Abstract
Coagulation factor XII (FXII) drives production of the inflammatory peptide bradykinin. Pathological mutations in the F12 gene, which encodes FXII, provoke acute tissue swelling in hereditary angioedema (HAE). Interestingly, a recently identified F12 mutation, causing a W268R substitution, is not associated with HAE. Instead, FXII-W268R carriers experience cold-inducible urticarial rash, arthralgia, fever, and fatigue. Here, we aimed to investigate the molecular characteristics of the FXII-W268R variant. We expressed wild type FXII (FXII-WT), FXII-W268R, and FXII-T309R (which causes HAE), as well as other FXII variants in HEK293 freestyle cells. Using chromogenic substrate assays, immunoblotting, and ELISA, we analyzed expression media, cell lysates, and purified proteins for FXII activation. Recombinant FXII-W268R forms increased amounts of intracellular cleavage products that are also present in expression medium and display enzymatic activity. The active site–incapacitated variant FXII-W268R/S544A reveals that intracellular fragmentation is largely dependent on autoactivation. Purified FXII-W268R is highly sensitive to activation by plasma kallikrein and plasmin, compared with FXII-WT or FXII-T309R. Furthermore, binding studies indicated that the FXII-W268R variant leads to the exposure of a plasminogen-binding site that is cryptic in FXII-WT. In plasma, recombinant FXII-W268R spontaneously triggers high-molecular-weight kininogen cleavage. Our findings suggest that the W268R substitution influences FXII protein conformation and exposure of the activation loop, which is concealed in FXII-WT. This results in intracellular autoactivation and constitutive low-grade secretion of activated FXII. These findings help to explain the chronically increased contact activation in carriers of the FXII-W268R variant.
Keywords: inflammation, serine protease, vascular biology, coagulation factor, enzyme mutation, autoinflammatory syndrome, bradykinin, factor XII, hereditary angioedema, kringle domain
Introduction
Factor XII (FXII)3 is a 78-kDa liver-expressed serine protease zymogen that circulates in blood plasma (30 μg/ml; 375 nm). It consists of five N-terminal domains that are connected to the C-terminal protease domain by a proline-rich region. The protease domain contains an activation loop with a cleavage motif for plasma kallikrein (PKa) and plasmin. Cleavage after arginine at position 353 of this sequence generates activated FXII (αFXIIa). This full-length two-chain disulfide-linked enzyme can be truncated to generate βFXIIa (∼28 kDa) (1). Both forms of FXIIa can activate plasma prekallikrein, as well as FXII zymogen (autoactivation). As a result of this reciprocal feedback, initial FXII activation is rapidly amplified. FXII is part of the plasma contact system. This system is activated by anionic surface materials, which can be artificial or of natural origin. When FXII binds to these surfaces, it changes conformation (2, 3). This promotes FXII activation through activation loop exposure. The interplay of FXII(a) with PK(a) depends on high-molecular-weight kininogen (HK); this protein is bound to PK in plasma and has surface-binding properties. The contact system is linked to the blood coagulation system but is also responsible for production of bradykinin. This peptide is liberated from its precursor HK by PKa and is a mediator of pain and vascular leakage (4, 5).
The connection between FXII activation, bradykinin release, and human pathology was first identified in hereditary angioedema (HAE). HAE is a vascular leakage disorder hallmarked by acute attacks of bradykinin-mediated tissue swelling. HAE often results from deficiency in C1-esterase inhibitor (C1-INH), the main plasma inhibitor of FXIIa and PKa. In addition, bradykinin-mediated HAE can also be caused by mutations in the F12 gene (encodes FXII). So far, five different mutations in FXII are associated with HAE (6–9). All of these are located in the proline-rich region, a 53-amino acid sequence that is unique to FXII. Three of these mutations alter the enzymatic processing of FXII during activation through introduction of cleavage sites that are sensitive to plasmin (10) and thrombin (11). The resulting truncated FXII products display an enhanced sensitivity for activation (10–12).
Recently, a new FXII mutation (FXII-W268R) was reported in the kringle domain of FXII (13, 14). Interestingly, FXII-W268R carriers do not present with acute symptoms of HAE. All four identified FXII-W268R carriers are first-degree family members. They suffer from an autoinflammatory syndrome characterized by cold-inducible urticaria, arthralgia, fever, and fatigue. Plasma from these subjects contains markedly increased biomarkers of contact activation. Furthermore, clinical symptoms improve upon administration of a bradykinin receptor antagonist. Strikingly, this pathogenic FXII mutation (W268R) is located only 9 amino acids before the proline-rich region begins, which harbors mutations that cause HAE. This raises the question how FXII-W268R influences protein function and how this differs from mutations that cause HAE. We here report the investigation on the molecular characteristics of FXII with the mutation W268R.
Results
FXII-W268R is spontaneously active
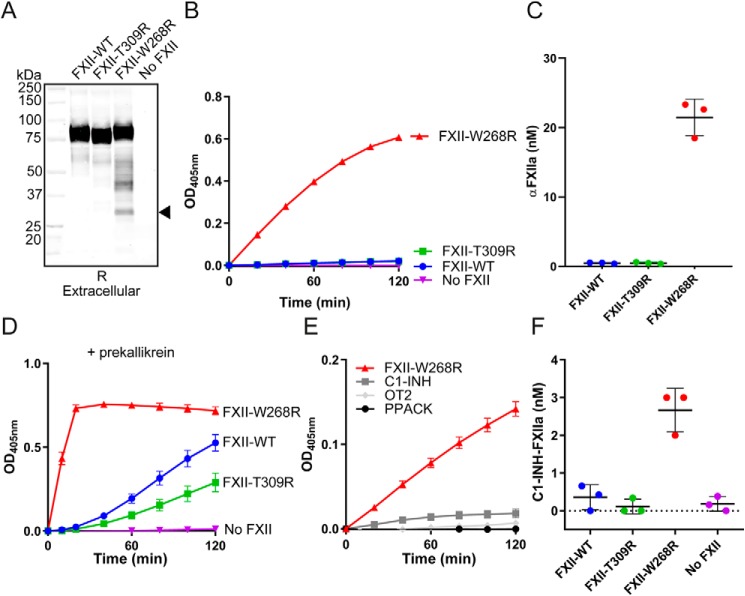
Western blotting analyses of expression medium containing FXII-W268R showed that it is partly fragmented (Fig. 1A). This fragmentation is predominantly seen under reducing conditions (nonreduced Western blotting in Fig. S1) and is much less pronounced during expression of FXII-WT or FXII-T309R (that causes HAE). The smallest fragment that FXII-W268R forms is ∼28 kDa, resembling the light chain of FXIIa (which contains the protease domain). Chromogenic substrate assays showed that expression medium containing FXII-W268R exhibits spontaneous enzyme activity that is equivalent to 21 ± 2.4 nm αFXIIa (progress curves in Fig. 1B, quantitation of FXIIa activity in Fig. 1C). Furthermore, incubation of FXII-W268R expression medium with plasma prekallikrein results in an accelerated development of enzyme activity, compared with expression medium containing FXII-WT or FXII-T309R (Fig. 1D). In a similar manner, prekallikrein activator activity is seen in expression medium containing FXII-W268R that was cloned without the purification tag (tagless FXII-W268R) but not in expression medium containing tagless FXII-WT (Fig. S2A). We subsequently found that the spontaneous enzymatic activity in FXII-W268R expression medium is sensitive to inhibition by the chemical serine protease inhibitor Phe-Pro-Arg-chloromethylketone (PPACK), C1-INH, and the FXIIa-inhibiting mAb OT2 (Fig. 1E) (15). Furthermore, enzymatic activity that develops in FXII-W268R expression medium in the presence of plasma prekallikrein is fully inhibited by PPACK and partially inhibited by C1-INH and OT2 (Fig. S3). When C1-INH is added to expression medium of the FXII-W268R mutant, FXIIa–C1-INH complexes form (Fig. 1F). These combined experiments show that FXII-W268R is already active upon production.
Figure 1.
Factor XII-W268R displays spontaneous activity during protein expression. A, Western blotting of expression medium under reducing (R) conditions. The closed arrow indicates the FXIIa light chain. The image is representative of three separate experiments. B, chromogenic substrate assay for FXIIa activity in expression medium. C, quantification of FXIIa activity in expression medium. Substrate conversion (linear) was monitored in the first 20 min of the assays and fitted to a standard curve of αFXIIa in expression medium. D, chromogenic substrate assay for plasma prekallikrein activator activity in expression medium. E, effects of PPACK, C1-INH, or antibody OT2 on FXIIa activity in expression medium. F, FXIIa–C1-INH complex formation in expression medium. After incubation with C1-INH, a sigmoidal 4PL fit model was used to interpolate concentrations from a standard curve. The figure panels represent means and S.D. of three repeated experiments.
FXII-W268R is prone to intracellular autoactivation
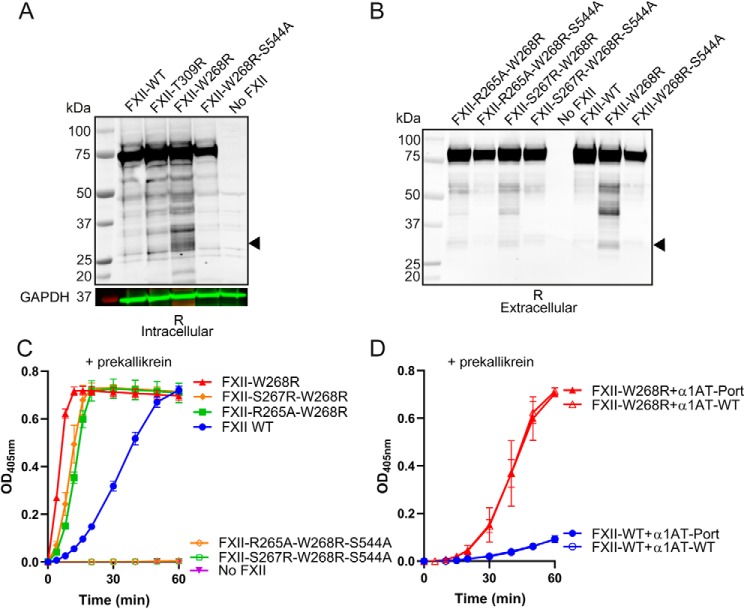
The spontaneous enzymatic activity in expression medium that is displayed by FXII-W268R led us to investigate the possibility of intracellular FXII-W268R activation. Hereto, we analyzed lysates of cells that express FXII-W268R by Western blotting. Increased levels of ∼28 kDa FXII fragments were found in lysates of cells that express FXII-W268R compared with those expressing FXII-WT or FXII-T309R (Fig. 2A). This suggests that FXII-W268R has an increased propensity towards intracellular cleavage and activation. To evaluate the contribution of autoactivation of FXII to the intracellular activation of FXII-W268R, we expressed an active site mutant, FXII-W268R/S544A. In this “silent” mutant, the active site serine is replaced by alanine. FXII-W268R/S544A is unable to autoactivate (i.e. cleave and activate other FXII molecules) and cannot activate plasma prekallikrein (Fig. S4). This FXII variant showed markedly less intracellular fragmentation (Fig. 2A), indicating that intracellular FXII-W268R cleavage predominantly reflects autoactivation.
Figure 2.
FXII-W268R is prone to intracellular autoactivation. A, Western blotting of transfected cell lysates under reducing (R) conditions. Cell lysates of mock-transfected cells were included as a control (No FXII). The closed arrow indicates FXIIa light chain. Where indicated, the FXII active site is incapacitated (S544A). GAPDH (in green; molecular weight marker in red) was used as a lane loading control. B, Western blotting (reduced; R) of expression medium containing FXII variants with altered putative proprotein convertase motifs. Where indicated, the FXII active site is incapacitated (S544A). Images are representative of three separate experiments. C, chromogenic substrate assay for plasma prekallikrein activator activity in expression medium of FXII variants with altered putative proprotein convertase motifs (closed symbols). Where indicated, the FXII active site is incapacitated (S544A; open symbols). D, chromogenic substrate assay for plasma prekallikrein activator activity in expression medium of FXII-WT and FXII-W268R after coexpression with WT α1AT (α1AT-WT) or α1AT-Portland (α1AT-Port). The data represent means and S.D. of three separate experiments.
Alternatively, we investigated whether other intracellular enzymes are involved in the intracellular misprocessing of FXII-W268R. Proprotein convertases are abundant in most cell types; nine subtypes are important for intracellular processing of secretory proteins, with furin as the archetypical example (16). In silico predictions suggest that the W268R mutation introduces an RXRXXR consensus motif for proprotein convertases in FXII (residues 263–270; Table S1). To question its functionality, we developed four FXII-W268R variants (S267R/W268R and R265A/W268R), respectively improving or disrupting this consensus motif and their active site-incapacitated counterparts (S267R/W268R/S544A and R265A/W268R/S544A; Table S2). Western blotting analyses of expression medium containing these FXII-W268R variants showed that FXII fragmentation occurs irrespective of disruption or improvement of the consensus motif for proprotein convertases (Fig. 2B). Disabling FXII autoactivation (S267R/W268R/S544A and R265A/W268R/S544A) prevents this FXII fragmentation to an extent that is comparable with that of FXII-WT and FXII-W268R/S544A (Fig. 2B). Furthermore, disruption or improvement of this motif did not influence plasma prekallikrein–activator activity (Fig. 2C), whereas plasma prekallikrein–activator potential was completely lost upon additional disabling of FXIIa enzymatic activity. It is therefore unlikely that the W268R mutation introduces a new functional proprotein convertase cleavage motif.
In addition, coexpression of FXII-W268R with α1-antitrypsin-Portland (α1AT-Portland; inhibits furin (17)) does not reduce plasma prekallikrein–activator activity (Fig. 2D), ruling out a role for furin. Together, these findings show that where FXII-WT is resistant to intracellular autoactivation during protein expression, FXII-W268R is not. We hypothesized that this is the result of inappropriate activation loop exposure by this pathogenic mutant.
Mutation W268R accelerates FXII activation
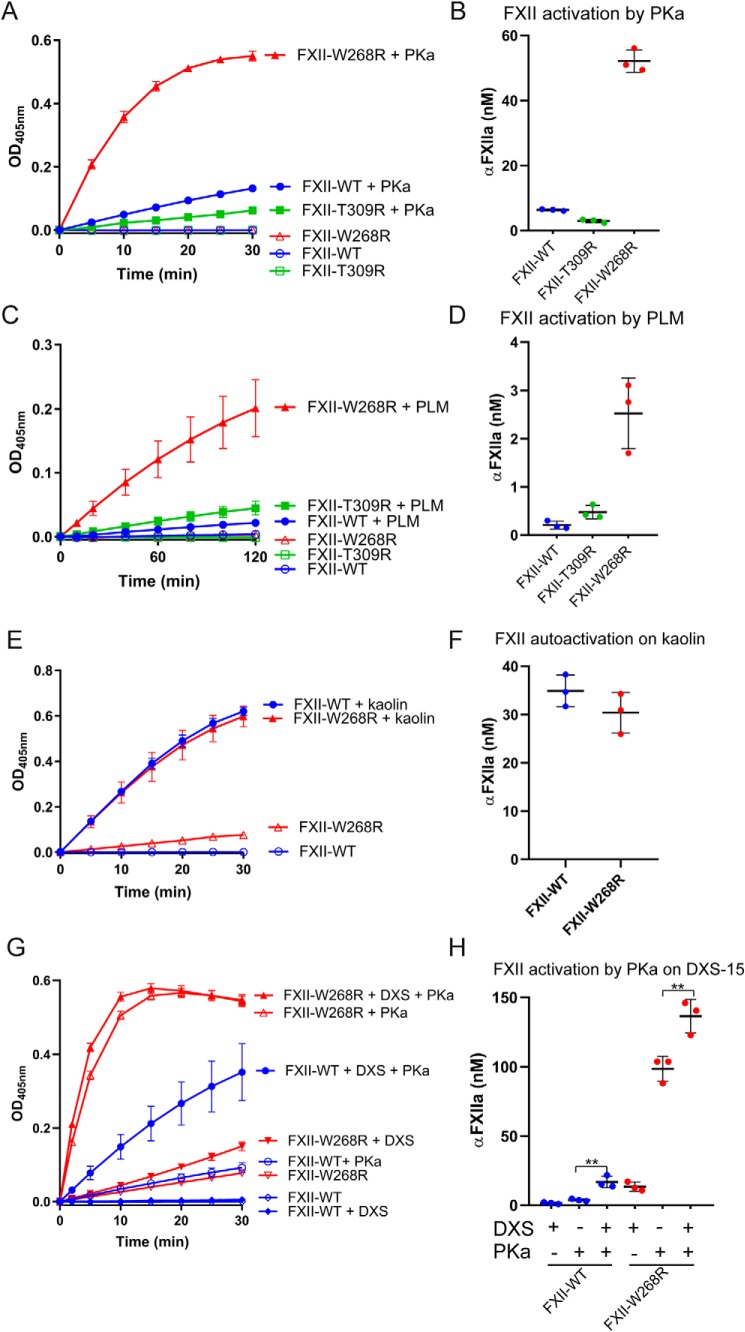
We next examined the sensitivity of FXII-W268R for activation by either PKa or plasmin. We first purified FXII-W268R via its N-terminal 2× Strep-tag. This removes active, truncated βFXIIa, lacking the purification tag (SDS-PAGE with Coomassie stain of purified FXII variants in Fig. S5). As a result, the spontaneous activity of purified FXII-W268R is substantially reduced (Fig. 3). Transient exposure to PKa activates both FXII-WT (6.4 ± 0.4 nm αFXIIa) and FXII-T309R (3.0 ± 0.6 nm αFXIIa) to a limited extent (progress curves in Fig. 3A, quantification of FXIIa activity in 3B). By comparison, FXII-W268R displays a strongly increased activity after exposure to PKa (52 ± 3.6 nm) (progress curves in Fig. 3A and quantification of FXIIa activity in 3B). Tagless versions of these FXII variants behaved highly similarly (Fig. S2, B and C). In a similar manner, transient exposure of FXII-W268R to plasmin generates enzyme activity that is equivalent to 2.5 ± 0.6 nm αFXIIa, ∼5-fold more than FXII-WT (0.2 ± 0.07 nm) and FXII-T309R (0.5 ± 0.2 nm) (progress curves in Fig. 3C and quantification of FXIIa activity in 3D). We next investigated the influence of surface binding on FXII-W268R activation. Anionic surfaces, like kaolin, enhance FXII (auto)activation. In contrast to activation in solution, FXII-WT and FXII-W268R develop similar enzymatic activity in the presence of kaolin (35 ± 3.0 and 30 ± 3.9 nm αFXIIa, respectively; progress curves in Fig. 3E and quantification of FXIIa activity in Fig. 3F).
Figure 3.
Mutation W268R accelerates FXII activation. A, chromogenic substrate assay for FXIIa activity of purified FXII variants in buffer after exposure to PKa (1 μg/ml; closed symbols) or buffer (open symbols). B, quantification of FXIIa activity. Substrate conversion (linear) was monitored in the first 5 min of the assays and fitted to a standard curve of αFXIIa (0–125 nm) in buffer. C, chromogenic substrate assay for FXIIa activity of purified FXII variants in buffer after exposure to plasmin (PLM; 25 μg/ml, closed symbols) or buffer (open symbols). D, quantification of FXIIa activity. Substrate conversion (linear) was monitored in the first 10 min of the assays and fitted to a standard curve of αFXIIa (0–25 nm). The figures represent means and S.D. of three repeated experiments. E, chromogenic substrate assay for FXIIa activity in buffer triggered by kaolin (closed symbols) or buffer (open symbols). F, quantification of kaolin-triggered FXIIa activity in buffer. Spontaneous background activity of FXII variants was subtracted before analyses. Substrate conversion (linear) was monitored in the first 5 min of the assays and fitted to a standard curve of αFXIIa (0–125 nm). G, chromogenic substrate assay for FXIIa activity after preincubation with dextran sulfate (DXS-15k) and subsequent exposure to PKa. FXII-WT and FXII-W268R are indicated by closed circles and closed triangles, respectively. Controls without DXS are indicated by open symbols. Activation of FXII-WT and FXII-W268R in the absence of PKa (autoactivation) is indicated by diamonds and inverted triangles, respectively. H, quantification of PKa-triggered FXIIa activity in the absence or presence of DXS-15k. Substrate conversion (linear) was monitored in the first 5 min of the assays and fitted to a standard curve of αFXIIa (0–150 nm). Spontaneous background activity of FXII variants was subtracted before analyses. The figures represent means and S.D. of three repeated experiments.
We next performed a similar experiment with short dextran sulfate polymers (DXS-15k; Mr = ∼15,000). Because of their short chain length (3), FXII autoactivation is limited (progress curves in Fig. 3G and quantification of FXIIa activity in Fig. 3H). However, the presence of DXS-15k does enhance activation of FXII-WT by PKa ∼4-fold (3.9 ± 0.74 nm αFXIIa without DXS; 16.9 ± 3.9 nm αFXIIa with DXS). Under the same conditions, FXII-W268R already becomes activated by PKa in the absence of DXS-15k (activity equivalent to 98.6 ± 8.1 nm αFXIIa; Fig. 3, G and H). Taken together, FXII-W268R behaves as if it is already in a surface-bound, unfolded conformation, exposing its activation loop where FXII-WT does not.
Mutation W268R enhances activating cleavage of FXII
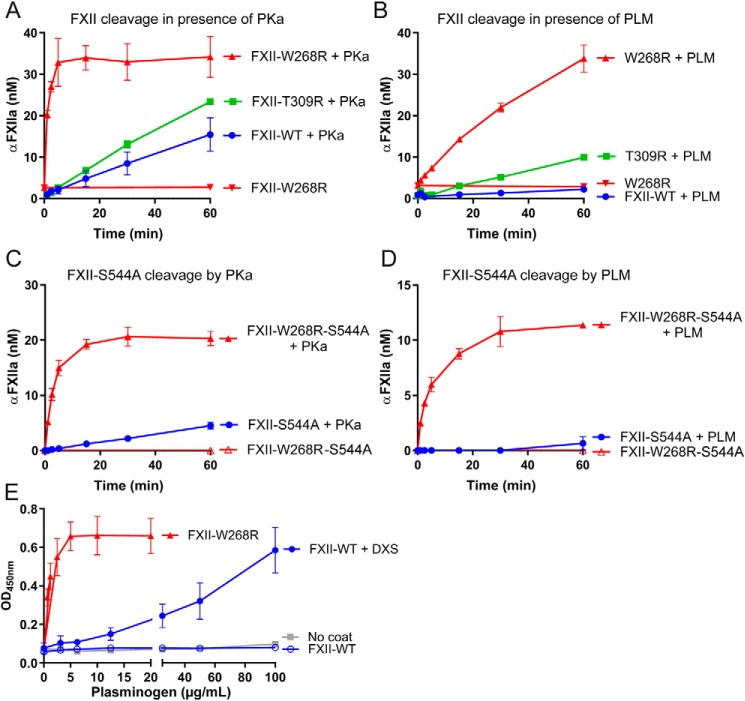
We next investigated FXII-W268R with an ELISA that recognizes two-chain FXIIa, which forms when FXII is cleaved after Arg-353. This ELISA does not require an operational active site for target recognition (18). In good agreement with our chromogenic substrate activity assays, purified FXII-W268R in buffer generates much more FXIIa in response to PKa than FXII-WT or FXII-T309R (Fig. 4A). Similarly, more FXIIa forms when this mutant is exposed to plasmin (Fig. 4B). To exclude a contribution of autoactivation, we next performed these experiment with active-site incapacitated mutants (S544A). As before, the W268R mutation accelerates formation of a two-chain form of FXII by PKa or plasmin (Fig. 4, C and D). These combined experiments suggest that cleavage site Arg-353 is cryptic in FXII-WT but uncovered by the W268R mutation, making it sensitive to activation in solution.
Figure 4.
Mutation W268R enhances activating cleavage of FXII. A and B, formation of two-chain FXIIa by purified FXII variants in buffer, monitored by ELISA during exposure to PKa (A) or plasmin (PLM, B). C and D, formation of two-chain FXII by active-site incapacitated FXII variants (S544A), monitored by ELISA during exposure to PKa (C) or plasmin (PLM, D). A sigmoidal 4PL fit model was used to interpolate FXIIa concentrations from a standard curve. E, plasminogen binding to immobilized purified FXII variants in the absence or presence of DXS. All data represent means and S.D. of three separate experiments.
In further experiments, we examined whether the W268R mutation influences the interaction of FXII with its natural binding partners. We were unable to detect a stable binding interaction between plasma prekallikrein and immobilized FXII-WT or FXII-W268R in direct binding studies (by ELISA; data not shown). In contrast, plasminogen binds to immobilized FXII-WT, but only when it is immobilized in the presence of DXS (Fig. 4E). This suggests that FXII contains a cryptic binding site for plasminogen, which is exposed by surface binding. Remarkably, plasminogen binds robustly to immobilized FXII-W268R without DXS (Fig. 4E), suggesting an open conformation. This behavior is specific for this mutation, because it was not the case for several FXII mutations that cause HAE (Fig. S6). Together, our results suggest that the conformation of FXII determines its activation rate, as well as the interaction with binding partners.
FXII-W268R activity in plasma overrides C1-INH function
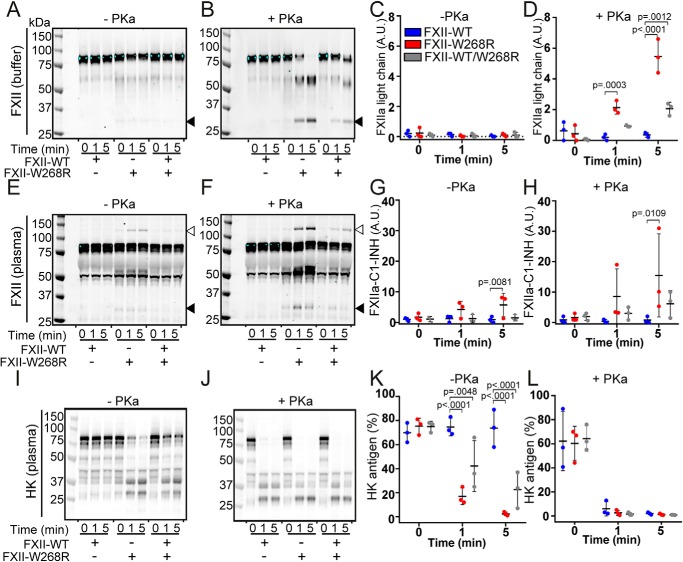
In physiology, FXIIa and PKa are inhibited by C1-INH. We next investigated how FXII-W268R interacts with the plasma contact system by Western blotting. We first examined FXII fragmentation in buffer in the absence or presence of PKa. Over a 5-min time span, 15 μg/ml FXII-WT, FXII-W268R, or a 1:1 combination of both (7.5 μg/ml each, to mimic heterozygous FXII-W268R carriers) did not display fragmentation (Fig. 5, A and C). In contrast, progressive FXII fragmentation was observed in the presence of PKa where FXII-W268R was present (Fig. 5, B and D).
Figure 5.
FXII-W268R activity in plasma overrides C1-INH activity. FXII and HK fragmentation were analyzed by Western blotting (reduced) in a buffered system or in FXII-immunodepleted plasma in the absence or presence of PKa. A, spontaneous FXII fragmentation in buffer. B, FXII cleavage by PKa in buffer. Black arrows indicate FXIIa light chain. C and D, densitometric quantification of FXIIa light chain. The y axis reflects the band intensity of A and B, respectively. E, spontaneous FXII fragmentation in plasma. F, FXII cleavage by PKa in plasma. Open arrows indicate the FXIIa–C1-INH complex. G and H, densitometric quantification of FXIIa–C1-INH complex bands. The y axis reflects band intensity of E and F, respectively. I, spontaneous HK cleavage in plasma in the presence of FXII variants. J, PKa-triggered HK cleavage in plasma in the presence of FXII variants. K and L, densitometric quantification of HK antigen. The y axis reflects band intensity of I and J, respectively. Conditions with FXII-WT were compared with FXII-W268R and FXII-WT/W268R using two-way analysis of variance. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Similarly, when we resupplemented FXII-immunodepleted plasma with FXII-W268R, we observed spontaneous formation of ∼130 kDa bands that correspond to FXIIa–C1-INH complexes (Fig. 5E, indicated by open arrows, and densitometric quantification in Fig. 5G), as well as FXIIa light chain (Fig. 5E, indicated by closed arrows). This was more apparent in the presence of PKa (Fig. 5, F and H).
Finally, we analyzed HK cleavage as a reflection of bradykinin production. Where resupplementation of FXII-immunodepleted plasma with FXII-WT did not provoke any HK cleavage, substantial HK cleavage was seen in the presence of FXII-W268R (Fig. 6, I and K). As expected, the direct addition of PKa to plasma leads to HK cleavage (Fig. 6, J and L). These combined observations suggest that the accelerated activation rate of FXII-W268R overwhelms the inhibitory capacity of C1-INH, triggering HK cleavage in plasma, and are consistent with observations in FXII-W268R carriers (13, 14).
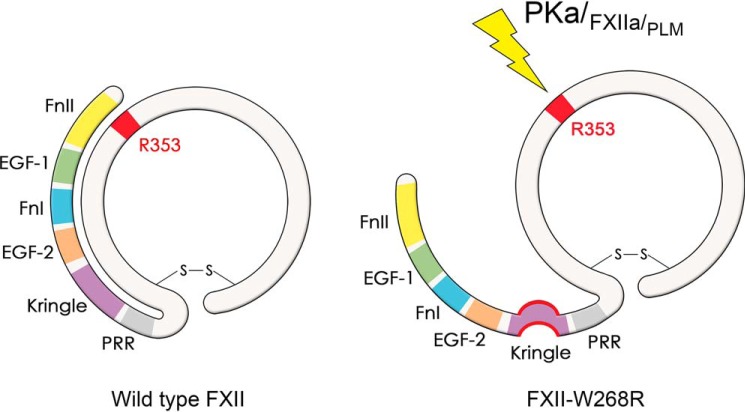
Figure 6.
Graphical representation of hypothesized molecular mechanism behind FXII-W268R. FnII, fibronectin type II domain; EGF-1, epidermal growth factor-like domain 1; FnI, fibronectin type I domain; EGF-2, epidermal growth factor-like domain 2; Kringle, kringle domain; PRR, proline-rich region.
Discussion
In the present study, we investigated the biochemical features of the FXII mutant FXII-W268R that was recently discovered in a family with autoinflammation (13, 14). We found that this pathogenic variant has a generally increased susceptibility for enzyme activation, unlike mutant FXII-T309R, a biochemically comparable mutation that causes HAE. This remarkable behavior of FXII-W268R is not restricted to the extracellular environment; activation already begins intracellularly. The finding that a fraction of FXII-W268R is secreted as an active enzyme and has a lowered threshold for activation enables it to override physiological C1-INH activity in plasma as a result of accelerated activation. This matches the clinical observations of excessive contact activation in plasma of FXII-W268R carriers.
Based on our findings, we hypothesize that the increased susceptibility for activation of FXII-W268R is related to its conformation (graphically summarized in Fig. 6). However, this might not be the only explanation for the observed clinical phenotype. It is becoming increasingly clear that leukocytes contain a distinct pool of FXII (19). The presence of intracellular FXIIa in these cells might trigger inflammasome activation, in turn leading to uncontrolled interleukin-1β production. Alternatively, this mutation might influence the nonproteolytic contributions of FXII to inflammation (20).
At the same time, there is a growing body of evidence that the conformation of FXII is paramount to its function. The binding of FXII to a negatively charged surface changes its conformation. This mediates (auto)activation and causes FXII to expose its activation loop (2, 3). In the present study, we show that the W268R mutation influences FXII in a strikingly similar way.
We previously proposed that FXII zymogen quiescence is determined by a “closed” protein conformation, in which the N terminus of FXII shields its activation loop (1). This concept is not completely novel; it is also described for the serine proteases plasminogen (21), Factor IX (22), prothrombin (23), and the metalloprotease ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) (24, 25). Each of these enzymes displays increased functional properties in an open conformation, which mostly is the result of binding to phospholipid surfaces, protein cofactors, or polymeric target proteins.
Although our data are consistent with the hypothesis that FXII adopts an open conformation as a result of the W268R mutation, we did not directly demonstrate this. Currently available data on FXII structure is limited to the protease domain (26), βFXIIa (27, 28), and the FnI-EGF-2 tandem domain (29).
The W268R mutation is located in the kringle domain of FXII, distant from the protease domain. The functional role of this domain in FXII has not been thoroughly investigated. Kringle domains are among others conserved in members of the plasminogen activation system, hepatocyte growth factors, and prothrombin (30). However, the kringle domain peptide sequence of FXII has low similarity to those of plasminogen, which are important for lysine-dependent binding to fibrin. Although FXII has fibrin-binding properties and is implicated in fibrinolysis, it remains unclear whether this depends on lysine-mediated binding of its kringle domain (31, 32). Interestingly, the two kringle domains of prothrombin are not only important for interactions with factor Va and factor Xa. Kringle 1 is involved in ensuring a closed prothrombin conformation through intramolecular binding to its serine protease domain (23). Disruption or reinforcement of this interaction directly influences the open/closed conformational equilibrium, which impacts prothrombin activation. Based on our current findings, one can speculate that the kringle domain of FXII has a role in regulation of the open/closed equilibrium of this enzyme.
Although our findings point toward the kringle domain as a regulator of zymogen quiescence, based on the identification of a human pathogenic mutation in this domain, it cannot be ruled out that other domains are involved. Indeed, although a mAb against the kringle domain enhances FXII activation by PKa (33), the same holds true for monoclonal antibodies against the adjacent EGF-2 domain (11) and the N-terminally positioned FnII domain (34).
This current work reveals the biomolecular differences between FXII-W268R and previously studied FXII-HAE mutations. Throughout our studies, we learned that FXII-W268R was extremely sensitive to activating cleavage but not as the result of inappropriate truncation. Moreover, FXII-W268R already activates intracellularly where FXII-T309R does not. Our binding studies show that FXII-W268R exposes a binding site for plasminogen that is cryptic in FXII-WT or FXII with HAE mutations. Furthermore, studies with active site–incapacitated FXII mutants and ELISAs show us that this mutant exposes its activation loop, whereas incapacitated FXII-WT does not. Together, these findings strongly suggest that mutation W268R forces FXII into an open conformation. This contrasts the FXII-HAE mutants that we studied earlier: these are in a quiescent state until they are truncated, which requires a specific enzymatic environment. We hypothesize that the continuously increased potential for activation matches the chronic, diffuse clinical picture of autoinflammatory urticaria, whereas the accelerated activation of the FXII-HAE mutations after truncation matches the acute clinical picture of HAE.
Our work presented here on the FXII-W268R mutation supports a pivotal role for the contact system and bradykinin in pathology beyond HAE. To our knowledge, this is the first study to suggest that a properly controlled conformation of FXII is decisive for the regulation of the contact system in human physiology.
Experimental procedures
Reagents
Aprotinin, benzamidine hydrochloride hydrate, bromphenol blue, d-desthiobiotin, DXS molecular (average) weight 500,000 and DXS molecular (average) weight 15,000 (DXS-15k), DTT, DNA oligonucleotides, EDTA, glycine, glycerol, hexadimethrine bromide (Polybrene), KCL, Dulbecco's PBS, skimmed milk powder, NaCl, Na2HPO4, NaH2PO4, soy bean trypsin inhibitor, BSA, and Tween 20 were from Sigma–Aldrich. Restriction enzymes BsmBI, HindIII, XmaI, BsaI, EcoRI, and NotI; T4 DNA polymerase; and CutSmart Buffer were from New England Biolabs (Ipswich, MA). 293fectin transfection reagent, CloneJET PCR cloning kit, pJET1.2/blunt vector, Gibco FreeStyle 293 expression medium, Nalgene single-use PETG Erlenmeyer flasks, PolySorp- and MaxiSorp microtiter plates, and PageBlue were from Thermo Scientific (Waltham, MA). Costar “V” vinyl microtiter plates were from (Corning, NY). Spectra/Por dialysis membranes (molecular mass cutoff, 3.5 kDa) were purchased from Spectrumlabs. Bolt 4–12% Bis-Tris Plus Gels, MOPS buffer, Alexa Fluor 680 donkey anti-sheep IgG, One-shot TOP10 chemically competent Escherichia coli were from Life Technologies, Inc. Immobilon-FL and ethanol were from Merck-Millipore. Sodium acetate and CaCl2 were from Merck. HEK293F cells were from ATCC (LGC Standards GmbH, Wesel, Germany), Tris-HCl was from Roche (Woerden, The Netherlands), and HEPES was from VWR International (Amsterdam, The Netherlands). Kaolin (light) was purchased from BDH (Poole, UK). DNA gel clean-up and plasmid purification kits were from Qiagen. Polyclonal affinity-purified FXII antibody CL20055 was from Cederlane (Burlington, Canada). Anti-human factor XII antibody clone OT2 and streptavidin–poly-HRP were from Sanquin Blood Supply (Division Reagents, Amsterdam, The Netherlands). Anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mAb (clone 4G5; sc-51906) was from Santa Cruz Biotechnology (Dallas, TX). Polyclonal affinity-purified HK antibody 1518BR1 was from Affinity Biologicals (Ancaster, Canada). Peroxidase-conjugated polyclonal rabbit anti-sheep antibodies were from Dako (Heverlee, Belgium). PKa, αFXIIa, and βFXIIa were from Enzyme Research Laboratories (South Bend, IN). Odyssey blocking reagent and donkey anti-mouse IRdye 800 antibody were from LI-COR (Hamburg, Germany). Streptokinase (Streptase) was purchased from CSL-Behring (Breda, The Netherlands), 3,3′,5,5′-tetramethyl-benzidine (TMB) was from Tebu Bio (Heerhugowaard, The Netherlands). PPACK was purchased from Hematologic Technologies (Essex Junction, VT) Ampicillin was obtained from Carl Roth GmbH (Karlsruhe, Germany), H-d-Pro-Phe-Arg-pNA (L2120) was from Bachem (Bubendorf, Switzerland). Plasma purified C1-INH was purchased from Alpha Diagnostics (San Antonio,TX). Blasticidin-S hydrochloride was from Enzo Life Sciences (Raamsdonksveer, The Netherlands). 2× yeast tryptone broth and agar capsules were from MP Biomedicals (Santa Ana, CA). Strep-Tactin–Sepharose beads were from IBA-Life Sciences (Goettingen, Germany). Gene fragments were ordered from Integrated DNA Technologies (Coralville, IA). FXII immunodepleted plasma was from Invitrogen. Plasminogen was purified from plasma as described (10) and converted to plasmin (2.17 mg/ml) by a 30-min incubation with 2000 IU/ml streptokinase in 10 mm HEPES, 150 mm NaCl, 1 mm MgSO4, 5 mm KCl, pH 7.4 (HBS), at 37 °C.
Factor XII cloning, protein expression, and purification
FXII-WT and HAE mutants T309K, T309R, and Del&Ins (c971_1018 + 24del72) were developed as described previously (10). For FXII-Dup (Pro279-Pro284dup), the two segments at both sides of the reported pathogenic duplication (7) were amplified by PCR (Table S3; fragment 1, N-Frag_For and Pro-279–Pro-284dup_P1_Rev; fragment 2, Pro-279–Pro-284dup_P2_For and C-Frag_Rev). Subsequently, the two PCR products were fused and amplified by PCR via overhanging primer sequences.
Mutants FXII-W268R and FXII-S544A were developed through two-step assembly. First, the insert encoding the FXII-WT cDNA sequence was amplified by PCR in three segments (annotated as N, M, and C, respectively; primer sequences in Table S3). Segments with the desired mutations were ordered as dsDNA fragments. First, the N and M segments were digested with XmaI. Subsequently, the fragments were fused by ligation. The fused product (NM) was amplified via PCR. Hereafter, the NM and C segments were digested with BsaI and fused together by ligation. For FXII mutants R265A/W268R, R265A/W268R/S544A, S267R/W268R, S267R/W268R/S544A, and W268R/S544A, the complete cDNA sequences were ordered as dsDNA fragments that were codon-optimized for expression in human cell lines. Fragments were first ligated into the pJET1.2/blunt vector according to the manufacturer's instructions. Inserts were next digested with EcoRI and NotI and ligated into the mammalian expression vector pSM2 encoding an N-terminal IgK secretion signal and 2× Strep-tags for affinity purification (based on pcDNA6/ V5-His A (10)). For FXII-WT and W268R expressed without the N-terminal 2× Strep-tags (tagless FXII), cDNA sequences were amplified by PCR from the FXII-WT or W268R cDNA sequence using the primers tagless_FXII_For and FXII_Rev Primer sequences in Table S3). The resulting cDNA fragment contains the N-terminal IgK secretion signal but lacks the 2× Strep-tags. The cDNA product was ligated into the pJET1.2/blunt vector according to the manufacturer's instructions. Inserts were digested with HindIII and NotI and ligated into the HindIII and NotI restricted mammalian expression vector pcDNA6/V5-His A. Sequences were verified by Sanger sequencing prior to transfection.
HEK293F cells at a concentration of 1.1 × 106 cells/ml were transfected with 293fectin and cultured in Gibco FreeStyle 293 expression medium with 2-ml cell suspensions in 6-well culture plates. After 4 days, cell suspensions were collected and centrifuged at 500 × g for analysis of expression medium and cell pellets. Stable cell lines were selected for resistance to 5 μg/ml blasticidin in the presence of penicillin and streptomycin. Resistant cells were expanded and grown in 2-liter Erlenmeyer flasks for cell culture, cells were centrifuged at 500 × g, and supernatant medium was harvested twice weekly. FXII inhibitor mix consisting of soy bean trypsin inhibitor (0.024 g/liter), benzamidine (0.174 g/liter), and Polybrene (0.056 g/liter) was added to the supernatant prior to storage at −20 °C. Supernatants were thawed and concentrated, and the buffer was exchanged (100 mm Tris-HCl, 150 mm NaCl, and 1 mm EDTA, pH 8.0, containing FXII inhibitor mix) on a 10-kDa dialysis membrane in a Quixstand benchtop system (GE Life Sciences). Strep-Tactin–Sepharose beads were used for purification of recombinant FXII via Strep-tag, and 12.5 μm PPACK was added to washing and elution buffers. Purified recombinant FXII was dialyzed against 4 mm sodium acetate-HCl and 150 mm NaCl (pH 5.5). Protein concentrations were routinely determined by absorption spectroscopy at 280 nm, Coomassie protein staining, or Western blotting (accompanied by densitometric analyses of bands) to ensure equal protein amounts in experiments. Table S2 displays an overview of the constructs used.
α1-Antitrypsin expression
α1AT-WT and α1AT-Portland (17) in pSM2 were cotransfected with FXII constructs where indicated. A SERPINA1 gene fragment was designed with two BsmBI restriction sites between reactive center loop (P4–P4′), allowing for later insertion of the desired reactive center loop DNA template; this SERPINA1-BsmBI fragment was ligated into pSM2 and digested via the BsmBI sites. DNA oligonucleotides encoding for amino acid sequences AIPMSIPP (P4–P4′ reactive center loop of α1AT-WT) or RIPRSIPP (P4-P4′ reactive center loop of α1AT-Portland) were ligated into the predigested SERPINA1-BsmBI template.
Western blotting: FXII in expression medium
Samples from expression medium were diluted in 3× sample buffer (30% glycerol, 190 mm Tris-HCl, 6% SDS, and 0.006% (m/v) bromphenol blue) with or without 25 mm DTT. HEK293F cell pellets of 2-ml expressions, seeded with 2.2 × 106 cells were washed, resuspended in Dulbecco's PBS, lysed by adding 3× sample buffer with 25 mm DTT, and heated for 10 min at 95 °C.
Purified FXII in plasma and buffer
For activation experiments in plasma, 10 μl of recombinant FXII-WT, FXII-W268R, or a 1:1 combination of both (final FXII concentration, 15 μg/ml) were added to 15 μl of FXII immunodepleted plasma or 15 μl of HBS containing 0.05% BSA (w/v). Directly after mixing, samples were placed at 37 °C for 2 min, and activation was started with 5 μl of PKa (final concentration, 12.5 μg/ml) or 5 μl of HBS as a control. Samples were taken after 1 and 5 min and diluted 20 times in 1× sample buffer containing 8.3 mm DTT. Prior to activation, t = 0 samples were created.
All samples were heated for 10 min at 95 °C and centrifuged for 5 min at 2000 × g. Samples were separated on 4–12% Bis-Tris gels at 165 V for 60 min in MOPS buffer, and proteins were transferred onto Immobilon-FL membranes at 125 V for 55 min in blotting buffer (14.4 g/liter glycine, 3.03 g/liter Tris-HCl, 20% ethanol). The membranes were blocked for 1–2 h at room temperature using Odyssey blocking reagent diluted 1:1 with Tris-buffered saline (TBS; 50 mm Tris-HCl and 150 mm NaCl, pH 7.4). Factor XII or HK were detected with affinity-purified polyclonal antibodies (1:2000) by overnight incubation at 4 °C. Next, primary antibodies were detected with Alexa Fluor 680–conjugated donkey anti-sheep IgG (1:5000). GAPDH in lane loading controls was detected with a mAb (1:4000) and a IRdye 800-conjugated donkey anti-mouse antibody (1:10000). The membranes were washed with TBSt and distilled water and analyzed on a near-IR Odyssey scanner (LI-COR).
FXIIa–C1-INH complex ELISA
Methods on VhH selection and production for this assay are described before (18). VhH 1B12 (5 μg/ml), a monoclonal VhH directed against complexed C1-INH, was coated overnight at 4 °C on 96-well MaxiSorp plates. The plates were tapped dry and blocked with HBS containing 0.05% (v/v) Tween 20 (HBSt) and 1% (w/v) skimmed milk powder (mHBSt). Plasma purified C1-INH was added to expression medium at a final concentration of 5 μg/ml. As a reference standard, βFXIIa was added to expression medium from untransfected HEK293F cells and incubated with 5 μg/ml C1-INH. After 10 min of incubation with purified C1-INH, 100 μl/sample was pipetted on the plate and incubated for 1 h at room temperature while shaking. The plates were washed with HBSt. The captured FXIIa–C1-INH complexes were detected by 5 μg/ml biotinylated polyclonal VhH targeted against βFXIIa in mHBSt and after rinsing by streptavidin-poly-HRP (1:2000) in mHBSt. Finally, plates were rinsed, and 50 μl/well TMB substrate was added. Substrate conversion was stopped by adding 25 μl/well H2SO4 (0.3 m), and absorbance was measured at 450 nm. GraphPad Prism 7.02 software was used to interpolate FXIIa–C1-INH concentrations, using a sigmoidal 4PL fit model to the βFXIIa–C1-INH reference standard.
FXIIa ELISA
VhH B7, a monoclonal VhH directed against FXIIa (18) was coated overnight at 4 °C on a 96-well MaxiSorp plates. The plates were tapped dry and blocked with mHBSt. 10 μg/ml recombinant FXII in HBSt was incubated with 1 μg/ml PKa or 25 μg/ml plasmin. At the indicated time points, the reactions were stopped by diluting samples five times in mHBSt, supplemented with 250 μm PPACK. Fifty μl per sample was pipetted on the blocked plates and incubated for 1 h at room temperature while shaking. A reference range of purified αFXIIa in mHBSt containing 200 μm PPACK was included as a standard. Captured FXIIa was detected by the biotinylated monoclonal VhH B2 targeted against βFXIIa (5 μg/ml). Further detection steps were performed as described above (FXIIa–C1-INH complex ELISA).
Protein-binding assays
To study binding interactions between FXII and plasminogen, 5 μg/ml recombinant FXII in HBS was coated on 96-well PolySorp plates (overnight at 4 °C) in the presence of 100 μm PPACK and in the presence of 1 μg/ml DXS, where indicated. The plates were tapped dry and blocked with 2% BSA in HBS (w/v). A concentration series of plasminogen was diluted in HBS containing 1% (w/v) BSA in the presence of 10 μm PPACK and incubated on plates for 1 h. After rinsing, bound plasminogen was detected using a polyclonal goat anti-human plasminogen antibody (1:2000) followed by a rabbit anti-sheep IgG–HRP antibody (1:4000) in the presence of 10 μm PPACK. Finally, the plates were rinsed, and 50 μl/well TMB substrate was added. Substrate conversion was stopped by adding 25 μl of H2SO4 (0.3 m), and absorbance was measured at 450 nm.
Chromogenic substrate assays
Chromogenic substrate H-d-Pro-Phe-Arg-pNA (0.5 mm) was used to detect FXIIa and/or PKa activity. 96-well Costar plates were blocked with HBS containing 1% BSA (w/v; filtered on 0.20-micron filters) for at least 20 min. All sample incubations were performed 37 °C. Substrate conversion was readout at 405 nm at 37 °C.
Spontaneous activity in medium was determined in 40 μl of expression medium containing FXII, to which 10 μl of chromogenic substrate H-d-Pro-Phe-Arg-pNA was added (final concentration, 0.5 mm). Amidolytic FXIIa activity was quantified by determining the initial (linear) slopes of substrate conversion for 20 min, which were fitted to a standard curve of αFXIIa in expression medium (0–25 nm). Where indicated, inhibitors PPACK (200 μm), C1-INH (25 μg/ml), or mAb OT2 (5 μg/ml) were added for 15 min prior to addition of chromogenic substrate H-d-Pro-Phe-Arg-pNA.
Plasma prekallikrein activator activity in 40 μl of expression medium containing FXII was measured by addition of 10 μl of chromogenic substrate H-d-Pro-Phe-Arg-pNA (final concentration, 0.5 mm) and plasma prekallikrein (5 μg/ml final concentration), after which PKa-like activity was measured. The effect of inhibitors PPACK (200 μm), C1-INH (25 μg/ml), and mouse mAb OT2 (5 μg/ml) was investigated by preincubation for 15 min, after which 10 μl of chromogenic substrate H-d-Pro-Phe-Arg-pNA (final concentration, 0.5 mm) and plasma prekallikrein (5 μg/ml) were added, and PKa-like activity was measured.
FXII activation of purified recombinant FXII variants by PKa or plasmin was investigated by preincubating FXII (10 μg/ml in HBS, 0.2% m/v BSA) with PKa (1 μg/ml, in HBS 0.2% BSA), plasmin (25 μg/ml in HBS 0.2% BSA), or buffer (HBS, 0.2% BSA) for 15 min at 37 °C, followed by a 5-min incubation with aprotinin for all conditions (100 KIU/ml in HBS 0.2% BSA) to inhibit PKa or plasmin. FXIIa activity was detected by conversion chromogenic substrate H-d-Pro-Phe-Arg-pNA (0.5 mm) and quantified by determining the initial (linear) slopes of substrate conversion for 5 min, which were fitted to a standard curve of αFXIIa (0–125 nm) in HBS, 0.2% BSA. Residual PKa activity (monitored as a control without FXII) was routinely subtracted where FXII activation by PKa was investigated.
FXII activation of purified recombinant FXII variants by kaolin was measured by preincubating FXII (10 μg/ml in HBS, 0.2% BSA) with kaolin (50 μg/ml) or buffer (HBS) at 37 °C for 15 min, after which chromogenic substrate H-d-Pro-Phe-Arg-pNA (0.5 mm) was added and FXIIa activity was detected. FXIIa activity was quantified by determining the initial (linear) slopes of substrate conversion for 5 min, which were fitted to a standard curve of αFXIIa (0–125 nm) in HBS, 0.2% BSA.
FXII activation of purified recombinant FXII variants by PKa in the presence of DXS-15 was measured by preincubating FXII (10 μg/ml in HBS, 0.2% BSA) with DXS-15 (10 μg/ml in HBS) or buffer (HBS) for 15 min at 37 °C after which PKa (1 μg/ml, in HBS 0.2% BSA) or buffer (HBS, 0.2% BSA) was added and incubated at 37 °C for 15 min, after which aprotinin (100 KIU/ml in HBS 0.2% BSA) was added and incubated at 37 °C for 5 min. Chromogenic substrate H-d-Pro-Phe-Arg-pNA (0.5 mm) was added, and FXIIa activity was quantified by determining the initial (linear) slopes of substrate conversion for 5 min, which were fitted to a standard curve of αFXIIa (0–156 nm) in HBS, 0.2% BSA. Residual PKa activity (monitored as a control without FXII) was routinely subtracted where FXII activation by PKa was investigated.
Statistical analysis
GraphPad Prism 7.02 software was used for statistical analysis were indicated. Two-way analysis of variance with Dunn's correction of multiple testing was used for comparison of repeated experiments with biological samples.
Author contributions
Z. L. M. H., K. K., N. A. M., J. S., C. E. H., S. d. M., and C. M. conceptualization; Z. L. M. H., C. C. C., W. S., A. N., N. M. J. P., M. Z., and S. d. M. data curation; Z. L. M. H. and C. C. C. formal analysis; Z. L. M. H. investigation; Z. L. M. H., W. S., S. d. M., and C. M. methodology; Z. L. M. H. and C. M. writing-original draft; Z. L. M. H., C. C. C., K.K., C.E.H., S. d. M., and C. M. writing-review and editing; W. S. resources; C. E. H., M. M., S. d. M., and C. M. supervision.
Supplementary Material
Acknowledgment
We thank Peter Boross for providing FXII immunodepleted plasma.
This work was supported by funds from the Landsteiner Foundation for Blood Transfusion Research Grant LSBR 1520 (to C. M.), The Netherlands Thrombosis Foundation Grant 2017-03 (to C. M.), the Alexandre Suermann Grant from the Utrecht University Medical Center (to Z. L. M. H.), and a grant from the Thai government (to W. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S6.
- FXII
- factor XII
- α1AT
- α1-antitrypsin
- αFXIIa
- active factor XII α-form
- βFXIIa
- active factor XII β-form
- C1-INH
- C1-esterase inhibitor
- DXS
- dextran sulfate
- FXIIa
- active factor XII
- FXII-HAE
- hereditary angioedema caused by FXII mutation
- HAE
- hereditary angioedema
- HK
- high-molecular-weight kininogen
- PPACK
- Phe-Pro-Arg-chloromethylketone
- VhH
- antigen-binding fragment of heavy chain only antibody
- PKa
- plasma kallikrein
- HRP
- horseradish peroxidase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TMB
- 3,3′,5,5′-tetramethyl-benzidine.
References
- 1. de Maat S., and Maas C. (2016) Factor XII: form determines function. J. Thromb. Haemost. 14, 1498–1506 10.1111/jth.13383 [DOI] [PubMed] [Google Scholar]
- 2. Samuel M., Pixley R. A., Villanueva M. A., Colman R. W., and Villanueva G. B. (1992) Human factor XII (Hageman factor) autoactivation by dextran sulfate: circular dichroism, fluorescence, and ultraviolet difference spectroscopic studies. J. Biol. Chem. 267, 19691–19697 [PubMed] [Google Scholar]
- 3. Citarella F., Wuillemin W. A., Lubbers Y. T., and Hack C. E. (1997) Initiation of contact system activation in plasma is dependent on factor XII autoactivation and not on enhanced susceptibility of factor XII for kallikrein cleavage. Br. J. Haematol. 99, 197–205 10.1046/j.1365-2141.1997.3513165.x [DOI] [PubMed] [Google Scholar]
- 4. Hall S. M., LeBaron L., Ramos-Colon C., Qu C., Xie J. Y., Porreca F., Lai J., Lee Y. S., and Hruby V. J. (2016) Discovery of stable non-opioid dynorphin A analogues interacting at the bradykinin receptors for the treatment of neuropathic pain. ACS Chem. Neurosci. 7, 1746–1752 10.1021/acschemneuro.6b00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Maat S., Hofman Z. L. M., and Maas C. (2018) Hereditary angioedema: the plasma contact system out of control. J. Thromb. Haemost. 16, 1674–1685 10.1111/jth.14209 [DOI] [PubMed] [Google Scholar]
- 6. Dewald G., and Bork K. (2006) Missense mutations in the coagulation factor XII (Hageman factor) gene in hereditary angioedema with normal C1 inhibitor. Biochem. Biophys. Res. Commun. 343, 1286–1289 10.1016/j.bbrc.2006.03.092 [DOI] [PubMed] [Google Scholar]
- 7. Kiss N., Barabás E., Várnai K., Halász A., Varga L. Á., Prohászka Z., Farkas H., and Szilágyi Á. (2013) Novel duplication in the F12 gene in a patient with recurrent angioedema. Clin. Immunol. 149, 142–145 10.1016/j.clim.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 8. Bork K., Wulff K., Meinke P., Wagner N., Hardt J., and Witzke G. (2011) A novel mutation in the coagulation factor 12 gene in subjects with hereditary angioedema and normal C1-inhibitor. Clin. Immunol. 141, 31–35 10.1016/j.clim.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 9. Gelincik A., Demir S., Olgaç M., Karaman V., Toksoy G., Çolakoğlu B., Büyüköztürk S., and Uyguner Z. O. (2015) Idiopathic angioedema with F12 mutation: is it a new entity? Ann. Allergy. Asthma Immunol. 114, 154–156 10.1016/j.anai.2014.11.018 [DOI] [PubMed] [Google Scholar]
- 10. de Maat S., Björkqvist J., Suffritti C., Wiesenekker C. P., Nagtegaal W., Koekman A., van Dooremalen S., Pasterkamp G., de Groot P. G., Cicardi M., Renné T., and Maas C. (2016) Plasmin is a natural trigger for bradykinin production in patients with hereditary angioedema with factor XII mutations. J. Allergy Clin. Immunol. 138, 1414–1423.e9 10.1016/j.jaci.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 11. Ivanov I., Matafonov A., Sun M.-F., Mohammed B. M., Cheng Q., Dickeson S. K., Kundu S., Verhamme I. M., Gruber A., McCrae K., and Gailani D. (2019) A mechanism for hereditary angioedema with normal C1 inhibitor: an inhibitory regulatory role for the factor XII heavy chain. Blood 133, 1152–1163 10.1182/blood-2018-06-860270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Maat S., Clark C. C., Boertien M., Parr N., Sanrattana W., Hofman Z. L. M., and Maas C. (2019) Factor XII truncation accelerates activation in solution. J. Thromb. Haemost. 17, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scheffel J., Mahnke N., Hofman Z., de Maat S., Wu J., Bonnekoh H., Pengelly R., Ennis S., Holloway J., Church M., Maurer M., Maas C., and Krause K. (2019) 10th Congress of International Society of Systemic Auto-Inflammatory Diseases (ISSAID). Pediatr. Rheumatol. 17, 18 10.1186/s12969-019-0313-x [DOI] [Google Scholar]
- 14. Mahnke N. A., Scheffel J., Wu J., de Maat S., Maas C., Hofman Z. L., Ennis S., Holloway J. W., Pengelly R. J., and Maurer M. K. K. (2018) 45th Annual Meeting of the Arbeitsgemeinschaft Dermatologische Forschung (ADF) Zurich, Switzerland, March 7–10, 2018. Exp. Dermatol. 27, e2–e106 10.1111/exd.13486 [DOI] [Google Scholar]
- 15. Dors D. M., Nuijens J. H., Huijbregts C. C., and Hack C. E. (1992) A novel sensitive assay for functional factor XII based on the generation of kallikrein–C1-inhibitor complexes in factor XII-deficient plasma by glass-bound factor XII. Thromb. Haemost. 67, 644–648 10.1055/s-0038-1648516 [DOI] [PubMed] [Google Scholar]
- 16. Seidah N. G., Sadr M. S., Chrétien M., and Mbikay M. (2013) The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J. Biol. Chem. 288, 21473–21481 10.1074/jbc.R113.481549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jean F., Stella K., Thomas L., Liu G., Xiang Y., Reason A. J., and Thomas G. (1998) α1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc. Natl. Acad. Sci. U.S.A. 95, 7293–7298 10.1073/pnas.95.13.7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Maat S., van Dooremalen S., de Groot P. G., and Maas C. (2013) A nanobody-based method for tracking factor XII activation in plasma. Thromb. Haemost. 110, 458–468 10.1160/TH12-11-0792 [DOI] [PubMed] [Google Scholar]
- 19. Stavrou E. X., Fang C., Bane K. L., Long A. T., Naudin C., Kucukal E., Gandhi A., Brett-Morris A., Mumaw M. M., Izadmehr S., Merkulova A., Reynolds C. C., Alhalabi O., Nayak L., Yu W.-M., et al. (2018) Factor XII and uPAR upregulate neutrophil functions to influence wound healing. J. Clin. Invest. 128, 944–959 10.1172/JCI92880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Renné T., and Stavrou E. X. (2019) Roles of factor XII in innate immunity. Front. Immunol. 10, 2011 10.3389/fimmu.2019.02011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xue Y., Bodin C., and Olsson K. (2012) Crystal structure of the native plasminogen reveals an activation-resistant compact conformation. J. Thromb. Haemost. 10, 1385–1396 10.1111/j.1538-7836.2012.04765.x [DOI] [PubMed] [Google Scholar]
- 22. Johnson D. J., Langdown J., and Huntington J. A. (2010) Molecular basis of factor IXa recognition by heparin-activated antithrombin revealed by a 1.7-A structure of the ternary complex. Proc. Natl. Acad. Sci. U.S.A. 107, 645–650 10.1073/pnas.0910144107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chinnaraj M., Planer W., and Pozzi N. (2018) Structure of coagulation factor II: molecular mechanism of thrombin generation and development of next-generation anticoagulants. Front. Med. (Lausanne) 5, 281 10.3389/fmed.2018.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. South K., Luken B. M., Crawley J. T., Phillips R., Thomas M., Collins R. F., Deforche L., Vanhoorelbeke K., and Lane D. A. (2014) Conformational activation of ADAMTS13. Proc. Natl. Acad. Sci. U.S.A. 111, 18578–18583 10.1073/pnas.1411979112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clark C. C., Mebius M. M., de Maat S., Tielens A. G. M., de Groot P. G., Urbanus R. T., Fijnheer R., Hazenberg B. P. C., van Hellemond J. J., and Maas C. (2018) Truncation of ADAMTS13 by plasmin enhances its activity in plasma. Thromb. Haemost. 118, 471–479 10.1055/s-0038-1627460 [DOI] [PubMed] [Google Scholar]
- 26. Pathak M., Wilmann P., Awford J., Li C., Hamad B. K., Fischer P. M., Dreveny I., Dekker L. V., and Emsley J. (2015) Coagulation factor XII protease domain crystal structure. J. Thromb. Haemost. 13, 580–591 10.1111/jth.12849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dementiev A., Silva A., Yee C., Li Z., Flavin M. T., Sham H., and Partridge J. R. (2018) Structures of human plasma β-factor XIIa cocrystallized with potent inhibitors. Blood Adv. 2, 549–558 10.1182/bloodadvances.2018016337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pathak M., Manna R., Li C., Kaira B. G., Hamad B. K., Belviso B. D., Bonturi C. R., Dreveny I., Fischer P. M., Dekker L. V., Oliva M. L. V., and Emsley J. (2019) Crystal structures of the recombinant β-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics. Acta Crystallogr. D Struct. Biol. 75, 578–591 10.1107/S2059798319006910 [DOI] [PubMed] [Google Scholar]
- 29. Beringer D. X., and Kroon-Batenburg L. M. (2013) The structure of the FnI-EGF-like tandem domain of coagulation factor XII solved using SIRAS. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 69, 94–102 10.1107/S1744309113000286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Castellino F. J., and Beals J. M. (1987) The genetic relationships between the kringle domains of human plasminogen, prothrombin, tissue plasminogen activator, urokinase, and coagulation factor XII. J. Mol. Evol. 26, 358–369 10.1007/BF02101155 [DOI] [PubMed] [Google Scholar]
- 31. Konings J., Govers-Riemslag J. W., Philippou H., Mutch N. J., Borissoff J. I., Allan P., Mohan S., Tans G., Ten Cate H., and Ariëns R. A. (2011) Factor XIIa regulates the structure of the fibrin clot independently of thrombin generation through direct interaction with fibrin. Blood 118, 3942–3951 10.1182/blood-2011-03-339572 [DOI] [PubMed] [Google Scholar]
- 32. Mitchell J. L., Lionikiene A. S., Georgiev G., Klemmer A., Brain C., Kim P. Y., and Mutch N. J. (2016) Polyphosphate colocalizes with factor XII on platelet-bound fibrin and augments its plasminogen activator activity. Blood 128, 2834–2845 10.1182/blood-2015-10-673285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ravon D. M., Citarella F., Lubbers Y. T., Pascucci B., and Hack C. E. (1995) Monoclonal antibody F1 binds to the kringle domain of factor XII and induces enhanced susceptibility for cleavage by kallikrein. Blood 86, 4134–4143 10.1182/blood.V86.11.4134.bloodjournal86114134 [DOI] [PubMed] [Google Scholar]
- 34. Citarella F., te Velthuis H., Helmer-Citterich M., and Hack C. E. (2000) Identification of a putative binding site for negatively charged surfaces in the fibronectin type II domain of human factor XII: an immunochemical and homology modeling approach. Thromb. Haemost. 84, 1057–1065 10.1055/s-0037-1614171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.