucleotides (Bio-ODN) were investigated respectively. TBA/Bio-ODN was stained with GelRed in gel. When the molecules of TBA/Bio-ODN and their targets are loaded into the opposite wells, they diffuse from their respective loading wells. The binding of aptamer-target will halt the each diffusion at their interface, thereby shortening the diffusion distance and even forming a precipitation line. Indeed, image analysis results showed that visible diffusion distance of target-bound aptamer/ODN were significantly shorter than that of target-free aptamer/ODN, even “precipitation lines” could be obviously observed in Bio-ODN and SA binding assay with dose-dependent manner (Fig. 2a & c). This is similar to double-immunodiffusion assay for detection of antibody-antigen interaction. Although none of “precipitation line” was found in thrombin and TBA binding groups, the visible diffusion distance of TBA to thrombin is consistently shorter than to BSA with dose-dependent manner (Fig. 2d & f). Double diffusion is reasonably easy to perform although its sensitivity is not very ideal. Whether appearance of “precipitation lines” or not could be dependent on the different conformations of complexes of thrombin/TBA and SA/Bio-ODN. Dissociation constant (Kd) for thrombin and TBA is 2–170 × 10− 6 M, but for SA and biotin is almost 10− 15 M [54, 55]. Detectable binding signal of protein target were observed ranging from 0.1 to 1.0 μg/μL by using the aptamer concentration of 10 μM in 5 μL volume system. With the increasing of length of the aptamer, the lower concentration such as 1 μM of 80 nt aptamer is also meet the demand for diffusion.
Fig. 2.
Characterization of oligodeoxynucleotides (ODN)-target binding by double-diffusion in mini-gels. Diffusion profiles of ODN modified with biotin (Bio-ODN) (a) and without biotin (control ODN) (b) during diffusion toward target streptavidin (SA) and non-target bovine serum albumin (BSA), respectively. Quantitative analysis of diffusion distance of ODN from the central point of different loading wells to their diffusion edge (c & f). Diffusion profiles of thrombin aptamer (TBA) (d) and non-aptamer ODN (control ODN) (e) when they diffused toward target thrombin and non-target BSA, respectively. Indication of binding: shortened diffusion distance due to stagnation of diffusion caused by the formation of binding complexes at the interface of the double-diffusion. Arrow heads in a: appearance of precipitation line of binding complexes at the interface of the double-diffusion. Thin arrow heads in d: interface of the double-diffusion of aptamer and its target; Data represent mean ± SEM. *P < 0.05, **P < 0.01, set control = 1.0. GelRed as the DNA indicator
For small molecules, two binding systems (acetamiprid and profenofos) were selected for monitoring by double diffusion. Image analysis results showed that there was no significant difference between target-bound and target-free aptamer using double diffusion approach (Additional file 5: Figure S4). When the molecular weight of small molecule targets is far lower than that of their corresponding aptamer, they are not heavy enough to affect the diffusion of aptamer. It suggests that a novel approach is demanded for detection of aptamer-target binding when the target molecule weight is very low.
Aptamer-based single diffusion
In single diffusion, the results showed that “precipitation ring” were easily observed in Bio-ODN and SA binding reaction (Additional file 6: Figure S5a). “Precipitation trace” rather than “precipitation ring” was found in thrombin-TBA binding groups (Additional file 6: Figure S5c) compared to the SA-Bio-ODN groups. This may be caused by the difference of affinity between TBA and Bio-ODN with theirs target molecules, which similar with double diffusion. Concentrations of protein target ranging from 0.5 to 1 μg/μL can cause the binding signals by using the aptamer concentration of 0.25 μM in gel. This kind of assay needs a larger volume system (almost 7 mL gels) with more aptamer to meet the requirement to form a visible diffusion difference. Though the sensitivity of this method is reasonable, not so suitable for its applications due to the large amount of aptamer required. Moreover, small molecules (i.e. acetamiprid) were used for testing the monitoring system, results showed that no significant difference was found (Additional file 7: Figure S6).
Aptamer-based chasing diffusion
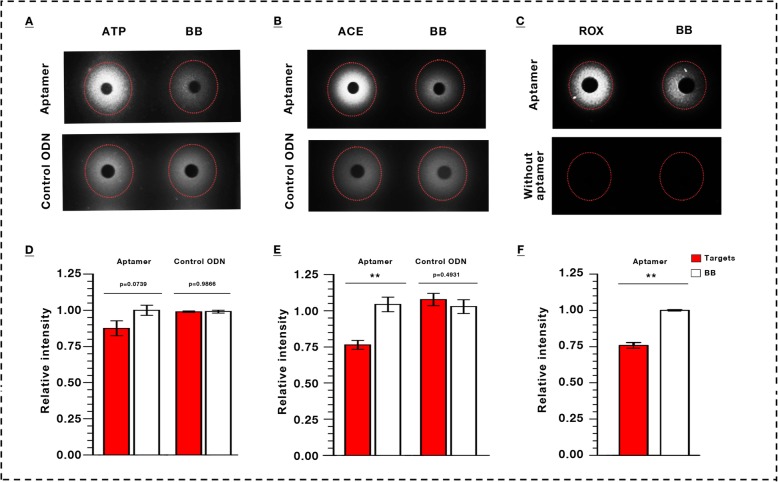
The conformation of aptamer could be changed resulting from binding of aptamer and its target. Consequently, the number of complementally binding base pair may be changed, i.e. double-stranded regions will be different [50]. In chasing diffusion, the significant signal difference between the target molecules and control molecules were found in this study (Fig. 3, Additional file 8: Figure S7). In our previous works, we applied an ssDNA library immobilized SELEX protocol to isolation of aptamers binding to roxithromycin (ROX). One novel candidate aptamer was identified that binds to ROX with high affinity by using SYBR Green I assay (data unpublished). To validate our method, we firstly performed the chasing diffusion to monitoring the binding of aptamer with ROX. The significant signal difference between the ROX molecules and control molecules were found in this study (Fig. 3c & f).
Fig. 3.
Characterization of aptamer-target binding by chasing diffusion in mini-gels. a, b, c, Binding signal of ATP, acetamiprid (ACE) and roxithromycin (ROX) with their aptamers or respective control oligodeoxynucleotides (ODN), respectively. d, e, f, Analysis of the intensity for a, b, c, respectively. Binding buffer (BB) as a negative control; Data represent mean ± SEM. **P < 0.01, set control = 1.0. SYBR Green I as the DNA indicator
The aptamer concentration of 10 μM in 5 μL volume system is enough for chasing diffusion. In principle, chasing diffusion used duplex-specific nucleic acid dye as indicator, which is same as the standard SGI assay [56]. Compared with standard SGI assay, the detection mode is changed from fluorescence to UV detection in our proposed gel-based method. This method does not require relatively sophisticated equipments and are user friendly with labor- and time- saving in experimental operation.
Monitoring the enrichment of the dynamic libraries in SELEX rounds
Various approaches for monitoring the SELEX process have been reported [42, 57–75]. The most straightforward monitoring method involves the assessment of sequence enrichment in RNA pools, a technique made possible by the advent of HTS technology [42, 57–61]. However, HTS is expensive, and sample preparation for HTS is time-consuming. Another effective way of monitoring is to assess the average affinity of the RNA pools to the target molecule [42, 64–74]. Several such assessment methods are available, including SPR, EMSA, and the filter-binding assay. Furthermore, fluorescence-activated cell sorting (FACS) has recently been applied for monitoring the evolution of nucleic acid [73, 74].
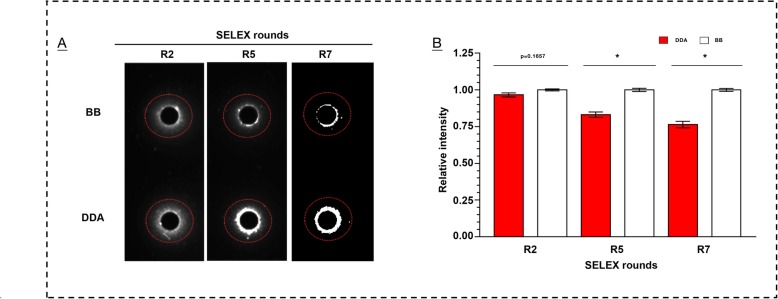
In the scenario that there is a significant difference of molecule weights between target (less than 500 Da) and aptamer, their diffusion rates must be significantly different. In many cases of double diffusion, the target molecules are too small to influence the diffusion distance of aptamer, even they bind each other with high affinity. Recently, we applied SELEX protocol to isolation of aptamers (almost 25.96 kD) bound to 2,2-bis(4-chlorophenyl) acetic acid (DDA) (281.134 Da). We firstly performed chasing diffusion to regularly monitor the enrichment of the library in SELEX process. In each round, the sample of library obtained from selection by candidate-target binding was loaded into the well first. After an optimized period, the target solution was then added into the well to chase the aptamer candidates. The significant signal difference between DDA molecules and control molecules were found since round 5. Three representative libraries were demonstrated in Fig. 4. It strongly implies the application potential of chasing diffusion. Real-time PCR also showed that the DDA specific elutions contain more DNAs than binding buffer washing controls since round 5 (Additional file 9: Figure S8b).
Fig. 4.
Monitoring the enriching situation of the dynamic libraries in SELEX process. a Binding signal of DDA with the libraries of the representative rounds; b analysis of the intensity. DDA: 2,2-bis(4-chlorophenyl) acetic acid; BB: binding buffer; Data represent mean ± SEM. *P < 0.05, set control =1.0. SYBR Green I as the DNA indicator
In selection of aptamers against DDA, the results are very encouraging [76]. One candidate aptamer bound to DDA with high affinity with low cross-binding activities on other small molecules were selected and detected by using SYBR Green I assay (Additional file 9: Figure S8c & d).
A experimental summary of the reported methods used in SELEX rounds
A table including the average duration time, requires of normal reagents, cost and the sensitivity was summarized to better compare the methods used in SELEX (Table 2). GBDM is a low-cost and easy performed method with reasonable sensitivity. In additional, the method focuses on the binding phase of the aptamer candidates and the target, which shows the real enrichment state in the SELEX process. The diffusion of our method does take time, in practical, it can be done overnight for each round of selection without additional labor cost.
Table 2.
Comparison of the reported methods used in SELEX rounds
| Monitoring methods | Average duration time | Normal reagents required | Average cost in reagents per assay | Instrument and its average cost | Sensitivity | Focus on phase |
|---|---|---|---|---|---|---|
| UV quantification | 0.5 h | ~ | ~ |
UV quantification instrument 15,000 USD |
ng | Eluting phase |
| GBDM | 6 to 9 h | Nucleic acid dye | Below 1 USD |
The user-defined mini gel cassette 15 USD UV imager 1500 USD |
μg or nM | Binding phase |
| Gel-shifting | 2 h | Nucleic acid dye | Below 1 USD |
Electrophoresis equipment 1800 USD UV imager 1500 USD |
μg | Binding phase |
| Agarose gel analysis | 0.5 h | PCR reaction reagent; Nucleic acid dye | Below 1.5 USD |
Electrophoresis equipment 1800 USD UV imager 1500 USD |
ng | Eluting phase |
| qPCR | 2 h | qPCR reaction reagent | 25 USD |
Real time PCR amplifier 30,000 USD |
pg | Eluting phase |
| Fluorescence quantification | 1 h | Fluorophore-labeled primer | 25 USD |
Fluorescence quantification instrument 70,000 to 150,000 USD |
pg | Eluting phase |
| Dot blotting | 7 h | Biotin labelled primer; NC membrane; SA-HRP reagents; ECL chemiluminescent solution | 30 USD |
Chemiluminescence apparatus 30,000 USD |
ng | Binding phase |
| EMSA | 7 h | Labeled aptamer (such asγ-32P and biotin); Nylon membrane; Detection reagents | 30 USD |
Electrophoresis equipment 1800 USD Imaging apparatus 50,000 USD |
μg | Binding phase |
| ELONA | 6 h | Biotin-labeled primer; ELISA plates; SA-HRP reagents; TMB solutions | 30 USD |
Microplate reader 20,000 USD |
ng | Binding phase |
| SPR | 6 h | Sensor chip | 90 USD |
SPR instrument 150,000 USD |
pg | Binding phase |
| Gel-elution assay | 7 h | Target-coupled column; PCR reaction reagent | N/A |
Electrophoresis equipment 1800 USD UV imager 1500 USD |
μg | Eluting phase |
Notes: The methods ranked by average cost in reagents per assay; ~ means none cost; N/A means not applicable
GBDM gel-based diffusion method, qPCR real-time quantitative PCR, EMSA Electrophoretic mobility shift assay, ELONA enzyme-linked oligonucleotide assay, SPR Surface plasmon resonance
These results indicate that GBDM, in particular, chasing diffusion is suitable for monitoring the interaction between enriched aptamer candidates and their targets. This developed method is easy performed with reasonable sensitivity. Moreover, we invented a device using proposed mechanism for its better application.
Materials & methods
All oligodeoxynucleotides (Additional file 10: Table S2) were synthesized and purified by High Performance Liquid Chromatography (HPLC, Sangon biotech, Shanghai, China). Thrombin (alpha) (Cat.No. HCT-0020, USA) was purchased from Haematologic Technologies Inc. (USA). Streptavidin (SA, Cat.No. S9170-1 mg), bovine serum albumin (BSA, Cat.No. A8020), goat anti-rabbit IgG (Cat.No. SPA134) and rabbit IgG (Cat.No. SP034) were purchased from Solarbio life sciences (Beijing, China). GelRed stain (Cat.No. KGM025R) was obtained from KeyGEN BioTECH (Nanjing, China). SYBR Green I (Cat.No. A502040–0500) was purchased from Sangon biotech (Shanghai, China). Acetamiprid (Cat.No. A109930-100 mg) was purchased from ALADDIN (Shanghai, China); Profenofos (Cat.No.1633000) was purchased from LGC Labor GmbH (Germany). Binding buffer A (20 mM Tris-HCL, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 5 mM KCl, 1 mM CaCl2), binding buffer B (0.1 M PBS with 1 mM Mg2+), binding buffer C (300 mM NaCl, 5 mM MgCl2, 20 mM Tris, pH 7.6) and binding buffer D (10 mM NaCl, 10 mM KCl, 10 mM MgCl2 and 50 mM Tris/HCl, pH 8.0) was prepared for these assays. All other chemical reagents were analytically pure grades and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Water used in all experiments was Elix water.
Preparation of mini gel cassette
The experimental models of gel cassette (Fig. 1) were designed by using Materialise 3-MATIC software (www.materialise.com) and fabricated by 3D printing technology (ZRapid Tech, iSLA550, China) using Stereo Lithography Apparatus (SLA) with transparent resin (ZRapid Tech, ZR580, China). Precision of printing was 0.1 mm.
In mini gel cassette, four parts could be assembled before making gel as the diagram in Fig. 1b. Briefly, the tray could be placed into the concave of the base, a smooth glass plate could be placed on the tray. Agarose with binding buffer were melted, mixed with GelRed or SYBR Green I stain (with aptamers for single diffusion) and then poured on the glass plate which was pre-assembled in the concave with tray. A suitable mold could be placed on the tray with melted agarose to make sample wells on the gel. After a half-hour setting, the mold was carefully removed off and the gel with sample wells was ready to be used for loading samples.
Double immunodiffusion (DID) tests by using our mini gel cassette
Immunodiffusion tests were performed in 2.0% agarose. Each agar plate contained a five-well pattern. The wells were 0.85 mm in diameter. Each of the 4 wells in the outer side was spaced 4.5 mm from the centre well (Additional file 1: Figure S1). Reagents were added to the wells in a consistent pattern. 5 μL of each reference antiserum (goat anti-rabbit IgG) and of each reference antigen (Rabbit IgG) and the negative control were added to the designated well. Slides were incubated in a moist atmosphere at 37 °C. Readings were made after overnight incubation (16 h).
GBDM for monitoring the binding of aptamer and target
A protocol for GBDM assay including a step-by-step guide to the procedure can be found in Additional file 11: Protocol.
Gel preparation
The 2.0–3.0% (m/v) agarose with binding buffer (pre-preparation refer to materials and methods, Additional file 10: Table S2) were melted, mixed with 1 × GelRed or 1× SYBR Green I and loaded into the device carefully. After a half-hour, gels are ready for monitoring after removing the hole-making molds carefully.
Aptamer-based double diffusion
Principle of double diffusion
There are three wells for each test. The middle one is for loading aptamer or control ODN sample. Both control materials and target molecules were simultaneously loaded in a designed well beside the aptamer/control ODN well in agarose gel respectively (Fig. 5). In the diffusion process, molecules including aptamer/control ODN, control material and target molecules are diffused freely in gel. Once target molecules meet with the aptamer, formation of aptamer-target complex reduces the diffusion speed. This difference can detect and visualize by UV-light system.
Fig. 5.
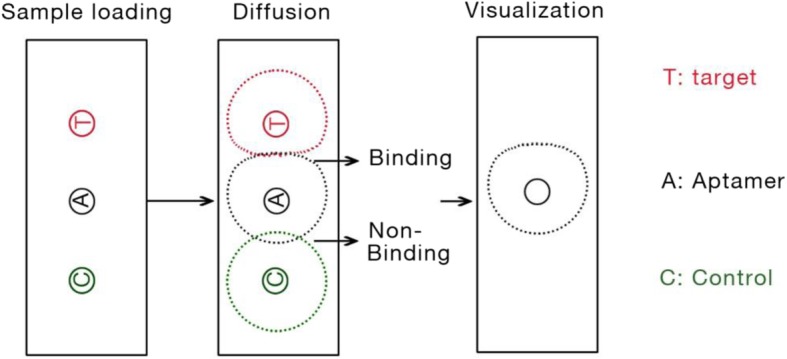
Schematic diagram of the aptamer-based double diffusion
Double diffusion
Optimal diffusion conditions were determined at room temperature. Details in thrombin and SA assays: Concentration of TBA and Bio-ODN: 10 μM. Targets concentration: 1.0, 0.5, and 0.1 μg/μL. Diffusion time was 6 h at room temperature. Gel concentration: 2.0% in binding buffer. Control ODN and bovine serum albumin (BSA) as negative controls. Sample volume: 5 μL for each well.
Aptamer-based single diffusion
Principle of single diffusion
For single diffusion, in gel preparation step, gels needed to be mixed with aptamer candidates or control ODN firstly. Then the target molecules and control materials were loaded for diffusion (Fig. 6). Once target molecules meet with the aptamer in gel, aptamer-target complex are constantly forming on the road. These aptamer-target complex can detect and visualize by UV-light system. Conversely, gels could be mixed with target molecules firstly.
Fig. 6.
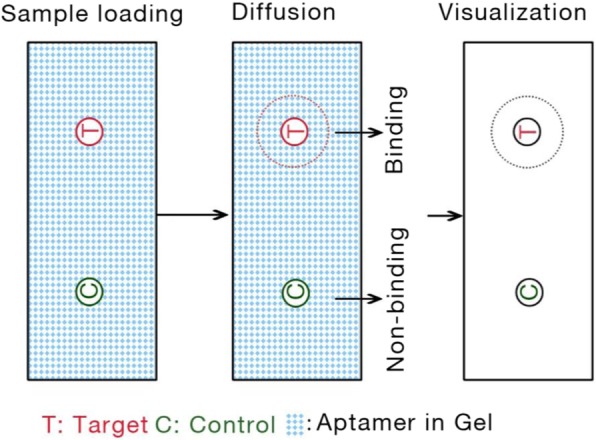
Schematic diagram of the aptamer-based single diffusion
Single diffusion
Optimal diffusion conditions were determined at room temperature. Gel concentration: 2.0% in binding buffer with aptamer candidates or control ODN. Details in thrombin and SA assays: Concentration of TBA and Bio-ODN: 0.25 μM. Targets concentration: 1.0, 0.5, and 0.1 μg/μL. Diffusion time was 16 h at room temperature. Control ODN and bovine serum albumin (BSA) as negative controls. Sample volume: 5 μL for each well.
Aptamer-based chasing diffusion
Principle of chasing diffusion
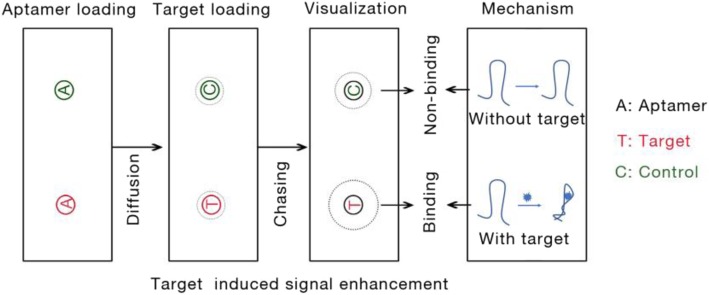
Preparation of low molecular weight target molecule or control material was loaded into the well where the aptamer preparation was previously loaded and allowed to diffuse for a definite time (Fig. 7). Once target molecules meet with the aptamer in gel, formation of aptamer-target complex could induce the signal enhancement or weakness. This difference can detect and visualize by UV-light system.
Fig. 7.
Schematic diagram of the aptamer-based chasing diffusion (signal enhancement)
Chasing diffusion
Optimal diffusion conditions for chasing diffusion were determined at room temperature. Concentration of Aptamer and Bio-ODN: 10 μM. Targets concentration: ATP, 10 mM; acetamiprid, 9.85 mM. Diffusion time was 9 h at room temperature including 30 min for aptamer diffusion. Control ODN or binding buffer only as a negative control. Gel concentration: 3.0% in binding buffer. Sample volume: 5 μL for each sample.
Image taken and analysis
Interaction of stained aptamer and target was visualized by photography of the gels with UV light using a gel imaging system (Tanon 4600SF, Shanghai, China). Images were processed and quantified by calculating distance or scanning densitometry using Image-pro plus (version 6.0) (www.mediacy.com) processing software and normalized to the signal intensity of control materials.
Monitoring the enriching situation of library in SELEX process
Selection of an aptamer against DDA was performed by using a variation of the SELEX protocol (Additional file 9: Figure S8). In brief, the synthetic ssDNA library (80 nt with 40 nt random sequence) was dissolved in binding buffer. Biotin-labeled library complementary sequence was used to capture the library onto the streptavidin magnetic beads. The magnetic beads were washed 6 times, and incubated for 90 min with DDA in binding buffer (200 μL) to a final concentration of 100 μM at room temperature. The supernatant was collected by magnetic separation and monitored by real-time PCR. Then the DDA-specific elution was amplified by PCR and library was generated for next selection round. In the last selection round, the library was sequencing. Characterization of candidate aptamers for DDA was conducted by SYBR Green I assay.
In this study, the library obtained from each round was sampled and monitored by chasing diffusion. Concentration of sub-libraties: 700 nM to 1 μM. Gel concentration: 3.0% in binding buffer. Targets concentration: 10 μM. Diffusion time was 9 h at room temperature. Binding buffer only as a negative control. Sample volume: 5 μL. Library samples of round 2, 5 and 7 were duplicated and demonstrated in this report.
Characterization of a novel aptamer against small molecule by using mini gel
To validate our concept of mini gel, a novel candidate aptamer against roxithromycin (ROX) was selected and monitored by mini gel. The candidate aptamer was newly selected and characterized by SYBR Green I assay with high affinity by our research group (data unpublished). Gel concentration: 3.0% in binding buffer. Candidate aptamer concentration: 5 μM; Targets concentration: 1 mM. Diffusion time was 9 h at room temperature. Binding buffer only as a negative control. Sample volume: 5 μL.
Statistical analysis
All experiments in this study were performed at least in triplicate for each control and treatment group. The numeric data are expressed as the mean ± SEM. Differences between groups were evaluated using Student’s t test. P < 0.05 was considered statistically significant. GraphPad Prism 7 (version 7.00) (www.graphpad.com) was used to display and analyze the data.
Conclusions
In summary, the GBDM described here is a kind of convenient aptamer-target binding assay. Both double diffusion and single diffusion are good to monitor the binding behavior of macromolecule target. Whereas, the developed chasing diffusion is recommended for monitoring the interaction between dynamic aptamer libraries and their targets, in particular for small molecule targets, round by round during the process of SELEX. Our study and theory will yield benefits in discovery of novel aptamers, the modern non-animal affinity recognition elements of aptasensors.
Supplementary information
Additional file 1: Figure S1. Excellent performance in double immunodiffusion (DID) experiment by using designed gel cassette. Stars: Precipitation line; Ag, rabbit IgG (10 mg/mL); Ab, goat anti-rabbit IgG (different ratio in PBS buffer); C, PBS as negative control; Diffusion time was 16 h at 37 °C.
Additional file 2: Table S1. Optimizations of the diameter of different well sizes and their maximum volumes for sample loading.
Additional file 3: Figure S2. Optimization (A) and analysis (B) of the diffusion distance by using streptavidin (SA) and biotinylated oligodeoxynucleotides (Bio-ODN) by double diffusion. Arrows: target-DNA binding complexes. Concentration of Bio-ODN: 5 μM. Targets concentration: 1.0 μg/μL. Diffusion time was 6 h at room temperature. Bovine serum albumin (BSA) as a negative control.
Additional file 4: Figure S3. There is no significant difference in diffusion situation of oligodeoxynucleotides (ODN) in non-horizontal placed gel. The gel was placed in erect direction after 5 min incubation for diffusion. It has little impact in the diffusion pattern of the ODN. Diffusion time was 6 h at room temperature.
Additional file 5: Figure S4. There are no binding signals in aptamer- profenofos (A) and -acetamiprid (B) by double diffusion, respectively. Concentration of Aptamers: 10 μM. Targets concentration: profenofos,10 mM; acetamiprid, 9.85 mM. Diffusion time was 6 h at room temperature. Binding buffer as a negative control.
Additional file 6: Figure S5. Characterization of aptamer-target binding by single diffusion in mini-gels. A and B, Binding signal of streptavidin (SA) and biotinylated oligodeoxynucleotides (Bio-ODN) (A) and control DNA (B). C, Analysis of diffusion radius centered at the centre of the well for SA. D and E, Binding signal of thrombin and its aptamer (TBA) (D) and control DNA (E). F, Analysis of diffusion radius centered at the centre of the well for thrombin. Arrow heads in A: diffusion ring; Thin arrow heads in D: diffusion trace. Data represent mean ± SEM. *P < 0.05, **P < 0.01, set control = 1.0. GelRed as the DNA indicator.
Additional file 7: Figure S6. There is no binding signal in aptamer-acetamiprid by single diffusion. Concentration of Apt: 0.25 μM. Targets concentration: 8.95 mM. Diffusion time was 16 h at room temperature. Binding buffer as a negative control. The abnormal high intensity spot at the top left well was caused by a bubble.
Additional file 8: Figure S7. Characterization of the binding of aptamer-profenofos and -atrazine by chasing diffusion. Concentration of Apt: 10 μM. Targets concentration: profenfos, 10 mM; atrazine, 20 mM; Diffusion time was 9 h at room temperature. Binding buffer only as a negative control.
Additional file 9: Figure S8. Generation and characterization of aptamers against DDA. A, SELEX protocol to isolation of aptamers bound to DDA; B, Enrichment of the dynamic libraries in SELEX rounds by real-time PCR; C and D, characterization of the affinity (C) and selectivity (D) of one aptamer against DDA by using SYBR Green I assay. BB:Binding buffer; DDA: 2,2-bis(4-chlorophenyl) Acetic Acid; DDT: 2,4′-DDT; PFOA: Perfluorooctanoic Acid; EP: Ethyl pyruvate; TBBPA: Tetrabromobisphenol A; Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Additional file 10: Table S2. Sequences of oligodeoxynucleotides (ODN) used in this study.
Additional file 11: PROTOCOL. A protocol for GBDM assay including a step-by-step guidance.
Acknowledgments
We thank Mr. Zhifa Zheng for his valuable advice in designing the device, and Dr. Jianguang Zhang on three view drawing. We also thank the lab-mates from Junsheng Research Group for their valuable advice and kind help in experiments.
Abbreviations
- Bio-ODN
Biotinylated oligodeoxynucleotides
- CE
Capillary electrophoresis
- DDA
2,2-bis(4-chlorophenyl) acetic acid
- DID
Double immunodiffusion
- ELONA
Enzyme-linked oligonucleotide assay
- EMSA
Electrophoretic mobility shift assay
- FACS
Fluorescence-activated cell sorting
- GBDM
Gel-based diffusion method
- HTS
High throughput sequencing
- ITC
Isothermal titration calorimetry
- ODN
Oligodeoxynucleotides
- QCM
Quartz crystal microbalance
- ROX
Roxithromycin
- SA
Streptavidin
- SELEX
Systematic Evolution of Ligands by EXponential enrichment
- SLA
Stereo Lithography Apparatus
- SPR
Surface plasmon resonance
- TBA
Thrombin binding aptamer
Authors’ contributions
JSL, QL and WZ conceived and drafted this manuscript. WZ, SC, and QL and performed the experiments. YZ performed 3D designing and printing. QL, WZ, ZZ, LJ and JSL discussed the data and revised the article. All the authors have discussed and approved the final version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (Grant No.2016YFE0101700), the Huaqiao University Grant (Grant No.13Y0391), the Promotion Program for Young and Middle-aged Teacher in Science and Technology Research of Huaqiao University (Grant No. ZQN-PY419), and the Subsidized Project for Cultivating Postgraduates’ Innovative Ability in Scientific Research of Huaqiao University (Grant No. 17011071006).
Availability of data and materials
All data generated or analyzed in this study are included in this published article, its supplementary information files, and additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qingxiu Liu and Wei Zhang contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13036-019-0223-y.
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science (New York, NY) 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344(6265):467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 4.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10(3):282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee J, Nilsen-Hamilton M. Aptamers: multifunctional molecules for biomedical research. J Mol Med (Berl) 2013;91(12):1333–1342. doi: 10.1007/s00109-013-1085-2. [DOI] [PubMed] [Google Scholar]
- 6.Hong KL, Sooter LJ. Single-stranded DNA aptamers against pathogens and toxins: identification and biosensing applications. Biomed Res Int. 2015;2015:419318. doi: 10.1155/2015/419318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Liu QX, Guo ZH, Lin JS. Practical application of aptamer-based biosensors in detection of low molecular weight pollutants in water sources. Molecules (Basel, Switzerland) 2018;23(2):344. doi: 10.3390/molecules23020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayat P, Nosrati R, Alibolandi M, Rafatpanah H, Abnous K, Khedri M, Ramezani M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie. 2018;154:132–155. doi: 10.1016/j.biochi.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Munzar JD, Ng A, Juncker D. Duplexed aptamers: history, design, theory, and application to biosensing. Chem Soc Rev. 2019;48(5):1390–1419. doi: 10.1039/C8CS00880A. [DOI] [PubMed] [Google Scholar]
- 10.Willner I, Zayats M. Electronic aptamer-based sensors. Angew Chem Int Ed Eng. 2007;46(34):6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 11.Tang Y, Ge B, Sen D, Yu HZ. Functional DNA switches: rational design and electrochemical signaling. Chem Soc Rev. 2014;43(2):518–529. doi: 10.1039/C3CS60264H. [DOI] [PubMed] [Google Scholar]
- 12.Labib M, Sargent EH, Kelley SO. Electrochemical methods for the analysis of clinically relevant biomolecules. Chem Rev. 2016;116(16):9001–9090. doi: 10.1021/acs.chemrev.6b00220. [DOI] [PubMed] [Google Scholar]
- 13.Wang RE, Zhang Y, Cai J, Cai W, Gao T. Aptamer-based fluorescent biosensors. Curr Med Chem. 2011;18(27):4175–4184. doi: 10.2174/092986711797189637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Acha N, Elosua C, Corres JM, Arregui FJ. Fluorescent sensors for the detection of heavy metal ions in aqueous media. Sensors (Basel) 2019;19(3):599. doi: 10.3390/s19030599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhouati A, Catanante G, Nunes G, Hayat A, Marty JL. Label-free aptasensors for the detection of mycotoxins. Sensors (Basel) 2016;16(12):2178. doi: 10.3390/s16122178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Dong S, Wang E. Homogeneous analysis: label-free and substrate-free aptasensors. Chem Asian J. 2010;5(6):1262–1272. doi: 10.1002/asia.200900660. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, Lu CH, Willner I. From cascaded catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem Rev. 2014;114(5):2881–2941. doi: 10.1021/cr400354z. [DOI] [PubMed] [Google Scholar]
- 18.Yu P, Zhang X, Xiong E, Zhou J, Li X, Chen J. A label-free and cascaded dual-signaling amplified electrochemical aptasensing platform for sensitive prion assay. Biosens Bioelectron. 2016;85:471–478. doi: 10.1016/j.bios.2016.05.047. [DOI] [PubMed] [Google Scholar]
- 19.Torres-Chavolla E, Alocilja EC. Aptasensors for detection of microbial and viral pathogens. Biosens Bioelectron. 2009;24(11):3175–3182. doi: 10.1016/j.bios.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alizadeh N, Memar MY, Mehramuz B, Abibiglou SS, Hemmati F, Samadi Kafil H. Current advances in aptamer-assisted technologies for detecting bacterial and fungal toxins. J Appl Microbiol. 2018;124(3):644–651. doi: 10.1111/jam.13650. [DOI] [PubMed] [Google Scholar]
- 21.Feng C, Dai S, Wang L. Optical aptasensors for quantitative detection of small biomolecules: a review. Biosens Bioelectron. 2014;59:64–74. doi: 10.1016/j.bios.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Bagheri E, Abnous K, Alibolandi M, Ramezani M, Taghdisi SM. Triple-helix molecular switch-based aptasensors and DNA sensors. Biosens Bioelectron. 2018;111:1–9. doi: 10.1016/j.bios.2018.03.070. [DOI] [PubMed] [Google Scholar]
- 23.Kim YS, Raston NH, Gu MB. Aptamer-based nanobiosensors. Biosens Bioelectron. 2016;76:2–19. doi: 10.1016/j.bios.2015.06.040. [DOI] [PubMed] [Google Scholar]
- 24.Razmi N, Baradaran B, Hejazi M, Hasanzadeh M, Mosafer J, Mokhtarzadeh A, de la Guardia M. Recent advances on aptamer-based biosensors to detection of platelet-derived growth factor. Biosens Bioelectron. 2018;113:58–71. doi: 10.1016/j.bios.2018.04.048. [DOI] [PubMed] [Google Scholar]
- 25.Toh SY, Citartan M, Gopinath SC, Tang TH. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens Bioelectron. 2015;64:392–403. doi: 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Liu M, Khan A, Wang Z, Liu Y, Yang G, Deng Y, He N. Aptasensors for pesticide detection. Biosens Bioelectron. 2019;130:174–184. doi: 10.1016/j.bios.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhou JH, Rossi J. Aptamers as targeted therapeutics: current potential and challenges (vol 16, pg 181, 2017) Nat Rev Drug Discov. 2017;16(6):440. doi: 10.1038/nrd.2017.86. [DOI] [PubMed] [Google Scholar]
- 28.Zhu C, Yang G, Ghulam M, Li L, Qu F. Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers. Biotechnol Adv. 2019;37:107432. doi: 10.1016/j.biotechadv.2019.107432. [DOI] [PubMed] [Google Scholar]
- 29.Boussebayle A, Groher F, Suess B. RNA-based capture-SELEX for the selection of small molecule-binding aptamers. Methods. 2019;161:10–15. doi: 10.1016/j.ymeth.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16(3):181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diao D, Qiao N, Wu X, Li J, Lou X. An efficient method to evaluate experimental factor influence on in vitro binding of aptamers. Anal Biochem. 2018;556:7–15. doi: 10.1016/j.ab.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem. 1993;36(7):831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 33.Gawande BN, Rohloff JC, Carter JD, von Carlowitz I, Zhang C, Schneider DJ, Janjic N. Selection of DNA aptamers with two modified bases. Proc Natl Acad Sci U S A. 2017;114(11):2898–2903. doi: 10.1073/pnas.1615475114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin JS, Kauff A, Diao Y, Yang H, Lawrence S, Juengel JL. Creation of DNA aptamers against recombinant bone morphogenetic protein 15. Reprod Fertil Dev. 2015;28:1164–71. [DOI] [PubMed]
- 35.Luo Z, He L, Wang J, Fang X, Zhang L. Developing a combined strategy for monitoring the progress of aptamer selection. Analyst. 2017;142(17):3136–3139. doi: 10.1039/C7AN01131H. [DOI] [PubMed] [Google Scholar]
- 36.Moreno M, Fernandez-Algar M, Fernandez-Chamorro J, Ramajo J, Martinez-Salas E, Briones C. A combined ELONA-(RT)qPCR approach for characterizing DNA and RNA aptamers selected against PCBP-2. Molecules (Basel, Switzerland) 2019;24(7):1213. doi: 10.3390/molecules24071213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Lu Q, Chen S, Wang F, Hou J, Xu Z, Meng C, Hu T, Hou Y. Selection and identification of novel aptamers specific for clenbuterol based on ssDNA library immobilized SELEX and gold nanoparticles biosensor. Molecules. 2018;23(9):2337. doi: 10.3390/molecules23092337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo M, Ahn JY, Lee J, Lee S, Hong SW, Yoo JW, Kang J, Dua P, Lee DK, Hong S, et al. Development of single-stranded DNA aptamers for specific Bisphenol a detection. Oligonucleotides. 2011;21(2):85–91. doi: 10.1089/oli.2010.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M, Guo X, Li H, Zuo X, Hao R, Song H, Aldalbahi A, Ge Z, Li J, Li Q, et al. Epitope binning assay using an electron transfer-modulated aptamer sensor. ACS Appl Mater Interfaces. 2018;10(1):341–349. doi: 10.1021/acsami.7b17324. [DOI] [PubMed] [Google Scholar]
- 40.Sedighian H, Halabian R, Amani J, Heiat M, Amin M, Fooladi AAI. Staggered target SELEX, a novel approach to isolate non-cross-reactive aptamer for detection of SEA by apta-qPCR. J Biotechnol. 2018;286:45–55. doi: 10.1016/j.jbiotec.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Yang KA, Pei RJ, Stojanovic MN. In vitro selection and amplification protocols for isolation of aptameric sensors for small molecules. Methods. 2016;106:58–65. doi: 10.1016/j.ymeth.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiga FM, Maietta P, Guiducci C. More DNA-aptamers for small drugs: a capture-SELEX coupled with surface plasmon resonance and high-throughput sequencing. ACS Comb Sci. 2015;17(5):326–333. doi: 10.1021/acscombsci.5b00023. [DOI] [PubMed] [Google Scholar]
- 43.Jia W, Li H, Wilkop T, Liu X, Yu X, Cheng Q, Xu D, Chen HY. Silver decahedral nanoparticles empowered SPR imaging-SELEX for high throughput screening of aptamers with real-time assessment. Biosens Bioelectron. 2018;109:206–213. doi: 10.1016/j.bios.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Huang Y, Duan N, Wu S, Xia Y, Ma X, Zhu C, Jiang Y, Wang Z. Screening and identification of DNA aptamers against T-2 toxin assisted by graphene oxide. J Agric Food Chem. 2014;62(42):10368–10374. doi: 10.1021/jf5032058. [DOI] [PubMed] [Google Scholar]
- 45.Stoltenburg R, Nikolaus N, Strehlitz B. Capture-SELEX: selection of DNA aptamers for aminoglycoside antibiotics. J Anal Methods Chem. 2012;2012:415697. doi: 10.1155/2012/415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Liu X, Zhang Q, Zhang C, Liu Y, Tu K, Tu J. Selection of DNA aptamers that bind to four organophosphorus pesticides. Biotechnol Lett. 2012;34(5):869–874. doi: 10.1007/s10529-012-0850-6. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Luo Y, Gao T, Yang L, Wang J, Pei R. In vitro selection of DNA aptamers for a small-molecule porphyrin by gold nanoparticle-based SELEX. J Mol Evol. 2019;87(7–8):231–239. doi: 10.1007/s00239-019-09905-4. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Bowser MT. Capillary electrophoresis-SELEX selection of catalytic DNA aptamers for a small-molecule porphyrin target. Anal Chem. 2013;85(3):1525–1530. doi: 10.1021/ac302721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang W, Yu H, Alkhamis O, Liu Y, Canoura J, Fu F, Xiao Y. In vitro isolation of class-specific oligonucleotide-based small-molecule receptors. Nucleic Acids Res. 2019;47:e71. doi: 10.1093/nar/gkz224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komarova N, Andrianova M, Glukhov S, Kuznetsov A. Selection, characterization, and application of ssDNA aptamer against furaneol. Molecules. 2018;23(12):3159. doi: 10.3390/molecules23123159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin JS, McNatty KP. Aptamer-based regionally protected PCR for protein detection. Clin Chem. 2009;55(9):1686–1693. doi: 10.1373/clinchem.2009.127266. [DOI] [PubMed] [Google Scholar]
- 52.Su X, He YT, Lin JS. Comparative analysis of detection methods for dissociation constant of aptamer and protein. Transducer Microsyst Technol. 2016;35(11):47–50. [Google Scholar]
- 53.Hornbeck P. Double-immunodiffusion assay for detecting specific antibodies (ouchterlony) Curr Protoc Immunol. 2017;116:2.3.1–2.3.4. doi: 10.1002/cpim.18. [DOI] [PubMed] [Google Scholar]
- 54.Trapaidze ABA, Brut M. Binding modes of thrombin binding aptamers investigated by simulations and experiments. Appl Phys Lett. 2015;106(4):043702. doi: 10.1063/1.4906594. [DOI] [Google Scholar]
- 55.Green NM. Avidin. 3. The nature of the biotin-binding site. Biochem J. 1963;89:599–609. doi: 10.1042/bj0890599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong L, Xu J, Xu Y, Xiang Y, Yuan R, Chai Y. A universal and label-free aptasensor for fluorescent detection of ATP and thrombin based on SYBR Green I dye. Biosens Bioelectron. 2013;42:193–197. doi: 10.1016/j.bios.2012.10.064. [DOI] [PubMed] [Google Scholar]
- 57.Zimmermann B, Gesell T, Chen D, Lorenz C, Schroeder R. Monitoring genomic sequences during SELEX using high-throughput sequencing: neutral SELEX. PLoS One. 2010;5(2):e9169. doi: 10.1371/journal.pone.0009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu G, Wang T, Yang Y, Xu X, Wang J. An improved SELEX-Seq strategy for characterizing DNA-binding specificity of transcription factor: NF-kappaB as an example. PLoS One. 2013;8(10):e76109. doi: 10.1371/journal.pone.0076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dupont DM, Larsen N, Jensen JK, Andreasen PA, Kjems J. Characterisation of aptamer-target interactions by branched selection and high-throughput sequencing of SELEX pools. Nucleic Acids Res. 2015;43(21):e139. doi: 10.1093/nar/gkv700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi M, Wu X, Ho M, Chomchan P, Rossi JJ, Burnett JC, Zhou J. High throughput sequencing analysis of RNA libraries reveals the influences of initial library and PCR methods on SELEX efficiency. Sci Rep. 2016;6:33697. doi: 10.1038/srep33697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ditzler MA, Lange MJ, Bose D, Bottoms CA, Virkler KF, Sawyer AW, Whatley AS, Spollen W, Givan SA, Burke DH. High-throughput sequence analysis reveals structural diversity and improved potency among RNA inhibitors of HIV reverse transcriptase. Nucleic Acids Res. 2013;41(3):1873–1884. doi: 10.1093/nar/gks1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanbrabant J, Leirs K, Vanschoenbeek K, Lammertyn J, Michiels L. reMelting curve analysis as a tool for enrichment monitoring in the SELEX process. Analyst. 2014;139(3):589–595. doi: 10.1039/C3AN01884A. [DOI] [PubMed] [Google Scholar]
- 63.Muller J, El-Maarri O, Oldenburg J, Potzsch B, Mayer G. Monitoring the progression of the in vitro selection of nucleic acid aptamers by denaturing high-performance liquid chromatography. Anal Bioanal Chem. 2008;390(4):1033–1037. doi: 10.1007/s00216-007-1699-8. [DOI] [PubMed] [Google Scholar]
- 64.Misono TS, Kumar PKR. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal Biochem. 2005;342(2):312–317. doi: 10.1016/j.ab.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 65.Horii K, Omi K, Yoshida Y, Imai Y, Sakai N, Oka A, Masuda H, Furuichi M, Tanimoto T, Waga I. Development of a sphingosylphosphorylcholine detection system using RNA aptamers. Molecules. 2010;15(8):5742–5755. doi: 10.3390/molecules15085742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshida W, Mochizuki E, Takase M, Hasegawa H, Morita Y, Yamazaki H, Sode K, Ikebukuro K. Selection of DNA aptamers against insulin and construction of an aptameric enzyme subunit for insulin sensing. Biosens Bioelectron. 2009;24(5):1116–1120. doi: 10.1016/j.bios.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 67.Wang CL, Yang G, Luo ZF, Ding HM. In vitro selection of high-affinity DNA aptamers for streptavidin. Acta Biochim Biophys Sin. 2009;41(4):335–340. doi: 10.1093/abbs/gmp022. [DOI] [PubMed] [Google Scholar]
- 68.Cao XX, Li SH, Chen LC, Ding HM, Xu H, Huang YP, Li J, Liu NL, Cao WH, Zhu YJ, et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009;37(14):4621–4628. doi: 10.1093/nar/gkp489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dausse E, Taouji S, Evade L, Di Primo C, Chevet E, Toulme JJ. HAPIscreen, a method for high-throughput aptamer identification. J Nanobiotechnol. 2011;9:25. doi: 10.1186/1477-3155-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mencin N, Smuc T, Vranicar M, Mavri J, Hren M, Galesa K, Krkoc P, Ulrich H, Solar B. Optimization of SELEX: comparison of different methods for monitoring the progress of in vitro selection of aptamers. J Pharmaceut Biomed. 2014;91:151–159. doi: 10.1016/j.jpba.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 71.Yufa R, Krylova SM, Bruce C, Bagg EA, Schofield CJ, Krylov SN. Emulsion PCR significantly improves nonequilibrium capillary electrophoresis of equilibrium mixtures-based aptamer selection: allowing for efficient and rapid selection of aptamer to unmodified ABH2 protein. Anal Chem. 2015;87(2):1411–1419. doi: 10.1021/ac5044187. [DOI] [PubMed] [Google Scholar]
- 72.Ohuchi SP, Shibuya M, Nakamura Y. The RNA aptamer inhibiting human vesicular endothelial growth factor receptor 1 without affecting cytokine binding. Biochemistry. 2013;52(13):2274–2279. doi: 10.1021/bi301669p. [DOI] [PubMed] [Google Scholar]
- 73.Fan L, Zhao G, Shi H, Liu M, Wang Y, Ke H. A femtomolar level and highly selective 17beta-estradiol photoelectrochemical aptasensor applied in environmental water samples analysis. Environ Sci Technol. 2014;48(10):5754–5761. doi: 10.1021/es405685y. [DOI] [PubMed] [Google Scholar]
- 74.Qu H, Csordas AT, Wang JP, Oh SS, Eisenstein MS, Soh HT. Rapid and label-free strategy to isolate aptamers for metal ions. ACS Nano. 2016;10(8):7558–7565. doi: 10.1021/acsnano.6b02558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amano R, Aoki K, Miyakawa S, Nakamura Y, Kozu T, Kawai G, Sakamoto T. NMR monitoring of the SELEX process to confirm enrichment of structured RNA. Sci Rep-Uk. 2017;7:283. doi: 10.1038/s41598-017-00273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang W, Liu Q, Lin JS. 3rd international caparica conference on pollutant toxic ions & molecules. Caparica: Proteomass Scientific Society (Portugal); 2019. In vitro selection and characterization of a single-stranded DNA aptamer probe for 2,2-bis(4-chlorophenyl) acetic acid (DDA) p. 166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Excellent performance in double immunodiffusion (DID) experiment by using designed gel cassette. Stars: Precipitation line; Ag, rabbit IgG (10 mg/mL); Ab, goat anti-rabbit IgG (different ratio in PBS buffer); C, PBS as negative control; Diffusion time was 16 h at 37 °C.
Additional file 2: Table S1. Optimizations of the diameter of different well sizes and their maximum volumes for sample loading.
Additional file 3: Figure S2. Optimization (A) and analysis (B) of the diffusion distance by using streptavidin (SA) and biotinylated oligodeoxynucleotides (Bio-ODN) by double diffusion. Arrows: target-DNA binding complexes. Concentration of Bio-ODN: 5 μM. Targets concentration: 1.0 μg/μL. Diffusion time was 6 h at room temperature. Bovine serum albumin (BSA) as a negative control.
Additional file 4: Figure S3. There is no significant difference in diffusion situation of oligodeoxynucleotides (ODN) in non-horizontal placed gel. The gel was placed in erect direction after 5 min incubation for diffusion. It has little impact in the diffusion pattern of the ODN. Diffusion time was 6 h at room temperature.
Additional file 5: Figure S4. There are no binding signals in aptamer- profenofos (A) and -acetamiprid (B) by double diffusion, respectively. Concentration of Aptamers: 10 μM. Targets concentration: profenofos,10 mM; acetamiprid, 9.85 mM. Diffusion time was 6 h at room temperature. Binding buffer as a negative control.
Additional file 6: Figure S5. Characterization of aptamer-target binding by single diffusion in mini-gels. A and B, Binding signal of streptavidin (SA) and biotinylated oligodeoxynucleotides (Bio-ODN) (A) and control DNA (B). C, Analysis of diffusion radius centered at the centre of the well for SA. D and E, Binding signal of thrombin and its aptamer (TBA) (D) and control DNA (E). F, Analysis of diffusion radius centered at the centre of the well for thrombin. Arrow heads in A: diffusion ring; Thin arrow heads in D: diffusion trace. Data represent mean ± SEM. *P < 0.05, **P < 0.01, set control = 1.0. GelRed as the DNA indicator.
Additional file 7: Figure S6. There is no binding signal in aptamer-acetamiprid by single diffusion. Concentration of Apt: 0.25 μM. Targets concentration: 8.95 mM. Diffusion time was 16 h at room temperature. Binding buffer as a negative control. The abnormal high intensity spot at the top left well was caused by a bubble.
Additional file 8: Figure S7. Characterization of the binding of aptamer-profenofos and -atrazine by chasing diffusion. Concentration of Apt: 10 μM. Targets concentration: profenfos, 10 mM; atrazine, 20 mM; Diffusion time was 9 h at room temperature. Binding buffer only as a negative control.
Additional file 9: Figure S8. Generation and characterization of aptamers against DDA. A, SELEX protocol to isolation of aptamers bound to DDA; B, Enrichment of the dynamic libraries in SELEX rounds by real-time PCR; C and D, characterization of the affinity (C) and selectivity (D) of one aptamer against DDA by using SYBR Green I assay. BB:Binding buffer; DDA: 2,2-bis(4-chlorophenyl) Acetic Acid; DDT: 2,4′-DDT; PFOA: Perfluorooctanoic Acid; EP: Ethyl pyruvate; TBBPA: Tetrabromobisphenol A; Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Additional file 10: Table S2. Sequences of oligodeoxynucleotides (ODN) used in this study.
Additional file 11: PROTOCOL. A protocol for GBDM assay including a step-by-step guidance.
Data Availability Statement
All data generated or analyzed in this study are included in this published article, its supplementary information files, and additional files.