Abstract
Deleted-in-liver cancer 1 (DLC1) exerts its tumor suppressive function mainly through the Rho-GTPase–activating protein (RhoGAP) domain. When activated, the domain promotes the hydrolysis of RhoA-GTP, leading to reduced cell migration. DLC1 is kept in an inactive state by an intramolecular interaction between its RhoGAP domain and the DLC1 sterile α motif (SAM) domain. We have shown previously that this autoinhibited state of DLC1 may be alleviated by tensin-3 (TNS3) or PTEN. We show here that the TNS3/PTEN-DLC1 interactions are mediated by the C2 domains of the former and the SAM domain of the latter. Intriguingly, the DLC1 SAM domain was capable of binding to specific peptide motifs within the C2 domains. Indeed, peptides containing the binding motifs were highly effective in blocking the C2-SAM domain-domain interaction. Importantly, when fused to the tat protein-transduction sequence and subsequently introduced into cells, the C2 peptides potently promoted the RhoGAP function in DLC1, leading to decreased RhoA activation and reduced tumor cell growth in soft agar and migration in response to growth factor stimulation. To facilitate the development of the C2 peptides as potential therapeutic agents, we created a cyclic version of the TNS3 C2 domain-derived peptide and showed that this peptide readily entered the MDA-MB-231 breast cancer cells and effectively inhibited their migration. Our work shows, for the first time, that the SAM domain is a peptide-binding module and establishes the framework on which to explore DLC1 SAM domain-binding peptides as potential therapeutic agents for cancer treatment.
Keywords: deleted in liver cancer 1 (DLC1); Ras homolog gene family, member A (RhoA); cell migration; cell proliferation; breast cancer; C2 domain; RhoGAP; SAM domain
Introduction
Deleted-in-liver cancer 1 (DLC1)4 is a tumor suppressor that was initially implicated in hepatocellular cancers (1–4). It is now known to be deregulated, via deletion or down-regulation by epigenetic mechanisms, in malignancies of the lung, stomach, colon, kidney, uterus, ovary, pancreas, prostrate, and breast (5). DLC1 has four known isoforms, α, β, γ, and 4i. The ubiquitous, 1091-residue DLC1α and the longer, 1528-residue DLC1β isoforms have been associated with focal adhesions (6, 7). Both isoforms harbor three structurally defined domains and an intervening, unstructured serine-rich (SR) region. An N-terminal sterile alpha motif (SAM) domain is separated from a Rho-GTPase activating protein (RhoGAP) domain by the SR region (6). The SR region is a hot spot for protein-protein interaction that plays important roles in the regulation of DLC1 function. Of note, the SR region has been shown to bind the 14-3-3 adaptor protein (8), tensins (9–11), the focal adhesion kinase and talin (12). The region has also been shown to assist in protein kinase A-induced dimerization of DLC1 (13). Moreover, the SR region undergoes phosphorylation mediated by the cyclin-dependent Ser/Thr kinase CDK5 (14). Recently, we identified a phosphorylation-mediated molecular interaction switch comprising DLC1, tensin-3 (TNS3), phosphatase and tensin homologue (PTEN), and phosphoinositide 3-kinase (PI3K) (10). We further showed that dynamic interactions of these proteins with each other in response to motility cues such as the epidermal growth factor (EGF) or platelet-derived growth factor (PDGF) played an important role in the migration of mammary epithelial cells and breast cancer cells. Remarkably, phosphorylation of specific Thr residues within the C2 domains of TNS3 and PTEN, following the growth factor stimulation, triggers the switch of binding partners for DLC1 and phosphoinositide 3-kinase to promote cell migration (10).
The function of DLC1 as a tumor suppressor and regulator of cell migration is primarily dependent on its RhoGAP domain, which catalyzes the hydrolysis of GTP-bound RhoA (2, 15). Although the RhoGAP activity may be regulated by phosphorylation of the SR region in DLC1 (14, 16), we have shown that a direct intramolecular interaction between the SAM and RhoGAP domains keeps DLC1 in an autoinhibited state (11, 17). The SAM domain is a versatile module shown to bind RNA (18), lipid (19), and tensins and tensin-like proteins (20). Although some SAM domains have been shown to dimerize or oligomerize (21, 22), there is no evidence to suggest that the DLC1-SAM undergoes oligomerization by itself (13, 23). Nevertheless, the DLC1 SAM domain can bind EF1A1 (24), TNS3 (tensin-3), and PTEN (10, 11, 25), in addition to the RhoGAP domain from the same protein (11, 17). Therefore, the DLC1 SAM domain is a protein interaction module capable of binding to a variety of different proteins. Intriguingly, binding of TNS3 to the DLC1 SAM domain blocks interaction of the latter with the RhoGAP domain, thereby promoting RhoGAP activity and resulting in decreased cellular RhoA-GTP levels (10). These studies suggest that targeting the SAM domain-mediated protein-protein interactions may be an attractive strategy to control cell migration by manipulating cellular RhoA-GTP levels via DLC1.
We report here that the DLC1 SAM domain binds directly to the TNS3 and PTEN C2 domains. Using peptide-walking arrays, we defined the regions in the two C2 domains that mediate SAM-binding and identified specific peptides that bound the SAM domain with micromolar affinities. Our work show, for the first time, the SAM domain is a peptide-interaction module that is capable of binding to multiple peptides derived from TNS3 and PTEN. Importantly, we provide evidence showing that the TNS3 or PTEN C2 domain-derived peptides can inhibit anchorage-independent cell growth and the migration of a variety of cancer cells in response to growth factor stimulation.
Results
The DLC1 SAM domain binds directly to the TNS3 and PTEN C2 domains
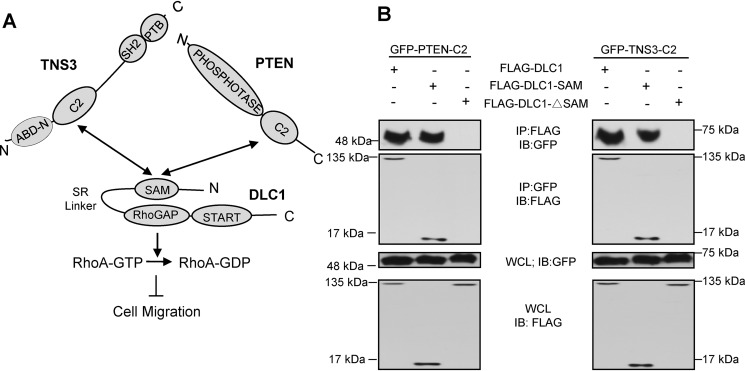
We have previously shown that the DLC1 SAM domain is capable of binding to PTEN and TNS3 through a homologous region (10, 11). Binding by TNS3 or PTEN may activate DLC1 by releasing the intramolecular interaction between the SAM and RhoGAP domains, thereby resulting in increased RhoGAP activity, decreased Rho-GTP level, and reduced cell migration (Fig. 1A). To exploit this mechanism to control cell migration, we first examined if the homologous C2 domains in TNS3 and PTEN would bind directly to the DLC1 SAM domain. To this end, we transfected HEK293 cells with expression constructs for the TNS3-C2 or PTEN-C2 domain fused to green fluorescence protein (GFP) together with the FLAG epitope-tagged DLC1, the SAM domain, or a DLC1 mutant in which the SAM domain was deleted (DLC1-ΔSAM). Immunoprecipitation of the GFP-C2 domains followed by Western blotting showed that both the full-length DLC1 and the SAM domain co-immunoprecipitated with the TNS3 or PTEN C2 domain, whereas DLC1-ΔSAM did not (Fig. 1B). This indicates that the SAM domain is required for DLC1 binding to the C2 domain.
Figure 1.
The SAM domain of DLC1 bound to its own RhoGAP domain and the C2 domain of TNS3 or PTEN. A, a schematic depicting the interactions between the DLC1 SAM domain and the TNS3 or PTEN C2 domain and how these interactions regulate cell migration through the RhoGAP-RhoA pathway. B, Western blots (WB) confirming the SAM-C2 domain interaction in cells. HEK293 cells co-expressing GFP-fused PTEN-C2 (left panel) or TNS3-C2 (right panel) and FLAG-tagged full-length DLC1, DLC1-SAM (SAM domain only), or DLC1-ΔSAM (SAM deletion mutant) were subjected to immunoprecipitation (IP) and immunoblotting (IB) using anti-GFP and anti-FLAG antibodies.
The DLC1 SAM domain recognizes specific peptides within the TNS3 or PTEN C2 domain
The SAM-C2 domain-domain interaction may be mediated by peptide motifs as is often the case for protein interaction modules (26). To explore this possibility, we synthesized peptide spot arrays representing the amino acid (aa) sequences of the PTEN and TNS3 C2 domains, respectively. Each peptide in the spot array is 12-aa in length with 2-aa overlap between neighboring peptides in the array (Fig. S1, A and B). The peptide arrays were subsequently probed for binding to the purified GST-SAM (Fig. 2A) or GST (for background binding, Fig. S2) and the bound protein visualized by anti-GST Western blots. This led to the identification of three distinct regions in the PTEN-C2 domain, with the 230GPTR, 257FFHK, and 334NRYF motifs, respectively, that were recognized by the DLC1-SAM domain (Fig. 2, A, upper panel, and B). In contrast, only a single motif on the TNS3-C2 domain, 244CYHK, was recognized by the SAM domain (Fig. 2, A, lower panel, and B). Intriguingly, this motif shares significant sequence identify to the 257FFHK motif in PTEN-C2 (Fig. 2B), suggesting similar mechanisms may be used by both C2 domains for SAM-binding. Although the C2 peptide arrays included phosphorylated versions of the bound peptides, none was found to bind the DLC1-SAM domain (white boxes in Fig. 2A; Fig. S1, A and B). The SAM-binding peptides mapped to specific β-strands or surface loops at one side of the PTEN-C2 structure (Fig. 2C). Although the TNS3-C2 domain structure is not yet available, it should be noted that the 244CYHK peptide from TNS3-C2 aligns with the 257FFHK motif of PTEN-C2 domain (Fig. 2, B and C).
Figure 2.
The DLC1 SAM domain bound to peptides derived from PTEN or TNS3 C2 domain. A, peptide-walking arrays of the PTEN-C2 (aa 174–403) or TNS3-C2 domain (aa 166–440) were probed with purified GST-SAM and the bound protein detected by anti-GST Western blotting. Peptides representing phosphorylated peptides, which showed no binding, are identified in white boxes. See Fig. S1 for the peptide array map and peptide identities. B, sequence alignment of the PTEN and TNS3 C2 domains with the SAM-binding peptides/motifs identified in A are shown in black boxes. C, SAM-binding peptides from B mapped onto the structure of the PTEN-C2 domain (PDB code 1D5R, aa 188–351; 230GPTR peptide in green, 257FFHK in red, and 334NRYF in blue). D and E, binding of the SAM domain to peptides from PTEN (230GPTRREDKFMYF) or TNS3 (244CYHKKYRSATRD) C2 domains. Shown are representative binding curves from fluorescence polarization (FP) analysis (n = 3) using the corresponding fluorescein-labeled C2 peptides. ΔFP, difference in fluorescence polarization.
To measure the binding affinities of the identified C2 peptides for the SAM domain in solution, we synthesized peptides representing the different motifs with a fluorescein tag. Fluorescence polarization binding assays were conducted for the peptide-SAM complexes and the equilibrium binding curves were used to derive the corresponding dissociation constants (KD) (Fig. 2, D and E; Table 1; Fig. S3). The strongest interaction was observed for the 230GPTR motif-containing peptide from PTEN-C2 (called the PTEN-C2 peptide for short hereafter) with KD of 0.87 μm (Fig. 2D; Table 1). In contrast, the 244CYHK motif-containing peptide (called the TNS3-C2 peptide hereafter) had a KD of 2.18 μm (Fig. 2E; Table 1), which is ∼7-fold stronger than the equivalent 257FFHK peptide from the PTEN-C2 domain (Fig. 2B; Table 1). A peptide containing the other PTEN-C2 motif, 334NRYF, was much weaker in binding, with a KD of 25.87 μm, (Table 1; Fig. S3A). To rule out that the interaction was disulfide bond-dependent, a 244SYHK motif-containing variant of the TNS3-C2 peptide was also synthesized and tested for binding, which showed a minor change in affinity (KD = 3.58 μm) compared with the 244CYHK peptide (Table 1; Fig. S3A).
Table 1.
Equilibrium dissociation constants for the SAM-C2 peptide interactions
All peptides contained a fluorescein tag coupled to the N terminus of the peptide via a 6-aminohexanoic acid spacer.
| Peptide (Sequence) | KDa |
|---|---|
| μm | |
| TNS3-C2 peptide (CYHKKYRSATRD) | 2.18 ± 0.45 |
| 244TNS3-C2 (Cys244-Ser) (SYHKKYRSATRD) | 3.58 ± 1.09 |
| PTEN-C2 peptide (GPTRREDKFMYF) | 0.87 ± 0.18 |
| PTEN-C2-FFHK (FFHKQNKMLKKD) | 14.05 ± 1.51 |
| PTEN-C2-NRYF (NRYFSPNFKVKLY) | 25.87 ± 2.87 |
a KD values were derived from fluorescence polarization measurements.
The importance of the 244CYHK and 230GPTR motifs for SAM-binding was investigated by Ala-scanning peptide spot arrays. The N-terminal half of each peptide was found to play an important role as substitution of any residue within this region led to a marked decrease or loss of SAM-binding (Fig. S4, A and B). To confirm this finding, we synthesized analogues of the C2 peptides in which the 2nd and 3rd residues in the two motifs were substituted by Ala. Fluorescence polarization assay showed that the resulting peptides, containing the GAAR or CAAK motif, lost binding to SAM (Fig. S4, C and D).
The C2 peptides disrupt the SAM domain-C2 domain interaction
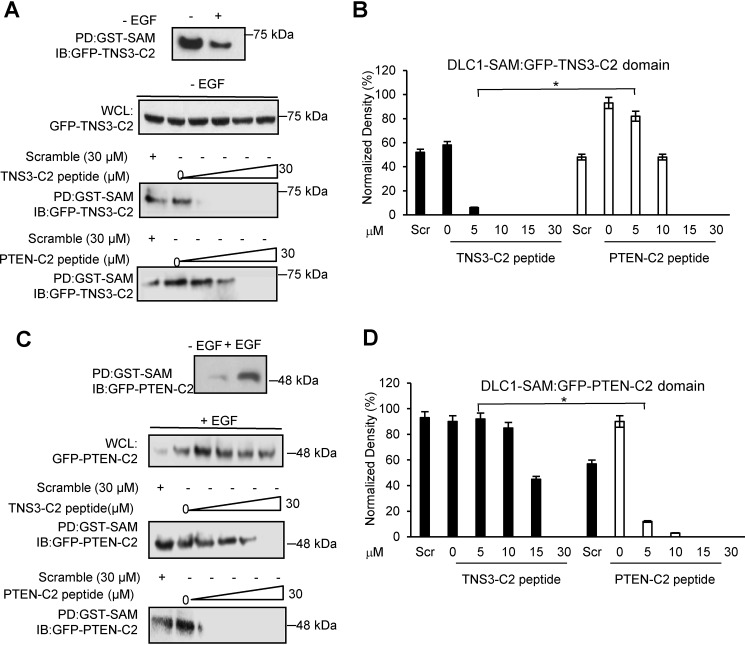
To find out if the two C2 peptides could inhibit the corresponding SAM-C2 interaction in cells, we resynthesized these peptides with an N-terminal, HIV-derived tat-motif (27) to facilitate cellular transduction and a fluorescein tag for easy detection. We also synthesized a scrambled version of the fluorescein-tat-TNS3-C2 peptide as control. The tat-peptides were able to transduce the MDA-MB-231 (breast cancer), HCC78 (lung cancer), and HEK293DLC1 (HEK293 overexpressing DLC1) cells with equal efficiency (Fig. S5) and the transduced peptides appeared as punctate structures that colocalized with actin (Fig. S6). With confirmation of cellular transduction, we then tested if the C2 peptides could block the interaction between GFP-TNS3-C2 or GFP-PTEN-C2 and GST-DLC1-SAM that were co-expressed in the HEK293DLC1 cells. To this end, we first established the conditions for binding between the SAM and two C2 domains. In agreement with previous findings (10), GST pulldown showed that the SAM domain bound more robustly to the TNS3-C2 domain in the absence, than in the presence, of EGF in serum-starved HEK293DLC1 cells (Fig. 3A). In contrast, the SAM-PTEN-C2 interaction was stronger in the presence of EGF stimulation (Fig. 3C). This discrepancy may be due to the distinct effect of phosphorylation of the two C2 domains on SAM-binding (10). To assess the efficacy of the C2 peptides in disrupting the SAM-C2 interaction, an incremental amount of a fluoscein-tat-C2 peptide was added to the cells in the absence or presence of EGF followed by GST pulldown. As shown in Fig. 3, A and B, the TNS3-C2 peptide blocked TNS3-C2 binding to the SAM domain at a concentration as low as 5 μm, whereas the PTEN-C2 peptide exhibited a significant inhibitory effect only at 10 μm or above (Fig. 3, A and B). Intriguingly, in the presence of EGF, the PTEN-C2 peptide disrupted SAM-binding by the PTEN-C2 domain at 5 μm, whereas the TNS3-C2 peptide was unable to completely inhibit the SAM-C2 domain-domain interaction even at 15 μm (Fig. 3, C and D). The scrambled TNS3-C2 peptide control, in contrast, was unable to block SAM binding to either C2 domain even at 30 μm regardless of EGF. These results indicate that the C2 peptides have distinct abilities in disrupting SAM-binding to the corresponding C2 domains. Nevertheless, at high peptide concentrations (e.g. 30 μm), both peptides are effective in blocking the SAM-C2 interactions.
Figure 3.
The C2 peptides disrupted the DLC1 SAM-PTEN/TNS3-C2 domain-domain interactions in cells. A and B, effect of the C2 peptides on DLC1-SAM binding to the TNS3-C2 domain in HEK293DLC1 cells without EGF stimulation. The TNS3 C2 domain bound more tightly to the DLC1 SAM domain in the absence of EGF stimulation; accordingly, the TNS3-C2–derived peptide inhibited this interaction more effectively than the PTEN C2–derived peptide. Quantification of the Western blots indicates a significantly different effect between the two C2 peptides at 5 μm (n = 3; *, p < 0.01, Student's t test). C and D, effect of the C2 peptides on DLC1-SAM binding to the PTEN C2 domain in HEK293 cells in the presence of EGF. The PTEN-C2 domain bound to the SAM domain more tightly with EGF stimulation (for 30 min). Accordingly, the PTEN C2–derived peptide blocked this interaction more effectively than the TNS3 C2–derived peptide in HEK293 cells with EGF. Quantification of Western blots (D) indicates a significantly different effect between the two C2 peptides at 5 μm peptide (n = 3; *, p < 0.01, Student's t test). Scr: scrambled TSN3-C2 peptide control, applied at 30 μm.
The C2 peptides inhibited RhoA activation and anchorage-independent cell growth
Because the TNS3-C2 domain can act as inhibitor of cell migration (10), apparently by binding to the SAM domain and mitigating its inhibition of DLC1-RhoGAP, we inquired whether the C2 peptides were sufficient to activate the RhoGAP and thereby, promoting RhoA-GTP hydrolysis (10, 11). To this end, the HEK293DLC1 cells were cultured in serum-free and EGF-containing medium with increasing concentrations of a C2 peptide (from 0 to 18 μm) or scrambled control (at 18 μm). Rhotekin-RBD beads were used to pulldown GTP-bound RhoA from the cell lysate followed by anti-RhoA Western blotting (10). Although both C2 peptides were capable of blocking RhoA activation at 18 μm, the PTEN-C2 peptide was more effective as it abolished cellular GTP-RhoA at 10 μm (Fig. 4A). This result is consistent with the finding above (Fig. 3, C and D) showing that the PTEN-C2 peptide inhibited the C2-SAM interaction more effectively than the TNS3-C2 peptide in the presence of EGF. To ascertain that the inhibitory effect of the C2 peptide on RhoA activation was not limited to the specific cell type or growth factor examined, we repeated the assay on MDA-MB-231 (a breast cancer line) under EGF stimulation and HCC78 (a lung cancer line) under the treatment of HGF (hepatocyte growth factor) and obtained essentially identical results (Fig. 4, B and C). It should be noted that the peptide treatment did not alter the cellular level of DLC1 in any of three cell lines tested (Fig. 4A; Fig. S7). Furthermore, the C2 peptides had no effect on Rac1 activation (Fig. S8), consistent with DLC1 being a Rho-specific GAP.
Figure 4.
The C2 peptides decreased RhoA activity in multiple cell lines and reduced colony formation by DLC1-expressing HEK293 cells. A, both TNS3 and PTEN C2 peptides were able to inhibit RhoA activation in the DLC1-overexpressing HEK293DLC1 cells in a concentration-dependent manner. B and C, the same RhoA-GTP inhibitory effect was observed for the C2 peptides in EGF-treated MDA-MB-231 cells (B) or HGF-treated HCC78 cells (C). D, colony-formation in soft agar for the HEK293DLC1 cells in the absence (no treatment) or presence of a C2 peptide or the scrambled TNS3-C2 peptide. E, quantify of colony-formation data in D; *, denotes a p value < 0.01 (n = 3), Student's t test; IB, immunoblotting.
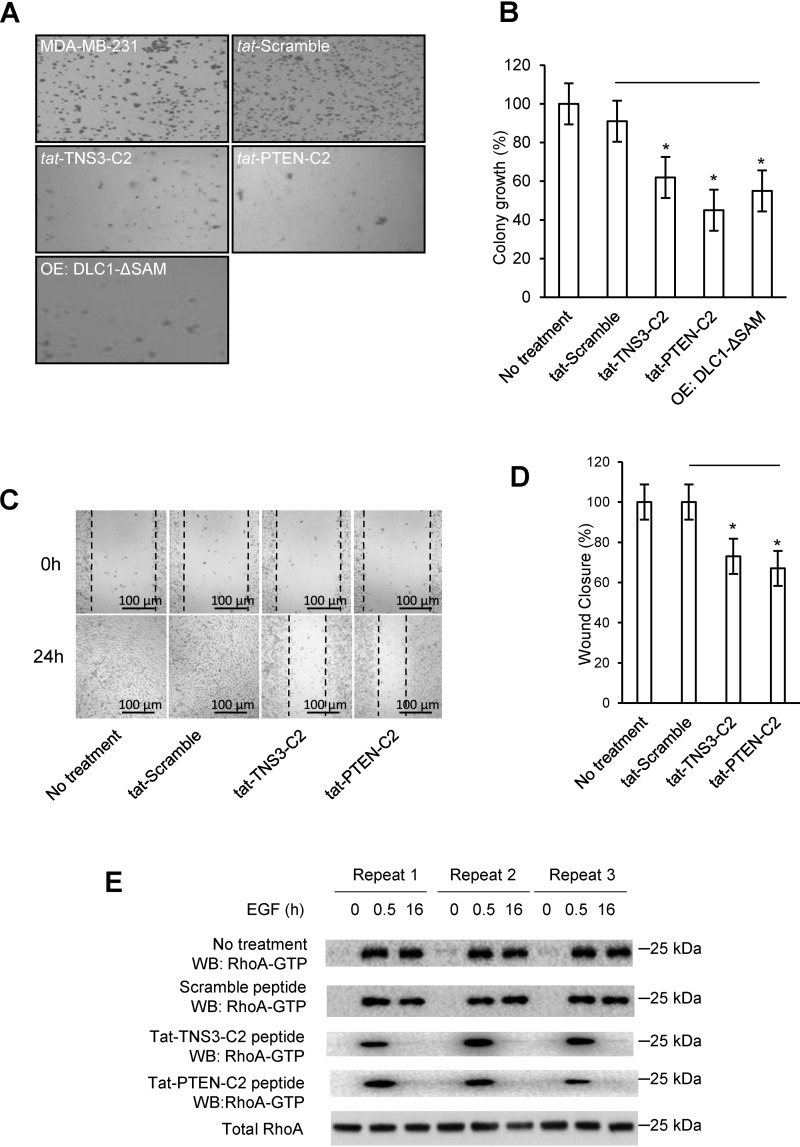
Because RhoA is known to play a role in tumorigenesis (4, 28, 29), we investigated if the corresponding tat-C2 peptides would inhibit cellular transformation using an anchorage-independent cell growth assay. To this end, the HEK293DLC1 cells were seeded on soft agar and allowed to grow in the absence or presence of a C2 peptide. Relative to the scrambled TNS3-C2 peptide control that showed no significant effect on colony formation, both tat-C2 peptides significantly (p < 0.01) reduced the number of colonies formed in soft agar. Moreover, the tat-PTEN-C2 peptide showed a more remarkable effect than the tat-TNS3-C2 peptide (Fig. 4, D and E), again reinforcing earlier results (Fig. 4A). It should be noted that the C2 peptides had no significant effect on cell proliferation or apoptosis (Figs. S9 and S10).
The C2 peptides reduced anchorage-independent growth and growth factor-induced migration of cancer cells
To corroborate the results obtained using HEK293DLC1 cells, we next tested the efficacy of the C2 peptides in inhibiting transformation and migration of MDA-MB-231, a triple negative breast cancer cell line with endogenous expression of DLC1 (10). To this end, we subjected the MDA-MB-231 cells to soft-agar growth assays in the presence of a C2 or control peptide. To ensure that the effect of C2 peptides was mediated by the SAM domain, we overexpressed the SAM domain deletion mutant of DLC1, DLC1ΔSAM, in the MDA-MB-231 cells. The two C2 peptides and DLC1ΔSAM caused a significant reduction in colony formation in soft agar (Fig. 5, A and B). Specifically, the tat-TNS3-C2 peptide reduced colony formation by 38%, whereas the tat-PTEN-C2 peptide reduced the colony number by 55% compared with the scramble control (Fig. 5, A and B). This result is in excellent agreement with that obtained using the HEK293DLC1 cells (Fig. 4, D and E).
Figure 5.
C2 peptides reduced anchorage-independent growth and EGF-dependent migration of breast cancer cells. A, colony formation by MDA-MB-231 cells treated with a C2 peptide or scrambled control (of tat-TNS3-C2) or transfected with DLC1-ΔSAM. B, quantification of data in A to show efficiency in colony formation for the above cells compared with the parent cells (set as 100%). C, representative images of wound-healing in peptide-treated MDA-MB-231 cells. D, quantification of the would-healing data in C. For panels B and D, * denotes p < 0.01 (compared with the scramble peptide, n = 3), Student's t test. E, Western blots of total and active RhoA in peptide-treated MDA-MB-231 cells in response to EGF treatment. Peptide concentration in C–E: 18 μm.
Because RhoA plays a critical role in cell migration (10, 29), we next determined the effect of the C2 peptides on cell migration using wound-healing assays. The C2 or control peptides were added, respectively, into the culture of serum-starved MDA-MB-231 under EGF stimulation (10). We found that both the tat-TNS3-C2 and tat-PTEN-C2 peptide significantly decreased the ability of the MDA-MB-231 cells to migration in response to EGF (Fig. 5, C and D). Rhotekin-RBD pulldown followed by Western blotting of the cell lysate harvested prior to the addition of EGF or at 0.5 or 16 h following EGF stimulation showed that both C2 peptides, but not the scrambled control, effectively inhibited RhoA activation at the 16th hour. These results suggest that the C2 peptides inhibited cell transformation and migration through inactivating RhoA. The C2 peptides showed the same inhibitory effect on the migration of the HCC78 and HEK293DLC1 cells (Fig. S11).
Cyclization of the TNS3-C2 peptide facilitated plasma membrane penetration without loss of anti-migration effect
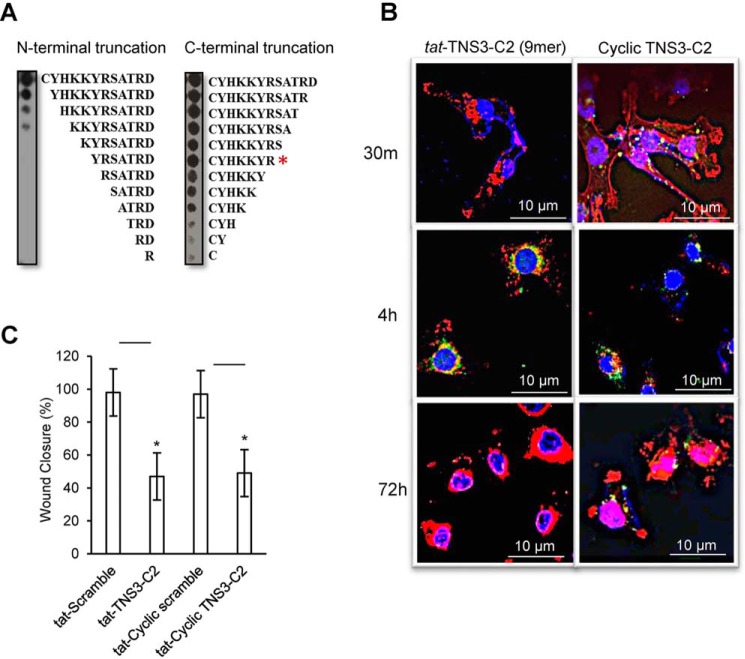
Because the target (DLC1) of the C2 peptides is intracellular, the inability of these peptides to cross the lipid bilayer of the plasma membrane greatly limits their therapeutic potential. Although we have shown that this limitation may be overcome by fusing the peptide with the tat sequence, we also wanted to test if cyclization of the C2 peptides themselves would provide an alternative for their intracellular delivery. Unlike linear peptides, cyclic peptides, exemplified by cyclosporine (30, 31), have been shown to penetrate the plasma membrane. Compared with the PTEN-C2 peptide that required all 12 aa for optimal binding, the TNS3 C2 peptide appeared to be less stringent in sequence requirement (Fig. S4, A and B). We therefore focused our cyclization of the latter. To optimize efficiency of cyclization, we first subjected the TNS3-C2 peptide to serial truncations to identify the minimal sequence for SAM-binding. Probing the peptide truncation arrays with GST-SAM allowed the identification of a 7-residue motif, CYHKKYR, which retained full binding ability of the parent peptide (Fig. 6A, red asterisk). To enable cyclization, we added the Gly-Cys dipeptide to the C terminus of this motif and used oxidization by air to promote the cyclization of the resulting peptide (now a 9-mer) through formation of an intramolecular Cys-Cys disulfide bond (32, 33). Ellman's reagent assay, HPLC separation, and MALDI-MS verified the cyclic peptide with a final yield of ∼52% (Fig. S12).
Figure 6.
Cyclization of the TNS3-C2 peptide led to increased stability without comprising its ability to inhibit MDA-MB-231 cell migration. A, identification of the minimal length for the active TNS3-C2 peptide by peptide truncation spot array. Peptide spot arrays representing N- or C-terminal truncation (or Ala-scanning analogues, Fig. S4A) of the TNS3-C2 peptides were probed with the GST-SAM domain. Based on the binding data, a 9-mer peptide (CYHKKYS-GC) was synthesized for cyclization. B, the cyclized C2 peptide showed increased cellular stability. MDA-MB-231 cells were transduced with fluorescein-labeled tat-TNS3-C2 9-mer linear peptide or the corresponding cyclized TNS3-C2 peptide (without tat label) and fixed at the indicated time points. The cells were stained with DAPI (blue for the nucleus) and phalloidin (red for actin) and imaged by confocal microscopy. The fluorescein-labeled peptides were shown in green punctate structures. C, serum-starved MDA-MB-231 cells were transduced with scrambled, linear, and cyclized TNS3 C2 peptides (each at 18 μm) and subjected to wound healing assays under EGF stimulation. Closed wound areas were measured at 18 h and are represented as percentages of wound area closed relative to the untreated cells; * denotes a p value < 0.01, n = 3; Student's t test.
Confocal microscopy suggested that the fluorescein-labeled TNS3-C2 cyclic peptide penetrated the MDA-MB-231 cells more efficiently than the corresponding linear peptide fused to the tat sequence as the former was detected intracellularly within 30 min, whereas the latter at 4 h (Fig. 6B). Moreover, the cyclic peptide, but not the linear counterpart, was detectable at 72 h, suggesting the cyclic peptide was more stable than the linear version (Fig. 6B). Importantly, the cyclic TNS3-C2 peptide showed a similar level of efficiency as the liner peptide in inhibiting the migration of the MDA-MB-231 cells triggered by EGF (Fig. 6C).
Discussion
DLC1 has been implicated in numerous cancers, including metastatic breast cancer (10, 11, 25, 34, 35). Recent studies have underscored the importance of DLC1 in actin skeleton re-organization and directional cell migration via its interactions with TNS3 and PTEN (10, 11, 36). TNS3 and PTEN act as alternate binding partners for DLC1 or P13K, depending on the phosphorylation states of the TNS3 and PTEN C2 domains (10). The binding partner-switch for DLC1 has been shown to regulate the spatiotemporal activation of Rac1 and RhoA under growth factor stimulation and govern directional cell migration fate (10). These previous studies suggest that the DLC1-TNS3/PTEN interactions are mediated by the C2 domains of TNS3 and PTEN and the SAM domain of DLC1 (10, 11, 25). Furthermore, these studies also indicate that the SAM domain plays a crucial role in the autoinhibition of the RhoGAP domain. Lowy and colleagues (12, 14, 16, 37) have shown that the autoinhibition of the DLC1-RhoGAP domain may also be mediated by Ser phosphorylation of the SR region by Akt1 and CDK5. It is therefore likely that either or both of the SAM domain and SR region may regulate the RhoGAP activity in DLC1, depending on cellular context.
The SAM domain is a small (60–90 residue) helical domain present in numerous proteins in the animal and plant kingdoms, including >200 in human proteins (17, 22). The modular domain primarily acts as scaffolding entity in larger, multidomain proteins involved in orchestrating cellular signaling. SAM domains display a remarkable promiscuity in binding macromolecules that include proteins, lipids, and RNA (18, 19, 22). There have been extensive investigations into the structure and function of the SAM domain of the EphA2 tyrosine kinase receptor (38–44). In comparison, the function of the DLC1 SAM domain is not fully understood despite a few early studies on the topic (17, 24, 45, 46). We have shown here that the DLC1 SAM domain binds directly to the TNS3 or PTEN C2 domain with micromolar affinities. Moreover, multiple peptides from the two C2 domains are capable of binding the DLC1 SAM domain. These findings have several important implications. First, our study shows, for the first time, that the DLC1 SAM domain is both a protein- and a peptide-interaction module. Although the SAM domain has been shown to form homo- or heterodimers, it has not been characterized for binding to other proteins or peptides. The versatile nature of the DLC1 SAM domain in protein/peptide binding greatly expands the repertoire of proteins that this modular domain may interact. It is likely that other SAM domains may have the same characteristics and therefore warrant further investigation. Second, our study suggests that the DLC1 SAM domain is capable of binding to peptides with distinct sequences. It would be interesting to find out how this versatility in ligand-binding is accomplished by such a small modular domain. Third, our study identifies the C2 domain as a protein-binding module. Like the SAM domain, the C2 domain has been found in numerous proteins (47). Studies to date have been focused on the calcium-dependent lipid-binding property of the C2 domain. In this regard, the PTEN/TNS3 C2 domains bind phospholipids, but in a calcium-independent manner (47, 48). Our work supports the notion that the C2 domain is also a protein-interaction module (49–51). It would be interesting to investigate if other C2 domains possess the same protein-binding capability and whether some C2 domains have the dual properties of lipid- and protein-binding.
Protein-protein interactions are frequently mediated by modular domains and targeting protein-protein interaction is an attractive approach for cancer therapeutics (50, 51). We explored this principle in the current study by identifying and developing peptides that block the SAM-C2 interaction. We have shown that peptides derived from the TNS3 or PTEN C2 domains are potent inhibitors of the SAM-C2 interaction in cells. These peptides, when fused to the tat protein transduction sequence (27, 52) or cyclized, effectively activated DLC1 RhoGAP in both DLC1-expressing HEK293 cells or cancer cells, including MDA-MB-231 and HCC78. The ensuing inactivation of RhoA led to markedly reduced tumorigenic potential for these cells as assessed by anchorage-independent growth and decreased migratory potential as measured by wound-healing assay. Although DLC1 expression can be silenced by epigenetic mechanisms, many cancers express DLC1 (53). Therefore, peptide-based strategies to activate DLC1 RhoGAP, as illustrated by this study, may be a feasible approach in cancer therapy. Our finding that both the linear tat-C2 peptide and the cyclic version were capable of effectively inhibiting tumorigenic and migratory potential of cancer cells raises the possibility that these peptide inhibitors may be exploited for potential therapeutic applications.
Experimental procedures
Antibodies
Rabbit anti-DLC1 (H-260; sc-32931) and mouse anti-DLC1 (C-12; sc-271915) were obtained from Santa Cruz Biotech Inc. Anti-RhoA (catalog number ARH03) was supplied in the RhoA pulldown kit from Cytoskeleton Inc. Anti-GST-HRP (catalog number A7340), rabbit anti-GFP (catalog number G1544), and mouse anti-FLAG (M2; catalog number F1804) were obtained from Sigma-Aldrich. Rabbit anti-Rac1 was obtained from GenScript (A00660). Goat anti-mouse IgG (H+L)-HRP (catalog number 170–6516) and anti-rabbit IgG (HþL)-HRP (catalog number 170–6515) conjugates were obtained from Bio-Rad Laboratories.
Expression constructs
DNA sequence encoding the DLC1-SAM (residues 13–78) was subcloned into the pGEX-2T vector (Addgene) for the expression of GST-SAM in Escherichia coli. FLAG-DLC1, FLAG-DLC1-SAM, FLAG-DLC1ΔSAM, GFP-TNS3-C2, and GFP-PTEN-C2 for mammalian cell expression were cloned as reported previously (10).
Cell culture and transfection
HEK293, MDA-MB-231, and HCC78 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured as monolayers as described (10). HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing antibiotics and 10% fetal bovine serum (FBS; Sigma-Aldrich). MDA-MB-231 cells were grown in DMEM/F-12 containing antibiotics and 10% FBS. HCC78 cells were grown in RPMI1640 medium containing antibiotics and 10% FBS. Serum-free/starved growth media contained no FBS and growth factor. EGF treatment medium contained 20 ng/ml of EGF, and EGF treatment was for 30 min at 37 °C in 5% CO2. Plasmids (2 μg) were transiently transfected using X-tremeGENE HP transfection reagent (Roche) at 70% confluence, using the manufacturer's protocols. Cultures were allowed to grow for an additional 16 h before further treatments.
Peptide arrays and soluble peptide synthesis
Cellulose spot peptide arrays and soluble peptides were synthesized on a Multipep synthesizer from Intavis AG Bioanalytical Instruments. Peptide arrays were printed on cellulose membranes and soluble peptides were synthesized on TFA-cleavable resins (Rink resin; AnaSpec Inc.) using Fmoc chemistry. Design, probing, and blotting (Far Westerns) of peptide arrays followed the same procedures as reported previously (54). Peptide walking arrays were designed with 2-amino acid residue overlaps. Truncation peptide arrays were designed to interrogate single-amino differences. N-hydroxysuccinimide (NHS) fluorescein (Pierce) was used to label the N-terminal of soluble peptides when required. 6-Aminohexanoic acid (AnaSpec Inc.) linkers separated the Tensin3 (TNS3) and PTEN C2 peptide motifs from the cell-penetrating peptide (HIV-based tat motif, GRKKRRQRRRPQ) and the NHS-fluorescein label, to avoid possible steric interference during protein-peptide interactions. Peptides were purified by repeated cold-ether precipitation and desalted on Sephadex® G10 (Sigma-Aldrich) columns or purified on HPLC (C18 columns; Waters). Peptide masses were validated by MALDI MS.
Determination of equilibrium dissociation constant (KD) by fluorescence polarization
Binding affinities of GST and the GST-DLC1-SAM domain for the fluorescein-labeled peptides from TNS3-C2 or PTEN-C2 were determined using a Multi-label Reader (PerkinElmer Life Sciences). Fluorescein-labeled peptides were diluted and incubated with increasing concentrations of DLC1-SAM in 20 mm Tris (pH 7.0), 150 mm NaCl2, 3 mm DTT. The FP data (experiments performed in triplicates) was fitted to a nonlinear regression model as described previously (55).
Pulldown assays for RhoA-GTP
Rhotekin-RBD beads pulldown and immunoblotting for Rho-GTP and total RhoA followed the manufacturer's protocol (Cytoskeleton Inc., BK036). For peptide penetration, tat-peptides (at concentrations of 0–18 μm) were incubated with the cell culture at 37 °C for 0.5 h before lysate preparation, pulldown, and immunoblotting. For the GFP-C2 domain pulldown assay, 0–30 μm tat-C2 peptides were used to treat the HEK293DLC1 cells. Western blots were quantified using ImageJ.
Pulldown assays for Rac1-GTP
PAK-PBD beads pulldown and immunoblotting for Rac1-GTP and total Rac1 followed the manufacturer's protocol (Cytoskeleton Inc., BK035S). For peptide penetration, tat-peptides (at concentrations of 0–18 μm) were incubated with the cell culture at 37 °C for 0.5 h before lysate preparation, pulldown, and immunoblotting. Western blots were quantified using ImageJ.
Fluorescence microscopy and confocal microscopy
MDA-MB-231 cells were grown in glass-bottom dishes until 20–40% confluence. Cultures were incubated for 30 min to 72 h with linear (9-mer) and cyclic (9-mer) fluorescein-tagged tat-labeled or nonlabeled peptides (18 μm) in serum-free media containing EGF. Cells were fixed by incubation in 3.7% formaldehyde for 5 min. The fixed cells were washed with 1× phosphate-buffered saline (PBS) twice and treated with 0.2% Triton X-100 for 10 min. The fixed cells were rhodamine phalloidin stained and washed with 1× PBS (twice). Cells were imaged using a LSM-510 Zeiss META/ConfoCor2 microscope or an Olympus FV1000 microscope, after adding VectaShield® mounting medium containing DAPI to the fixed cells.
Anchorage-independent growth (soft-agar) assay
HEK293 and MDA-MB-231 cells (70% confluence) were transfected with plasmids encoding DLC1-SAM or treated with a tat-peptide (30 μm). After incubation with peptides for 4 h, the cells were trypsinized and plated at a density of 5 × 104 cells in 0.30% agarose in DMEM (10% FBS), on a layer of 0.6% agarose suspended in DMEM (10% FBS) in 6-well-plates (in triplicates). A small aliquot of cells was saved for ascertaining cell counts using a hemocytometer, post-peptide treatment (4 h) or transfection (16 h). The soft agar bilayers, with proliferating colonies, were replenished with DMEM (10% FBS) containing ±tat-peptides and EGF, twice a week. Colonies were allowed to grow in 37 °C in 5% CO2 for 22 days. Soft agar wells were stained for 1 h with crystal violet (Sigma-Aldrich) and washed in PBS. Colonies were counted and the numbers normalized to untreated cell cultures grown in soft-agar. Colonies >100 μm in diameter were counted. Representative micrographs were taken using an Infinity Capture Imaging system (Lumenera Corporation) mounted on a Motic AE31 inverted microscope (Matic Microscope).
Wound-healing assay
Cell monolayers at ∼100% confluence in serum-free medium were scratched using a 200-μl pipette tip and cell debris were rinsed off with PBS. Wounded monolayers were incubated for 24 h in ±serum, ±EGF, and tat-peptides (18 μm) or cyclic peptides (30 μm). Images were captured at 0 and 24 h using the Infinity Capture Imaging System (Lumenera Corporation) mounted on a Motic AE31 inverted microscope (Matic Microscope). The cell migration front was established as one that had a continuous line of live migrating cells. As all wounds created were not the exact size between wells in which the cells were allowed to migrate, the wound areas at the end points were calculated by normalizing it to the wound area at 0 h for each well. Wound area dimensions were then determined using the analyze and measure functions of the ImageJ software and are expressed as a percentage.
Optimization of TNS3-C2 peptide
The 7-mer motif of TNS3-C2 was used as the core sequence for synthesizing cyclic peptides with two additional amino residues (Gly-Cys) at the C-terminal end to provide a linker and a disulfide-bond partner. The 9-mer linear peptide was synthesized as described before and all further cyclization steps were done on-resin. The peptide was allowed to cyclize, via disulfide bond formation at room temperature by incubating the peptide-resin in 18% DMSO at pH 6.0 for 48 h (32, 33, 56). The side chain protection groups were removed in a sequential manner, with the cysteine side chains de-protected first before cyclization. The reduction of free-sulfhydryl groups were tracked by spectrometer readings, using Ellman's reagent (5,5′-dithiobis-(2-nitrobenzoic acid); Sigma-Aldrich) at 412 nm. Monomers and multimers were separated by HPLC and the peaks analyzed by MALDI-MS. Total yield and purity of cyclic peptides were calculated from the volume of the reaction mixture loaded onto the HPLC column, the area under the HPLC peaks, and from a cysteine-Ellman's reagent assay-based standard curve. The cyclic peptides were labeled with fluorescein after 20% piperidine treatment of the peptides to remove the Fmoc moiety and before the cleavage of the peptides from resin with 95% TFA.
Flow cytometry
Cells were culture until 70% confluence. After 16 h serum starvation, cells were incubated with 18 μm peptides for 24 h. Subsequently, cells were harvested and resuspended in annexin binding buffer (10 mm Hepes (pH 7.4), 140 mm NaCl, and 2.5 mm CaCl2) and stained with annexin V-FITC (Biolegend) and SYTOXTM AADvanced (ThermoFisher Scientific). All flow cytometry samples were analyzed using LSRII flow cytometer (BD Biosciences) and FlowJo V10 (FlowJo LLC). A minimum of 20,000 events was recorded.
Cell proliferation assay
Cells were cultured in 100 μl of medium in 96-well-plates. After 16 h serum starvation, cells were incubated with 18 μm peptides for 24 h. Subsequently, cell number was evaluated using Sigma WST-8 ((2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt). 10 μl of WST-8 solution was directly added to the 100 μl of medium. Cells were then returned to the incubation chamber for 30 min. The 96-well-plates were then read for absorbance at 460 nm.
Peptide transfection efficiency detection by fluorescein absorbance measurement
Cells were cultured in 96-well-plates. After 16 h serum starvation, cells were incubated with or without 18 μm peptides for 4 h. After washing twice with PBS, 200 μl of RIPA buffer (25 mm Tris, 75 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, pH 7.6) was used to lyse the cells. Intracellular fluorescence was detected by 480 and 535 nm for excitation and emission, respectively. For comparison, the absorbance of the scramble peptide was set as 100% (Fig. S5).
Statistical analysis
All statistical analyses were based on paired Student's t test using Excel and GraphPad®. A p value of <0.01 was considered to be statistically significant.
Author contributions
R. J., L. Q., and S. S.-C. L. data curation; R. J. and L. Q. software; R. J. and L. Q. formal analysis; R. J., L. Q., X. C., S. Z., C. V., and W. M. investigation; R. J. and L. Q. methodology; R. J. and S. S.-C. L. writing-original draft; L. Q. conceptualization; L. Q. validation; L. Q. and S. S.-C. L. writing-review and editing; W. M. and S. S.-C. L. supervision; S. S.-C. L. resources; S. S.-C. L. funding acquisition; S. S.-C. L. project administration.
Supplementary Material
This work was supported, in part, by grants from the Canadian Institute of Health Research (to S. S. C. L.), the Canadian Cancer Society (to S. S. C. L.), and the Natural Science Foundation of Hubei Province, China Grant 2016CFA053 (to X. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S12.
- DLC1
- deleted-in-liver cancer 1
- SR
- serine-rich
- SAM
- sterile α motif
- Rho-GTP
- Rho-GTPase–activating protein
- TNS3
- tensin-3
- PTEN
- phosphatase and tensin homologue
- EGF
- epidermal growth factor
- PDGF
- platelet-derived growth factor
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- aa
- amino acid
- GST
- glutathione S-transferase
- HRP
- horseradish peroxidase
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- DAPI
- 4′,6-diamidino-2-phenylindole
- HGF
- hepatocyte growth factor
- WST-8
- 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt.
References
- 1. Zhou X., Thorgeirsson S. S., and Popescu N. C. (2004) Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene 23, 1308–1313 10.1038/sj.onc.1207246 [DOI] [PubMed] [Google Scholar]
- 2. Wong C. C., Wong C. M., Ko F. C., Chan L. K., Ching Y. P., Yam J. W., and Ng I. O. (2008) Deleted in liver cancer 1 (DLC1) negatively regulates Rho/ROCK/MLC pathway in hepatocellular carcinoma. PLoS ONE 3, e2779 10.1371/journal.pone.0002779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xue W., Krasnitz A., Lucito R., Sordella R., Vanaelst L., Cordon-Cardo C., Singer S., Kuehnel F., Wigler M., Powers S., Zender L., and Lowe S. W. (2008) DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 22, 1439–1444 10.1101/gad.1672608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou X., Zimonjic D. B., Park S. W., Yang X. Y., Durkin M. E., and Popescu N. C. (2008) DLC1 suppresses distant dissemination of human hepatocellular carcinoma cells in nude mice through reduction of RhoA GTPase activity, actin cytoskeletal disruption and down-regulation of genes involved in metastasis. Int. J. Oncol. 32, 1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Durkin M. E., Avner M. R., Huh C. G., Yuan B. Z., Thorgeirsson S. S., and Popescu N. C. (2005) DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 579, 1191–1196 10.1016/j.febslet.2004.12.090 [DOI] [PubMed] [Google Scholar]
- 6. Braun A. C., and Olayioye M. A. (2015) Rho regulation: DLC proteins in space and time. Cell Signal. 27, 1643–1651 10.1016/j.cellsig.2015.04.003 [DOI] [PubMed] [Google Scholar]
- 7. Ko F. C., Yeung Y. S., Wong C. M., Chan L. K., Poon R. T., Ng I. O., and Yam J. W. (2010) Deleted in liver cancer 1 isoforms are distinctly expressed in human tissues, functionally different and under differential transcriptional regulation in hepatocellular carcinoma. Liver Int. 30, 139–148 10.1111/j.1478-3231.2009.02133.x [DOI] [PubMed] [Google Scholar]
- 8. Scholz R. P., Regner J., Theil A., Erlmann P., Holeiter G., Jähne R., Schmid S., Hausser A., and Olayioye M. A. (2009) DLC1 interacts with 14-3-3 proteins to inhibit RhoGAP activity and block nucleocytoplasmic shuttling. J. Cell Sci. 122, 92–102 10.1242/jcs.036251 [DOI] [PubMed] [Google Scholar]
- 9. Chan L. K., Ko F. C., Ng I. O., and Yam J. W. (2009) Deleted in liver cancer 1 (DLC1) utilizes a novel binding site for Tensin2 PTB domain interaction and is required for tumor-suppressive function. PLoS ONE 4, e5572 10.1371/journal.pone.0005572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao X., Kaneko T., Li J. S., Liu A. D., Voss C., and Li S. S. (2015) A phosphorylation switch controls the spatiotemporal activation of Rho GTPases in directional cell migration. Nat. Commun. 6, 7721 10.1038/ncomms8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao X., Voss C., Zhao B., Kaneko T., and Li S. S. (2012) Differential regulation of the activity of deleted in liver cancer 1 (DLC1) by tensins controls cell migration and transformation. Proc. Natl. Acad. Sci. U.S.A. 109, 1455–1460 10.1073/pnas.1114368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li G., Du X., Vass W. C., Papageorge A. G., Lowy D. R., and Qian X. (2011) Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin and focal adhesion kinase (FAK). Proc. Natl. Acad. Sci. U.S.A. 108, 17129–17134 10.1073/pnas.1112122108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ko F. C., Chan L. K., Sze K. M., Yeung Y. S., Tse E. Y., Lu P., Yu M. H., Ng I. O., and Yam J. W. (2013) PKA-induced dimerization of the RhoGAP DLC1 promotes its inhibition of tumorigenesis and metastasis. Nat. Commun. 4, 1618 10.1038/ncomms2604 [DOI] [PubMed] [Google Scholar]
- 14. Tripathi B. K., Qian X., Mertins P., Wang D., Papageorge A. G., Carr S. A., and Lowy D. R. (2014) CDK5 is a major regulator of the tumor suppressor DLC1. J. Cell Biol. 207, 627–642 10.1083/jcb.201405105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim T. Y., Vigil D., Der C. J., and Juliano R. L. (2009) Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev. 28, 77–83 10.1007/s10555-008-9167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tripathi B. K., Grant T., Qian X., Zhou M., Mertins P., Wang D., Papageorge A. G., Tarasov S. G., Hunter K. W., Carr S. A., and Lowy D. R. (2017) Receptor tyrosine kinase activation of RhoA is mediated by AKT phosphorylation of DLC1. J. Cell Biol. 216, 4255–4270 10.1083/jcb.201703105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim T. Y., Healy K. D., Der C. J., Sciaky N., Bang Y. J., and Juliano R. L. (2008) Effects of structure of Rho GTPase-activating protein DLC-1 on cell morphology and migration. J. Biol. Chem. 283, 32762–32770 10.1074/jbc.M800617200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aviv T., Lin Z., Lau S., Rendl L. M., Sicheri F., and Smibert C. A. (2003) The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 10, 614–621 10.1038/nsb956 [DOI] [PubMed] [Google Scholar]
- 19. Barrera F. N., Poveda J. A., González-Ros J. M., and Neira J. L. (2003) Binding of the C-terminal sterile alpha motif (SAM) domain of human p73 to lipid membranes. J. Biol. Chem. 278, 46878–46885 10.1074/jbc.M307846200 [DOI] [PubMed] [Google Scholar]
- 20. Thanos C. D., Goodwill K. E., and Bowie J. U. (1999) Oligomeric structure of the human EphB2 receptor SAM domain. Science 283, 833–836 10.1126/science.283.5403.833 [DOI] [PubMed] [Google Scholar]
- 21. Peterson A. J., Kyba M., Bornemann D., Morgan K., Brock H. W., and Simon J. (1997) A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell Biol. 17, 6683–6692 10.1128/MCB.17.11.6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qiao F., and Bowie J. U. (2005) The many faces of SAM. Sci. STKE 2005, re7 [DOI] [PubMed] [Google Scholar]
- 23. Yang X. Y., Guan M., Vigil D., Der C. J., Lowy D. R., and Popescu N. C. (2009) p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth-suppressing activities. Oncogene 28, 1401–1409 10.1038/onc.2008.498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhong D., Zhang J., Yang S., Soh U. J., Buschdorf J. P., Zhou Y. T., Yang D., and Low B. C. (2009) The SAM domain of the RhoGAP DLC1 binds EF1A1 to regulate cell migration. J. Cell Sci. 122, 414–424 10.1242/jcs.027482 [DOI] [PubMed] [Google Scholar]
- 25. Heering J., Erlmann P., and Olayioye M. A. (2009) Simultaneous loss of the DLC1 and PTEN tumor suppressors enhances breast cancer cell migration. Exp. Cell Res. 315, 2505–2514 10.1016/j.yexcr.2009.05.022 [DOI] [PubMed] [Google Scholar]
- 26. Li S. S. (2005) Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem. J. 390, 641–653 10.1042/BJ20050411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bechara C., and Sagan S. (2013) Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 587, 1693–1702 10.1016/j.febslet.2013.04.031 [DOI] [PubMed] [Google Scholar]
- 28. Moscow J. A., He R., Gnarra J. R., Knutsen T., Weng Y., Zhao W. P., Whang-Peng J., Linehan W. M., and Cowan K. H. (1994) Examination of human tumors for rhoA mutations. Oncogene 9, 189–194 [PubMed] [Google Scholar]
- 29. Ullmannova-Benson V., Guan M., Zhou X., Tripathi V., Yang X. Y., Zimonjic D. B., and Popescu N. C. (2009) DLC1 tumor suppressor gene inhibits migration and invasion of multiple myeloma cells through RhoA GTPase pathway. Leukemia 23, 383–390 10.1038/leu.2008.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Myrdal P. B., Karlage K. L., Stein S. W., Brown B. A., and Haynes A. (2004) Optimized dose delivery of the peptide cyclosporine using hydrofluoroalkane-based metered dose inhalers. J. Pharm. Sci. 93, 1054–1061 10.1002/jps.20025 [DOI] [PubMed] [Google Scholar]
- 31. Ahlbach C. L., Lexa K. W., Bockus A. T., Chen V., Crews P., Jacobson M. P., and Lokey R. S. (2015) Beyond cyclosporine A: conformation-dependent passive membrane permeabilities of cyclic peptide natural products. Future Med. Chem. 7, 2121–2130 10.4155/fmc.15.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. White C. J. Y., AK. (2011) Contemporary strategies for peptide macrocylization. Nat. Chem. 3, 509–524 10.1038/nchem.1062 [DOI] [PubMed] [Google Scholar]
- 33. Andreu D., Albericio F., Sole N. A., Munson M. C., Ferrer M, and Barany G. (1994) Formation of disulphide bonds in synthetic peptides and proteins. in Methods in Molecular Biology Peptide Synthesis Protocols, Vol. 35, pp. 91–169, Humana Press, Totowa, NJ: 10.1385/0-89603-273-6:91 [DOI] [PubMed] [Google Scholar]
- 34. Ullmannova V., and Popescu N. C. (2007) Inhibition of cell proliferation, induction of apoptosis, reactivation of DLC1, and modulation of other gene expression by dietary flavone in breast cancer cell lines. Cancer Detect. Prev. 31, 110–118 10.1016/j.cdp.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang X., Popescu N. C., and Zimonjic D. B. (2011) DLC1 interaction with S100A10 mediates inhibition of in vitro cell invasion and tumorigenicity of lung cancer cells through a RhoGAP-independent mechanism. Cancer Res. 71, 2916–2925 10.1158/0008-5472.CAN-10-2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katz M., Amit I., Citri A., Shay T., Caravalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jocob-Hirsch J., et al. (2007)) A reciprocal tensin-3–cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 9, 961–969 10.1038/ncb1622 [DOI] [PubMed] [Google Scholar]
- 37. Tripathi B. K., and Lowy D. R. (2017) DLC1: a tumor suppressor that regulates Rho signaling. Oncotarget 8, 27674–27675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leone M., Cellitti J., and Pellecchia M. (2008) NMR studies of a heterotypic Sam-Sam domain association: the interaction between the lipid phosphatase Ship2 and the EphA2 receptor. Biochemistry 47, 12721–12728 10.1021/bi801713f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mercurio F. A., Costantini S., Di Natale C., Pirone L., Guariniello S., Scognamiglio P. L., Marasco D., Pedone E. M., and Leone M. (2017) Structural investigation of a C-terminal EphA2 receptor mutant: does mutation affect the structure and interaction properties of the Sam domain? Biochim. Biophys. Acta 1865, 1095–1104 10.1016/j.bbapap.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 40. Mercurio F. A., and Leone M. (2016) The Sam domain of EphA2 receptor and its relevance to cancer: a novel challenge for drug discovery? Curr. Med. Chem. 23, 4718–4734 10.2174/0929867323666161101100722 [DOI] [PubMed] [Google Scholar]
- 41. Mercurio F. A., Marasco D., Pirone L., Pedone E. M., Pellecchia M., and Leone M. (2012) Solution structure of the first Sam domain of Odin and binding studies with the EphA2 receptor. Biochemistry 51, 2136–2145 10.1021/bi300141h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mercurio F. A., Pirone L., Di Natale C., Marasco D., Pedone E. M., and Leone M. (2018) Sam domain-based stapled peptides: structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2. Bioorg. Chem. 80, 602–610 10.1016/j.bioorg.2018.07.013 [DOI] [PubMed] [Google Scholar]
- 43. Park J. E., Son A. I., Hua R., Wang L., Zhang X., and Zhou R. (2012) Human cataract mutations in EPHA2 SAM domain alter receptor stability and function. PLoS ONE 7, e36564 10.1371/journal.pone.0036564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi X., Hapiak V., Zheng J., Muller-Greven J., Bowman D., Lingerak R., Buck M., Wang B. C., and Smith A. W. (2017) A role of the SAM domain in EphA2 receptor activation. Sci. Rep. 7, 45084 10.1038/srep45084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H., Fung K. L., Jin D. Y., Chung S. S., Ching Y. P., Ng I. O., Sze K. H., Ko B. C., and Sun H. (2007) Solution structures, dynamics, and lipid-binding of the sterile α-motif domain of the deleted in liver cancer 2. Proteins 67, 1154–1166 10.1002/prot.21361 [DOI] [PubMed] [Google Scholar]
- 46. Kwan J. J., and Donaldson L. W. (2007) The NMR structure of the murine DLC2 SAM domain reveals a variant fold that is similar to a four-helix bundle. BMC Struct. Biol. 7, 34 10.1186/1472-6807-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang D., and Aravind L. (2010) Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene 469, 18–30 10.1016/j.gene.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- 49. Kaneko T., Joshi R., Feller S. M., and Li S. S. (2012) Phosphotyrosine recognition domains: the typical, the atypical and the versatile. Cell Commun. Signal. 10, 32 10.1186/1478-811X-10-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fry D. C., and Vassilev L. T. (2005) Targeting protein-protein interactions for cancer therapy. J. Mol. Med. (Berl.) 83, 955–963 10.1007/s00109-005-0705-x [DOI] [PubMed] [Google Scholar]
- 51. Biron E., and Bédard F. (2016) Recent progress in the development of protein-protein interaction inhibitors targeting androgen receptor-coactivator binding in prostate cancer. J. Steroid Biochem. Mol. Biol. 161, 36–44 10.1016/j.jsbmb.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 52. Howl J., and Jones S. (2009) Transport molecules using reverse sequence HIV-Tat polypeptides: not just any old Tat? (WO200808225). Expert Opin. Ther. Pat. 19, 1329–1333 10.1517/17530050902824829 [DOI] [PubMed] [Google Scholar]
- 53. Low J. S., Tao Q., Ng K. M., Goh H. K., Shu X. S., Woo W. L., Ambinder R. F., Srivastava G., Shamay M., Chan A. T., Popescu N. C., and Hsieh W. S. (2011) A novel isoform of the 8p22 tumor suppressor gene DLC1 suppresses tumor growth and is frequently silenced in multiple common tumors. Oncogene 30, 1923–1935 10.1038/onc.2010.576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jia C. Y., Nie J., Wu C., Li C., and Li S. S. (2005) Novel Src homology 3 domain-binding motifs identified from proteomic screen of a Pro-rich region. Mol. Cell Proteomics 4, 1155–1166 10.1074/mcp.M500108-MCP200 [DOI] [PubMed] [Google Scholar]
- 55. Kaneko T., Huang H., Cao X., Li X., Li C., Voss C., Sidhu S. S., and Li S. S. (2012) Superbinder SH2 domains act as antagonists of cell signaling. Sci. Signal. 5, ra68 [DOI] [PubMed] [Google Scholar]
- 56. Isidro-Llobet A., Alvarez M., and Albericio F. (2009) Amino acid-protecting groups. Chem. Rev. 109, 2455–2504 10.1021/cr800323s [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.