Abstract
Considering the role of proto-oncogene c-Met (c-Met) in oncogenesis, we examined the effects of the metastasis suppressor, N-myc downstream-regulated gene-1 (NDRG1), and two NDRG1-inducing thiosemicarbazone-based agents, Dp44mT and DpC, on c-Met expression in DU145 and Huh7 cells. NDRG1 silencing without Dp44mT and DpC up-regulated c-Met expression, demonstrating that NDRG1 modulates c-Met levels. Dp44mT and DpC up-regulated NDRG1 by an iron-dependent mechanism and decreased c-Met levels, c-Met phosphorylation, and phosphorylation of its downstream effector, GRB2-associated binding protein 1 (GAB1). However, incubation with Dp44mT and DpC after NDRG1 silencing or silencing of the receptor tyrosine kinase inhibitor, mitogen-inducible gene 6 (MIG6), decreased c-Met and its phosphorylation, suggesting NDRG1- and MIG6-independent mechanism(s). Lysosomal inhibitors rescued the Dp44mT- and DpC-mediated c-Met down-regulation in DU145 cells. Confocal microscopy revealed that lysosomotropic agents and the thiosemicarbazones significantly increased co-localization between c-Met and lysosomal-associated membrane protein 2 (LAMP2). Moreover, generation of c-Met C-terminal fragment (CTF) and its intracellular domain (ICD) suggested metalloprotease-mediated cleavage. In fact, Dp44mT increased c-Met CTF while decreasing the ICD. Dp44mT and a γ-secretase inhibitor increased cellular c-Met CTF levels, suggesting that Dp44mT induces c-Met CTF levels by increasing metalloprotease activity. The broad metalloprotease inhibitors, EDTA and batimastat, partially prevented Dp44mT-mediated down-regulation of c-Met. In contrast, the ADAM inhibitor, TIMP metallopeptidase inhibitor 3 (TIMP-3), had no such effect, suggesting c-Met cleavage by another metalloprotease. Notably, Dp44mT did not induce extracellular c-Met shedding that could decrease c-Met levels. In summary, the thiosemicarbazones Dp44mT and DpC effectively inhibit oncogenic c-Met through lysosomal degradation and metalloprotease-mediated cleavage.
Keywords: cancer therapy, cancer biology, molecular pharmacology, receptor tyrosine kinase, hepatocyte growth factor (HGF), c-Met, hepatocyte growth factor receptor, MET proto-oncogene receptor tyrosine kinase (MET), N-myc downstream-regulated gene-1 (NDRG1), thiosemicarbazone
Introduction
Receptor tyrosine kinases (RTKs)5 are important regulators of proliferation, differentiation, cell survival, cell metabolism, cell migration, and cell-cycle progression (1). Despite their critical roles in normal physiology, these molecules have been shown to potently drive cancer progression when they are either mutated or amplified in tumor cells (2, 3).
The hepatocyte growth factor receptor (also termed MET or c-Met) has been shown to regulate multiple oncogenic pathways, including signal transducer and activator of transcription 3 (STAT3), Ras/Raf/MEK/ERK, PI3K/AKT/mTOR, and NF-κB pathways, leading to cancer cell migration and invasion (4, 5), proliferation (5), and survival (6, 7). Studies have shown that c-Met is involved in cross-talk with several key oncogenic pathways that synergistically drive cancer progression and development of drug resistance (8, 9). Considering the crucial role c-Met plays in cancer, it is vital to target this molecule for effective management of malignant tumors, particularly drug-resistant cancers.
The regulation of c-Met expression is complex, with the protein being reported to be processed by internalization, endocytosis, and lysosomal degradation (10), but also via the proteasomal pathway (11). More recently, it has been suggested that membrane-bound metalloproteinases of the a disintegrin and metalloproteinase (ADAM) family (12) could be involved in c-Met degradation at the extracellular level (13). This is because these proteases are major mediators of cell-surface protein shedding during tumor progression, which interferes with intracellular signaling (13). In fact, ADAM10 is the protease regulating spontaneous c-Met shedding, as its knockdown leads to increased cell-surface levels of c-Met in vitro (14). ADAM17 has also been proposed as another mediator involved in shedding (15). Additionally, c-Met has been recently been described to be cleaved by metalloproteases both intracellularly (e.g. in lysosomes) and also at the extracellular level at the plasma membrane by metalloproteases (e.g. ADAM10, ADAM17, etc.), leading to the generation of a C-terminal fragment (CTF) (14–17).
Novel agents of the di-2-pyridylketone thiosemicarbazone (DpT) series, namely di-2-pyridylketone 4,4-dimethyl-3-thiosemicarbazone (Dp44mT) and di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC; Fig. 1A), are members of a class of clinically trialed anti-cancer agents (18) that potently suppress tumor growth and metastasis in vitro and in vivo in a variety of cancer types (19–25). Our recent studies have demonstrated that these agents effectively down-regulate EGFR and other ErbB family receptors (i.e. HER2 and HER3) and inhibit their activities in pancreatic and colon cancers (26, 27). Furthermore, these agents inhibit STAT3 (28), Ras/ERK (29), AKT (30, 31), and NF-κB (30, 32) pathways, which are downstream signaling pathways of c-Met (3). These findings suggest the novel DpT analogues may modulate c-Met protein levels in cancer.
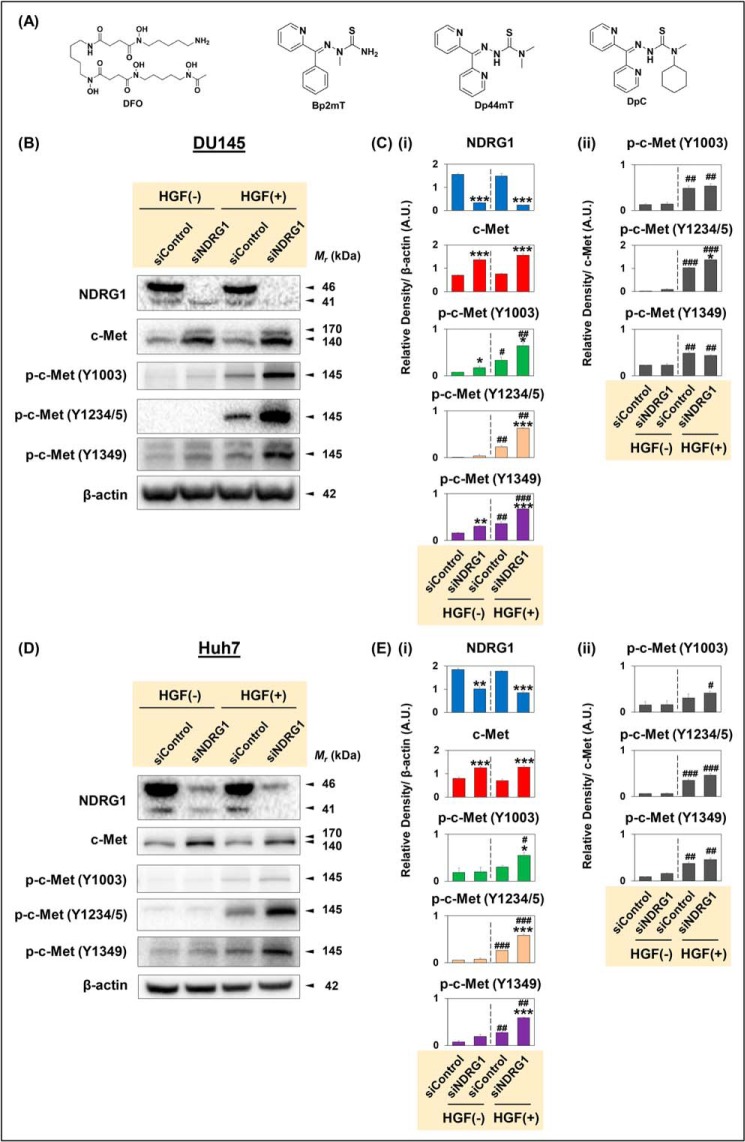
Figure 1.
A, line drawings of the chemical structures of the agents used in this investigation, and B–E, NDRG1 silencing increases c-Met protein expression and its phosphorylation levels at multiple sites (Y1003, Y1234/5, and Y1349). A, line drawings of the chemical structures of DFO, Bp2mT, Dp44mT, and DpC. DU145 (B and C) and Huh7 (D and E) cells were incubated with Opti-MEM media containing nontargeting, negative control siRNA (siControl) or siRNA specific for NDRG1 (siNDRG1) as per “Experimental procedures.” The cells were then incubated in normal media alone without hepatocyte growth factor (HGF(−)) or media containing HGF (50 ng/ml; HGF (+)) for 15 min at 37 °C. Densitometric analyses of Western blottings for DU145 (C) and Huh7 (E) cells are mean ± S.E. (n = 3) normalized to β-actin, which was used as a protein-loading control. Phosphorylation levels for c-Met are presented both relative to β-actin and as a ratio of total c-Met protein levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to the respective siControl. #, p < 0.05; ## p < 0.01; and ###, p < 0.001 relative to the HGF(−)-treated cells.
In this investigation, we examined the effect of the DpT agents on c-Met expression in prostate cancer and hepatocellular carcinoma (HCC) cells. Previous investigations have shown c-Met exerts oncogenic effects in both prostate cancer (33, 34) and HCC (35). A putative metastasis suppressor, N-myc downstream-regulated gene-1 (NDRG1), is an important molecular target of the DpT class of agents (20, 36). Therefore, we initially aimed to examine whether NDRG1 affects c-Met expression in these latter cancer types and the molecular nature of the effect.
Herein, for the first time, we demonstrate that Dp44mT and DpC, which are effective metal chelators (37–39) and desferrioxamine (DFO; Fig. 1A), a well-characterized iron chelator (40), potently down-regulate c-Met expression in prostate cancer and HCC cells. Furthermore, silencing NDRG1 in cancer cells significantly increased c-Met protein expression and phosphorylation, but it did not inhibit the chelator-mediated down-regulation of c-Met. Three lysosomotropic agents, which interrupt lysosomal degradation, significantly inhibited the ability of the chelators to decrease c-Met levels, suggesting the role of lysosomal processing. Incubation with Dp44mT increased the levels of c-Met CTF, a metalloprotease cleavage product of c-Met (14–16), indicating that Dp44mT may induce higher rates of c-Met shedding in cancer cells. However, no increase in extracellular shedding of c-Met was observed after incubation with Dp44mT, suggesting a role for intracellular shedding and lysosomal degradation.
Results
NDRG1 silencing increases c-Met protein levels in prostate cancer and HCC cells
The initial aim of this investigation was to examine the effect of NDRG1 silencing on c-Met expression in cancer cells. To elucidate these aims, the NDRG1 gene was silenced in c-Met-expressing DU145 and Huh7 cells using siNDRG1 and compared with the nontargeting, negative control siRNA (siControl). Then, cells were either incubated in media alone or media containing HGF (50 ng/ml) for 15 min at 37 °C, which induces c-Met phosphorylation (41). Protein levels of c-Met and its phosphorylated forms were then examined using Western blotting (Fig. 1, B–E).
First, in DU145 cells, upon silencing NDRG1, NDRG1 protein levels were significantly reduced relative to the siControl in the presence or absence of HGF (Fig. 1, B and Ci). Two major bands were detected for NDRG1 protein at 41 and 46 kDa in DU145 cells, as reported previously (42). Notably, only the top NDRG1 band was silenced after treatment with siNDRG1, as demonstrated previously by our laboratory (27, 31, 43). A second NDRG1 siRNA (i.e. NDRG1 II) demonstrated the same effect (Fig. S1, A and B). These two NDRG1 bands are the result of differential processing (44) and represent NDRG1 truncated at the N terminus and the full-length NDRG1 isoform, respectively (42, 43). Notably, HGF stimulation did not significantly affect NDRG1 expression relative to nonstimulated HGF(−) cells (Fig. 1, B and Ci).
Examining the c-Met protein, two major bands were detected at ∼140 and 170 kDa (Fig. 1B). The 170-kDa band represents the single-chain c-Met precursor, whereas the ∼140-kDa band corresponds to the β-subunit of the mature 190-kDa c-Met protein (45, 46). Both the 140- and 170-kDa bands of c-Met were significantly increased in siNDRG1 cells versus siControl cells in the presence or absence of HGF (Fig. 1, B and Ci). Furthermore, HGF stimulation did not significantly change the total c-Met protein levels when compared with nonstimulated HGF(−) cells (Fig. 1, B and Ci).
Assessing c-Met phosphorylated at Y1003, Y1234/5, or Y1349, as expected, HGF stimulation significantly increased phosphorylation of c-Met at all sites compared with nonstimulated DU145 cells (Fig. 1, B and Ci). The phosphorylation at Y1003 results in recruitment of casitas b-lineage lymphoma leading to c-Met ubiquitination and endocytosis and its degradation via a lysosome-mediated pathway (10). In contrast, phosphorylation at Y1234/5 and Y1349 is involved in recruitment and activation of downstream effector molecules, such as growth factor receptor-bound protein 2 (Grb2), Grb2-associated-binding protein 1 (Gab1), phospholipase C, and Src (3). Importantly, NDRG1 silencing significantly increased phosphorylation of c-Met at Y1003, Y1234/5, and Y1349 relative to the respective siControl in the presence of HGF (Fig. 1, B and Ci).
Investigating the effect of NDRG1 silencing on c-Met expression in Huh7 HCC cells, NDRG1 protein was significantly reduced in siNDRG1 cells relative to siControl cells in the presence or absence of HGF (Fig. 1, D and Ei). Similarly to DU145 cells, NDRG1 silencing significantly increased c-Met expression in Huh7 cells (Fig. 1, D and Ei). Upon HGF stimulation of siControl cells, there was a significant increase in phosphorylation at Y1234/5 and Y1349, and a slight increase at Y1003 versus the respective HGF(−) conditions (Fig. 1, D and Ei). Notably, NDRG1 silencing significantly increased c-Met phosphorylation at all sites relative to the respective siControl in the presence of HGF (Fig. 1, D and Ei). Similarly, the second NDRG1 siRNA (i.e. NDRG1 II) demonstrated the same effect on increasing c-Met phosphorylation at all sites in DU145 and Huh7 cells (Fig. S1, A and B).
There are two potential mechanisms regarding how silencing NDRG1 could increase c-Met phosphorylation: 1) by increasing total c-Met protein levels; and/or 2) by increasing phosphorylation at the specific sites examined. To clarify which of the above mechanisms is responsible for the increase in c-Met phosphorylation, ratios of phosphorylated c-Met to the total c-Met were calculated (Fig. 1, Cii and Eii). Indeed, the ratios were generally significantly increased for all phosphorylation sites by HGF stimulation under both siControl and siNDRG1 conditions in DU145 and Huh7 cells relative to unstimulated cells (Fig. 1, Cii and Eii). In both cell types, the phosphorylated to total c-Met ratio at all sites was generally not significantly altered after siNDRG1 relative to the siControl after HGF stimulation (Fig. 1, Cii and Eii). The only exception to this was for the p-c-Met Y1234/5 to total c-Met ratio in DU145 cells, where a slight but significant increase was observed for siNDRG1 versus the siControl (Fig. 1Cii). Thus, in general, the increased total c-Met after siNDRG1 treatment observed with HGF stimulation could be responsible for the increased phosphorylation at Y1003, Y1234/5, and Y1349 found under these conditions in both cell types (Fig. 1, Ci and Ei). Together, the results in Fig. 1 demonstrate that NDRG1 silencing markedly increased c-Met expression and phosphorylation at Y1003, Y1234/5, and Y1349 in DU145 or Huh7 cells.
Thiosemicarbazones down-regulate c-Met protein in prostate cancer and HCC cells
Our laboratory has demonstrated that novel anti-cancer agents of the DpT class (i.e. Dp44mT or DpC; Fig. 1A) that potently up-regulate NDRG1 and its phosphorylation (21) can inhibit oncogenic pathways mediated by tyrosine kinases via both NDRG1-dependent and -independent mechanism(s) depending upon the thiosemicarbazone examined (26, 28). Therefore, it was important to examine whether the DpT agents could affect c-Met expression and activation in DU145 and Huh7 cells (Fig. 2).
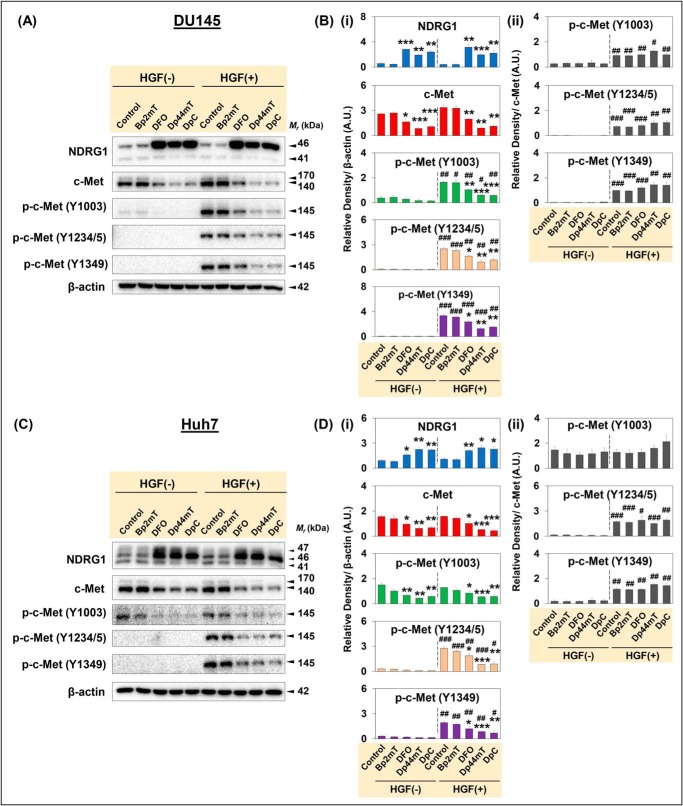
Figure 2.
Thiosemicarbazones, Dp44mT and DpC, markedly down-regulate c-Met protein expression and its phosphorylation. DU145 (A and B) and Huh7 (C and D) cells were incubated with Control medium or medium containing Bp2mT (5 μm; negative control), DFO (100 μm), Dp44mT (5 μm), or DpC (5 μm) for 24 h at 37 °C. These cells were then incubated in media alone without HGF (HGF −) or media containing HGF (50 ng/ml; HGF +) for 15 min at 37 °C. Densitometric analyses of Western blottings for DU145 (Bi and ii) and Huh7 (Di and ii) cells are mean ± S.E. (n = 3) normalized to the protein-loading control, β-actin. Phosphorylation levels for c-Met are presented both relative to β-actin and as a ratio of total c-Met protein levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to the respective Control. #, p < 0.05; ##, p < 0.01; ###, p < 0.001 relative to the HGF(−)-treated cells.
In these studies, the DpT analogue, 2-benzoylpyridine 2-methyl-3-thiosemicarbazone (Bp2mT; Fig. 1A; 5 μm), was used as a negative control, as it has a similar chemical structure to Dp44mT and DpC (Fig. 1A), but it was specifically designed not to bind metal ions (47). Cells were incubated with Bp2mT (5 μm), Dp44mT (5 μm), DpC (5 μm), or DFO (100 μm; Fig. 1A) for 24 h at 37 °C. Notably, DFO was used at a higher concentration due to its poor membrane permeability compared with the lipophilic, highly membrane-permeable agents, Dp44mT and DpC (19, 30, 48, 49). Following this incubation, cells were either incubated with medium containing HGF (50 ng/ml) or medium alone for 15 min at 37 °C.
DU145 cells
Examining DU145 prostate cancer cells, DFO, Dp44mT and DpC markedly and significantly up-regulated NDRG1 expression compared with the Control in the presence or absence of HGF (Fig. 2, A and Bi). Incubation of cells with the chelators, DFO, Dp44mT, or DpC, up-regulated the 46-kDa full-length NDRG1 isoform, but not the 41-kDa truncated form (Fig. 2, A and Bi), as shown previously (21). Upon examining c-Met expression in the presence or absence of HGF, DFO significantly decreased the 170-kDa c-Met precursor and the 140-kDa β-chain of c-Met (Fig. 2, A and Bi). Furthermore, Dp44mT and DpC were more potent than DFO and markedly and significantly reduced levels of the 170- and 140-kDa c-Met versus the Control (Fig. 2, A and Bi).
Considering the down-regulation of total c-Met with the agents, which could be due to increased degradation, it was considered important to also examine c-Met phosphorylation at Y1003, which is involved in lysosome-mediated degradation of c-Met (10) and also Y1234/5 and Y1349 that are involved in downstream c-Met signaling (3) (Fig. 2, A and B). Of note, phosphorylation at Y1003, Y1234/5, and Y1349 was significantly increased by HGF stimulation in DU145 cells relative to all the corresponding HGF(−) treatments (Fig. 2, A and Bi). However, DFO and particularly Dp44mT and DpC significantly reduced c-Met phosphorylation at all three sites examined versus the HGF-stimulated Control (Fig. 2, A and Bi).
To clarify which mechanism is responsible for the reduction in c-Met phosphorylation (i.e. decreased c-Met total protein or a specific inhibition of phosphorylation), ratios of phosphorylated c-Met to the total c-Met were calculated (Fig. 2Bii). The ratios were significantly increased for the three phosphorylation sites by HGF stimulation under all conditions versus the unstimulated cells (Fig. 2Bii). This was expected for the HGF-incubated Control cells, as the total c-Met level remained similar after HGF stimulation relative to the HGF(−) Control (Fig. 2, A and Bii). However, the phosphorylated to total c-Met ratio was not significantly decreased in DFO, Dp44mT, or DpC incubated cells relative to the Control (Fig. 2Bii). This indicates that the decreased c-Met phosphorylation in the presence of these agents could be due to a reduction of total c-Met protein levels rather than a specific inhibition of phosphorylation.
Huh7 cells
Similar effects of the agents on c-Met expression were also observed in Huh7 cells (Fig. 2, C and Di and ii). In fact, DFO, Dp44mT, or DpC markedly and significantly up-regulated NDRG1 in the presence or absence of HGF (Fig. 2, C and Di). By examining c-Met levels in Huh7 cells, incubation with DFO, Dp44mT, or DpC induced a significant decrease in c-Met protein levels relative to the Control in the presence or absence of HGF (Fig. 2, C and Di).
Of note, HGF stimulation had no significant effect on c-Met Y1003 phosphorylation in Huh7 cells relative to their untreated counterparts (Fig. 2, C and Di). Considering Y1003 phosphorylation is important for endocytosis and lysosomal degradation (10, 50), this observation in Huh7 cells may indicate perturbed c-Met regulation and trafficking, irrespective of ligand binding (50). However, DFO, Dp44mT, and DpC significantly decreased Y1003 phosphorylation relative to the Control in both nonstimulated and HGF-stimulated cells (Fig. 2, C and Di).
Phosphorylation at Y1234/5 and Y1349 was significantly increased with HGF stimulation under all conditions relative to the HGF(−) treatment (Fig. 2, C and Di). However, DFO, Dp44mT, and DpC markedly and significantly reduced their levels versus the HGF-stimulated Control (Fig. 2, C and Di). The ratio of c-Met phosphorylated at Y1003 to the total c-Met was not significantly altered by HGF stimulation in Huh7 cells (Fig. 2Dii). In contrast, the ratios for Y1234/5 and Y1349 were significantly increased with HGF incubation relative to the nonstimulated cells under all conditions (Fig. 2, C and Dii). Similarly to DU145 cells, DFO, Dp44mT, and DpC did not significantly reduce the ratios for all three phosphorylation sites relative to the HGF-stimulated Control in Huh7 cells (Fig. 2Dii). This suggests the decrease in c-Met phosphorylation in Huh7 cells is due to the decrease in total c-Met induced by the agents, rather than specific inhibition of phosphorylation.
Together, these results in Fig. 2 demonstrate that DFO, Dp44mT, and DpC markedly decrease c-Met levels in DU145 and Huh7 cells. Phosphorylation of c-Met at Y1003, Y1234/5, and Y1349 was also markedly reduced by these latter agents in both cell types. This decrease in phosphorylation was due to the reduction in total c-Met induced by DFO, Dp44mT, and DpC.
Considering these studies using DU145 prostate cancer and Huh7 hepatoma cells, four additional cell types were investigated, including SCC25 oral squamous cell carcinoma, Hep3B hepatoma, PANC-1 pancreatic carcinoma, and MDA-MB-231 breast cancer cells (Fig. S1, C–F). For all cell types, Dp44mT and DpC decreased c-Met protein expression, suggesting these agents act similarly in a variety of tumor cell types (Fig. S1, C–F).
Thiosemicarbazones inhibit phosphorylation of Gab1, a downstream effector of c-Met
After binding to HGF, c-Met undergoes auto-phosphorylation at its C-terminal bidentate substrate-binding site, which is composed of Y1349 and Y1356 (51). Various downstream effector molecules are then recruited to these latter phosphorylation sites to activate downstream signaling (51, 52). One of these downstream effectors is Gab1, which is phosphorylated upon binding to activated c-Met (53).
Phosphorylated Gab1 bound to c-Met further recruits docking proteins, such as PI3K, Src homology 2 domain-containing phosphatase 2 (SHP2), etc. (5). These docking proteins together with various downstream effectors induce multiple oncogenic signaling pathways, such as Ras/Raf/MEK/ERK (54), PI3K/AKT (55), and cell division control protein 42 (Cdc42)/Rac1-mediated cytoskeletal reorganization (56). Hence, Gab1 protein levels and its phosphorylation were examined after incubation of cells with the agents (Fig. S2).
Similarly to Fig. 2, DU145 and Huh7 cells were incubated with the chelators DFO (100 μm), Dp44mT (5 μm), or DpC (5 μm) for 24 h at 37 °C, and Bp2mT (5 μm) was used as a negative control. Following this incubation, cells were incubated with medium containing HGF (50 ng/ml) or medium alone for 15 min at 37 °C (Fig. S2).
Examining total Gab1 expression in DU145 and Huh7 cells, its levels were not significantly affected by any of the agents nor HGF stimulation (Fig. S2, A–D). In the absence of HGF, DFO, Dp44mT, and DpC only slightly decreased Y307-phosphorylated Gab1 (p-Gab1 Y307) and the p-Gab1 Y307/Gab1 ratio relative to the Control. In contrast, p-Gab1 Y307 levels were markedly and significantly increased by HGF in the Control- and Bp2mT-treated DU145 and Huh7 cells versus the nonstimulated cells (Fig. S2, A–D). In both cell types in the presence of HGF, incubation with DFO, Dp44mT, or DpC, significantly decreased p-Gab1 (Y307) levels versus the Control, whereas Bp2mT had no significant effect. The ratio of p-Gab1 (Y307) to total Gab1 also demonstrated a significant increase by HGF in the Control and Bp2mT-treated DU145 and Huh7 cells (Fig. S2, B–D). However, this ratio was significantly reduced by DFO, Dp44mT, or DpC, relative to the respective HGF-stimulated Control cells. These data indicated that these latter agents decreased Gab1 Y307 phosphorylation, rather than total Gab1 levels.
Binding of cellular iron is partially required for Dp44mT and DpC to down-regulate c-Met protein
One of the major mechanisms of action of Dp44mT and DpC is via their potent binding to iron in tumor cells (19, 39). Hence, it was important to investigate whether the effect of Dp44mT and DpC on c-Met expression was due to their ability to bind cellular iron.
In these studies, DU145 cells were incubated with either DFO (100 μm), Dp44mT (5 μm), DpC (5 μm), or Bp2mT (5 μm) as a negative control (Fig. S3A) for 24 h at 37 °C. For comparison, cells were also incubated with the Fe complexes of these chelators, namely DFO–Fe (1:1), Dp44mT–Fe (1:2) and DpC–Fe (1:2), as well as Bp2mT (5 μm) together with FeCl3 (2.5 μm). Additionally, Fe(III) (as FeCl3) was also implemented as a relevant control at the same Fe(III) concentrations that were used to prepare the Fe(III) complex of DFO ([Fe] = 100 μm) relative to the Fe(III) complexes of Dp44mT or DpC ([Fe] = 2.5 μm). These stoichiometries of ligand to iron take account of their coordination properties (37, 39, 48). This ensures saturation of the coordination sites of DFO and the thiosemicarbazones with Fe(III), which is using ligand/iron ratios of 1:1 and 2:1, respectively (Fig. S3A) (37, 39, 48).
By examining NDRG1 expression, incubation of DU145 cells with DFO, Dp44mT, and DpC significantly up-regulated the 46-kDa band (Fig. S3, A and B). In contrast to either DFO, Dp44mT, or DpC, their respective iron complexes (i.e. DFO–Fe, Dp44mT–Fe, and DpC–Fe) did not have any significant effect on NDRG1 expression (Fig. S3, A and B). This is because the agents can no longer bind cellular iron pools, as their ligating sites are occupied with the metal ion, preventing depletion of intracellular iron. These results demonstrate the regulation of NDRG1 by iron deprivation is expected, as NDRG1 is well-known to be iron-regulated in vitro and in vivo (20, 36, 57, 58) and acts as a positive control for cellular Fe levels.
The levels of c-Met were not affected by Bp2mT, whereas DFO, Dp44mT, or DpC again significantly reduced its expression (Fig. S3, A and B). Incubation with iron alone or iron bound to Bp2mT or DFO did not significantly alter c-Met expression (Fig. S3, A and B). However, whereas the addition of Fe to Dp44mT and DpC to form their Fe complexes prevented the up-regulation of NDRG1 observed with the ligands alone, the down-regulation of c-Met was partially attenuated (Fig. S3, A and B). In fact, although Dp44mT–Fe and DpC–Fe still induced a significant decrease in c-Met versus the Control, their effects were less marked than the ligands. Similar results to those observed above in DU145 cells were also obtained using Huh7 cells (data not shown).
In conclusion, the results in Fig. S3, A and B indicate DFO, Dp44mT, or DpC down-regulates c-Met, and in the case of DFO, this could be effectively rescued by the addition of Fe to the ligand to form a complex to prevent chelation of cellular Fe pools. In contrast, the Dp44mT–Fe and DpC–Fe complexes still possessed some activity at down-regulating c-Met. This may be related to the fact that these Fe complexes are also redox-active (19, 37, 38, 47), with redox species potentially playing a role in the activity observed. In contrast, the DFO–Fe complex is redox-inactive (48, 59), with only the ability of the ligand to deplete cellular iron being active in decreasing c-Met levels (Fig. S3, A and B).
To assess whether the effects of Dp44mT and DpC on c-Met were mediated by their redox activity upon binding metal ions (37, 39), further studies were performed using the anti-oxidant, N-acetylcysteine (NAC), which supplements GSH and markedly prevents the anti-proliferative effects of these agents (38). Using both cell types, the incubation of NAC (5 mm) with the ligands for 24 h at 37 °C, which inhibits redox stress induced by these agents (38, 47), had no significant effect on rescuing c-Met versus the ligands alone (Fig. S3, A–F). This finding suggested that generation of redox species by the Fe complexes formed within cells was not key to the ability of the ligands to down-regulate c-Met (Fig. S3, C–F).
Thiosemicarbazones up-regulate inhibitors of c-Met, namely NDRG1 and MIG6, but down-regulate c-Met by NDRG1- and MIG6-independent mechanism(s)
The studies above (Fig. 2) demonstrate that DFO and the thiosemicarbazones markedly up-regulate NDRG1, as shown previously (20, 36). Our laboratory has recently demonstrated that NDRG1 can down-regulate the RTK, EGFR, through a mechanism mediated by its ability to bind to, and stabilize, the tumor suppressor, mitogen-inducible gene-6 (MIG6), which is an EGFR inhibitor that mediates its degradation (27). Considering these facts and that MIG6 has been demonstrated to decrease c-Met activity by an indirect mechanism (60), it was hypothesized that NDRG1 and MIG6 could play a role in the observed chelator-mediated down-regulation of c-Met. This was intriguing, because like NDRG1 (36), MIG6 is also potently induced after cellular Fe chelation (27) potentially via a hypoxia-inducible factor-α–dependent mechanism (27, 61).
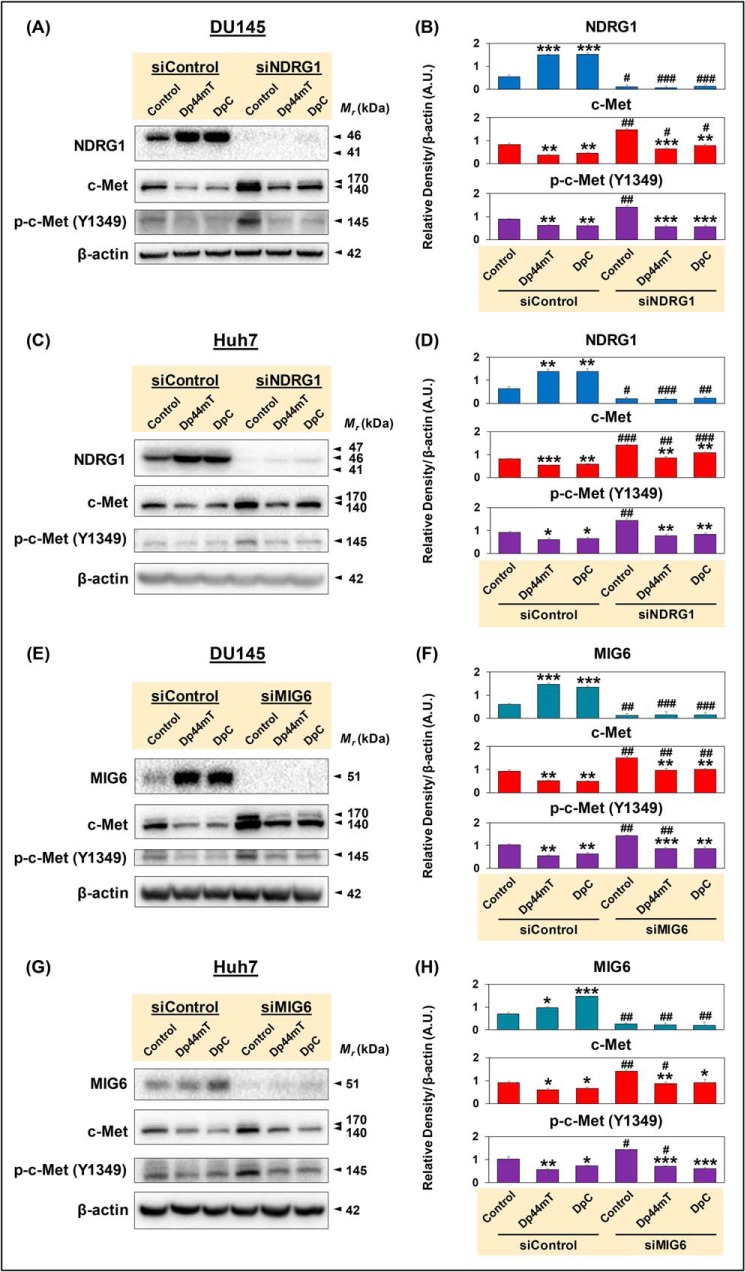
To investigate the role of NDRG1 and MIG6 in the chelator-mediated down-regulation of c-Met, DU145 or Huh7 cells were treated with NDRG1 siRNA (Fig. 3, A–D) or MIG6 siRNA (Fig. 3, E–H) for 48 h at 37 °C and then incubated with Control medium or this medium containing Dp44mT or DpC at 5 μm for 24 h at 37 °C. Western blot analysis was then performed. Examining the siControl-treated cells, Dp44mT and DpC significantly increased NDRG1 (Fig. 3, A–D) and MIG6 (Fig. 3, E–H) versus the Control in both cell types. In contrast, after silencing NDRG1 or MIG6, there was a pronounced and significant decrease in their levels under all conditions, with the thiosemicarbazones failing to up-regulate expression.
Figure 3.
Thiosemicarbazones up-regulate inhibitors of c-Met, namely NDRG1 and MIG6, but down-regulate c-Met by NDRG1- and MIG6-independent mechanism(s). DU145 or Huh7 cells were incubated with Opti-MEM media containing nontargeting, negative control siRNA (siControl) or siRNA specific for NDRG1 (siNDRG1; A–D) or MIG6 (siMIG6; E–H) in the absence of HGF and then incubated with Control medium, or this medium containing Dp44mT or DpC at 5 μm for 24 h at 37 °C. Western blot analysis was then performed. Densitometric analyses are mean ± S.E. (n = 3) normalized to the protein-loading control, β-actin. *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to the respective Control. #, p < 0.05; ##, p < 0.01; ###, p < 0.001 relative to siControl samples.
Assessing c-Met and its phosphorylation after siControl treatment (Fig. 3, A–H) demonstrated that the thiosemicarbazones significantly decreased total c-Met and p-c-Met (Y1349) versus the Control, as demonstrated in Fig. 2. After siNDRG1 or siMIG6 treatment, it was notable that Control levels of both c-Met and p-c-Met were significantly increased relative to the Control in the siControl treatment (Fig. 3, A–H). This suggested that the metastasis suppressor and tumor-suppressive roles of NDRG1 and MIG6 expression, respectively, are involved in down-regulating c-Met levels and activation under Control conditions.
However, irrespective of either NDRG1 or MIG6 siRNA treatment, the thiosemicarbazones still significantly decreased both c-Met and p-c-Met relative to the NDRG1 or MIG6 siRNA-treated Controls (Fig. 3, A–H). The fact that the thiosemicarbazones could still significantly down-regulate c-Met when either NDRG1 or MIG6 was silenced indicates a mechanism(s) independent of these molecules. To examine this, the decrease in c-Met or p-c-Met was expressed as a percentage of the relevant control after incubation with the thiosemicarbazones. Irrespective of siNDRG1 or siMIG6 treatment, there was no significant difference in the effect of the thiosemicarbazones to that observed after incubation with the siControl. Similar results were obtained when the cells were incubated with HGF (data not shown). As described previously for NDRG1 silencing using a second NDRG1 siRNA (i.e. NDRG1 II siRNA; Fig. S1A, B), a second MIG6 siRNA (siMIG6 II) resulted in very similar results to those demonstrated for MIG6 siRNA in Fig. 3, E–H, i.e. up-regulation of c-Met and p-c-Met (Y1349; Fig. S4, A and B). These controls argue against off-target effects of the siRNAs.
Examining the effect of the thiosemicarbazones on Gab1 and p-Gab1 (Y307) in the presence and absence of NDRG1 and MIG6 silencing in DU145 and Huh7 cells, it was demonstrated that there was no appreciable effect on total Gab1 under any conditions relative to the Control (Fig. S5, A–H). Assessing p-Gab1 (Y307) in the siControl treatments, Dp44mT and DpC slightly decreased its levels in DU145 cells, whereas Dp44mT had no appreciable effect, and DpC increased p-Gab1 (Y307) in Huh7 cells relative to the Control (Fig. S5, A–H). For DU145 or Huh7 cells incubated with siNDRG1, Dp44mT or DpC resulted in no marked effect on p-Gab1 (Y307) relative to the control (Fig. S5, A–D). Incubation of DU145 cells or Huh7 cells with siMIG6 followed by treatment with Dp44mT and DpC led to a decrease of p-Gab1 (Y307) in DU145 cells; although there was a slight increase in p-Gab1 (Y307) levels with Dp44mT and no change using DpC in Huh7 cells, relative to the Control (Fig. S5, A–H).
In summary, the thiosemicarbazones were able to down-regulate c-Met and p-c-Met even when either MIG6 or NDRG1 was markedly silenced (Fig. 3, A–H), implicating another mechanism independent of these molecules.
Thiosemicarbazones do not modulate mRNA levels of HSP90AA1 but decrease MET mRNA in Huh7 cells
Next, as an initial assessment of the mechanism involved in the down-regulation of c-Met protein levels after incubation with thiosemicarbazones, studies then examined MET mRNA expression (Fig. 4, A–D). In these studies, NDRG1 mRNA levels were examined as a positive control for cellular iron depletion along with HSP90AA1. The expression of the latter was assessed as it was demonstrated by others that cellular iron chelation decreases HSP90AA1 mRNA expression (62). The HSP90AA1 mRNA translates to HSP90, which is known to act as a chaperone for c-Met protein (63). Furthermore, alterations in HSP90 activity can modulate c-Met expression (63). Hence, we hypothesized that the down-regulation of c-Met after incubation of cells with chelators (Figs. 2 and 3) could be due to the effect of these agents on HSP90 expression at the mRNA and/or protein levels.
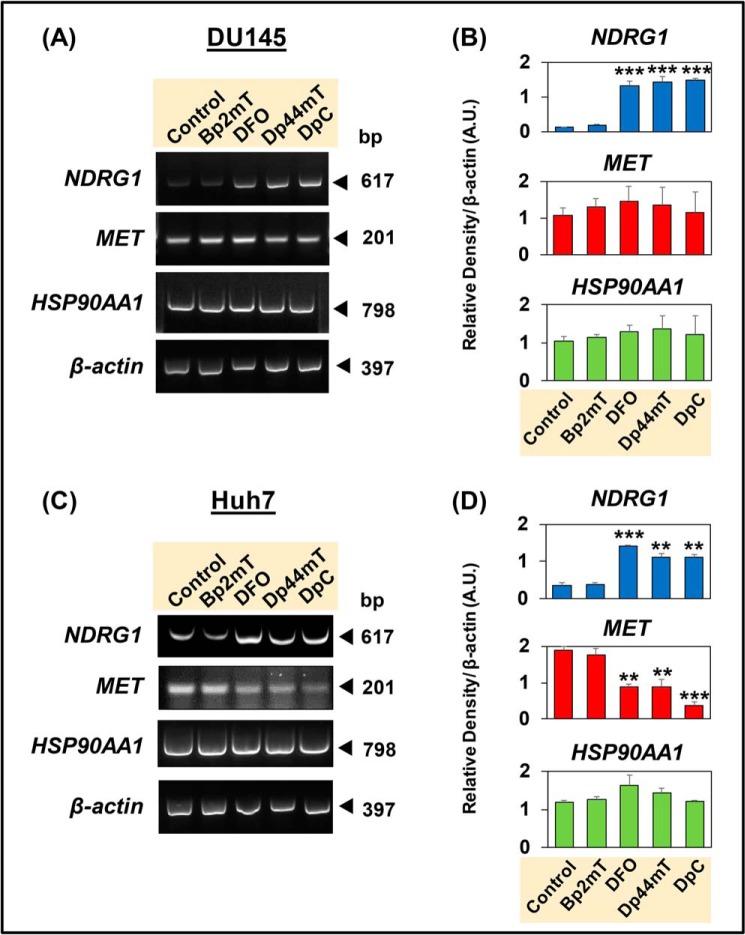
Figure 4.
Thiosemicarbazones, Dp44mT and DpC, decrease MET mRNA levels in Huh7 cells, but not in DU145 cells. DU145 (A and B) and Huh7 (C and D) cells were incubated with Control medium or this medium containing either Bp2mT (5 μm; negative control), DFO (100 μm), Dp44mT (5 μm), or DpC (5 μm) for 24 h at 37 °C. Densitometric analyses of NDRG1, MET, and HSP90AA1 mRNA levels from RT-PCR are shown as the mean ± S.E. (n = 3) normalized to β-actin mRNA. *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to the Control.
In these experiments, DU145 cells (Fig. 4, A and B) and Huh7 cells (Fig. 4, C and D) were incubated for 24 h at 37 °C with either Control medium or medium containing Bp2mT (5 μm), DFO (100 μm), Dp44mT (5 μm), or DpC (5 μm) in the absence of HGF. HGF was not implemented, as the effect of chelators on c-Met protein expression was independent of incubation with this cytokine (Fig. 2). Examining NDRG1 mRNA levels in DU145 and Huh7 cells, incubation with DFO, Dp44mT, or DpC led to a significant increase relative to the Control, whereas the negative control, Bp2mT, had no significant effect (Fig. 4, A–D). The up-regulation of NDRG1 confirms cellular iron depletion by DFO, Dp44mT, and DpC and acts as a positive control (20, 36). On the other hand, MET mRNA levels were not decreased in DU145 cells by Dp44mT or DpC relative to the Control (Fig. 4, A and B). In contrast, for Huh7 cells, there was a significant reduction in MET mRNA levels by these agents compared with the Control (Fig. 4, C and D). In both cell types, none of the compounds had a significant effect on HSP90AA1 mRNA levels relative to the Control (Fig. 4, A–D). In summary, the results in Fig. 4 indicate that down-regulation of c-Met protein by the chelators in Huh7 cells (Fig. 2, C and D) could be partially due to a reduction in MET mRNA levels.
Considering that HSP90, the protein encoded by HSP90AA1, acts as a chaperone for c-Met, its levels were also examined after incubation of cells with the chelators (Fig. S6). Assessing HSP90 protein levels in DU145 and Huh7 cells after incubation with the agents, its levels were not significantly altered in both cell types (Fig. S6). These results agree with the mRNA data (Fig. 4, A–D), indicating that DFO, Dp44mT, or DpC do not modulate HSP90 expression at the mRNA and protein levels, and thus do not decrease c-Met through this mechanism.
Dp44mT and DpC increase lysosomal co-localization of c-Met protein, with the decrease in c-Met levels being rescued by three lysosomotropic agents in DU145 cells
As the effect of the thiosemicarbazones was due, in part, to cellular iron chelation (Fig. S3, A and B), it was critical to understand the mechanism of how these agents decrease c-Met expression (Figs. 2 and 3). The decrease in c-Met protein levels has been attributed to changes in either lysosomal (64) or ubiquitin-dependent proteasomal degradation (11). First, regarding lysosomal processing, HGF binding to c-Met activates and phosphorylates it at Y1003 (Fig. 2, A and B), which then recruits casitas b-lineage lymphoma, leading to internalization and lysosome-mediated degradation of c-Met (10). Second, internalized c-Met could also be degraded by the proteasome-mediated pathway (64). These two mechanisms appear to function together, as inhibiting proteasomal activity has been reported to block c-Met internalization after HGF stimulation (64). Hence, internalization and degradation of c-Met could involve multiple mechanisms.
Both Dp44mT and DpC, which markedly down-regulate c-Met (Figs. 2 and 3), are known to affect lysosomes in cancer cells (18, 38, 65). Hence, it was hypothesized the thiosemicarbazones may enhance c-Met degradation via a lysosome-mediated mechanism. To investigate this, the cellular co-localization of the classical late endosomal/lysosomal marker, LAMP2 (66), with c-Met was investigated using confocal immunofluorescence microscopy (Fig. 5, A–D). In these studies, the well-established lysosomotropic agents that disrupt lysosomal acidification and degradation, namely ammonium chloride (NH4Cl), methylamine (MA), and chloroquine (CLQ) (67–69), were also utilized to examine their ability to rescue c-Met levels after incubation with Dp44mT and DpC (Fig. 5, A–D).
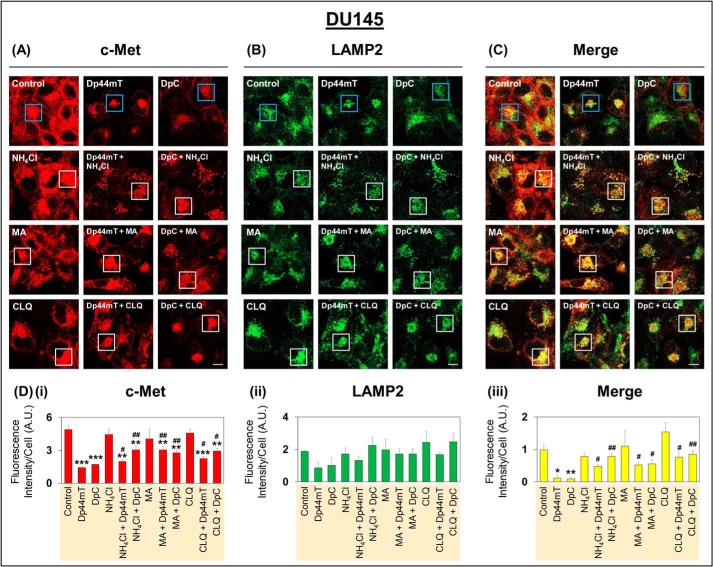
Figure 5.
Confocal immunofluorescence microscopy demonstrates that incubation of DU145 cells with lysosomotropic agents rescues down-regulation of c-Met expression mediated by Dp44mT and DpC. A–C, DU145 cells were incubated with Control medium or this medium containing either Dp44mT (5 μm), DpC (5 μm), NH4Cl (15 mm), MA (15 mm), CLQ (50 μm), alone or in combination for 24 h at 37 °C. Confocal immunofluorescence microscopy was then performed examining the following: A, c-Met (red); B, LAMP2 (green); and C, merge between A and B. Blue boxes in images indicate Control, Dp44mT, and DpC relative to when these conditions were also treated with lysosomotropic agents, which are indicated by white boxes. All images were taken with a ×63 objective and at the same acquisition settings using Zeiss Zen Blue software. Images are representative from three experiments. D, quantitative analyses of pixel intensities of c-Met (i), LAMP2 (ii), and co-localization of c-Met and LAMP2 (iii) were performed using ImageJ software. All quantitative analyses are mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to the Control. #, p < 0.05, and ##, p < 0.01 relative to either Dp44mT or DpC alone. Pixel intensity and co-localization analysis utilized a total of 20–30 cells. Scale bar represents 10 μm and is the same for all images.
To test the involvement of the lysosome-mediated degradation pathway, DU145 cells were incubated with either Control medium or medium containing Dp44mT (5 μm) or DpC (5 μm) in the absence or presence of NH4Cl (15 mm), MA (15 mm), or CLQ (50 μm) for 24 h at 37 °C (Figs. 5 and 6). These concentrations of lysosomotropic agents have been previously shown to be effective in terms of inhibiting lysosomal acidification and biological mechanisms dependent on this process (27, 68, 69). Optimization experiments demonstrated that the lysosomotropic agents were not cytotoxic over the incubation used, as demonstrated by cellular morphology (Fig. 5, A–C).
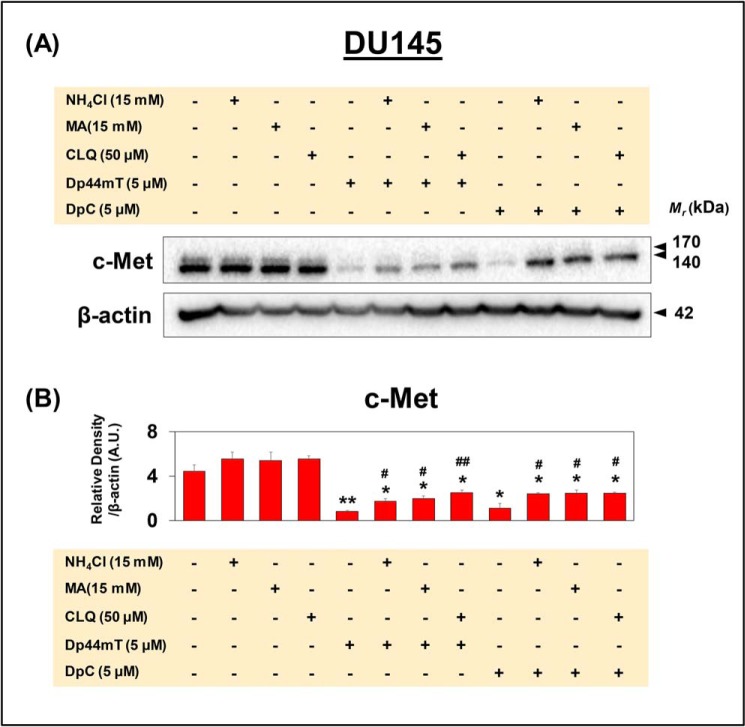
Figure 6.
Western blot analysis demonstrates that incubation of DU145 cells with lysosomotropic agents partially rescues down-regulation of c-Met expression mediated by Dp44mT and DpC. A and B, DU145 cells were incubated as in Fig. 5, and Western blot analysis for c-Met and β-actin was performed. B, densitometric analyses are mean ± S.E. (n = 3) normalized to the protein-loading control, β-actin. *, p < 0.05; and **, p < 0.01 are relative to the Control. #, p < 0.05, and ##, p < 0.01 are relative to either Dp44mT or DpC.
Examining c-Met expression by confocal microscopy demonstrated that incubation of cells with either of the three lysosomotropic agents in the absence of Dp44mT or DpC had no significant effect on its levels (Fig. 5, A and Di). Blue boxes in the images in Fig. 5, A–C, indicate Control, Dp44mT, and DpC relative to when these conditions were also treated with lysosomotropic agents, which are indicated by white boxes. As shown previously (Figs. 2 and 3), c-Met levels were significantly decreased by Dp44mT and DpC relative to the Control (Fig. 5, A and Di). It was also of note that Dp44mT and DpC resulted in tighter intracellular clustering of c-Met and LAMP2 into foci (Fig. 5, A and B), suggesting altered intracellular trafficking. Assessing cells incubated with Dp44mT or DpC and also the lysosomotropic agents, this resulted in significantly higher c-Met levels relative to cells treated with thiosemicarbazones alone (Fig. 5, A and Di). These results suggested that inhibition of lysosomal activity by the lysosomotropic agents at least partly reversed the effect of thiosemicarbazones on down-regulating c-Met. Co-localization of c-Met was then examined relative to the LAMP2-defined late endosomal/lysosomal compartment (Fig. 5B) (66).
Because confocal microscopy images cells on a single lateral focal plane, only proteins in the same location are co-localized (70). Co-localization analysis between c-Met (red) and LAMP2 (green) demonstrated that incubation of cells with either Dp44mT or DpC significantly decreased the co-localization of these two proteins (yellow) in the merge (Fig. 5, C and Diii). This effect is probably due to the markedly decreased c-Met levels mediated by these agents after a 24-h incubation (Fig. 5Di). Upon incubation of DU145 cells with either of the three lysosomotropic agents alone, there was no significant change observed in co-localization compared with the Control. However, co-incubation of cells with either Dp44mT or DpC and either of the three lysosomotropic agents induced a significant increase in co-localization between c-Met and LAMP2 when compared with cells incubated with Dp44mT or DpC alone (Fig. 5, C and Diii). These latter results suggested the lysosomotropic agents inhibited c-Met degradation by the thiosemicarbazones, leading to increased co-localization with LAMP2. Furthermore, Dp44mT and DpC induced c-Met lysosomal localization for its degradation.
Assessing c-Met expression in the studies with lysosomotropic agents and thiosemicarbazones by Western blot analysis (Fig. 6, A and B) resulted in similar results to those demonstrated by confocal microscopy (Fig. 5, A and Di). Indeed, c-Met levels were significantly decreased by Dp44mT or DpC, whereas cells incubated with Dp44mT or DpC together with the lysosomotropic agents expressed significantly higher c-Met versus the thiosemicarbazones alone (Fig. 6, A and B).
Collectively, these results using confocal and Western blot analysis of DU145 cells indicated that inhibition of lysosome activity at least partially reversed the effect of these agents on decreasing c-Met levels. This finding suggested an additional mechanism also contributed to the decrease of c-Met in DU145 cells after incubation with thiosemicarbazones.
In contrast to DU145 cells, incubation of Huh7 cells with these lysosomotropic agents did not significantly rescue the ability of Dp44mT or DpC to decrease c-Met, as demonstrated by both confocal microscopy and Western blot analysis (Fig. S7, A–C). This indicated another processing pathway in Huh7 cells was responsible for the thiosemicarbazone-mediated decrease in total c-Met. This could be explained by the decreased MET mRNA levels observed after incubation with these agents (Fig. 4, C and D). Of interest, a difference in c-Met trafficking between DU145 and Huh7 cells was also suggested by the differential response of c-Met phosphorylation at Y1003 between these cell types (cf. Fig. 2, A and B with C and D). This phosphorylation mediates c-Met endocytosis and lysosome-mediated degradation (10).
Proteasomal inhibition up-regulates levels of the 55-kDa C-terminal fragment of c-Met, which is increased in the presence of Dp44mT
The investigations above in Figs. 5 and 6 demonstrate some role of lysosomal degradation in the decrease of c-Met expression after incubation of DU145 cells with Dp44mT and DpC, but not Huh7 cells. Considering this, other mechanisms of c-Met processing were then examined, namely the proteasomal pathway and c-Met shedding (10). It has been demonstrated that c-Met can be ubiquitinated and directly degraded by the proteasome (11). Additionally, c-Met can be shed by intracellular (e.g. in lysosomes) and extracellular cleavage via the metalloproteases, such as ADAM10 and ADAM17, leading to the 55-kDa c-Met CTF (14–17). The c-Met CTF is then cleaved by the γ-secretase complex to generate a 50-kDa c-Met intracellular domain (c-Met ICD), which is then degraded by the proteasome (10, 15).
Previous studies have demonstrated that inhibition of the proteasome using the well-characterized inhibitor, MG132, resulted in increased levels of the 50-kDa c-Met ICD and the 55-kDa c-Met CTF (15, 71). Furthermore, agents that increase c-Met cleavage up-regulate the 55-kDa c-Met CTF (15, 71). To examine the effect of proteasomal inhibition in the presence and absence of Dp44mT on c-Met expression, DU145 and Huh7 cells were incubated for 8 h at 37 °C with either Control medium alone or medium containing either Dp44mT (5 μm), MG132 (2.5 or 5 μm), EDTA (1 mm), or these agents in combination. Notably, EDTA was used as a potent metalloprotease inhibitor, which is known to inhibit ADAM10 and the activity of other metalloproteases (72–74).
As shown previously, Dp44mT significantly decreased the expression of the mature and precursor isoforms of c-Met at 140- and 170-kDa relative to the Control (Fig. 7, Ai and B). Upon longer exposures of the blots (Fig. 7, Aii and iii), the 50- and 55-kDa c-Met ICD and CTF bands, respectively, became visible under all conditions and were significantly decreased by Dp44mT relative to the Control (Fig 7, Aiii, C, and D). The low levels of these two later bands in the Control and Dp44mT-treated groups suggests they may be rapidly processed relative to the high-molecular-weight precursor and mature forms (10). Upon incubation with MG132 (2.5 μm) alone, there was a significant decrease in the mature and precursor c-Met isoforms and also a pronounced and significant increase in the c-Met ICD and c-Met CTF isoforms at 50- and 55-kDa, respectively (Fig. 7, A–D), as shown previously (15).
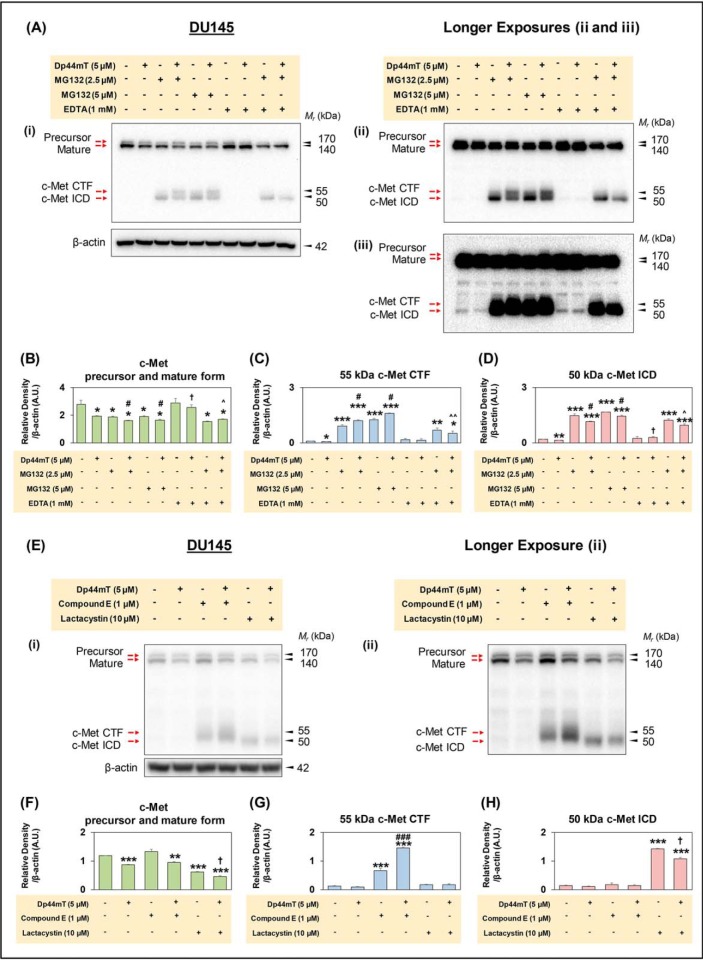
Figure 7.
Proteasome inhibition prevents processing and degradation of c-Met CTF and c-Met ICD, and Dp44mT increases the levels of c-Met CTF in DU145 cells. A, DU145 cells were incubated with Control medium or this medium containing either Dp44mT (5 μm), MG132 (2.5 or 5 μm), EDTA (1 mm) alone, or in combination for 8 h at 37 °C. Western blot analysis was then performed, and three images of the blots taken at different exposure times (Ai–Aiii) are shown for optimal presentation of different c-Met isoforms, namely the c-Met precursor, mature form, c-Met CTF, and c-Met ICD. Densitometric analyses of the c-Met precursor and mature form (B), 55-kDa c-Met CTF (C), and 50-kDa c-Met ICD (D) are mean ± S.E. (n = 3) normalized to a protein-loading control, β-actin. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 relative to the Control. #, p < 0.05 relative to cells incubated with MG132 (2.5 or 5 μm). †, p < 0.05 relative to cells incubated with Dp44mT. ∧, p < 0.05; ∧∧, p < 0.01 relative to cells incubated with MG132 and Dp44mT. E, DU145 cells were incubated with Control medium or this medium containing either Dp44mT (5 μm), compound E (1 μm), lactacystin (10 μm) alone, or in combination for 8 h at 37 °C. Ei–Eii are shown for optimal presentation of the different c-Met isoforms, namely the c-Met precursor, mature form, c-Met CTF, and c-Met ICD. Densitometric analyses of c-Met precursor and mature form (F), 55-kDa c-Met CTF (G), and 50-kDa c-Met ICD (H). Results are mean ± S.E. (n = 3) normalized to the protein-loading control, β-actin. **, p < 0.01, and ***, p < 0.001 relative to the Control. #, p < 0.05, and ###, p < 0.01 relative to cells incubated with Compound E (1 μm). †, p < 0.05 relative to cells incubated with lactacystin (10 μm) alone.
Incubation of cells with another proteasomal inhibitor, lactacystin, had a similar effect on the mature and precursor c-Met isoforms (Fig. 7, E and F). Co-incubation of cells with both Dp44mT and MG132 resulted in a further slight but significant decrease in the mature and precursor isoforms of c-Met relative to MG132 alone (Fig. 7, Ai and B). Furthermore, co-incubation with Dp44mT and MG132 resulted in a significant increase in the 55-kDa c-Met CTF and a significant decrease in the c-Met ICD relative to MG132 alone (Fig. 7, Ai, ii, C, and D). Hence, this observation suggests that Dp44mT induces increased c-Met CTF fragment levels, potentially by increased c-Met cleavage by metalloproteases, such as ADAM10 and ADAM17 (14–17). Similar results to MG132 (2.5 μm) in the absence and presence of Dp44mT were also observed at an MG132 concentration of 5 μm, except the levels of the c-Met CTF and c-Met ICD were higher (Fig. 7, Ai, ii, and B).
Hence, Dp44mT increased the c-Met CTF in the presence of the proteasomal inhibitor versus the inhibitor alone (Fig. 7, A and C), suggesting Dp44mT-generated CTF is degraded by the proteasome. However, this did not occur upon incubation of DU145 cells with the lysosomotropic agents (Fig. S8). In fact, the addition of Dp44mT or DpC to the lysosomotropic agents decreased CTF versus the lysosomotropic agents alone (Fig. S8). In contrast, the lysosomotropic agents alone without Dp44mT markedly increased c-Met CTF when compared with the untreated Control. This indicates c-Met CTF generated under Control conditions is degraded by the lysosome (Fig. S8), as demonstrated by others (17).
Broad metalloprotease inhibitor, EDTA, potently rescues the ability of Dp44mT to decrease c-Met expression
The CTF is generated by increased c-Met cleavage by metalloproteases (14–17). As such, studies next examined the effect of incubation of DU145 cells with the broad-spectrum metalloprotease inhibitor EDTA (72–74) in the presence and absence of Dp44mT (Fig. 7, A–D). Incubation with EDTA alone had no significant effect on the expression of the mature, precursor, c-Met CTF and c-Met ICD isoforms of c-Met relative to the Control (Fig. 7, A–D). In contrast, EDTA co-incubated with Dp44mT totally and significantly prevented the loss of the mature and precursor isoforms of c-Met relative to that observed with Dp44mT alone (Fig. 7, Ai and B). Furthermore, c-Met CTF levels were not significantly changed, although the c-Met ICD levels were slightly but significantly increased after incubation with Dp44mT and EDTA relative to Dp44mT alone (Fig. 7, Aiii, C, and D).
Co-incubation of EDTA and MG132 (2.5 μm) had a similar effect as MG132 alone, significantly decreasing the mature and precursor c-Met isoforms (Fig. 7, Ai and B) and significantly (p < 0.001–0.01) increasing the c-Met CTF and c-Met ICD levels (Fig. 7, Aii, C, and D) relative to the Control. Co-incubation of EDTA, MG132 (2.5 μm), and Dp44mT resulted in significantly higher levels of c-Met mature and precursor isoforms relative to MG132 (2.5 μm) and Dp44mT (Fig. 7, Ai and B). In contrast, c-Met CTF and c-Met ICD were significantly lower after co-incubation of EDTA, MG132 (2.5 μm), and Dp44mT versus MG132 (2.5 μm) and Dp44mT (Fig. 7, Aii, C, and D). This latter observation suggests that EDTA may prevent the cleavage of c-Met via inhibition of metalloprotease activities (e.g. ADAM10 and -17, etc.), which are known to cleave c-Met resulting in the CTF (14–16) and then the ICD via γ-secretase processing (10, 15).
Similar results to those found in DU145 cells (Fig. 7, A–D) were also observed in Huh7 cells (Fig. S9), where down-regulation of the mature and precursor forms of c-Met and up-regulation of c-Met ICD isoforms were demonstrated after incubation with these agents. It was also of interest that when EDTA was added with Dp44mT it significantly rescued the loss of c-Met precursor and mature forms when compared with Dp44mT alone (Fig. S9, A and B). Again, as for DU145 cells (Fig. 7, A and B), this suggested some involvement of metalloprotease activity in the Dp44mT-induced decrease of c-Met levels.
In contrast to DU145 cells where two well-defined c-Met ICD and CTF bands were identified (Fig. 7, A and B), only a single well-defined band at ∼50-kDa was observed in Huh7 cells (Fig. S9, A and C), which could represent the c-Met ICD. Only a shadow at ∼55 kDa was present in Huh7 cells (Fig. S9, A–C), which may represent low levels of the c-Met CTF. However, this could not be reliably quantitated. Upon the addition of MG132 that prevents CTF and ICD degradation, a clear band representative of the CTF was still not apparent (Fig. S9A). This was in contrast to the marked increase in CTF and ICD observed in DU145 cells (Fig. 7, A, C, and D). Together, these findings indicate that in Huh7 cells the generation of c-Met CTF and ICD appeared less than that of DU145 cells.
In conclusion, the results in Fig. 7, A–D, demonstrate that Dp44mT decreases the mature and precursor isoforms and concurrently leads to increased c-Met CTF levels in DU145 cells, suggesting cleavage by a metalloprotease (e.g. ADAM10, -17, etc.) resulting in c-Met shedding (14, 16). Using Huh7 cells (Fig. S9), Dp44mT induced similar down-regulation of the mature and precursor forms of c-Met, although metalloprotease degradation appeared less marked. As such, further studies examining metalloprotease activity on c-Met processing were focused on the DU145 cell type.
Compound E, a γ-secretase inhibitor, increases the 55-kDa c-Met CTF and this is increased in the presence of Dp44mT
To further assess the ability of Dp44mT to enhance c-Met CTF levels, observed in the presence of MG132 (Fig. 7, A and C), a γ-secretase inhibitor, Compound E, and a proteasomal inhibitor, lactacystin, were utilized (Fig. 7, E–H). In fact, it was previously demonstrated by others that Compound E selectively increases c-Met CTF (17). This occurs through the ability of Compound E to inhibit γ-secretase from processing c-Met CTF to ICD, whereas proteasomal inhibitors, such as MG132 or lactacystin, increase c-Met ICD levels via proteasomal inhibition (17). In this study, DU145 cells were incubated with Compound E (1 μm) or lactacystin (10 μm) and the absence or presence of Dp44mT (5 μm) for 8 h at 37 °C.
Examining the levels of c-Met precursor and the mature form, Dp44mT, significantly reduced their levels, whereas Compound E alone had no significant effect when compared with the Control (Fig. 7, Ei and ii, and F). Notably, incubation with both Compound E and Dp44mT did not rescue the decrease in c-Met observed with Dp44mT alone. However, a significant decrease in the c-Met precursor and mature form was observed in cells treated with lactacystin alone, which was further decreased by the addition of Dp44mT (Fig. 7, Ei and ii, and F).
Assessing the 55-kDa c-Met CTF levels, there was a significant increase in DU145 cells incubated with Compound E relative to the control. Intriguingly, in cells incubated with Dp44mT and Compound E, there was a marked and significant increase in c-Met CTF levels compared with cells incubated with Compound E alone (Fig. 7, Eii and G). These results indicate that Dp44mT induced the generation of c-Met CTF potentially through increased metalloprotease activity, strongly supporting the results in Fig. 7, A and C. In contrast, the intensity of the CTF was very low in cells treated with lactacystin (Fig. 7, Ei and ii, and G), as reported previously (17), and also after lactacystin and Dp44mT.
Furthermore, investigating the 50-kDa c-Met ICD, its levels were not affected by either Dp44mT, Compound E, or Dp44mT and Compound E (Fig. 7, Ei and ii, and H). In contrast, lactacystin markedly and significantly increased the c-Met ICD relative to the Control. Co-incubation of cells with lactacystin and Dp44mT also caused a pronounced and significant increase in the ICD relative to the Control (Fig. 7, Ei and ii, and H). However, the extent of the increase was significantly decreased relative to cells incubated with lactacystin alone (Fig. 7, Ei and ii, and H). This latter effect was similar to the response observed in Fig. 7, A and D, where c-Met ICD was reduced by Dp44mT in the presence of MG132.
Overall, these results from Fig. 7, E–H, support and extend the data presented in Fig. 7, A–D. Indeed, Dp44mT markedly increased c-Met CTF levels in the presence of the γ-secretase inhibitor, Compound E, suggesting increased metalloprotease-mediated shedding of c-Met by Dp44mT.
Metalloprotease inhibitor, batimastat, limits the Dp44mT-mediated decrease in cellular c-Met levels, whereas TIMP-3 has no effect
Considering that the broad-spectrum metalloprotease inhibitor, EDTA (75), markedly rescued c-Met expression in the presence of Dp44mT (Fig. 7, A and B, and Fig. S9, A and B), another well-characterized metalloprotease inhibitor, batimastat (76, 77), was used to examine its effect on c-Met levels. Incubation of DU145 cells with Dp44mT (5 μm) for 8 h at 37 °C led to a significant reduction in c-Met expression relative to the Control, whereas batimastat (5 μm) alone had no appreciable effect (Fig. 8, A and B). In contrast, co-incubation of Dp44mT (5 μm) and batimastat (5 μm), largely prevented the down-regulation of c-Met, leading to significantly higher c-Met levels compared with Dp44mT alone (Fig. 8, A and B). These results suggest the ability of batimastat to inhibit metalloprotease activity prevents the loss of c-Met.
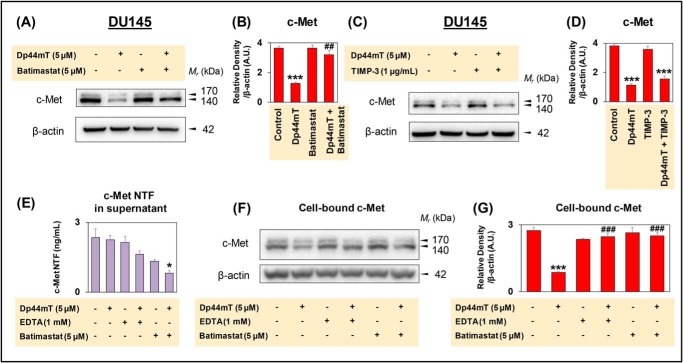
Figure 8.
Broad spectrum metalloprotease inhibitor, batimastat, rescues c-Met expression in DU145 cells. A, DU145 cells were incubated with Control medium or this medium containing either Dp44mT (5 μm) or batimastat (5 μm) alone, or in combination for 8 h at 37 °C. B, densitometric analyses for c-Met as mean ± S.E. (n = 3) normalized to β-actin. ***, p < 0.001 relative to the Control. ##, p < 0.01 relative to cells incubated with Dp44mT alone. C, DU145 cells were incubated with Control medium or this medium containing either Dp44mT (5 μm) or TIMP-3 (1 μg/ml) alone or in combination for 8 h at 37 °C. D, densitometric analyses for c-Met as mean ± S.E. (n = 3) normalized to β-actin. ***, p < 0.001 relative to the Control. E and F, DU145 cells were incubated with Control medium or this medium containing either Dp44mT (5 μm), EDTA (1 mm), or batimastat (5 μm) alone, or in combination for 8 h at 37 °C. E, extracellular culture medium was collected, and an ELISA was performed, as described under “Experimental procedures,” for the measurement of c-Met NTF levels. Results are presented as mean ± S.E. (n = 3). *, p < 0.05 relative to the Control (F and G). Cell lysates were prepared from the same cells in E for the measurement of cell-bound c-Met levels via Western blot analysis. G, densitometric analyses of the data in F are presented as mean ± S.E. (n = 3) normalized to β-actin. ***, p < 0.001 relative to the Control; ###, p < 0.001 relative to cells incubated with Dp44mT alone.
Studies then examined the effect of TIMP-3, which has been demonstrated to be an inhibitor of metalloprotease activity that potently inhibits c-Met shedding by the antibody, DN30, via blocking ADAM10 proteolytic activity (16). Incubation of DU145 cells for 8 h at 37 °C with TIMP-3 at a concentration previously shown to markedly inhibit ADAM10 activity (i.e. 1 μg/ml (16)) had no significant effect on c-Met expression relative to the Control (Fig. 8, C and D). Furthermore, co-incubation of TIMP-3 (1 μg/ml) and Dp44mT (5 μm) for 8 h at 37 °C did not prevent the ability of Dp44mT to reduce c-Met expression (Fig. 8, C and D). These results suggested that ADAM10 may not mediate the c-Met down-regulation by Dp44mT. Collectively, these data in Figs. 7, A and B, and 8, A–D, and Fig. S9, A and B suggest that Dp44mT decreases c-Met expression via inducing metalloprotease activity that can be inhibited by batimastat.
Dp44mT does not induce extracellular shedding of c-Met
The results in Fig. 7, A–D, demonstrated that Dp44mT increases c-Met CTF in the presence of MG132, suggesting Dp44mT induces extracellular cleavage by metalloproteases resulting in shedding of the N-terminal fragment (NTF) of c-Met. Considering these results, it was deemed important to examine the shedding of c-Met into the extracellular medium, and in this case, an ELISA method was utilized that detects c-Met NTF (78).
In these studies, DU145 cells were incubated for 8 h at 37 °C with either Dp44mT (5 μm), EDTA (1 mm), batimastat (5 μm), Dp44mT (5 μm), and EDTA (1 mm) or Dp44mT (5 μm), and batimastat (5 μm; Fig. 8, E–G). In contrast to our expectation, incubation of cells with Dp44mT alone did not increase release of the c-Met NTF into the extracellular medium, with a slight decrease being observed relative to the Control (Fig. 8E). This was unexpected considering that the corresponding total cellular levels of c-Met decreased after Dp44mT (Fig. 8, F and G), as observed previously (Figs. 2, A and B, and 3, A, B, E, and F). Hence, c-Met NTF was not being shed extracellularly, despite that c-Met CTF was being generated after incubation with Dp44mT (Fig. 7, A and E). As c-Met CTF is known to be generated after shedding (14, 16), this therefore must occur intracellularly, and because lysosomes are involved in c-Met internalization and degradation (Figs. 5 and 6) (64), this may potentially occur in these vesicles. This would account for the observed decrease in total cellular c-Met, the lack of c-Met NTF release into the extracellular medium, and the generation of the c-Met CTF that occurs via intracellular or extracellular shedding (14–17).
Examining EDTA and batimastat, both agents decreased c-Met NTF release into the extracellular medium potentially due to their ability to prevent metalloprotease activity (Fig. 8E). Moreover, EDTA and batimastat did not have any significant effect on corresponding total cellular levels of c-Met relative to the Control (Fig. 8, F and G). The addition of Dp44mT to EDTA or batimastat further decreased release of c-Met NTF into the extracellular medium (Fig. 8E) and significantly prevented the corresponding loss of total cell c-Met induced by Dp44mT alone (Fig. 8, F and G).
In conclusion, the results in Fig. 8, E–G, demonstrate in Control DU145 cells that c-Met NTF is released into the extracellular medium, and this can be decreased by the metalloprotease inhibitors EDTA or batimastat. Whereas Dp44mT decreases total cellular c-Met, this is not due to increased extracellular c-Met NTF shedding. This finding implies that the decrease in total cellular c-Met by Dp44mT is mediated by increased intracellular processing. Because c-Met CTF generation can also occur after metalloprotease cleavage in intracellular vesicles (17), and because Dp44mT stimulated both c-Met CTF levels (Fig. 7, A and C) and lysosomal degradation of c-Met (Figs. 5 and 6), it can be suggested that the shedding and processing of c-Met occurs in lysosomes. The decrease in c-Met NTF release observed with EDTA and batimastat after Dp44mT treatment could be due to their potent inhibition of metalloprotease cleavage stimulated by incubation of cells with Dp44mT.
However, for Huh7 cells, the results suggest that lysosomal activity (Fig. S7) is not involved in the c-Met down-regulation by the thiosemicarbazones. The decrease observed in MET mRNA after Dp44mT treatment (Fig. 4, C and D) and, to a lesser extent, the metalloprotease-mediated intracellular processing of c-Met after Dp44mT (Fig. S9) could be important mediators of the decrease of c-Met in Huh7 cells.
Multiple mechanisms of c-Met down-regulation observed in DU145 and Huh7 cells are also differentially identified in four other tumor cell types
The studies above have identified three major mechanisms of down-regulation of c-Met in DU145 and Huh-7 cells induced by thiosemicarbazones, namely c-Met lysosomal degradation, metalloprotease-induced c-Met cleavage, and decreased MET mRNA expression. To investigate whether these mechanisms were also responsible for the down-regulation of c-Met by Dp44mT observed in four other tumor cell types (Fig. S1, C–F), the following studies were performed, as per the protocols used previously. In this case, PANC-1 pancreatic cancer, SCC25 oral squamous cell carcinoma, MDA-MB-231 breast cancer cells, and Hep3B hepatoma cells were compared with DU145 and Huh7 cells as relative controls (Fig. S10). In these studies, cells were incubated with Dp44mT (5 μm) in the presence or absence of the lysosomotropic agent, CLQ (50 μm/24 h at 37 °C), to inhibit lysosomal c-Met degradation (Fig. S10A) or EDTA (1 mm/8 h at 37 °C) to prevent metalloprotease activity (Fig. S10B). The effect on NDRG1 mRNA and MET mRNA levels was also assessed in all these cell types after incubation with Dp44mT for 24 h at 37 °C (Fig. S10C).
In these studies, incubation with Dp44mT decreased c-Met expression in all cell types relative to the control, with CLQ rescuing c-Met expression in DU145 cells, but not Huh7 cells (Fig. S10A), as observed previously (Fig. 6 and Fig. S7). For the additional cell types examined, CLQ rescued c-Met expression in PANC-1 and SCC25 cells, but not in the others (Fig. S10A). This suggests that CLQ can prevent lysosomal degradation of c-Met in DU145, PANC-1, and SCC25 cells. Examining the effect of EDTA on c-Met expression again demonstrated that it rescued the down-regulation mediated by Dp44mT in DU145 and Huh7 cells (Fig. S10B), as shown previously (Fig. 7, A and B, and Fig. S9, A and B). Furthermore, EDTA also rescued the decrease in c-Met mediated by Dp44mT in all other cell types (Fig. S10B).
Assessing Dp44mT activity at the mRNA level, the agent markedly up-regulated NDRG1 mRNA demonstrating its ability to up-regulate a key target (positive control) in all cell types assessed relative to control medium alone (Fig. S10C). By investigating the effect of Dp44mT on MET mRNA expression, again no down-regulation was observed for DU145 cells versus the control, although it was markedly decreased in Huh7 cells (Fig. S10C), as shown previously (Fig. 4). In terms of the other cell types, Dp44mT had no effect on MET mRNA levels in PANC-1 cells, while a decrease was observed in all other cell types examined (Fig. S10C). In conclusion, as found for DU-145 and Huh7 cells, Dp44mT induced multiple mechanisms of c-Met degradation that were differentially observed depending on the cell type examined.
Discussion
A major mechanism that dictates the pharmacological activity of thiosemicarbazones is their binding to iron (19, 39, 48). In fact, saturation of the iron-binding site of these agents largely prevented their ability to down-regulate c-Met, suggesting a role for iron chelation in this effect. Considering this, our laboratory has previously demonstrated that both NDRG1 and MIG6 are up-regulated by thiosemicarbazones by mechanisms involving cellular iron depletion and the up-regulation of HIF-1α (27, 36). In fact, we previously reported that the down-regulation of EGFR was mediated by NDRG1 through its ability to directly associate with the EGFR inhibitor, MIG6, after genetic up-regulation of NDRG1 or after incubation of cells with thiosemicarbazones, which pharmacologically induce NDRG1 (26, 27). Indeed, NDRG1 markedly increased the half-life of the EGFR inhibitor, MIG6, resulting in the lysosomal degradation of EGFR (26, 27).
In contrast to the activity of the thiosemicarbazones on down-regulating EGFR via MIG6, which was NDRG1-dependent (26, 27), this study demonstrates that the decrease in the level of c-Met was mediated by an NDRG1-independent mechanism for both agents (Fig. 3, A–D). In fact, in the presence of the thiosemicarbazones, effective silencing of NDRG1 or MIG6 did not rescue the ability of these agents to decrease c-Met and its phosphorylation. However, under control conditions, silencing NDRG1 or MIG6 increased c-Met expression and its phosphorylation, suggesting that this metastasis (79) and tumor suppressor (80), respectively, acts to regulate c-Met expression.
To dissect the mechanism involved in the decrease in c-Met induced by these agents, the effect of the thiosemicarbazones on different c-Met processing pathways was examined. Studies progressed to examining the role of the lysosome in the thiosemicarbazone-mediated increase in c-Met degradation. This was important, as lysosomal processing plays a significant role in c-Met degradation (10). Confocal microscopy studies using DU145 cells indicated that association of the late endosome/lysosomal marker, LAMP2, with c-Met was increased by co-incubation with Dp44mT or DpC and three lysosomotropic agents, relative to the thiosemicarbazones alone.
The lysosomotropic agents also inhibited the ability of Dp44mT and DpC to induce c-Met degradation by DU145 cells. In contrast, using Huh7 cells, no rescue of c-Met was found upon incubation with the lysosomotropic agents (Fig. S7, A–C). As such, a different mechanism was active in Huh7 cells, with the thiosemicarbazones decreasing MET mRNA levels (Fig. 4, C and D), which could account for the decrease in c-Met protein in this cell type. No such down-regulation of MET mRNA by these agents was observed in DU145 cells (Fig. 4, A and B), indicating a post-transcriptional mechanism of regulation, which was then further assessed in this later cell type. Examining DU145 cells, the proteasomal inhibitors did not prevent the ability of Dp44mT to decrease c-Met expression and, in fact, further decreased levels of the precursor and mature forms. Intriguingly, inhibition of the proteasome by MG132 also increased the c-Met CTF and ICD levels, as demonstrated by others (10, 15). The CTF c-Met isoform is known to be generated by extracellular or intracellular c-Met shedding mediated by metalloproteases (10, 14–16). Importantly, in the presence of MG132, Dp44mT markedly increased c-Met CTF levels, suggesting increased c-Met shedding (Fig. 7, A and C).
Furthermore, incubation of DU145 cells with a γ-secretase inhibitor, Compound E, that blocks c-Met CTF degradation to the ICD markedly increased CTF levels, as demonstrated by others (17), and its levels were further increased in the presence of Dp44mT (Fig. 7, E and G). These data strongly support the hypothesis that Dp44mT induced metalloprotease cleavage of c-Met that could lead to its shedding and could, in part, explain the decrease in the precursor and mature forms of c-Met. The ability of Dp44mT to down-regulate the c-Met precursor and mature forms was largely abrogated by EDTA and batimastat, both of which are well-characterized, broad metalloproteinase inhibitors (72–74). These results with the inhibitors suggested that Dp44mT induces shedding of the NTF of c-Met, via stimulating the plasma membrane bound-metalloproteinases, such as ADAM10 or ADAM17 (81).
During this shedding process, the c-Met NTF would be liberated into the extracellular medium along with the generation of a c-Met CTF (Fig. 9A) (10, 15). To test this possibility, c-Met NTF levels were measured in the extracellular culture medium after incubation of cells with Dp44mT. Although extracellular shedding of the c-Met NTF was detected in the Control, contrary to our initial hypothesis, there was no significant increase in c-Met NTF levels in the extracellular medium after incubation of DU145 cells with Dp44mT alone (Fig. 8E). In fact, Dp44mT decreased c-Met NTF levels in the extracellular medium in the presence of the metalloprotease inhibitors, batimastat or EDTA (Fig. 8E), that prevent the decrease in total c-Met (Fig. 8F). Hence, the decrease in total cellular c-Met in the presence of Dp44mT alone (Fig. 8F) was not due to increased extracellular shedding of c-Met NTF. This observation suggests that intracellular shedding and c-Met degradation results in the c-Met CTF and the decreased total levels of cellular c-Met.
Figure 9.
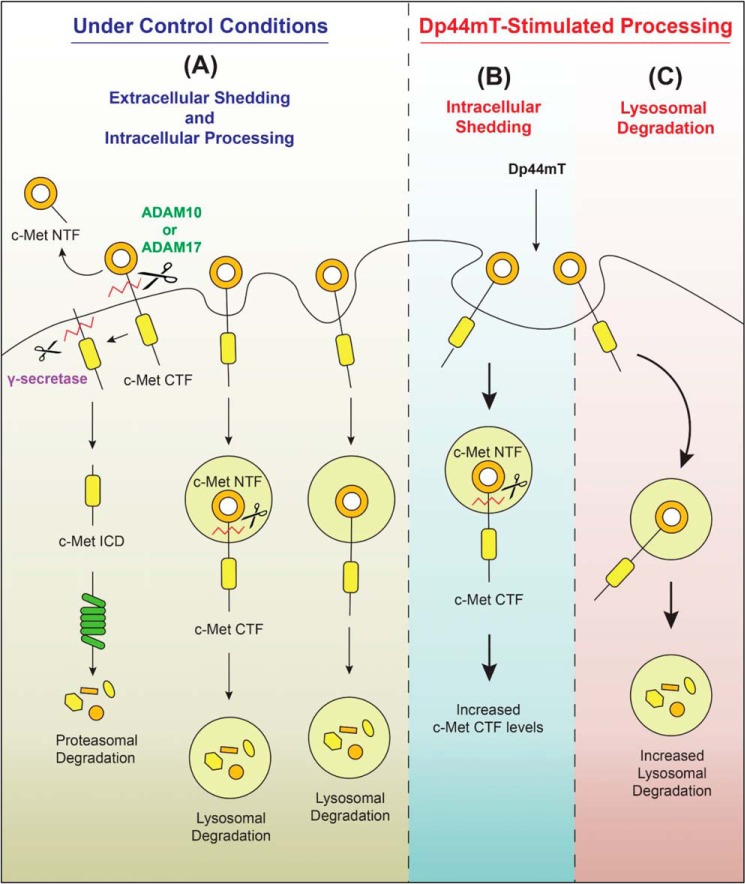
Schematic demonstrating that thiosemicarbazones decrease expression of c-Met by two mechanisms involving enhanced lysosomal degradation and intracellular shedding. The post-transcriptional processing of c-Met (mature and precursor 140 and 170 kDa) in DU145 cells is complex and consists of the following: A, extracellular shedding and intracellular processing (including intracellular shedding (17)) under Control Conditions mediated by metalloproteases, e.g. ADAM 10, 17, etc., followed by the generation of a c-Met CTF (55 kDa). The c-Met CTF is then processed by γ-secretase to generate a c-Met ICD (50 kDa). Lysosomal processing and proteasomal degradation are also involved in a concerted manner to degrade c-Met under Control Conditions (15, 16, 17). B, in contrast, after Dp44mT-stimulated processing there is enhanced c-Met CTF generation that can be demonstrated in the presence of the γ-secretase inhibitor, compound E, followed by decreased generation of the ICD. There was a decrease in the release of the c-Met NTF after Dp44mT treatment indicating potential intracellular shedding and degradation. C, three lysosomotropic agents (NH4Cl, methylamine, and chloroquine) prevent lysosomal degradation. The extracellular shedding of c-Met (A) was decreased after incubation with Dp44mT and is due to an increase in process (B and C) that is enhanced by treatment with the thiosemicarbazones.
Considering the findings and reasoning above, an alternative hypothesis could be suggested that the Dp44mT-induced increase in c-Met metalloprotease cleavage may also occur inside cells within vesicles, such as lysosomes (Fig. 9B). For example, this may occur once c-Met is internalized by the ligand-independent trafficking of the protein via endocytosis (15, 17). In fact, intracellular shedding of c-Met has been reported, with c-Met being shed inside cellular vesicles to produce the c-Met CTF and NTF (17). Moreover, both c-Met CTF and NTF were subsequently degraded by the lysosome (17).
In our studies using DU145 cells, Dp44mT increased the c-Met CTF in the presence of the proteasomal inhibitor (Fig. 7, A and C), but not the lysosomotropic agents (Fig. S8), suggesting Dp44mT-generated CTF is degraded by the proteasome. In contrast, the lysosomotropic agents alone without Dp44mT markedly increased c-Met CTF when compared with the untreated Control. This indicates c-Met CTF generated under Control conditions is degraded by the lysosome (Fig. S8). Overall, these results suggest that after incubation with Dp44mT, it could induce intracellular c-Met shedding to produce the c-Met CTF, which is then degraded by the proteasome (Fig. 7, A and C).
It has been described that extracellular and intracellular c-Met shedding could involve a number of different metalloproteases (17). This hypothesis also agrees with our data, where TIMP-3, an endogenous inhibitor of ADAM10 and -17 (16, 82, 83), did not prevent c-Met loss in the presence of Dp44mT. In contrast, the broad metalloproteinase inhibitors, EDTA and batimastat (72–74), prevented the Dp44mT-induced down-regulation of c-Met. These results suggest metalloprotease(s) inhibited by EDTA and batimastat, but not by TIMP-3, could be involved in Dp44mT-induced down-regulation of c-Met (Fig. 8, A–G).
From the results of this investigation, we propose a model in DU145 cells (Fig. 9) where under control conditions there is endocytosis of c-Met and lysosomal degradation, the extracellular and intracellular shedding of c-Met NTF, and the generation of c-Met CTF and ICD (Fig. 9A). However, after incubation with Dp44mT, two mechanisms are stimulated: 1) there is enhanced intracellular shedding, due to increased metalloprotease cleavage that results in increased c-Met CTF (Fig. 9B); and 2) there is enhanced lysosomal processing of c-Met (Fig. 9C). As such, the Dp44mT-induced enhancement of intracellular shedding (Fig. 9B) and lysosomal degradation of c-Met (Fig. 9C) result in decreased total cellular c-Met.
The ability of Dp44mT to activate these latter processes may be related to its known efficacy of depleting intracellular iron and up-regulating iron-sensitive genes in vitro and in vivo (19). This could act as a stress, and indeed Dp44mT activates the catabolic enzyme AMP-activated protein kinase, which induces lysosomal processing and autophagy (84, 85). In fact, it has been reported that a range of stress stimuli, including Dp44mT, that induce catabolic autophagy (84, 85), increase internalization of plasma membrane proteins into early endosomes and then into the lysosomal compartment (65, 86). Hence, we propose this mechanism may be important for the initial increased internalization via endocytosis of c-Met into lysosomes mediated by these agents, which leads to increased lysosomal processing. This is key to Dp44mT-induced intracellular c-Met shedding (Fig. 9B) and the lysosomal degradation of c-Met (Fig. 9C).
In summary, this investigation provides an integrated model (Fig. 9, A–C), which demonstrates the molecular mechanisms of how thiosemicarbazones decrease oncogenic cellular c-Met. Studies in multiple cell types demonstrated that Dp44mT decreases c-Met via a unique combined mechanism involving lysosomal degradation, shedding by a metalloprotease-sensitive mechanism, and also MET mRNA down-regulation. Various cell types utilize combinations of these mechanisms to down-regulate c-Met. This investigation also demonstrates that c-Met is another key molecular target of the clinically trialed DpT group of thiosemicarbazones (18) that explains their potent and selective anti-cancer efficacy in vitro and in vivo (19–23, 26, 39).
Experimental procedures
Cell culture
Human DU145 prostate cancer cells, PANC-1 pancreatic cancer cells, SCC25 oral squamous cell carcinoma, MDA-MB-231 breast cancer, and Hep3B hepatoma cells were purchased from the American Type Culture Collection (Manassas, VA). Human Huh7 hepatocellular carcinoma cells were kindly provided by Dr. D. Seth (University of Sydney). DU145 cells were cultured in RPMI 1640 medium (Sigma) supplemented with 10% (v/v) fetal bovine serum (Sigma), 1% (v/v) nonessential amino acids, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1% (v/v) sodium pyruvate (Sigma). Huh7, HepG2, Hep3B, MDA-MB-231, and PANC-1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) with the same supplements added to RPMI 1640 medium. The SCC25 cell line was grown in DMEM/F12 with the same supplements as with DMEM and RPMI 1640 medium. All cells were maintained at 37 °C in a humidified atmosphere of 5% CO2.
Gene silencing by small interfering RNA (siRNA)
Two siRNAs for NDRG1 and MIG6 were implemented, namely siNDRG1 (catalog no. 4392422; Life Technologies, Inc.) and siNDRG1 II (catalog no. s20334; Life Technologies, Inc.), and siMIG6 (catalog no. AM16708; Ambion), and siMIG6 II (catalog no. 4392420; Life Technologies, Inc.). These were compared with a nontargeting negative control siRNA (siControl; Life Technologies, Inc.; catalog no. AM4635). The siRNAs were diluted in Opti-MEMTM (Thermo Fisher Scientific, Waltham, MA) with Lipofectamine RNAiMAX (Invitrogen), as per the manufacturer's instructions. Then, DU145 and Huh7 cells at ∼50% confluence were incubated in this medium containing siRNA (75 nm) for 6 h at 37 °C, and then the media were removed and replaced with Control medium alone for a further 66 h at 37 °C. After incubation with siRNA, cells were harvested, and protein extraction and immunoblotting were performed, as described below.
Reagents and treatments
Desferrioxamine (DFO) was purchased from Sigma. The thiosemicarbazones, 2-benzoylpyridine 2-methyl-3-thiosemicarbazone (Bp2mT), Dp44mT, and DpC were synthesized and characterized, as described previously (37, 39, 47). Of note, Bp2mT was used as a negative control for Dp44mT and DpC. Briefly, at ∼80% confluence, cells were incubated with the compounds at the following concentrations: DFO (100 μm), Bp2mT (5 μm), Dp44mT (5 μm), and DpC (5 μm) for 24 h in 37 °C. In all studies, DFO was dissolved directly in media, while Bp2mT, Dp44mT, and DpC were freshly prepared in dimethyl sulfoxide (DMSO) at concentrations of 10 mm and then diluted in media (final [DMSO], ≤0.1%).
After incubation with the compounds, cells were serum-starved for 3 h at 37 °C, followed by 15 min at 37 °C treatment with HGF (50 ng/ml; Life Technologies Inc.; catalog no. PHG0324), which was dissolved in 0.01% BSA/sterile water and diluted in fresh media. The lysosomotropic agents, NH4Cl, methylamine, and chloroquine, and the proteasomal inhibitor, MG132, were purchased from Sigma. The broad metalloprotease inhibitor, EDTA, was also purchased from Sigma, and batimastat and TIMP-3 were purchased from Tocris and Sigma, respectively. Compound E and lactacystin were purchased from Life Technologies, Inc., and Santa Cruz Biotechnology, respectively.
Protein extraction and Western blot analysis
Total protein was extracted using standard procedures in our laboratory (58). Western blot analysis was performed, as described previously (87). The primary antibody used against human NDRG1 was from Abcam Inc. (amino acids 382–394; goat; 1:2000; catalog no. ab37897; Abcam Inc., Cambridge, UK). The following antibodies were purchased from Cell Signaling Technology (Danvers, MA) and were used at a 1:1000 dilution: rabbit monoclonal anti-Met XP® (catalog no. 8198), rabbit monoclonal anti-phospho-Met (Y1003; catalog no. 3135), rabbit monoclonal anti-phospho-Met (Y1234/1235; catalog no. 3077), rabbit monoclonal anti-phospho-Met (Y1349; catalog no. 3133), anti-Gab1 (catalog no. 3232), anti-phospho-Gab1 (Y307; catalog no. 3234), and anti-HSP90 (catalog no. 4877). An antibody against β-actin (mouse; 1:10,000; catalog no. A5441; Sigma) was used as a protein-loading control.
RNA extraction and reverse transcription PCR (RT-PCR)
The total RNA from cells was extracted using the TRI Reagent® solution (catalog no. AM9738; Thermo Fisher Scientific, Waltham, MA) following the manufacturer's protocol. The RNA concentration was measured using a NanoDrop® 1000 spectrophotometer (Thermo Fisher Scientific). RT-PCR was performed following standard procedures (87). The primers used are as follows: for NDRG1, 5′-TCACCCAGCACTTTGCCGTCT-3′ (forward) and 5′-GCCACAGTCCGCCATCTT-3′ (reverse); for MET, 5′-CAGGCAGTGCAGCATGTAGT-3′ (forward) and 5′-GATGATTCCCTCGGTCAGAA-3′ (reverse); for HSP90AA1, 5′-GGGACCAAAGCGTTCA-3′ (forward) and 5′-AGGGATTAGCTCCTCACA-3′ (reverse); and for β-actin, 5′-CCCGCCGCCAGCTCACCATGG-3′ (forward) and 5′-AAGGTCTCAAACATGATCTGGGTC-3′ (reverse). As an internal control, the housekeeping gene β-actin was amplified from the same samples. RT-PCR was semi-quantitative, as demonstrated by an optimization protocol showing it was in the log-phase of amplification.
Immunofluorescence and confocal microscopy
Cells were seeded into 24-well plates (Sigma) containing sterile glass coverslips (12 mm diameter; catalog no. G401-12; ProSciTech, Queensland, Australia) in media and were grown to ∼50% confluence. These cells were then incubated with Dp44mT (5 μm), DpC (5 μm), NH4Cl (15 mm), MA (15 mm), and CLQ (50 μm), alone or in combination for 24 h at 37 °C. The media were then discarded, and the cells were fixed with 4% (w/v) paraformaldehyde (Sigma) for 10 min at room temperature.
Cells were then washed with PBS (three times for 5 min) followed by permeabilization with 0.2% (v/v) Triton X-100/PBS for 5 min at room temperature. Then, another set of washes with PBS was performed. A blocking solution, 5% (w/v) BSA/PBS, was then applied for 1 h at room temperature and washed off with PBS (two times for 10 min), and then the primary antibodies against Met XP® (rabbit; 1:100; catalog no. 8198; Cell Signaling Technology) and LAMP2 (mouse; 1:100; catalog no. ab25631; Abcam Inc.) in 1% (w/v) BSA/PBS were applied with agitation at 4 °C overnight.
The primary antibody was then removed, and the coverslips were washed with PBS (three times for 10 min), followed by incubation with secondary anti-mouse Alexa Fluor® 488 conjugate (catalog no. 4408; Cell Signaling Technology) and anti-rabbit Alexa Fluor® 594 conjugate (catalog no. 8889; Cell Signaling Technology) diluted in 1% (w/v) BSA/PBS for 1 h/room temperature. After another set of washes with PBS, coverslips were mounted on glass microscope slides (Menzel Glaser; Braunschweig, Germany) using ProLong Gold anti-fade mounting solution with 4′6-diamidino-2-phenylindole (catalog no. P36935; Life Technologies, Inc.).
Cells were then visualized using a Zeiss LSM 510 Meta spectral confocal microscope (Carl Zeiss Microscopy, Jena, Germany; Advanced Microscopy Facility, Bosch Institute, University of Sydney) at ×63 magnification. Images were taken and processed using LSM 510 Meta software (Carl Zeiss Microscopy). Z-stack imaging was performed according to the manufacturer's instructions.
ELISA for the measurement of c-Met NTF levels in the culture medium
Cells were incubated with the agents Dp44mT (5 μm), EDTA (1 mm), and batimastat (5 μm) alone or in combination for 8 h at 37 °C. Culture media were then collected and centrifuged for 1000 × g for 5 min at 4 °C to remove cellular debris. These media samples were then filtered using a 0.22-μm filter unit (Merck Millipore) to remove cellular debris. The soluble c-Met/c-Met NTF levels in the filtered media were measured using an ELISA kit (catalog no. KHO2031, Invitrogen) following the manufacturer's instructions. Culture media incubated in Petri dishes without cells were used as appropriate blanks for the measurements. All samples were processed on ice.
Densitometry
Densitometric analysis was performed to quantify data obtained from immunoblotting, RT-PCR, and immunofluorescence. For quantifying the intensities of bands from immunoblotting and RT-PCR, ChemiDoc Image Lab software (Bio-Rad) and Quantity One software (Bio-Rad) were used, respectively. Intensities of target protein bands relative to β-actin are presented in arbitrary units (A.U.).
Statistical analysis
Results are presented as mean ± S.E. Data were compared using Student's paired t test. Results were considered statistically significant when p < 0.05. To quantify co-localization, Pearson's coefficients were calculated using ImageJ software (National Institutes of Health).
Author contributions
K. C. P., P. J. J., Z. K., and D. R. R. conceptualization; K. C. P. and B. G. data curation; K. C. P., P. J. J., Z. K., and D. R. R. formal analysis; K. C. P., B. G., and L. Y. W. L. validation; K. C. P., B. G., L. Y. W. L., J. P., S. C., P. J. J., and D. R. R. investigation; K. C. P. and D. R. R. visualization; K. C. P., J. P., S. C., P. J. J., Z. K., and D. R. R. methodology; K. C. P. and D. R. R. writing-original draft; K. C. P., B. G., L. Y. W. L., J. P., S. C., P. J. J., Z. K., and D. R. R. writing-review and editing; Z. K. and D. R. R. resources; Z. K. and D. R. R. supervision; Z. K. and D. R. R. funding acquisition.
Supplementary Material
Acknowledgments
We sincerely appreciate the comments regarding this manuscript from Dr. Danuta Kalinowski and Dr. Vera Richardson of the Molecular Pharmacology and Pathology Program, Dept. of Pathology and Bosch Institute, University of Sydney.
This work was supported by National Health and Medical Research Council of Australia (NHMRC) Project Grant 1060482 (to D. R. R. and Z. K.) and Priority-driven Collaborative Cancer Research Scheme Grant 1086449 co-funded by the Cure Cancer Australia Foundation of Australia and Cancer Australia) (to Z. K.). This research was also supported by NHMRC/PdCCRS Cancer Australia Project Grant co-Funded by NBCF (NHMRC APP1146599) and National Breast Cancer Foundation (NBCF) grant support IIRS-19–048 (to D. R. R. and P. J. J.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S10.
- RTK
- receptor tyrosine kinase
- Bp2mT
- 2-benzoylpyridine 2-methyl-3-thiosemicarbazone
- CTF
- C-terminal fragment
- DFO
- desferrioxamine
- Dp44mT
- di-2-pyridylketone 4,4-dimethyl-3-thiosemicarbazone
- DpC
- di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone
- DpT
- di-2-pyridylketone thiosemicarbazone
- EGFR
- epidermal growth factor receptor
- ICD
- intracellular domain
- NH4Cl
- ammonium chloride
- NTF
- N-terminal fragment
- HGF
- hepatocyte growth factor
- HCC
- hepatocellular carcinoma
- DMEM
- Dulbecco's modified Eagle's medium
- MA
- methylamine
- CLQ
- chloroquine
- NAC
- N-acetylcysteine
- PI3K
- phosphatidylinositol 3-kinase.
References
- 1. Lemmon M. A., and Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arteaga C. L., and Engelman J. A. (2014) ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 25, 282–303 10.1016/j.ccr.2014.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gherardi E., Birchmeier W., Birchmeier C., and Vande Woude G. (2012) Targeting MET in cancer: rationale and progress. Nat. Rev. Cancer 12, 89–103 10.1038/nrc3205 [DOI] [PubMed] [Google Scholar]
- 4. Nabeshima K., Shimao Y., Inoue T., Itoh H., Kataoka H., and Koono M. (1998) Hepatocyte growth factor/scatter factor induces not only scattering but also cohort migration of human colorectal-adenocarcinoma cells. Int. J. Cancer 78, 750–759 [DOI] [PubMed] [Google Scholar]
- 5. Birchmeier C., Birchmeier W., Gherardi E., and Vande Woude G. F. (2003) Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 4, 915–925 10.1038/nrm1261 [DOI] [PubMed] [Google Scholar]
- 6. Xiao G. H., Jeffers M., Bellacosa A., Mitsuuchi Y., Vande Woude G. F., and Testa J. R. (2001) Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc. Natl. Acad. Sci. U.S.A. 98, 247–252 10.1073/pnas.98.1.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan S., Ma Y. X., Gao M., Yuan R. Q., Meng Q., Goldberg I. D., and Rosen E. M. (2001) The multisubstrate adapter Gab1 regulates hepatocyte growth factor (scatter factor)–c-Met signaling for cell survival and DNA repair. Mol. Cell. Biol. 21, 4968–4984 10.1128/MCB.21.15.4968-4984.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mueller K. L., Hunter L. A., Ethier S. P., and Boerner J. L. (2008) Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 68, 3314–3322 10.1158/0008-5472.CAN-08-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., et al. (2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- 10. Lefebvre J., Ancot F., Leroy C., Muharram G., Lemière A., and Tulasne D. (2012) Met degradation: more than one stone to shoot a receptor down. FASEB J. 26, 1387–1399 10.1096/fj.11-197723 [DOI] [PubMed] [Google Scholar]
- 11. Jeffers M., Taylor G. A., Weidner K. M., Omura S., and Vande Woude G. F. (1997) Degradation of the Met tyrosine kinase receptor by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 17, 799–808 10.1128/MCB.17.2.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edwards D. R., Handsley M. M., and Pennington C. J. (2008) The ADAM metalloproteinases. Mol. Aspects Med. 29, 258–289 10.1016/j.mam.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murphy G. (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer 8, 929–941 10.1038/nrc2459 [DOI] [PubMed] [Google Scholar]
- 14. Kopitz C., Gerg M., Bandapalli O. R., Ister D., Pennington C. J., Hauser S., Flechsig C., Krell H. W., Antolovic D., Brew K., Nagase H., Stangl M., von Weyhern C. W., Brücher B. L., Brand K., et al. (2007) Tissue inhibitor of metalloproteinases-1 promotes liver metastasis by induction of hepatocyte growth factor signaling. Cancer Res. 67, 8615–8623 10.1158/0008-5472.CAN-07-0232 [DOI] [PubMed] [Google Scholar]
- 15. Foveau B., Ancot F., Leroy C., Petrelli A., Reiss K., Vingtdeux V., Giordano S., Fafeur V., and Tulasne D. (2009) Down-regulation of the met receptor tyrosine kinase by presenilin-dependent regulated intramembrane proteolysis. Mol. Biol. Cell 20, 2495–2507 10.1091/mbc.e08-09-0969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schelter F., Kobuch J., Moss M. L., Becherer J. D., Comoglio P. M., Boccaccio C., and Krüger A. (2010) A disintegrin and metalloproteinase-10 (ADAM-10) mediates DN30 antibody-induced shedding of the met surface receptor. J. Biol. Chem. 285, 26335–26340 10.1074/jbc.M110.106435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ancot F., Leroy C., Muharram G., Lefebvre J., Vicogne J., Lemiere A., Kherrouche Z., Foveau B., Pourtier A., Melnyk O., Giordano S., Chotteau-Lelievre A., and Tulasne D. (2012) Shedding-generated Met receptor fragments can be routed to either the proteasomal or the lysosomal degradation pathway. Traffic 13, 1261–1272 10.1111/j.1600-0854.2012.01384.x [DOI] [PubMed] [Google Scholar]
- 18. Jansson P. J., Kalinowski D. S., Lane D. J., Kovacevic Z., Seebacher N. A., Fouani L., Sahni S., Merlot A. M., and Richardson D. R. (2015) The renaissance of polypharmacology in the development of anti-cancer therapeutics: inhibition of the “Triad of Death” in cancer by di-2-pyridylketone thiosemicarbazones. Pharmacol. Res. 100, 255–260 10.1016/j.phrs.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 19. Yuan J., Lovejoy D. B., and Richardson D. R. (2004) Novel di-2-pyridyl–derived iron chelators with marked and selective antitumor activity: in vitro and in vivo assessment. Blood 104, 1450–1458 10.1182/blood-2004-03-0868 [DOI] [PubMed] [Google Scholar]
- 20. Whitnall M., Howard J., Ponka P., and Richardson D. R. (2006) A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. U.S.A. 103, 14901–14906 10.1073/pnas.0604979103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kovacevic Z., Chikhani S., Lovejoy D. B., and Richardson D. R. (2011) Novel thiosemicarbazone iron chelators induce up-regulation and phosphorylation of the metastasis suppressor N-myc downstream-regulated gene 1: a new strategy for the treatment of pancreatic cancer. Mol. Pharmacol. 80, 598–609 10.1124/mol.111.073627 [DOI] [PubMed] [Google Scholar]
- 22. Liu W., Xing F., Iiizumi-Gairani M., Okuda H., Watabe M., Pai S. K., Pandey P. R., Hirota S., Kobayashi A., Mo Y. Y., Fukuda K., Li Y., and Watabe K. (2012) N-myc downstream-regulated gene 1 modulates Wnt-β-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol. Med. 4, 93–108 10.1002/emmm.201100190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo Z. L., Richardson D. R., Kalinowski D. S., Kovacevic Z., Tan-Un K. C., and Chan G. C. (2016) The novel thiosemicarbazone, di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC), inhibits neuroblastoma growth in vitro and in vivo via multiple mechanisms. J. Hematol. Oncol. 9, 98 10.1186/s13045-016-0330-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Y. X., Zeng M. L., Yu D., Ren J., Li F., Zheng A., Wang Y. P., Chen C., and Tao Z. Z. (2018) In vitro assessment of the role of DpC in the treatment of head and neck squamous cell carcinoma. Oncol. Lett. 15, 7999–8004 10.3892/ol.2018.8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li P., Zheng X., Shou K., Niu Y., Jian C., Zhao Y., Yi W., Hu X., and Yu A. (2016) The iron chelator Dp44mT suppresses osteosarcoma's proliferation, invasion and migration: in vitro and in vivo. Am. J. Transl. Res. 8, 5370–5385 [PMC free article] [PubMed] [Google Scholar]
- 26. Kovacevic Z., Menezes S. V., Sahni S., Kalinowski D. S., Bae D. H., Lane D. J., and Richardson D. R. (2016) The metastasis suppressor, N-myc downstream-regulated gene-1 (NDRG1), down-regulates the ErbB Family of receptors to inhibit downstream oncogenic signaling pathways. J. Biol. Chem. 291, 1029–1052 10.1074/jbc.M115.689653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Menezes S. V., Kovacevic Z., and Richardson D. R. (2019) The metastasis suppressor NDRG1 down-regulates the epidermal growth factor receptor via a lysosomal mechanism by up-regulating mitogen-inducible gene 6. J. Biol. Chem. 294, 4045–4064 10.1074/jbc.RA118.006279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lui G. Y., Kovacevic Z., V Menezes S., Kalinowski D. S., Merlot A. M., Sahni S., and Richardson D. R. (2015) Novel thiosemicarbazones regulate the signal transducer and activator of transcription 3 (STAT3) pathway: inhibition of constitutive and interleukin 6-induced activation by iron depletion. Mol. Pharmacol. 87, 543–560 10.1124/mol.114.096529 [DOI] [PubMed] [Google Scholar]
- 29. Dixon K. M., Lui G. Y., Kovacevic Z., Zhang D., Yao M., Chen Z., Dong Q., Assinder S. J., and Richardson D. R. (2013) Dp44mT targets the AKT, TGF-β, and ERK pathways via the metastasis suppressor NDRG1 in normal prostate epithelial cells and prostate cancer cells. Br. J. Cancer 108, 409–419 10.1038/bjc.2012.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xi R., Pun I. H., Menezes S. V., Fouani L., Kalinowski D. S., Huang M. L., Zhang X., Richardson D. R., and Kovacevic Z. (2017) Novel thiosemicarbazones inhibit lysine-rich CEACAM1 co-isolated (LYRIC) and the LYRIC-induced epithelial-mesenchymal transition via up-regulation of N-myc downstream-regulated gene 1 (NDRG1). Mol. Pharmacol. 91, 499–517 10.1124/mol.116.107870 [DOI] [PubMed] [Google Scholar]
- 31. Kovacevic Z., Chikhani S., Lui G. Y., Sivagurunathan S., and Richardson D. R. (2013) The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid. Redox Signal. 18, 874–887 10.1089/ars.2011.4273 [DOI] [PubMed] [Google Scholar]
- 32. Menezes S. V., Fouani L., Huang M. L. H., Geleta B., Maleki S., Richardson A., Richardson D. R., and Kovacevic Z. (2019) The metastasis suppressor, NDRG1, attenuates oncogenic TGF-β and NF-κB signaling to enhance membrane E-cadherin expression in pancreatic cancer cells. Carcinogenesis 40, 805–818 10.1093/carcin/bgy178 [DOI] [PubMed] [Google Scholar]
- 33. Ghatak S., Hascall V. C., Markwald R. R., and Misra S. (2010) Stromal hyaluronan interaction with epithelial CD44 variants promotes prostate cancer invasiveness by augmenting expression and function of hepatocyte growth factor and androgen receptor. J. Biol. Chem. 285, 19821–19832 10.1074/jbc.M110.104273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Humphrey P. A., Zhu X., Zarnegar R., Swanson P. E., Ratliff T. L., Vollmer R. T., and Day M. L. (1995) Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am. J. Pathol. 147, 386–396 [PMC free article] [PubMed] [Google Scholar]
- 35. Ogunwobi O. O., Puszyk W., Dong H. J., and Liu C. (2013) Epigenetic upregulation of HGF and c-Met drives metastasis in hepatocellular carcinoma. PLoS ONE 8, e63765 10.1371/journal.pone.0063765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Le N. T., and Richardson D. R. (2004) Iron chelators with high antiproliferative activity up-regulate the expression of a growth inhibitory and metastasis suppressor gene: a link between iron metabolism and proliferation. Blood 104, 2967–2975 10.1182/blood-2004-05-1866 [DOI] [PubMed] [Google Scholar]
- 37. Richardson D. R., Sharpe P. C., Lovejoy D. B., Senaratne D., Kalinowski D. S., Islam M., and Bernhardt P. V. (2006) Dipyridyl thiosemicarbazone chelators with potent and selective antitumor activity form iron complexes with redox activity. J. Med. Chem. 49, 6510–6521 10.1021/jm0606342 [DOI] [PubMed] [Google Scholar]
- 38. Lovejoy D. B., Jansson P. J., Brunk U. T., Wong J., Ponka P., and Richardson D. R. (2011) Antitumor activity of metal-chelating compound Dp44mT is mediated by formation of a redox-active copper complex that accumulates in lysosomes. Cancer Res. 71, 5871–5880 10.1158/0008-5472.CAN-11-1218 [DOI] [PubMed] [Google Scholar]
- 39. Lovejoy D. B., Sharp D. M., Seebacher N., Obeidy P., Prichard T., Stefani C., Basha M. T., Sharpe P. C., Jansson P. J., Kalinowski D. S., Bernhardt P. V., and Richardson D. R. (2012) Novel second-generation di-2-pyridylketone thiosemicarbazones show synergism with standard chemotherapeutics and demonstrate potent activity against lung cancer xenografts after oral and intravenous administration in vivo. J. Med. Chem. 55, 7230–7244 10.1021/jm300768u [DOI] [PubMed] [Google Scholar]
- 40. Ceci A., Felisi M., De Sanctis V., and De Mattia D. (2003) Pharmacotherapy of iron overload in thalassaemic patients. Expert Opin. Pharmacother. 4, 1763–1774 10.1517/14656566.4.10.1763 [DOI] [PubMed] [Google Scholar]
- 41. Xu K. P., and Yu F. S. (2007) Cross-talk between c-Met and epidermal growth factor receptor during retinal pigment epithelial wound healing. Invest. Ophthalmol. Vis. Sci. 48, 2242–2248 10.1167/iovs.06-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ghalayini M. K., Dong Q., Richardson D. R., and Assinder S. J. (2013) Proteolytic cleavage and truncation of NDRG1 in human prostate cancer cells, but not normal prostate epithelial cells. Biosci. Rep. 33, e00042 10.1042/BSR20130042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park K. C., Menezes S. V., Kalinowski D. S., Sahni S., Jansson P. J., Kovacevic Z., and Richardson D. R. (2018) Identification of differential phosphorylation and sub-cellular localization of the metastasis suppressor, NDRG1. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2644–2663 10.1016/j.bbadis.2018.04.011 [DOI] [PubMed] [Google Scholar]
- 44. Sahni S., Park K. C., Kovacevic Z., and Richardson D. R. (2019) Two mechanisms involving the autophagic and proteasomal pathways process the metastasis suppressor protein, N-myc downstream-regulated gene 1. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1361–1378 10.1016/j.bbadis.2019.02.008 [DOI] [PubMed] [Google Scholar]
- 45. Giordano S., Di Renzo M. F., Narsimhan R. P., Cooper C. S., Rosa C., and Comoglio P. M. (1989) Biosynthesis of the protein encoded by the c-met proto-oncogene. Oncogene 4, 1383–1388 [PubMed] [Google Scholar]
- 46. Bottaro D. P., Rubin J. S., Faletto D. L., Chan A. M., Kmiecik T. E., Vande Woude G. F., and Aaronson S. A. (1991) Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 251, 802–804 10.1126/science.1846706 [DOI] [PubMed] [Google Scholar]
- 47. Stacy A. E., Palanimuthu D., Bernhardt P. V., Kalinowski D. S., Jansson P. J., and Richardson D. R. (2016) Structure-activity relationships of di-2-pyridylketone, 2-benzoylpyridine, and 2-acetylpyridine thiosemicarbazones for overcoming Pgp-mediated drug resistance. J. Med. Chem. 59, 8601–8620 10.1021/acs.jmedchem.6b01050 [DOI] [PubMed] [Google Scholar]
- 48. Kalinowski D. S., and Richardson D. R. (2005) The evolution of iron chelators for the treatment of iron overload disease and cancer. Pharmacol. Rev. 57, 547–583 10.1124/pr.57.4.2 [DOI] [PubMed] [Google Scholar]
- 49. Liu W., Yue F., Zheng M., Merlot A., Bae D. H., Huang M., Lane D., Jansson P., Lui G. Y., Richardson V., Sahni S., Kalinowski D., Kovacevic Z., and Richardson D. R. (2015) The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget 6, 8851–8874 10.18632/oncotarget.3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abella J. V., Peschard P., Naujokas M. A., Lin T., Saucier C., Urbé S., and Park M. (2005) Met/hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol. Cell. Biol. 25, 9632–9645 10.1128/MCB.25.21.9632-9645.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ponzetto C., Bardelli A., Zhen Z., Maina F., dalla Zonca P., Giordano S., Graziani A., Panayotou G., and Comoglio P. M. (1994) A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 77, 261–271 10.1016/0092-8674(94)90318-2 [DOI] [PubMed] [Google Scholar]
- 52. Weidner K. M., Di Cesare S., Sachs M., Brinkmann V., Behrens J., and Birchmeier W. (1996) Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature 384, 173–176 10.1038/384173a0 [DOI] [PubMed] [Google Scholar]
- 53. Gual P., Giordano S., Williams T. A., Rocchi S., Van Obberghen E., and Comoglio P. M. (2000) Sustained recruitment of phospholipase C-γ to Gab1 is required for HGF-induced branching tubulogenesis. Oncogene 19, 1509–1518 10.1038/sj.onc.1203514 [DOI] [PubMed] [Google Scholar]
- 54. Ridley A. J., Comoglio P. M., and Hall A. (1995) Regulation of scatter factor/hepatocyte growth factor responses by Ras, Rac, and Rho in MDCK cells. Mol. Cell. Biol. 15, 1110–1122 10.1128/MCB.15.2.1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Potempa S., and Ridley A. J. (1998) Activation of both MAP kinase and phosphatidylinositide 3-kinase by Ras is required for hepatocyte growth factor/scatter factor-induced adherens junction disassembly. Mol. Biol. Cell 9, 2185–2200 10.1091/mbc.9.8.2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Royal I., Lamarche-Vane N., Lamorte L., Kaibuchi K., and Park M. (2000) Activation of cdc42, rac, PAK, and rho-kinase in response to hepatocyte growth factor differentially regulates epithelial cell colony spreading and dissociation. Mol. Biol. Cell 11, 1709–1725 10.1091/mbc.11.5.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kovacevic Z., Fu D., and Richardson D. R. (2008) The iron-regulated metastasis suppressor, Ndrg-1: identification of novel molecular targets. Biochim. Biophys. Acta 1783, 1981–1992 10.1016/j.bbamcr.2008.05.016 [DOI] [PubMed] [Google Scholar]
- 58. Chen Z., Zhang D., Yue F., Zheng M., Kovacevic Z., and Richardson D. R. (2012) The iron chelators Dp44mT and DFO inhibit TGF-β–induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J. Biol. Chem. 287, 17016–17028 10.1074/jbc.M112.350470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chaston T. B., Lovejoy D. B., Watts R. N., and Richardson D. R. (2003) Examination of the antiproliferative activity of iron chelators: multiple cellular targets and the different mechanism of action of triapine compared with desferrioxamine and the potent pyridoxal isonicotinoyl hydrazone analogue 311. Clin. Cancer Res. 9, 402–414 [PubMed] [Google Scholar]
- 60. Pante G., Thompson J., Lamballe F., Iwata T., Ferby I., Barr F. A., Davies A. M., Maina F., and Klein R. (2005) Mitogen-inducible gene 6 is an endogenous inhibitor of HGF/Met-induced cell migration and neurite growth. J. Cell Biol. 171, 337–348 10.1083/jcb.200502013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saarikoski S. T., Rivera S. P., and Hankinson O. (2002) Mitogen-inducible gene 6 (MIG-6), adipophilin and tuftelin are inducible by hypoxia. FEBS Lett. 530, 186–190 10.1016/S0014-5793(02)03475-0 [DOI] [PubMed] [Google Scholar]
- 62. Sidarovich V., Adami V., Gatto P., Greco V., Tebaldi T., Tonini G. P., and Quattrone A. (2015) Translational down-regulation of HSP90 expression by iron chelators in neuroblastoma cells. Mol. Pharmacol. 87, 513–524 10.1124/mol.114.095729 [DOI] [PubMed] [Google Scholar]
- 63. Bachleitner-Hofmann T., Sun M. Y., Chen C. T., Liska D., Zeng Z., Viale A., Olshen A. B., Mittlboeck M., Christensen J. G., Rosen N., Solit D. B., and Weiser M. R. (2011) Antitumor activity of SNX-2112, a synthetic heat shock protein-90 inhibitor, in MET-amplified tumor cells with or without resistance to selective MET Inhibition. Clin. Cancer Res. 17, 122–133 10.1158/1078-0432.CCR-10-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hammond D. E., Urbé S., Vande Woude G. F., and Clague M. J. (2001) Down-regulation of MET, the receptor for hepatocyte growth factor. Oncogene 20, 2761–2770 10.1038/sj.onc.1204475 [DOI] [PubMed] [Google Scholar]
- 65. Seebacher N. A., Lane D. J., Jansson P. J., and Richardson D. R. (2016) Glucose modulation induces lysosome formation and increases lysosomotropic drug sequestration via the P-glycoprotein drug transporter. J. Biol. Chem. 291, 3796–3820 10.1074/jbc.M115.682450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huynh K. K., Eskelinen E. L., Scott C. C., Malevanets A., Saftig P., and Grinstein S. (2007) LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 26, 313–324 10.1038/sj.emboj.7601511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Seglen P. O., Grinde B., and Solheim A. E. (1979) Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. Eur. J. Biochem. 95, 215–225 10.1111/j.1432-1033.1979.tb12956.x [DOI] [PubMed] [Google Scholar]
- 68. Richardson D. R., and Baker E. (1994) Two saturable mechanisms of iron uptake from transferrin in human melanoma cells: the effect of transferrin concentration, chelators, and metabolic probes on transferrin and iron uptake. J. Cell. Physiol. 161, 160–168 10.1002/jcp.1041610119 [DOI] [PubMed] [Google Scholar]
- 69. Morgan E. H. (1981) Inhibition of reticulocyte iron uptake by NH4Cl and CH3NH2. Biochim. Biophys. Acta 642, 119–134 10.1016/0005-2736(81)90143-7 [DOI] [PubMed] [Google Scholar]
- 70. Dunn K. W., Kamocka M. M., and McDonald J. H. (2011) A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. 300, C723–C742 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Galvani A. P., Cristiani C., Carpinelli P., Landonio A., and Bertolero F. (1995) Suramin modulates cellular levels of hepatocyte growth factor receptor by inducing shedding of a soluble form. Biochem. Pharmacol. 50, 959–966 10.1016/0006-2952(95)00219-P [DOI] [PubMed] [Google Scholar]
- 72. Grantcharova E., Furkert J., Reusch H. P., Krell H. W., Papsdorf G., Beyermann M., Schulein R., Rosenthal W., and Oksche A. (2002) The extracellular N terminus of the endothelin B (ETB) receptor is cleaved by a metalloprotease in an agonist-dependent process. J. Biol. Chem. 277, 43933–43941 10.1074/jbc.M208407200 [DOI] [PubMed] [Google Scholar]
- 73. Lohi J., Wilson C. L., Roby J. D., and Parks W. C. (2001) Epilysin, a novel human matrix metalloproteinase (MMP-28) expressed in testis and keratinocytes and in response to injury. J. Biol. Chem. 276, 10134–10144 10.1074/jbc.M001599200 [DOI] [PubMed] [Google Scholar]
- 74. Peng M., Guo S., Yin N., Xue J., Shen L., Zhao Q., and Zhang W. (2010) Ectodomain shedding of Fcα receptor is mediated by ADAM10 and ADAM17. Immunology 130, 83–91 10.1111/j.1365-2567.2009.03215.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Codony-Servat J., Albanell J., Lopez-Talavera J. C., Arribas J., and Baselga J. (1999) Cleavage of the HER2 ectodomain is a pervanadate-activable process that is inhibited by the tissue inhibitor of metalloproteases-1 in breast cancer cells. Cancer Res. 59, 1196–1201 [PubMed] [Google Scholar]
- 76. Prenzel N., Zwick E., Daub H., Leserer M., Abraham R., Wallasch C., and Ullrich A. (1999) EGF receptor transactivation by G-protein–coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature 402, 884–888 10.1038/47260 [DOI] [PubMed] [Google Scholar]
- 77. Wojtowicz-Praga S. M., Dickson R. B., and Hawkins M. J. (1997) Matrix metalloproteinase inhibitors. Invest. New Drugs 15, 61–75 10.1023/A:1005722729132 [DOI] [PubMed] [Google Scholar]
- 78. Copin M. C., Lesaffre M., Berbon M., Doublet L., Leroy C., Tresch E., Porte H., Vicogne J., Cortot B. A., Dansin E., and Tulasne D. (2016) High-MET status in non-small cell lung tumors correlates with receptor phosphorylation but not with the serum level of soluble form. Lung Cancer 101, 59–67 10.1016/j.lungcan.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 79. Park K. C., Paluncic J., Kovacevic Z., and Richardson D. R. (2019) Pharmacological targeting and the diverse functions of the metastasis suppressor, NDRG1, in cancer. Free Radic. Biol. Med. S0891–5849(19)30256–4 10.1016/j.freeradbiomed.2019.05.020 [DOI] [PubMed] [Google Scholar]
- 80. Zhang Y. W., Staal B., Su Y., Swiatek P., Zhao P., Cao B., Resau J., Sigler R., Bronson R., and Vande Woude G. F. (2007) Evidence that MIG-6 is a tumor-suppressor gene. Oncogene 26, 269–276 10.1038/sj.onc.1209790 [DOI] [PubMed] [Google Scholar]
- 81. Chalupsky K., Kanchev I., Zbodakova O., Buryova H., Jirouskova M., Korinek V., Gregor M., and Sedlacek R. (2013) ADAM10/17-dependent release of soluble c-Met correlates with hepatocellular damage. Folia Biol. 59, 76–86 [PubMed] [Google Scholar]
- 82. Amour A., Knight C. G., Webster A., Slocombe P. M., Stephens P. E., Knäuper V., Docherty A. J., and Murphy G. (2000) The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 473, 275–279 10.1016/S0014-5793(00)01528-3 [DOI] [PubMed] [Google Scholar]
- 83. Amour A., Slocombe P. M., Webster A., Butler M., Knight C. G., Smith B. J., Stephens P. E., Shelley C., Hutton M., Knäuper V., Docherty A. J., and Murphy G. (1998) TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 435, 39–44 10.1016/S0014-5793(98)01031-X [DOI] [PubMed] [Google Scholar]
- 84. Sahni S., Bae D. H., Lane D. J., Kovacevic Z., Kalinowski D. S., Jansson P. J., and Richardson D. R. (2014) The metastasis suppressor, N-myc downstream-regulated gene 1 (NDRG1), inhibits stress-induced autophagy in cancer cells. J. Biol. Chem. 289, 9692–9709 10.1074/jbc.M113.529511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Krishan S., Richardson D. R., and Sahni S. (2016) The anticancer agent, di-2-pyridylketone 4,4-dimethyl-3-thiosemicarbazone (Dp44mT), up-regulates the AMPK-dependent energy homeostasis pathway in cancer cells. Biochim. Biophys. Acta 1863, 2916–2933 10.1016/j.bbamcr.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 86. Al-Akra L., Bae D. H., Sahni S., Huang M. L. H., Park K. C., Lane D. J. R., Jansson P. J., and Richardson D. R. (2018) Tumor stressors induce two mechanisms of intracellular P-glycoprotein-mediated resistance that are overcome by lysosomal-targeted thiosemicarbazones. J. Biol. Chem. 293, 3562–3587 10.1074/jbc.M116.772699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gao J., and Richardson D. R. (2001) The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents, IV: the mechanisms involved in inhibiting cell-cycle progression. Blood 98, 842–850 10.1182/blood.V98.3.842 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.