Abstract
Rapid detection of nucleic acids is integral for applications in clinical diagnostics and biotechnology. We have recently established a CRISPR-based diagnostic platform that combines nucleic acid pre-amplification with CRISPR-Cas enzymology for specific recognition of desired DNA or RNA sequences. This platform, termed Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK), allows for multiplexed, portable, and ultra-sensitive detection of RNA or DNA from clinically relevant samples. Here we provide step-by-step instructions for setting up SHERLOCK assays with recombinase-mediated polymerase pre-amplification of DNA or RNA, and subsequent Cas13- or Cas12-mediated detection via fluorescent and colorimetric readouts that provide results in less than 1 hour. We also include guidelines for designing efficient CRISPR RNA and isothermal amplification primers, and discuss important considerations for multiplex and quantitative SHERLOCK detection assays.
INTRODUCTION
Rapid nucleic acid detection is an important part of many applications in human health and biotechnology, including the sensing of infectious agents, agricultural pathogens, or circulating DNA or RNA associated with disease. Standard methods to amplify nucleic acids for detection (such as PCR) are effective, but require instrumentation that is not portable, precluding their deployment in the field. Demand for instrument-free nucleic acid detection technologies has driven the development of multiple methods for isothermal amplification. However, common approaches for isothermal amplification, such as recombinase polymerase amplification (RPA)1, require optimization and cannot typically discriminate between single base pair differences in target sequences, a distinction that can have important consequences for pathogenicity. Recently, enzymes from CRISPR-Cas systems have been adapted for the specific, rapid, sensitive, and portable sensing of nucleic acids.2–4 These approaches rely on Cas135,6 or Cas127, which exhibit non-specific endonuclease activity after binding to a specific target via programmable CRISPR RNAs (crRNA)6,8,9. By combining the programmable specificity of Cas13 or Cas12 with a reporter molecule that is activated upon target recognition, these enzymes result in specific and sensitive indication of the presence or quantity of a nucleic acid.
Here we describe a protocol for SHERLOCK nucleic acid detection (using RPA and CRISPR-Cas13) with instructions on reagent preparation, including recombinant Leptotrichia wadei Cas13 (LwaCas13a) protein expression and purification, as well as in vitro transcription of crRNAs and sample extraction from various starting materials. Experimental design and downstream analysis considerations are also discussed.
Development of the Protocol
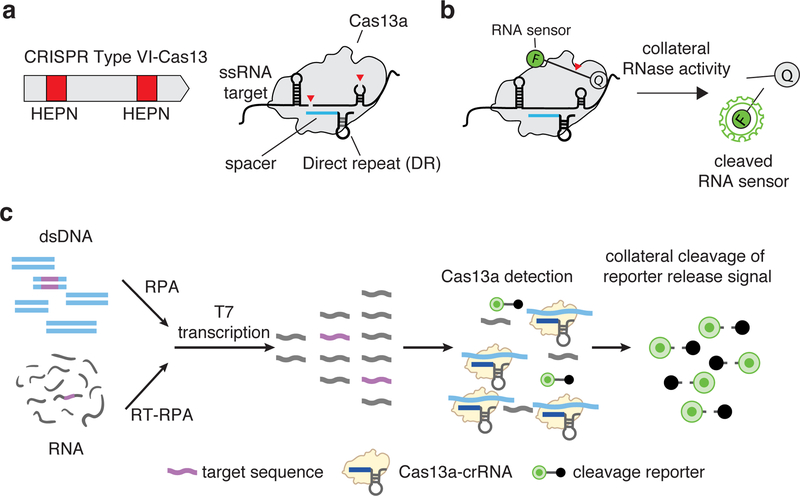
Cas13 (which encompasses four divergent family members, Cas13a-d) is an RNA-guided RNase that produces multiple cleavage sites in single-stranded areas of an RNA target with specific base preferences.5,6,10–13 (Figure 1a). Cas13 also exhibits target-dependent promiscuous RNase activity, leading to trans cleavage of bystander RNA molecules, an effect termed “collateral activity”6. It has recently been discovered that Cas effectors from other types of CRISPR-Cas systems also display collateral activity, including subfamilies of Cas12 (Box 1)8,9. The discovery of collateral activity, which links highly specific target recognition to a subsequent general RNase activity that can cleave a reporter molecule (Figure 1b), made it possible to develop a new approach for nucleic acid detection6,10,14.
Figure 1: Cas13 complex and collateral activity.
(a) CRISPR-Cas13 RNA targeting complex components. CRISPR-Cas13 enzymes are programmed by a crRNA, which is composed of a direct repeat sequence (DR) flanked by a target complementary spacer sequence (shown in blue). RNA cleavage is mediated by two Higher eukaryotic and prokaryotic nuclease domains (HEPN) domains (shown as red boxes) within a typical Type VI-A Cas13.
(b) Reporter unlocking via CRISPR-Cas13 collateral RNase activity. The CRISPR-Cas13-RNA complex is activated by binding to a complementary target RNA. The activation triggers collateral cleavage of a nonspecific RNA sensor in trans. The fluorescently labeled sensor is quenched when intact and emits fluorescence when cleaved by activated CRISPR-Cas13 complex.
(c) SHERLOCK detection assay Schematic of SHERLOCK assay steps, starting with pre-amplification of either a DNA or RNA target input. Amplified targets are converted to RNA via T7 transcription and are then detected by Cas13:crRNA complexes, which activate and cleave fluorescent RNA sensors. For Cas12 detection, the T7 transcription step is omitted, allowing for direct detection of amplified targets.
Box 1. Cas13 and Cas12 orthologs for CRISPR diagnostics.
To date, both the Cas13 and Cas12 protein families of CRISPR systems have been shown to have collateral activity, making them useful for nucleic acid detection applications6,9. Many of the Cas13 subtypes and orthologs have different base preferences, cleaving at specific dinucleotide motifs16. Additionally, Cas13 subtypes differ in CRISPR RNA (crRNA) structure, direct repeat sequence, and size (see Table below). While Cas13 has a PAM-like sequence motif called the protospacer flanking site (PFS) that restricts activity to only certain target sites, there are a number of very active Cas13 orthologs, such as LwaCas13a, that show no PFS. Lack of a PFS is a distinguishing feature of these orthologs that enables them to target any possible sequence or mutation.
Cas12a has weak collateral activity, enabling nucleic acid detection with low sensitivity (low nanomolar range)9,16. When combined with pre-amplification, Cas12a-mediated detection can detect concentrations of 2 aM14,16.
Comparison of Cas13 and Cas12 orthologs
| LwaCas13a | LbaCas13a | CcaCas13b | PsmCas13b | AsCas12a | |
|---|---|---|---|---|---|
| Organism | Leptotrichia wadei | Lachnospiraceae bacterium NK4A179 | Capnocytophaga canimorsus | Prevotella sp. MA2016 | Acidaminococcus sp. BV3L6 |
| Target | ssRNA | ssRNA | ssRNA | ssRNA | ssDNA/dsDNA |
| DR orientation | 5’ | 5’ | 3’ | 3’ | 5’ |
| Motif preference | Poly U/AU | Poly A/AC | Poly U/UA/UC | Poly A/GA | N/A |
| Spacer length | 28 | 28 | 30 | 30 | 20 |
| Sensitivity | ~5e5 aM16 | ~1e9 aM31 | ~5e6 aM16 | ~5e8 aM16 | ~5e8 aM16 |
Nucleic acid detection with collateral activity can be achieved using a variety of reporter molecules and methods, including: 1) gel-based detection with fluorescently-labeled reporters6, 2) fluorimeter detection with quenched fluorescent reporters10, 3) visual detection through modulation of solution turbidity via liquid-liquid phase separation15 and 4) lateral flow detection with antigen-labeled reporters16. Although Cas13 orthologs have been identified that can detect as little as 50 fM of target RNA input (~600,000 molecules)14, they cannot reach single molecule detection, a necessity for many clinical applications. In addition, a versatile detection platform for clinical diagnostics would need to detect DNA as well as RNA.
Several platforms have been developed to boost Cas13 detection. We focus here on SHERLOCK, which includes a pre-amplification step, typically RPA, that can amplify either RNA or DNA and introduce a T7 RNA polymerase promoter, allowing RNA transcription and subsequent detection by Cas13 (Figure 1c)14. Building on the original SHERLOCK technology, we developed additional features (including multiplex fluorescence-based detection and visual readout on lateral flow strips) to create SHERLOCKv216. These additional features are also described in the protocol.
Overview of the Procedure
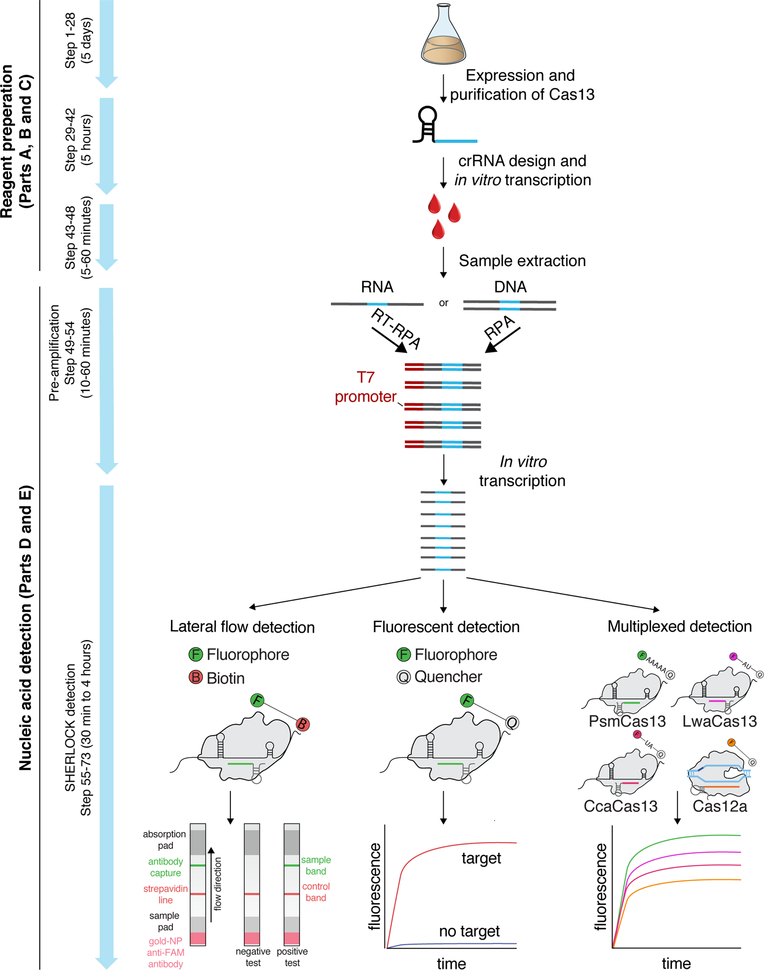
The four main stages of the SHERLOCK procedure are reagent preparation (steps 1–41), sample extraction (step 42), isothermal target nucleic acid pre-amplification (steps 43–48), and CRISPR-Cas13 nucleic acid detection (steps 49–52), as outlined in Figure 2. The first two stages (LwaCas13a protein purification and crRNA in vitro transcription) are optional as both products can be purified by a contract research organization (CRO)or commercially obtained. While this protocol focuses mostly on LwaCas13a purification, it can also be used and adapted for Cas13b purification. The next two stages are the pre-amplification reaction, most often with RPA, and the CRISPR collateral detection reaction, which cleaves a nucleic acid reporter and generates a detectable signal (Figure 1c). These two reactions can either be run sequentially, with a transfer step in between (two-step detection procedure; steps 49–52 of main procedure), or as a single combined mixture (one-pot detection procedure; Box 2).
Figure 2: Complete SHERLOCK experimental workflow.
First, LwaCas13a is recombinantly expressed and purified in E. coli (steps 1–28). After crRNA design and in vitro transcription (steps 29–41), sample extraction is performed to yield target nucleic acid (step 42). This sample is then used for RPA-based pre-amplification (steps 43–48) and detection by Cas13 (steps 49–52). Detection can be performed as a single-plex colorimetric lateral flow reaction (step 52 option A), or as fluorescence-based single or multiplex SHERLOCK reactions (step 52 option B). Multiple targets can be detected within the same reaction using CRISPR-Cas13 enzymes with orthogonal cleavage preferences or by combining Cas13 with Cas12 in the same assay.
Box 2: One-pot Cas13-SHERLOCK nucleic acid detection (1–4 hours).
Critical: The recommended procedures for RNA or DNA pre-amplification are nearly identical. However, important distinctions are summarized in the Experimental Design section. If multiplexed nucleic acid detection or quantification are desired, the reader should also follow the guidance provided in the Experimental Design section.
Pre-heat the fluorescence plate reader to 37°C.
Prepare the pre-amplification area by wiping down the work surface and pipettors with RNase away.
Thaw normalized crRNA (at 300 ng/μL), SHERLOCK RNase Alert Reporter (2 μM), and Cas13 protein aliquot (2 mg/mL, 14.44 μM) on ice, covered with aluminum foil to protect from light exposure.
Dilute LwaCas13 to 50x (348 ng/mL) by adding 19 μL of protein storage buffer (SB) to 4 μL of LwaCas13a (2 mg/mL).
Dilute the crRNA to 25 ng/μL by adding 137.5 μL of Ultragrade water to 12.5 μL of crRNA (300 ng/μL).
- Prepare the following Cas13-SHERLOCK master mix by adding the following components to an Eppendorf tube in top-down order. The master mix below is enough for one sample with 3–4 technical replicates. Scale up proportionally to the number of samples.
Component per reaction F primer (100μM) 0.48 μL R primer (100μM) 0.48 μL Rehydration Buffer, RPA kit 59 μL LwCas13a in SB (348 ng/mL) 2 μL crRNA (12.5 ng/mL) 0.17 μL RNase Alert v2 (2 μM) 6.83 μL Murine RNase Inhibitors, 40 U/μL 2.73 μL rNTPs 8 μL T7 RNA polymerase, 5 U/μL 2 μL MgCl2, 1M 0.5 μL MgAc, RPA kit 5 μL Ultragrade water 7.82 μL Total 95 μL -
Add 95 μL of master mix without sample to a single pellet aliquot, and carefully resuspend the mixture on ice. Then, transfer the entire reconstituted reaction back to the initial Eppendorf or PCR master mix tube.
Critical. One pellet yields roughly three individual 20 μL one-pot reactions. If a larger number of reactions is needed, simply scale the master mix volume and pellets up accordingly. When doing so, it is critical not to change the pipette in between transfers of resuspended pellets, as the stickiness of RPA reaction components could result in a significant loss of reaction enzymes.
Transfer 71.25 μL of master mix into a PCR-strip tube or 96-well PCR plate. This is the amount for one condition and three technical replicates.
Spin down the samples, carefully open the tubes, and transfer 3.75 μL of each sample to 71.25 μL aliquoted master mix on ice. Briefly vortex and spin down for 15 seconds in a plate to collect the entire reaction mix in the well.
Carefully open the reaction tubes and transfer 20 μL per technical replicate and condition into a 384-well, round black-well clear-bottom bio-assay plate. To group technical replicates, pipette the remaining technical replicates beside and below, to form a square of 2×2 grouped reactions. Note the lower right quadrant of this grid will be empty as the one-pot reactions have only three technical replicates.
-
Briefly spin down the bio-assay plate to remove potential bubbles and place into a 37°C pre-heated Biotek plate reader, or equivalent.
Critical. Following bio-assay plate preparation, quickly place into the fluorescence plate reader as SHERLOCK Cas13 reactions may begin as soon as they are removed from ice.
-
Start data acquisition by monitoring fluorescence over 3 hours at 37°C with a 5-minute interval between well-data acquisitions.
TROUBLESHOOTING
SHERLOCK is compatible with multiple readouts, either fluorescent detection or lateral flow detection, depending on reporter molecule choice. While either Cas12 or Cas13 enzymes can be used for detection, this protocol focuses on Cas13, which requires the introduction of a T7 RNA polymerase promoter during pre-amplification and a T7 RNA polymerase during the detection reaction to generate RNA for Cas13 collateral activation (Figure 1c). For fluorescent detection, detection can be performed either as an endpoint readout or in real time using a plate reader or other compatible fluorometer. Lateral flow detection is an endpoint assay, with lateral flow strips exposed to the reaction mixture post-incubation.
Alternative Methods
SHERLOCK joins a large array of other portable and isothermal methods for nucleic acid detection, including traditional RPA1, loop-mediated isothermal amplification (LAMP)17, and helicase dependent amplification (HDA)18. These methods all provide varying degrees of speed, sensitivity, portability, and ease-of-design. In the case of traditional RPA, risk of non-specific amplification requires the screening of numerous primer sets to find robust pairs. Alternatively, RPA can be combined with probe-based methods, such as exo-probes, to increase the stringency of detection. However, these probes require multiple modifications, such as fluorophores and quenchers, and are expensive to order. LAMP suffers from similar limitations, although a higher temperature of operation (65°C) can reduce non-specific effects. However, this high temperature can make heating difficult in situations without electricity. Furthermore, primer design with LAMP requires four different primer sets, leading to a substantial degree of complexity that limits the programmability of LAMP for certain users. In the case of HDA, commercial methods require high temperatures, long incubation times, and often fail to reach single-molecule detection thresholds.
In addition, portable versions of more conventional methods for nucleic acid detection, such as PCR (e.g. Taqman qPCR) and next generation sequencing (NGS), have been developed for rapid or point-of care-detection19–21. However, these methods require specialized instrumentation that may not be practical in many low-resource settings. While some portable NGS devices are commercially available, they are better suited for unbiased nucleic acid discovery, rather than sensitive and targeted detection of known targets. Thus, portable next generation sequencing and SHERLOCK are complementary approaches in nucleic acid diagnostics.
Since details of SHERLOCK were first published, additional CRISPR-Dx platforms have been developed, including DETECTR and HOLMES9,22. Whereas SHERLOCK refers to the method using a CRISPR enzyme for collateral detection with any pre-amplification of RNA or DNA, DETECTR refers to the specific instance of using Cas12a collateral detection after pre-amplification by RPA9. The HOLMES method, similar to DETECTR, uses Cas12a for DNA or RNA detection after pre-amplification with rapid PCR amplification22. HOLMESv2 is a modified version of HOLMES that allows for one-pot detection using thermophilic Cas12b with loop-mediated isothermal amplification (LAMP)23. Catalytically inactive Cas9 (dCas9) has also been harnessed for a digital chip-based nucleic acid detection platform24, enabling femtomolar detection without pre-amplification, but this approach, referred to as CRISPR-Chip, requires bespoke instrumentation (a graphene field-effect transistor on which Cas9 has been immobilized) that is currently not widely available.
Advantages and Limitations
SHERLOCK is highly sensitive and specific. It is capable of single molecule detection in 1uL sample volumes (2 aM) of both DNA and RNA targets. Additionally, by scaling up the pre-amplification volume, it is possible to achieve single-molecule detection in large sample input volumes up to 540μL (8 zM)16. SHERLOCK leverages the specificity of Cas136,14,25 and Cas12 enzymes7,26,27 to enable single nucleotide mismatch distinction in target sequences. Cas13 does not catalytically activate when there are two or more mismatches in the crRNA:target duplex. As a result, sequences of similar viruses, such as Dengue virus and Zika virus, can easily be distinguished by SHERLOCK14. In cases where detection of the target requires single-base distinction, such as viral and human genotyping or the detection of cancer-associated mutations in circulating nucleic acids, the specificity of Cas13 can be enhanced by the introduction of a “synthetic mismatch” in the crRNA14,16. The Cas13 enzyme used in SHERLOCK does not require strict sequence preferences at the target site, while Cas12 enzymes require a protospacer adjacent motif for cleavage. This allows for more flexibility and broader target range of SHERLOCK compare to DETECTR and HOLMES.
An attractive feature of SHERLOCK is the rapid nature of the assay. Usually, RPA is performed for 5–10 minutes as an initial reaction, and part of this solution is transferred to the Cas13 detection reaction as a two-step reaction, which can then detect the target in as little as 5 minutes. As with sensitivity, RPA primer and crRNA design are important determinants for the kinetics of target detection. It is important to screen designs, as choosing the right primer and crRNA combination can drastically increase speed of detection. RPA can also be combined in the same reaction as Cas13 detection for a one-pot assay, although the viscosity of the mixture can reduce robustness of detection leading to a potentially higher false-negative rate and less sensitivity (Table 1).14 Analogous to one and two-step RT-qPCR, combining pre-amplification and detection in a one-pot assay has both benefits and disadvantages (Table 1). One-pot SHERLOCK is faster and simpler than two-step SHERLOCK. By combining the pre-amplification and Cas13/Cas12 detection into a single one-pot reaction, it is possible to achieve detection in as little as 10–15 minutes for targets in the femtomolar concentration range and in less than an hour for targets in the attomolar concentration range. As well as being faster, there is also less risk of contamination and it is easier to obtain quantitative results (owing to real-time detection) with one-pot SHERLOCK. However, it is also less sensitive than two-step SHERLOCK and experiment optimization is often more challenging. In general, one-pot SHERLOCK is more useful for time-sensitive, high-throughput or quantitative applications, as well as applications with increased contamination risk (e.g. repeated testing, large target amounts), whereas two-step SHERLOCK is more useful for applications with challenging sample inputs (e.g. quick extractions from body fluids such as urine or saliva).
Table 1:
Comparison of two-step and one-pot formats of SHERLOCK
| Two-step SHERLOCK | One-pot SHERLOCK | |
|---|---|---|
| Experimental time | 30–60 minutes | 15–30 minutes |
| Protocol optimization | Easy (each step can be individually optimized) | Difficult (more challenging compared to two-step) |
| Contamination risk | increased risk of surface contamination with pre-amplification reaction | decreased risk due to single tube reaction |
| Quantitative | Difficult (depends on finding time-point where separation is possible) | Easy (owing to real-time detection). |
| Sensitivity | zeptomolar range (single molecule per mL) | femto to attomolar range (1000–1 molecule(s) per μL). Variability in sensitivities have been observed which we largely attribute to the viscous RPA sample composition that can influence Cas13 detection. |
| Appropriate applications |
|
|
|
|
|
|
Another advantage of the SHERLOCK platform over other detection platforms (such as Taqman qPCR) is the low cost of components. A typical single-plex reaction is estimated to cost as little as $0.6014. This is partly because the quencher has a fixed sequence and does not need to be redesigned and ordered for every new target. Additionally, the crRNA is a short RNA sequence that is cheap to synthesize, and the enzyme components can be produced in bulk. Since the majority of reagents are shared between SHERLOCK, DETECTR and HOLMES, the associated costs are similar.
Despite its advantages to existing detection technologies, SHERLOCK has several caveats that can make it unacceptable for certain use cases. SHERLOCK currently involves the preparation and testing of reaction components, some of which require expertise in protein purification and RNA biology. Moreover, pre-designed assays, including reaction mixtures and RNA/DNA oligonucleotides, are currently not commercially available for SHERLOCK. Since validated target assays exist for established PCR-based methods, routine laboratory applications might benefit from such simplified solutions and can minimize the optimization process compared to SHERLOCK. In addition, applications that do not demand the speed or portability of SHERLOCK, such as oncology assays, may be better suited for more standard detection technologies.
Another potential limitation of SHERLOCK is the multi-step nucleic acid amplification process, which may affect precise target quantification. While we recently demonstrated the quantitative detection of nucleic acids with SHERLOCK, absolute digital quantification such as in digital droplet PCR are currently not possible, and small differences in target quantity (<2x changes) have not been demonstrated. SHERLOCK may therefore currently be less suited for accurate gene expression profiling.
Applications
As the SHERLOCK assay described in this protocol forms the basis of a reprogrammable amplification system, it can be applied in any situation that requires sensitive detection of a DNA or RNA target. SHERLOCK has been used for the detection genotyping of bacterial and viral infectious disease agents, including distinguishing single-nucleotide variants and finding antibiotic resistance genes14. For example, SHERLOCK has been applied to detect Zika and Dengue virus directly from patient urine and serum samples, demonstrating the field-deployable nature of the technology. Furthermore, SHERLOCK can distinguish between Zika strains from Honduras, the Dominican Republic, and the United States, even in regions that differ only by a single-nucleotide28. The single-nucleotide specificity of SHERLOCK has also been applied to genotyping of patient samples and detection of cancer-associated mutations from circulating cell-free DNA, even in samples where the target at an abundance of 0.1% compared to background14,16.
Experimental Design
Sample format
An initial consideration for choosing a nucleic acid amplification or detection method is the concentration of RNA or DNA, which is dependent on sample extraction and removal of any inhibitory or confounding components. The appropriate method of extraction depends on the abundance of nucleic acids in the sample, the difficulty of releasing DNA or RNA from any cells or viral particles, and the presence of amplification inhibitors or nucleases that would interfere with downstream reactions. In resource-limited settings such as point-of-care applications, even “standard” laboratory equipment (such as centrifuges) may be unavailable. Thus, selecting a method for isolating nucleic acids is context-dependent and may have to be optimized for special cases. Common commercial methods for isolating and concentrating DNA or RNA from samples include column- or bead-based purification methods, which allow for extraction of pure concentrated nucleic acids but require equipment and power, as well as significant time investment from trained operators. Simpler methods, which do not extract pure nucleic acids, but instead rely on mechanical force or chemical disruption to free DNA or RNA for detection are alternatives in low-resource settings. However, raw nucleic acid extractions may result in contamination by nucleases or PCR/amplification inhibitors. Nucleases can degrade the sample and result in false negative signal, while nuclease carryover to SHERLOCK reactions may degrade the sensor and lead to a false positive signal. For RNA samples, nucleases may also degrade the target RNA, leading to false negative signals. The addition of RNase Inhibitor is therefore essential to prevent such unwanted source of variation. Additionally, pre-treatments such as HUDSON have developed to inactive nuclease commonly found in body fluids28. Other commonly known PCR/amplification inhibitors such as ethanol, detergents, strong acids or bases may lead to failed SHERLOCK reactions and false negatives. For purified nucleic acids, target detection with Cas13 or Cas12 is almost identical for RNA and DNA, as both inputs are pre-amplified as DNA first. However, in the case of RNA, a reverse transcriptase needs to be included during pre-amplification.
SHERLOCK is compatible with many different sample formats, including crude biological matrices14,28. However, it is imperative to remove or inactivate all resident RNases, as they will cleave reporter molecules and contribute to background signal. One approach, termed HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases)28, relies on heating and reducing reagents to prepare samples for input into SHERLOCK reactions. Heating viral particles releases enclosed nucleic acids, while additional treatment with the reducing agent TCEP inactivates RNases that are present in the sample matrix. HUDSON is compatible with human blood, serum, urine, and saliva, and allows instrument-free direct sampling of viruses28.
Multiplexing
Multiplexing with SHERLOCK can be performed either spatially, allowing up to 384 small reactions to be performed in a single reaction plate, or spectrally, allowing for simultaneous detection of up to four targets in a single reaction. The most basic form of SHERLOCK with LwaCas13a, an active ortholog selected from a screen of Cas13a orthologs in bacterial cells, allows for detection of only a single target per reaction via a fluorescence channel or a line on a lateral flow strip. Characterization of additional Cas13 and Cas12 orthologs has allowed for multiplexed detection of up to four targets in a single reaction in separate fluorescent color channels16. Biochemical profiling of these orthologs showed diverse cleavage preferences, allowing for collateral cleavage activity of different enzymes to be read out by reporters with different nucleotide motifs and attached fluorophores. Due to spectral limitations, it may be difficult to multiplex more than four target and enzyme pairs, but so far five orthogonal cleavage preferences have been discovered for Cas12a from Acidaminococcus sp. (AsCas12a) and Cas13s from Leptotrichia wadei (LwaCas13a), Capnocytophaga canimorsus Cc5 (CcaCas13b), Prevotella sp. MA2016 (PsmCas13b), and Lachnospiraceae bacterium NK4A179 (LbaCas13a) (Box 1)16. To combine multiplexed Cas13 and Cas12 detection with pre-amplification, multiple primer pairs must be designed, unless universal primers are being used for a variable insert site or multiple targets along a longer amplicon are being sensed. Owing to inherent base bias of the recombinases in RPA for specific primer sequences, designing compatible primer pairs can be difficult. Multiple primer pairs must be designed, and upon experimental testing, the primer concentrations must be adjusted depending on these biases to allow for even amplification of all the amplicons. Alternative pre-amplification strategies could also be employed that have less primer bias, such as helicase dependent amplification or nicking enzymatic amplification reactions29. However, RPA has been demonstrated to be more robust and more sensitive than other isothermal amplification methods such as NASBA14. The reaction temperature of RPA also allows for the combination of pre-amplification with detection in one-pot SHERLOCK assays.
Primer Design
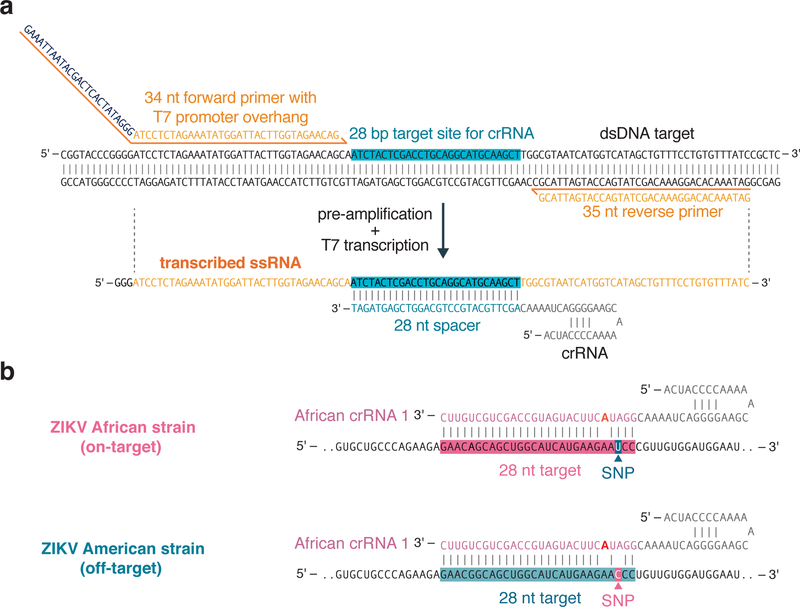
For pre-amplification with RPA, two primers (forward and reverse) must be designed (Figure 2a). To maximize amplification success, the primer lengths should be around 25–35 nt, and the total amplicon size should be 80–140 bp1. Typical primers are designed with melting temperatures between 54°C and 67°C. Primers can be designed with NCBI Primer-BLAST30, using the custom parameters discussed above. Although the RPA manufacturer recommends designing around 96 primer pairs in order to find an optimal pair that is specific, only a few primer pairs need to be designed and tested for SHERLOCK, as specificity is encoded by the crRNA and most primer pairs will result in high amplification rates. Due to the additional specificity from the CRISPR detection, it is not necessary to evaluate primer sets using gel-electrophoresis to look for specific amplicon amplification. For detection reactions involving Cas13, a T7 RNA polymerase promoter must be appended onto the 5’ end of one of the primers to allow for T7 transcription (Figure 3a). It is recommended to append the promoter sequence to the forward primer and design crRNAs complementary to the sense strand (Figure 3a).
Figure 3: Considerations for primer and crRNA design.
(a) RPA primer and LwaCas13a crRNA design for target detection. Schematic of LwaCas13 crRNA detection of target RNA transcribed from an RNA amplicon. Sequences of the RPA primers and crRNA are shown to highlight how these should be designed relative to the target DNA and transcribed RNA. A T7 promoter should be added to the 5´ end of the forward RPA primer and the crRNA sequence should be the reverse complement of the target site in the transcribed RNA. The direct repeat of the LwaCas13a crRNA is on the 5énd of the spacer sequence.
(b) Synthetic mismatch crRNA design for single-nucleotide specificity. Schematic of crRNA design for sensing a single-nucleotide mismatch difference between African and American strains of the Zika virus. The single-nucleotide polymorphism (SNP) site should be placed in the 3rd position of the spacer sequence and the synthetic mismatch (highlighted in red) should be placed in the 5th position of the spacer sequence.
crRNA Design
The most important consideration of crRNA design for Cas13 detection is the lack of overlap with RPA primers, as crRNA overlap with primers may result in detection of the primer sequence in off-target amplification products, increasing background (Figure 3a). The crRNA sequence should be the reverse complement of the target site in the transcribed RNA. A spacer sequence (28-nt in the case of LwaCas13a) can be placed anywhere within the resulting pre-amplified RPA product, and avoidance of targeting target RNA secondary structure has also been shown to increase RNA targeting with LwaCas13a{Abudayyeh, 2017 #611}. The spacer sequence is then joined with a 5’ direct repeat (DR) sequence to generate a complete crRNA.
crRNA can be generated from DNA templates via in vitro transcription (IVT) or produced as chemically synthesized oligonucleotides via phosphoramidite synthesis providers. IVT methods are typically less expensive and can be used to produce large amounts of crRNA at scale, but the generation of crRNA and subsequent purification takes longer. In addition, chemically synthesized crRNAs are purer, resulting in a stronger detection signal and increased sensitivity. When generating crRNA via IVT, an additional T7 RNA polymerase promoter is placed upstream of the crRNA sequence to allow for T7 transcription, and the entire sequence is ordered as the reverse complement.
When enzymes other than LwaCas13a are used (Box 1), more considerations apply. For the use of additional Cas13 variants, such as Cas13b orthologs16, the spacer length must be changed (Cas13b orthologs use spacer lengths of 30 nt), the DR sequence must be modified to correspond to the ortholog, and the position of the DR may need to be moved from 5’ to 3’ end of the spacer, as Cas13b crRNAs have a 3’ DR. When using Cas12a for detection, the spacer length (20 nt for AsCas12a) and DR position (5’) should be considered. When Cas12a targets ssDNA, it does not require a protospacer adjacent motif (PAM), but when targeting dsDNA, such as in combination with RPA, the crRNA must be placed next to a PAM (commonly 5’ TTTV for AsCas12a)9.
Owing to the inherent specificity of Cas13, which will not cleave sequences with two or more mismatches, SHERLOCK is capable of specific sequence detection. To design crRNA for single-base pair specificity, the base to be distinguished is placed in the 3rd position of the crRNA, and an additional synthetic mismatch is placed in either the 4th or 5th position of the crRNA (Figure 2b). This synthetic mismatch generates a bubble of 2 base pairs when the base pair of interest is not present, resulting in more stringent discrimination at the targeted position. Additionally, by placing the region where a small deletion occurs in the seed region of the crRNA (positions ~5–15), deletions can be sensed16.
Readout formats
Typically, SHERLOCK reaction readout uses a fluorescent sensor involving a short RNA oligonucleotide with a fluorophore on one end and a quencher on the other14. These assays are easy to set up in a plate reader and can be measured using any standard fluorescence plate reader or qPCR machine. Given the right setup, these reactions can also be monitored using handheld fluorimeters, allowing for portable fluorescent detection. Fluorescent readout also allows for continuous monitoring, enabling real-time quantitation.
For even simpler instrument-free and portable detection, lateral flow readouts can be used16. The lateral flow sensor works by using an RNA sensor flanked by a fluorescein and biotin on separate ends. On the lateral flow strips, a line of streptavidin will bind to biotin, capturing all the intact probe. Anti-fluorescein antibodies labeled with gold nanoparticles will bind the fluorescein end of the sensor and form a dark purple color at this first line. When RNA sensors are cleaved due to target presence and collateral activity, gold nanoparticle labeled antibodies will flow over to a second line of anti-species secondary antibody, which will capture all the antibodies. This will form a dark purple color at the second line, indicating the presence of target. The lateral flow readout only takes a few minutes and is usually performed after 5–10 minutes of pre-amplification and 20–30 minutes of Cas13 detection.
Quantitation
For many nucleic acid detection applications, it is sufficient to simply distinguish between the presence or absence of a target, but some situations, such as quantifying the fraction of cancer-associated cell-free DNA, or determining viral load, may call for quantitation of target levels. The activation of Cas13 is quantitative with respect to RNA target abundances, and increased levels of target will result in increased activation and faster accumulation of signal when measured by fluorescence output. In cases where RPA and T7 transcription are used, these additional steps add complexities to quantitation. T7 transcription is a linear amplification and does not introduce many barriers to quantitation, but RPA, as with many isothermal amplification methods, undergoes asynchronous amplification with the potential to saturate, which can prevent accurate quantitation14. To overcome this barrier, the concentration of primer in the RPA reaction can be tuned to find a quantitative window. In published experiments, a primer concentration of 240nM resulted in quantitative SHERLOCK activity over multiple orders of magnitude16, but quantitation should be optimized for each target. Generally, we recommend to optimize primer concentration and pre-amplification times for individual targets, in both single-plex and multiplex detection formats. Synthetic versions of the desired nucleic acid target may help to establish robust reaction conditions.
Trial run
Readers should attempt a trial run on synthesized DNA or RNA, or in vitro transcribed RNA. Example sequences for American ZIKV synthetic RNA or synthetic DNA1 can be found in Table 2. This will familiarize the experimenter with the procedure without potentially wasting extracted nucleic acid material. It also provides a quality control assessment of reagents, as SHERLOCK nucleic acid detection is performed with RNA probes that undergo enzymatic hydrolysis and therefore require sensitive handling.
Table 2:
Example sequences for testing SHERLOCK
| Name | Sequence | Purpose |
|---|---|---|
| American ZIKV synthetic RNA | ||
| Zika American synthetic RNA | gacaccggaacuccacacuggaacaacaaagaagcacugguagaguucaaggacgcacaugccaaaaggcaaacugucgugguucuagggagucaagaaggagcaguucacacggcccuugcuggagcucuggaggcugagauggauggugcaaagggaaggcuguccucuggc | American ZIKV synthetic RNA |
| Zika American strain RPA forward primer | gaaatTAATACGACTCACTATAGGGCGGAACTCCACACTGGAACAACAAA | RPA primer for pre-amplification of American ZIKV |
| Zika American strain RPA reverse primer | TGCACCATCCATCTCAGCCTCCAGAGCTCC | RPA primer for pre-amplification of American ZIKV |
| Zika American LwaCas13a crRNA | gauuuagacuaccccaaaaacgaaggggacuaaaacacucccuagaaccacgacaguuugccuu | American ZIKV LwaCas13 detection crRNA |
| Zika American LwaCas13a crRNA IVT template | aaggcaaactgtcgtggttctagggagtgttttagtccccttcgtttttggggtagtctaaatcccctatagtgagtcgtattaatttc | American ZIKV LwaCas13 detection crRNA IVT template |
| Synthetic DNA 1 | ||
| Synthetic DNA 1 | ggccagtgaattcgagctcggtacccggggatcctctagaaatatggattacttggtagaacagcaatctactcgacctgcaggcatgcaagcttggcgtaatcatggtcatagctgtttcctgtgtttatccgctcacaattccacacaacatacgagccggaagcataaag | synthetic DNA 1 |
| synthetic DNA 1 RPA forward primer | AATTCTAATACGACTCACTATAGGGATCCTCTAGAAATATGGATTACTTGGTAGAACAG | RPA primer for pre-amplification of DNA 1 |
| synthetic DNA 1 RPA reverse primer | GATAAACACAGGAAACAGCTATGACCATGATTACG | RPA primer for pre-amplification of DNA 1 |
| synthetic DNA 1 LwaCas13a crRNA | gauuuagacuaccccaaaaacgaaggggacuaaaaccgccaagcuugcaugccugcaggucgag | DNA 1 LwaCas13 detection crRNA |
| synthetic DNA 1 LwaCas13a crRNA IVT template | ctcgacctgcaggcatgcaagcttggcggttttagtccccttcgtttttggggtagtctaaatcccctatagtgagtcgtattaatttc | DNA 1 LwaCas13 detection crRNA IVT template |
| T7–3G IVT primer | gaaatTAATACGACTCACTATAGGG | T7 promoter annealing oligo for IVT |
MATERIALS & REAGENTS
Protein expression
Rosetta™ 2(DE3)pLysS Singles Competent Cells (Millipore cat. no. 71401–3)
TwinStrep-SUMO-LwaCas13a expression plasmid (Addgene: pC013 - Twinstrep-SUMO-huLwCas13a, Plasmid # 90097)
SOC Outgrowth Medium 25 mL (New England BioLabs cat. no. B9020S)
Luria Broth (LB), Ready-Made Powder 250 g, (Thermo Scientific cat. no. J75854.30
TB Broth powder (VWR™ cat. no. 89126–196)
Glycerol ≥99.5% (Sigma-Aldrich cat. no. G9012–1GA)
Ampicillin, sodium salt (AmericanBio, cat. no. AB00115–00025; CAS 69–52-3)
Isopropyl β-D-1-thiogalactopyranoside (IPTG), Sigma-Aldrich cat. no. I6758
Sodium chloride (Sigma-Aldrich cat. no. S7653)
DL-Dithiothreitol solution, BioUltra, for molecular biology, 1 M in H2O (Sigma Aldrich cat. no. 43816–50ML)
Tris(2-carboxyethyl)phosphine (TCEP) hydrochloride solution 0.5M, pH 7.0 (Sigma-Aldrich cat. no. 646547–10X1ML)
-
β-Mercaptoethanol (Sigma-Aldrich cat. no. M6250–100ML)
CAUTION: β-Mercaptoethanol is considered toxic. Wear gloves and work in a well-ventilated area.
1M Tris-HCI Buffer, pH 8.0 (Invitrogen™ UltraPure™, cat. no. 15–568-025)
1M Tris-HCI Buffer, pH 7.5 (Invitrogen™ UltraPure™, cat. no. 15–568-027)
cOmplete™ ULTRA Tablets, Mini, EDTA-free (Sigma-Aldrich, cat. no. 05892791001)
Lysozyme from chicken egg white (Sigma-Aldrich, cat. no. L6876–10G)
Benzonase® Nuclease (Sigma-Aldrich cat. no. E1014–25KU)
Bolt™ 4–12% Bis-Tris Plus Gels for SDS-Page Electrophoresis (ThermoFisher Scientific cat. no. NW04125BOX)
10X Bolt™ Sample Reducing Agent (ThermoFisher Scientific cat. no. B0004)
4X Bolt™ LDS Sample Buffer (ThermoFisher Scientific cat. no. B0007)
20X Bolt™ MES SDS Running Buffer (ThermoFisher Scientific cat. no. B000202)
-
Strep-Tactin Superflow Plus suspension, 50% (Qiagen cat. no. 30004)
CRITICAL: We have found superior performance of Strep-Tactin Superflow resin from Qiagen compared to other vendors.
SUMO Protease, 250 units, ThermoFisher Scientific cat. no. 12588018
Octylphenoxy poly(ethyleneoxy)ethanol, branched (IGEPAL® CA-630), Sigma-Aldrich cat. no. I8896–50ML or NP-40 Surfact-Amps™ Detergent Solution (ThermoFisher Scientific, cat. no. 85124, 50 mL)
4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) powder (Sigma-Aldrich cat no. H4034–1KG)
1 M HEPES buffer (ThermoFisher Scientific, cat. no.15630–080)
0.2 μM filtered 1 M MgCl2 (ThermoFisher Scientific, cat. no. AM9530G)
5 mL HiTrap SP HP cation exchange column (GE Healthcare Life Sciences, cat. no. 17115101)
Superdex® 200 Increase 10/300 GL (GE Healthcare Life Sciences, cat. no. 28990944)
HiScribe™ T7 Quick High Yield RNA Synthesis Kit (New England Biolabs, cat. no. E2050S)
-
Sodium Hydroxide 10M solution (Sigma-Aldrich, cat. no. 72068–100ML)
CAUTION: Sodium Hydroxide is a strong base and should be handled with appropriate protective wear.
EDTA (0.5 M), pH 8.0, RNase-free (Invitrogen, cat. no. AM9260G)
NTP Buffer Mix, T7 RNA Polymerase Mix (part of HiScribe™ T7 Quick High Yield RNA Synthesis Kit, New England Biolabs, cat. no. E2050S)
Standard Taq Buffer 10X (New England Biolabs cat. no. B9014S),
UltraPure™ DNase/RNase-Free Distilled water (Invitrogen cat. no. 10977015)
Agencourt® RNAClean® XP Kit (Beckman-Coulter part. no. A63987)
-
Absolute ethanol, anhydrous, 200 proof/100% (VWR cat. no. 89125–170)
CAUTION: Absolute Ethanol is highly flammable. Keep away from open flames and work in well-ventilated area.
Novex™ TBE-Urea Gels, 10% (ThermoFisher Scientific cat. no. EC68755BOX)
SYBR™ Gold Nucleic Acid Gel Stain 10,000X Concentrate in DMSO (ThermoFisher Scientific cat. no. S11494)
TwistAmp® Basic (TwistDX cat. no. TABAS03KIT)
TwistAmp® Basic RT (TwistDX cat. no. TABARTS01kit)
RNase Inhibitor, Murine (New England Biolabs cat. no. M0314S)
NxGen® T7 RNA Polymerase 125000U (Lucigen cat. No. 30223–2 30223–2)
RNaseAlert™ Lab Test Kit v2 (Invitrogen cat. No. 4479768)
Ribonucleotide Solution Mix (New England Biolabs cat. No. N0466L)
Milenia HybriDetect 1 (TwistDx™ cat. no. MILENIA01)
EQUIPMENT
Constant temperature incubator (e.g., Quincy Lab, Inc Incubator Model 12–140E)
100 mm × 15 mm Not TC-Treated Bacteriological Petri Dish (Corning, cat. no. 351029)
Culture flasks, 1 L (e.g., Corning® 5100–1L PYREX® Wide Mouth flask)
Biological shaker (e.g., Eppendorf Innnova S44i)
Fisherbrand™ Disposable Cuvettes; Polystyrene (cat. no. 14–955-127)
Floor centrifuge with 4 L capacity (e.g., Beckman-Coulter, Avanti J-E)
Gel electrophoresis tank and power source (e.g., Invitrogen™ Mini Gel Tank; VWR Power Source 250 V cat. no. 93000–746)
LM20 Microfluidizer® from Microfluidics
Corning™ Polypropylene Centrifuge Tubes, 250 mL (cat. no. 05–538-53)
Tube rotator (e.g., Scilogex MX-RL-Pro, cat. no. 824222019999)
Plate shaker (e.g., VWR® Mini Shaker)
Chromatography column (e.g., Bio-Rad Glass Econo-Column®)
eStain® L1 Protein Staining System (GenScript®) or Coomassie Stain (Coomassie Brilliant Blue R-250, cat. no. 1610400, Bio-Rad) and destain solution (40% methanol, 10% acetic acid)
FPLC (e.g., ÄKTA pure by GE Healthcare Life Sciences)
LightCycler® 480 Instrument II (Roche Diagnostics, product no. 05015243001)
QX200™ droplet generator (Bio-Rad, cat. no. 186–4002)
QX200™ droplet reader (Bio-Rad, cat. no. 186–4003)
Benchtop centrifuge (e.g., Beckman Coulter Allegra X-15)
Axygen™ 8-Strip PCR tubes (Fisher Scientific, cat. no. 14–222-250)
Axygen™ PCR plates, 96 well (VWR, cat. no. PCR-96M2-HS-C)
384-well optical plate (e.g., Roche, LightCycler 480 Multiwell plate 384, cat. no. 5102430001)
Axygen 1.5 mL Boil-Proof Microcentrifuge Tubes, (VWR, cat. no. 10011–702)
Falcon tubes, polypropylene, 15 mL (Corning cat. no. 352097)
Falcon tubes, polypropylene, 50 mL (Corning, cat. no. 352070)
Eppendorf Safe-lock microcentrifuge tubes, 1.5 mL (Eppendorf, cat. no. T9661–1000EA)
Filtered sterile pipette tips (e.g., Rainin)
Corning bottle-top vacuum filter system, 0.22 μM (Sigma-Aldrich, cat. no. CLS431098)
Stericup filter unit, 0.45 μM (Millipore, cat. no. SCHVU02RE)
Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-50 membrane (Sigma-Aldrich, cat. no. UFC905024)
Thermocycler with programmable temperature stepping functionality, 96 well (e.g., Applied Biosystems Veriti, cat. no. 4375786)
Desktop microcentrifuges (e.g., Eppendorf, cat. no. 5424)
Digital gel imaging system (GelDoc EZ, Bio-Rad, cat. no. 170–8270) and white sample tray (Bio-Rad, cat. no. 170–8272)
Gel quantification software (Bio-Rad, ImageLab or open-source ImageJ from the National Institutes of Health (NIH), USA, available at http://rsbweb.nih.gov/ij/)
UV spectrophotometer (e.g., NanoDrop 2000c, Thermo Scientific)
Plate spectrophotometer (e.g., Synergy H4 Hybrid Multi-Mode Microplate Reader, BioTek)
REAGENT SETUP
Bacterial transformation and large-scale protein expression
Large LB agar plates (10 cm Petri dish, ampicillin):
Reconstitute the LB Broth with agar at a concentration of 35 g L−1 in deionized water and swirl to mix. Autoclave to sterilize at 121.0°C for 20 minutes. Allow the LB agar to cool to 55°C before adding ampicillin to a final concentration of 100 μg mL−1 and swirl to mix. On a sterile bench area, pour ~20 mL of LB agar per 10 cm plate. Place the lids on the plates and allow them to cool at room temperature (22°C) for 30–60 min until solidified. Invert the plates and let sit for several more hours or overnight. Agar plates can be stored in plastic bags or sealed with parafilm at 4°C for up to 3 months.
Terrific Broth – TB Media (1 L, ampicillin):
Reconstitute TB media by adding 50.8 g of TB powder to a 2 L flask, add 4 mL of 100% Glycerol, and fill up with deionized water to 1 L final volume. Heat at 50°C with repeated stirring to dissolve completely, then autoclave at 121.0°C for 15 minutes. Let cool for several hours, then add Ampicillin to 100 μg mL−1 final concentration. TB Media is stable at 4°C for 6 months, or for 1 month at room temperature.
IPTG:
Dissolve 1.19 g of IPTG by adding to 8 mL of deionized water and then vortexing. Add deionized water to 10 mL and filter sterilize with a 0.22 μm syringe filter. Store at −20°C for up to six months.
Protein Purification
Lysis Buffer:
Combine 40ml Tris-HCl (pH 8.0, 1M), 200ml NaCl (5M) and 2ml DTT (1M) and make up to 2L final volume with ultragrade water. Use within 48 hours of preparation. For longer storage, prepare the buffer without the reducing agent at keep at 4°C for up to 2 weeks.
Supplemented Lysis Buffer:
Add 12 cOmplete Ultra EDTA-free tablets, 600mg lysozyme (KU) and 6 μL Benzonase (U/mL) to 600ml lysis buffer. Prepare fresh and use within 24 hours following preparation. Keep at 4°C when not in use.
Critical.
The lysis buffer contains the reducing agent DTT, which can be substituted with TCEP or beta-mercaptoethanol (BME). DTT and BME are less stable in solution, and so the lysis buffer should be freshly prepared for optimal results.
CAUTION: Follow the handling instructions in the MSDS to minimize risk when using hazardous reducing agents.
SUMO protease cleavage solution:
Supplement 15 mL of Lysis Buffer with 250 μL of SUMO protease and 22.5 uL of NP-40.
FPLC
Ion exchange buffer A:
Combine 40ml Tris-HCl (pH 7.5, 1M), 200ml glycerol (50% w/v) and 2ml DTT (1M) and make up to 2L final volume with ultragrade water. Filter through a 0.22 μm vacuum filter and store at 4°C for up to 48 hours. For storage up to two weeks, prepare the buffer without the reducing agent and add this fresh when performing FPLC.
Ion exchange buffer B:
Combine 20ml Tris-HCl (pH 7.5, 1M), 100ml glycerol (50% w/v), 400ml NaCl (5M) and 1ml DTT (1M) and make up to 1L final volume with ultragrade water. Filter through a 0.22μm vacuum filter and store at 4°C for up to 48 hours. For storage up to two weeks, prepare the buffer without the reducing agent and add this fresh when performing FPLC.
S200 size exclusion buffer:
Combine 10ml HEPES (pH 7, 1M), 5ml MgCl2 (1M), 200ml NaCl (5M) and 2ml DTT (1M) and make up to 1L final volume with ultragrade water. Filter through a 0.22μm vacuum filter and store at 4°C for up to 48 hours. For storage up to two weeks, prepare the buffer without the reducing agent and add this fresh when performing FPLC.
LwaCas13 protein storage buffer:
Combine 2.5ml Tris-HCl (pH 7.5, 1M), 6ml NaCl (5M), 2.5ml glycerol and 100 μL DTT (1M), and make up to 50ml final volume with ultragrade water. Filter through a 0.22 μm 50ml vacuum filter and store at 4°C for up to 48 hours. For storage up to 1 year, aliquot buffer and store at −20°C. Avoid repeated freeze-thaw cycles of the buffer.
CRITICAL: Prepare buffer under Nuclease-free conditions.
SDS-PAGE sample buffer:
Add 4 μL 10X Bolt™ Sample Reducing Agent and 10 μL 4X Bolt™ LDS Sample Buffer to 16 μL Ultragrade water. The buffer mixture can be stored for up to two weeks at 4°C
When running SDS-PAGE, 10 μL of sample is added to the above mixture and heated to 95°C for 5 minutes. If the sample volume needs to be changed, adjust the sample buffer component volumes accordingly to obtain a final volume of 40 μL, including the sample.
Optional reagents
Plant extraction buffer:
Prepare 10 mL of alkaline plant extraction buffer my adding 0.5 mL 10M NaOH and 0.2 mL 0.5M EDTA to 9.3 mL of Nuclease free water. Aliquot 1 mL of buffer in 1.5 mL plastic Eppendorf tubes and store at −20°C for up to 6 months.
PROCEDURE
Recombinant expression and purification of LwaCas13a (5 days)
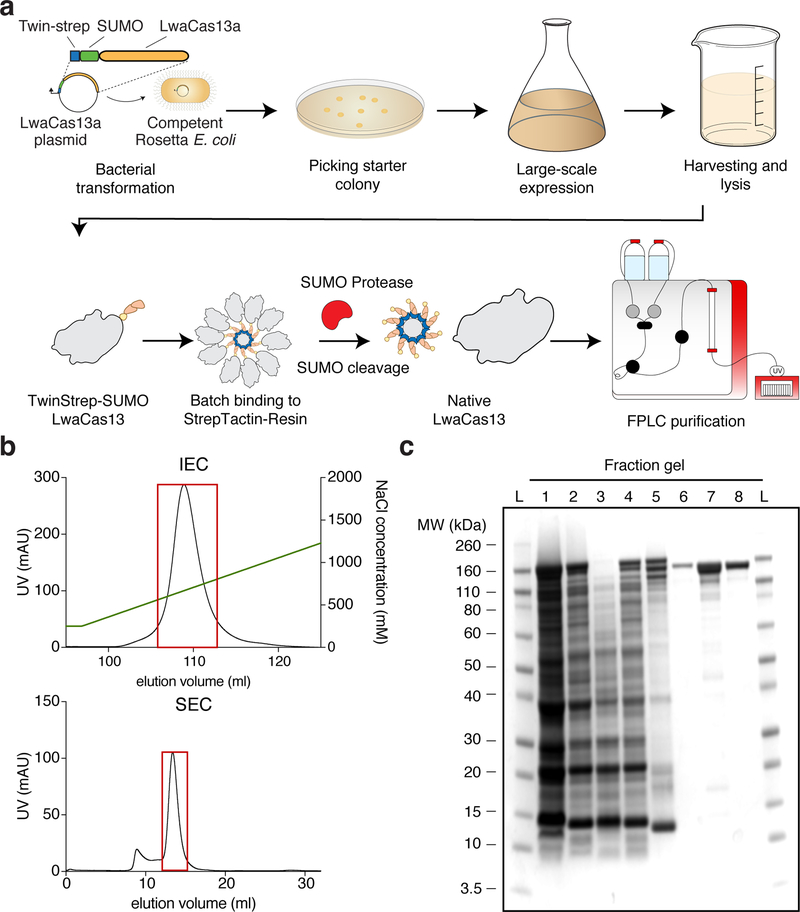
CRITICAL The workflow for transforming bacteria with the appropriate expression constructs to large scale expression and protein purification is shown in Figure 4a
Figure 4: Protein purification workflow and expected results.
(a) LwaCas13a protein expression and purification workflow. The TwinStrep-SUMO-LwaCas13a protein expression plasmid is first transformed into competent Rosetta E. Coli cells. After antibiotic selection and initial growth, expression is induced by IPTG. Following growth, cells are harvested and lysed. The recombinant protein is subsequently enriched from the total cell protein by affinity Streptactin purification. The TwinStrep-SUMO tag is then cleaved by SUMO protease to obtain native LwaCas13 protein. The enzyme is further purified by ion-exchange and size exclusion chromatography on an FPLC system.
(b) FPLC Chromatograms. The graph shows a representative chromatogram of ion-exchange (IEC, top image) and size-exclusion chromatogram (SEC, bottom image) for LwaCas13a. The UV-absorbance in mAU is plotted against the elution volume in mL. The NaCl gradient for ion-exchange chromatography is shown by the green line. The red boxes indicate the protein-containing fractions that were pooled and concentrated.
(c) Coomassie stained SDS-PAGE gel of protein fraction The progress of protein purification is shown on a Coomassie stained SDS-PAGE gel. The fractions are L: Ladder, 1: Cell lysate, 2: Cleared cell lysate, 3: Cell pellet after clearing of lysate, 4: Flow-through following StrepTactin batch-binding, 5: StrepTactin resin before SUMO protease cleavage, 6: Eluted fraction post SUMO protease cleavage, 7: Concentrated sample post ion-exchange chromatography, 8: Final product after size-exclusion chromatography.
Large-scale expression of LwaCas13 (3 days)
-
1
Thaw one vial of RosettaTM 2 (DE3) pLysS competent cells on ice for 30 minutes, then add 1 μL of 50 ng/μL LwaCas13 expression plasmid. Incubate on ice for 5 minutes.
-
2
Heat shock cells by placing the vial into a 42°C pre-heated water bath for 45 seconds, then cold-shock cells on ice for 2 minutes.
-
3
Add 200 μL of SOC media to cells, and plate 100 μL of cell suspension on a pre-warmed LB-Agarose plate containing 100 μg/mL Ampicillin. Incubate the plate overnight in 37°C incubator.
TROUBLESHOOTING
-
4
The next day, inoculate 25 mL of TB media containing 100 μg/mL Ampicillin with a single colony and incubate overnight at 37°C on rotary shaker at 300 RPM.
-
5
Inoculate 4–12 L TB media, containing 100 μg/mL Ampicillin, with 5 mL/L starter culture and determine the optical density (OD, 600nm). The amount of starter culture depends on the downstream expression scale. It is recommended to start with 5 mL of starter culture for every 1 L of large-scale culture. Then shake at 37°C, 300 RPM.
-
6
Monitor OD every hour until cells reach an optical density of 0.4 – 0.6, then transfer flasks to 4°C for 30 min to allow flasks to cool prior to induction. Take an aliquot of uninduced culture for SDS-PSGE analysis.
Critical step: For optimal expression, it is important to strictly follow the indicated OD of 0.4–0.6 at the time-point of induction.
-
7
Induce expression by adding 1 mL/L 0.5M IPTG and shake cultures for 16 hours on rotary shaker at 300 RPM, in a pre-chilled 21°C incubator.
-
8
Harvest cells by spinning the culture down at 5000 RPM for 15 minutes at 4°C. Take a small aliquot and resuspend in 500 μL P1 Buffer (Qiagen). Run together with the uninduced culture aliquot on a Bolt™ 4–12% Bis-Tris Plus SDS-PAGE gel, in 1x Bolt™ MES Buffer for 20 minutes at 200V.
Pause point. Cells can be directly used for purification, or stored at −80°C for up to 1 year. Cells are routinely stored as spread paste in Ziploc bags, which enables future expression testing and aliquoting by breaking frozen paste.
TROUBLESHOOTING
Purification of LwaCas13 (2 days)
Critical. Perform all steps at 4°C and do not let the resin run dry. In steps where a working environment of 4°C cannot be achieved, try to keep the sample near 4°C by cooling on ice.
-
9
Crush and resuspend frozen pellet in 4x (w/v) supplemented lysis buffer (e.g. 20g of pellet in 80mL of buffer) by stirring the mixture at 4°C for 30 minutes on a magnetic stir table.
Critical step. To ensure optimal lysis downstream, monitor resuspension progress until a homogenous mixture is obtained.
-
10
Lyse cells by passing the cell suspension once through a pre-chilled LM20 Microfluidizer system at 27k PSI. Alternatively, cells can be ruptured on ice using sonication, with an amplitude setting of 100% for 1 second on and 2 seconds off, in a total of 10-minute sonication time is recommended to avoid heat induced denaturation of lysed protein. Collect a 100 μL fraction for SDS-PAGE analysis.
-
11
Clear Lysate by centrifugation for 1 hour at 10k RPM at 4°C.
-
12
Decant cleared supernatant into a conical 250 mL tube and collect a 100 μL fraction for SDS-PAGE analysis. With a 1000 μL pipette tip, streak an aliquot of insoluble fraction and resuspend in 100 μL of lysis buffer for SDS-PAGE.
-
13
Add 5 mL of Superflow Streptactin resin to the supernatant. Batch-bind the recombinant protein to the resin for 2 hours by gentle shaking at 4°C.
-
14
Meanwhile, prepare a 50 mL Bio-Rad Glass Econo-Column® by washing the column with 2x 50 mL of cold lysis buffer, then add 20 mL of cold lysis buffer to equilibrate the column bed. Drain the column immediately prior to sample application.
Critical step. Do not use supplemented lysis buffer, as the presence of protease inhibitor will affect downstream cleavage of Streptactin-SUMO tag.
-
15
Pour the resin-sample suspension over the prepared column and collect the flow-through. Collect a 100 μL fraction for SDS-PAGE analysis. Then wash the collected resin three times with 25 mL cold lysis buffer. With a 200 μL pipette tip, take a small aliquot of resin and resuspend in 100 μL lysis buffer for SDS-PAGE analysis.
-
16
Add 15 mL of SUMO protease cleavage solution to the resin and allow for SUMO protease cleavage overnight at 4°C under gentle shaking.
Critical step. Avoid vigorous shaking to prevent foam formation and extensive coating of the column glass surface with protein-bound resin.
-
17
The next day, drain the column and collect the cleavage solution into a separate 50-mL Falcon tube. Then wash the remaining sample three times with 5 mL of lysis buffer to ensure complete transfer of cleaved protein. Collect a 100 μL aliquot of cleaved fraction for SDS-PAGE analysis.
Critical step. The resin bound with Twin-Strep-SUMO tag will remain in the column, while the collected fraction should contain untagged, native LwaCas13a. To ensure cleavage is completed, take a small aliquot of resin with a 200 μL pipette and resuspend in 100 μL lysis buffer for SDS-PAGE analysis.
-
18Perform SDS-PAGE analysis of collected protein fractions to assure successful cleavage by SUMO protease. To do so, add 10 μL of sample to 30 μL of SDS-PAGE sample buffer, heat to 95°C for 5 minutes and run SDS-PAGE on a Bolt™ 4–12% Bis-Tris Plus SDS-PAGE gel, in 1x BoltTM MES Buffer for 20 minutes at 200V.
Step Sample 10 Cell lysate 12 Cleared cell lysate 12 Cell pellet after clearing of lysate 15 Flow-through following StrepTactin batch-binding 15 StrepTactin resin before SUMO protease cleavage 17 Eluted fraction post SUMO protease cleavage Critical step. While the sample taken at stage 17 might appear as a clear, single band, it is advised that the experimenter further purify the enzyme via FPLC tandem purification (steps 19–28) to remove unwanted contaminants, such as SUMO protease and nucleases.
TROUBLESHOOTING
-
19
Using an ÄKTA-Pure or similar FPLC system, wash a 5 mL HP SP column with 5 column volumes (25 mL) of Buffer B followed by 5 column volumes of Buffer A (25 mL) using a system flow-rate of 5 mL/min. Then equilibrate the column with 5 column volumes of a mixture of 12.5% Buffer B in A (250 mM NaCl) at a flow rate of 5 mL/min.
Critical step. Use freshly prepared and filtered buffers for operating the FPLC to avoid column or system damage.
-
20
Dilute the collected sample two-fold by adding 30 mL of Buffer A.
Critical step. The dilution step is important to lower the NaCl concentration to 250 mM, to efficiently bind the sample to the equilibrated column during sample application.
-
21Start sample application and cation exchange chromatography by running below program at 5 mL/min.
Step Program Volume 1 Sample application (continuous application until entire volume is run through column) 2 Column wash: 12.5% Buffer B 5 column volumes 3 Gradient elution: 12.5% Buffer B to 100% Buffer B 10 column volumes 4 Column wash: 100% Buffer B 10 column volumes 5 Equilibration: 12.5% B 5 column volumes -
22
Collect 100 μL of peak-containing fractions for SDS-PAGE analysis. Example FPLC chromatogram of the ion exchange chromatography result is shown in Figure 4b.
TROUBLESHOOTING
-
23
Pool fractions containing LwaCas13 protein into a 15-mL centrifugal spin filter (50 MWCO) and concentrate to <1 mL final volume by spinning for 15 min at 4000 g centrifugation.
CRITICAL STEP. Sample concentration is important to reduce the sample loading volume for down-stream size exclusion chromatography, which is done to ensure a high-resolution separation of potential protein aggregates and other proteins from monomeric LwaCas13.
TROUBLESHOOTING
-
24
Equilibrate an S200 Increase 10/300 GL size exclusion column with 5 column volumes of S200 buffer at 0.75 mL/min.
-
25Apply sample and start size-exclusion chromatography by running below program at 0.75 mL/min.
Step Program Volume 1 Sample application 0.5 mL or 2 mL sample loop 2 Elution 1 column volume 3 Wash 1 column volumes 4 Equilibration 2 column volumes -
26
Collect 100 μL of peak-containing fractions for SDS-PAGE analysis. Example FPLC chromatogram of the size exclusion chromatography result is shown in Figure 4b. Protein gel results for samples from steps leading up to the size exclusion chromatography are shown in Figure 4c.
TROUBLESHOOTING
-
27
Pool and concentrate fractions containing LwaCas13a by centrifugation at 4000 g for 15 min in a 15-mL (50 MWCO) centrifugal spin filter. Then add protein storage buffer to 15 mL into the same centrifugal filter and repeat the centrifugation step.
Critical step. Buffer exchange into protein storage buffer is very important for storage and downstream SHERLOCK. Therefore, for optimal results, do not change the formulation of protein storage buffer.
TROUBLESHOOTING
-
28
Determine the final protein concentration, dilute to 2 mg/mL in protein storage buffer, and store as 5 μL aliquots at −80°C until use. Aliquots are stable for at least six months at −80°C.
Pause point. The purified protein should be high quality and ready for Cas13-based nucleic acid detection. A 5 μL aliquot is sufficient for roughly 50 individual SHERLOCK reactions, or 12.5 individual SHERLOCK conditions with 4 technical replicates for each condition.
In vitro transcription and purification of Cas13 crRNAs (5 hours or overnight)
Critical. crRNA quality is instrumental to the performance of CRISPR-diagnostics. An RNase-free bench and nuclease free reagents and consumables are therefore strongly recommending for in vitro transcription of crRNAs.
-
29Prepare the following annealing reactions in a PCR tube:
Component Volume to add crRNA template, 100 μM 1 μL T7–3G oligonucleotide, 100 μM 1 μL Standard Taq Buffer, 10x 1 μL Ultragrade water 7 μL -
30
Anneal the crRNA template and T7–3G oligonucleotide by performing a 5-minute denaturation, followed by slowly cooling the reaction to 4°C in a PCR thermocycler. For slow-cooling, adjust the ramp rate to 0.08°C/sec (or 2.5% ramp rate on most thermocyclers with 3.2°C/sec maximum ramp rate).
-
31Add the following in vitro transcription mix to the annealing reactions:
Component Volume to add Annealing reaction 10 μL NTP buffer mix 10 μL T7 Polymerase mix 2 μL Ultragrade water 17 μL -
32
Incubate the reaction for 4 hours at 37°C, then cool to 4°C in a PCR thermocycler until purification.
Pause point. In vitro transcription reactions can either be stored at - 20°C (for up to 2 weeks) or directly purified.
-
33
Move Ampure XP RNA beads to room temperature.
Critical. Ampure XP RNA beads must be brought to room temperature for optimal use. To do this, let the entire bottle or an aliquot rest on the bench for 30 minutes.
-
34
Per 30 μL of sample, add 100 μL of Ampure XP RNA magnetic beads and 90 μL of isopropanol.
-
35
Mix by pipetting up and down five times, then incubate the mixture for 5 minutes at room temperature.
-
36
Place the reaction on a magnetic stand for 5 minutes. The beads will accumulate near the magnetic surface.
-
37
While keeping the reaction on the magnetic stand, pipette off the clear supernatant and discard. Then add 200 μL of 85% EtOH to the beads, wait for 30 seconds and then discard the ethanol solution. Repeat the washing step and discard the ethanol solution.
-
38
Remove the reaction from the magnetic stand, and air-dry for 5 minutes at room temperature.
-
39
Add 50 μL of Ultragrade water to the dry beads and incubate for 5 minutes at room temperature.
-
40
Place the reaction on a magnetic stand and wait 5 minutes to allow for separation of the beads from eluted crRNA sample.
-
41
Transfer the clear supernatant into a new nuclease free tube and determine concentration by nanodrop. To ensure the correct size, run an aliquot on a 10% TBE Urea gel and stain with SYBR Gold to visualize nucleic acid species.
Critical. Try not to transfer any magnetic beads to the final eluant, as this can inhibit downstream reactions. Using this protocol, roughly 50–100 μg of RNA is made See the troubleshooting section if the result substantially deviates from this (Table 3).
TROUBLESHOOTING
Table 3.
| Step(s) | Problem | Possible reason | Solution |
|---|---|---|---|
| 3 | No colonies | Wrong antibiotic | Check if the correct antibiotic has been used (Ampicillin for LwaCas13a plasmid) |
| 3 | No colonies | Inefficient heat-shock | Repeat transformation procedure |
| 8 | No visible difference between induced and uninduced | Poor expression | Optimize temperature and IPTG induction conditions |
| 18 | No specific protein band in final elute | Protein did not bind to beads or precipitated | Check flowthrough for recombinant protein, try re-batch binding of flow-through with more resin |
| 18 | Two protein bands or wrong size | Insufficient sumo protease cleavage | Add more SUMO protease, incubate for 4–12 hours, and monitor cleavage by SDS-PAGE |
| 22 | No protein peak | Sample did not bind the column | Salt too high, check FPLC waste for protein and change initial buffer A/B mix ratio |
| 23, 27 | Slow spin-filter sample concentration | Glycerol sedimentation or protein precipitation | Carefully pipette up and down and centrifuge again |
| 26 | No protein peak | Sample lost during application | Check FPLC waste or spin-filter flow-through |
| 41 | Low crRNA concentration | Wrong crRNA design | Check crRNA design for T7 promoter sequence |
| 41 | Low crRNA concentration | Ethanol concentration in washing solution too low | Prepare fresh 85% EtOH solution |
| 52A-vii, 52B-vii Box 2 - 12 | No signal | No amplification during RPA, wrong primer design | Redesign primers |
| 52A-vii, 52B-vii Box 2 - 12 | No signal | No amplification during RPA, contaminants in sample | Re-purify or check presence of nucleic acid by other technologies such as PCR |
| 52A-vii, 52B-vii Box 2 - 12 | Signal in all samples including negative control | RNase contamination | Incubate reaction without sample, and remove suspected components to determine if signal is reduced |
| 52A-vii, 52B-vii Box 2 - 12 | Signal in all samples including negative control | DNA template contamination in reagent | Use new reagents and primer aliquots |
Sample extraction (5 min to 1 hour)
CRITICAL: SHERLOCK detection is compatible with both pure and crude sample extractions, as long as contaminating nucleases are inactivated. For pure extraction of genomic DNA or RNA, it is recommended to use the Qiagen QIAamp® DNA Blood Mini Kit (DNA) for isolating genomic DNA from human serum, saliva, or blood, as well as genomic DNA from bacterial cultures. For viral RNA detection from urine and tissue culture, it is recommended to use the QIAamp Viral RNA minikit (Qiagen) and RNARNeasy Plus Mini Kit (RNA) kit. The manufacturer’s instructions for the above mentioned extractions are sufficient.
-
42
Follow the procedures for crude DNA extraction from human saliva (option A) or plant material (option B).
(A) Human crude DNA extraction from human saliva (15 min)
Collect 2 mL of human saliva from patients or volunteers after appropriate IRB approval is in place.
Add 400 μL of phosphate buffered saline (PBS) to 100 μL of saliva, and centrifuge for 5 minutes at 1800 g.
Carefully remove the supernatant and resuspend the pellet in 30 μL PBS, supplemented with 0.2 % Triton X-100. Denature the sample for 5 minutes at 95°C.
Centrifuge for 5 minutes at 1800 g and use 1 μL of the supernatant as sample input for recombinase-polymerase amplification (RPA).
(B) Crude extraction from plant material (5 minutes)
To 20 mg of crushed seeds, leaf, or root tissue, add 200 μL of plant extraction buffer (500 mM NaOH, 10 mM EDTA). Vortex the sample for 30 seconds and incubate at room temperature for 1 minute. The raw alkaline extraction can be stored at −20°C for up to two weeks.
-
Transfer a 5 uL aliquot to a new tube and dilute 1:20 with Ultragrade water. Then subject 1 μL of the sample to RPA. The remaining dilution can be stored at −20°C for up to two weeks.
Critical: Reaction inhibitors might still be present after dilution, and we recommend that the sample volume in the reaction does not exceed 10% of the final volume.
RPA of nucleic acid (30 min to 1 hour)
Critical. The recommended procedures for RNA or DNA pre-amplification are nearly identical. However, important distinctions are summarized in the Experimental Design section. Here we provide a protocol for single target identification using two-step SHERLOCK. For one-pot SHERLOCK detection, please follow the instructions provided in Box 2. If multiplexed nucleic acid detection or quantification are desired, the reader should follow the guidance provided in the Experimental Design section. We also recommend to perform positive and negative controls alongside tested samples. A negative could include a water-only input sample for RPA, or a sample where the target molecules is known to be absent. Positive controls include synthetic RNA or DNA targets, or purified DNA/RNA where the target in known to be present.
-
43
Prepare the pre-amplification area by wiping down the work surface and pipettors with RNase Away.
Critical step. RPA is an isothermal amplification technology that produces large quantities of amplified product within minutes. This can lead to sample contamination of reagents and the work area, generating a false positive signal. It is therefore highly recommended to set up pre- and post-amplification work areas. Laminar flow hoods used for standard tissue culture provide an optimal pre-amplification work environment, and primer aliquots, rather than stocks, are recommended as another measure to prevent contamination spread.
-
44On ice, prepare the following master mix sufficient for four replicates in a 1.5 mL Eppendorf or PCR tube.
Component Volume Forward Primer 10 μM 2.4 μL Reverse Primer 10 μM 2.4 μL TwistAmp Rehydration Buffer 29.5 μL Ultragrade water 8.65 μL -
45
Add 40 μL of master mix to a single pellet aliquot, and carefully resuspend the mixture on ice. Then, transfer the entire reconstituted reaction back to the initial Eppendorf or PCR master mix tube.
Critical step. One pellet yields roughly four individual RPA reactions. If a larger number of reactions are needed, simply scale the master mix volume and pellets up accordingly. When doing so, it is critical not to change the pipette in between transfers of resuspended pellets, as the stickiness of RPA reaction components could result in a significant loss of reaction enzymes when using multiple pipette tips.
-
46
Add 2.5 μL of 280 mM TwistAmp magnesium acetate to the reaction, then briefly vortex and quickly spin down for 2–3 seconds.
Critical step. Addition of magnesium acetate to the reconstituted RPA pellet activates the enzymes and may lead to strong primer dimer formation. It is therefore important to quickly proceed to sample addition.
-
47
Aliquot 10 μL of reconstituted RPA reaction into PCR strip tubes or PCR plates, then add 1 μL of extracted sample to the reaction. Mix by carefully pipetting up and down. Include an Ultragrade water only negative control.
-
48
Run the reaction at 37–42°C in a pre-heated PCR thermocycler or incubator for 10–30 min. The reaction time can be extended to 1 hour for highly sensitive reactions.
Pause point. After the run is completed, you can either immediately proceed to Cas13 SHERLOCK detection or store the reaction at 4°C for 2–3 days. Alternatively, reactions can be stored at −20°C for up to several weeks.
Cas13-SHERLOCK nucleic acid detection (1–4 hours)
Critical: Here (in the main Procedure), we provide details for two-step detection. For details of one-pot detection, please follow the instructions in Box 2.
Critical. SHERLOCK is a highly sensitive RNA-based detection assay and requires RNase-free post-amplification. As with RPA, we recommend you wipe down the surface and pipettors with RNase Away. We also recommend to perform positive controls and negative controls alongside. Suitable positive and negative controls are water-only RPA reactions, or RPA reactions where RPA. To out rule Cas13 detection component contamination, also perform a water-only negative control instead of adding RPA pre-amplified reaction.
-
49
Thaw sufficient amounts of normalized crRNA (300 ng/μL), RNase Alert Reporter (2 μM, Part of RNaseAlert™ Lab Test Kit v2), and Cas13 protein aliquot (2 mg/mL, 14.44 μM) on ice, covered with aluminum foil to protect from light exposure. Sufficient amounts are calculated based on the number of desired reactions with a minimum 25% excess.
-
50
Dilute LwaCas13 to 10x concentration (63.3 μg/mL) by adding 153 μL of protein storage buffer (SB) to 5 μL of LwaCas13a at 2 mg/mL.
Critical step. Reactions are performed in 60 mM NaCl by adding 2 μL of diluted protein per 20 μL SHERLOCK reaction mixture. If multiplexed SHERLOCK is performed, dilute proteins in storage buffer accordingly. For example, for dual target multiplexing, dilute each CRISPR enzyme to 20x and add 1 μL of each per 20 μL SHERLOCK reaction. For triplex detection, dilute to 30x and add 0.67 μL of each enzyme. For quadruplex detection, dilute to 40x and add 0.5 μL of each enzyme to the reaction. For crRNAs, add 1μL of each ortholog’s crRNA at 10 ng/μL to the reaction. For sensors, add 1.25 μL of each ortholog’s sequence-specific sensor at 2 μM to the reaction. The sensors are specific to each ortholog and are as follows: LwaCas13a (/5TEX615/T*A*rArUG*C*/3IAbRQSp/), CcaCas13b (/5Cy5/T*A*rUrAG*C*/3IAbRQSp/), PsmCas13b (/56-FAM/rArArArArA/3IABkFQ/), and AsCas12a (commercial DNase alert from IDT).
-
51
Dilute the crRNA to 10 ng/μL by adding 145 μL of Ultragrade water to 5 μL of crRNA (300 ng/μL).
-
52
Perform fluorescence-based detection assay using a fluorescence plate reader (option A) or colorimetric lateral flow detection assay (option B).
(A) Fluorescence-based detection assay
-
i
Pre-heat the fluorescence plate reader to 37°C.
-
iiPrepare the Cas13-SHERLOCK master mix by adding the following components to an Eppendorf tube in the order they are listed, starting with water.
Component Volume per reaction Volume for four replicates Ultragrade water 11.27 μL 56.35 μL HEPES pH 6.8, 1M 0.4 μL 2 μL MgCl2, 1M 0.18 μL 0.9 μL rNTP solution mix, 25 mM each 0.8 μL 4 μL LwaCas13 in SB, 63.3 μg/mL 2 μL 10 μL Murine Rnase Inhibitors, 40 U/μL 1 μL 5 μL T7 RNA polymerase, 5 U/μL 0.5 μL 2.5 μL crRNA (10 ng/μL) 1 μL 5 μL RNase Alert v2 (2 μM) 1.25 μL 6.25 μL Total 19 μL 95 μL Critical step. It is recommended to routinely perform four technical replicates per SHERLOCK condition. When working with multiple conditions (e.g., if adding positive and negative controls or additional samples), scale up the reaction accordingly and adjust for pipetting errors by adding a 15% excess volume. For example, the following calculation would be required for an experiment with one sample, a positive and a negative control: amount per four technical replicates × 3 *x1.15.
-
iii
For one condition and four technical replicates, transfer 87.4 μL of master mix into a PCR-strip tube or 96-well PCR plate. Scale up according to total number of conditions and replicates.
-
iv
Spin down the RPA pre-amplification reaction sample, carefully open the eppendorf tube, and transfer 4.6 μL of RPA-reaction mix to 87.4 μL aliquoted master mix on ice. Briefly vortex and spin down for 15 seconds in a 96-well plate to collect the entire reaction mix in the well.
-
v
Carefully open each eppendorf tube and transfer 20 μL per technical replicate and condition into a 384-well, round black-well clear-bottom bio-assay plate on ice. To group technical replicates, we recommend you routinely pipette the remaining technical replicates beside and below, to form a square of 2×2 grouped reactions. When transferring reaction from a 96-well to a 384-well plate, we recommend using a 8-well or 12-well multichannel pipette for quicker set-up, which will also load every second well of a 384-well plate. We therefore recommend a 2×2 set up to avoid pipetting mistakes and better grouping of replicates.
-
vi
Briefly spin down the bio-assay plate to remove potential bubbles and place into a pre-heated Biotek plate reader, or equivalent.
Critical step. After bio-assay plate preparation, samples should be placed into the fluorescence plate reader quickly as SHERLOCK Cas13 reactions may begin as soon as they are removed from ice.
-
vii
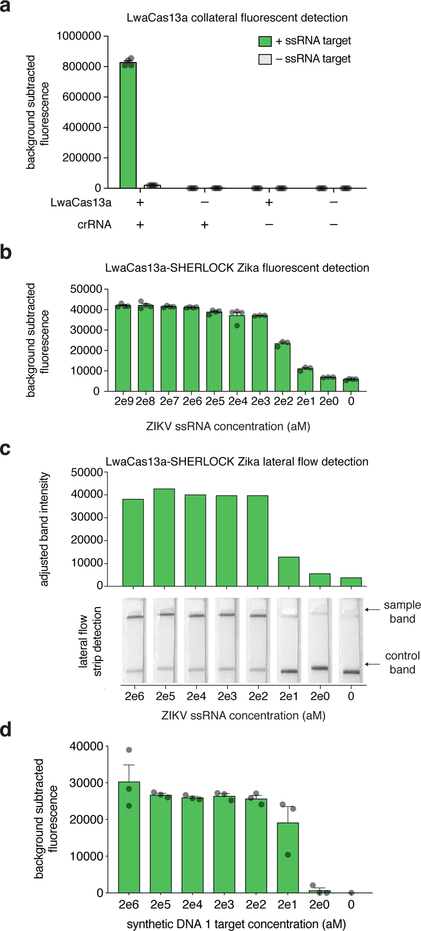
Start data acquisition by monitoring fluorescence over 3 hours at 37°C with a 5-minute interval between well-data acquisitions. Please see anticipated results section and Figure 5a–b for representative results.
TROUBLESHOOTING
Figure 5: Anticipated fluorescence and lateral flow results.
(a) LwaCas13 detection of ssRNA 1 is dependent on the formation of a protein-crRNA-target complex. Reactions contain LwaCas13a and crRNA, LwaCas13 or crRNA alone, or neither protein nor crRNA, in the presence or absence of a single ssRNA 1 target. Bar graphs indicate mean ± SEM of background-subtracted fluorescence measured from four technical replicates; each individual replicate is represented as a dot.
(b) Fluorescence detection for a synthetic RNA version of Zika virus with decreasing input concentrations with a two-step SHERLOCK reaction. Each bar represents the detected collateral cleavage activity for a given input concentration. Bar graphs indicate mean ± SEM of background-subtracted fluorescence measured from either three or four technical replicates; each individual replicate is represented as a dot.
(c) Lateral flow detection for a synthetic RNA version of Zika virus with decreasing input concentrations with a two-step SHERLOCK reaction. Samples were detected with a 30 minute RT-RPA incubation followed by a 1 hour LwaCas13 reaction prior to lateral flow strip detection. Adjusted band intensities (determined from lateral flow strips) are shown, as well as individual lateral flow strips, with positive and control sample lines indicated with arrows16.
(d) Fluorescence detection of synthetic DNA 1 target with decreasing input concentrations using a one-pot SHERLOCK reaction combining the RPA, T7, and LwaCas13 detection steps. Bar graphs indicate mean ± SEM of background-subtracted fluorescence measured from either three technical replicates; each individual replicate is represented as a dot14.
(B) Colorimetric-based lateral-flow detection assay
- Prepare the Cas13-SHERLOCK master mix by adding the following components in an Eppendorf tube in the order they are listed, starting with water.
Component Volume per reaction Volume for four replicates Ultragrade water 12.32 μL 61.6 μL HEPES pH 6.8, 1M 0.4 μL 2 μL MgCl2, 1M 0.18 μL 0.9 μL rNTP solution mix, 25 mM each 0.8 μL 4 μL LwaCas13 in SB (63.3 μg/mL) 2 μL 10 μL Murine RNase Inhibitors, 40 U/μL 1 μL 5 μL T7 RNA polymerase, 5 U/μL 0.5 μL 2.5 μL crRNA (10 ng/μL) 1 μL 5 μL LF-RNA sensor 1 (100 μM) 0.2 μL 1 μL Total 19 μL 95 μL For one condition and four technical replicates, transfer 87.4 μL of master mix into a PCR-strip tube or 96-well PCR plate. Scale up according to total number of conditions and replicates.
Spin down the RPA pre-amplification reaction mix, carefully open the eppendorf tube, and transfer 4.6 μL of RPA-reaction mix to 87.4 μL aliquoted master mix on ice. Briefly vortex and spin down for 15 seconds in a 96-well plate to collect the entire reaction mix in the well.
Carefully open the Eppendorf tubes for each reaction mix and transfer 20 μL per technical replicate and condition into a 96-well plate. Incubate reaction for 1 hour at 37°C in a PCR thermocycler or incubator.
Transfer the 20 μL reaction mix into a 2 mL Eppendorf tube, and add 100 μL of HybriDetect 1 assay buffer. A nuclease-free 2 mL 96-well block can be used instead of Eppendorf tubes.
Perform lateral flow detection by placing a HybriDetect 1 lateral flow strip into diluted reactions and waiting 1–2 minutes for the development of the colored readout.
-
After completion, remove lateral flow strip and place on a white background for visual inspection. See anticipated results and Figure 5c for positive and negative reaction. Alternatively, for quantitative assessment, remove the lateral flow strip and place on a white background for image acquisition with conventional smart-phone cameras or a gel imaging system. Using a gel image or image processing software, measure band intensity for the bands and determine positive detection against a positive control dilution series.
Critical step. It is important to perform positive and negative controls alongside tested samples to verify a functional reaction and rule out false positives and false negatives.
Troubleshooting
TIMING
Step 1–28, Recombinant expression and purification of LwaCas13a (5 days)
Step 29–41, In vitro transcription and purification of crRNA (5 hours)
Step 42, Sample extraction (5 minutes to 1 hour)
Step 43–48, RPA pre-amplification (30 minutes to 1 hour)
Step 49–52, Cas13-SHERLOCK detection (1 to 4 hours)
Box 2, Step 1–12, One-Pot Cas13-SHERLOCK nucleic acid detection (1 to 4 hours)
ANTICIPATED RESULTS
The presented protocol for expression and purification of LwaCas13a yields roughly 0.5 – 1 mg of pure protein. A recommended culture volume of 4 liters therefore yields enough protein to perform thousands of individual Cas13 SHERLOCK reactions from the same batch. Moreover, the enzyme is free of contaminating nucleases as water input SHERLOCK controls do not result in background fluorescence (Fig 1D, Figure 5a–b).
Nucleic acid detection as described above can detect DNA and RNA with attomolar sensitivity. Figure 5b shows the result from a Cas13-SHERLOCK fluorescence detection of a synthetic RNA dilution series down to 2 aM. The same dilution series was used to detect RNA with a Cas13-SHERLOCK lateral flow colorimetric readout with similar sensitivity (Figure 5c). Combining the RPA, T7, and Cas13 steps in a single one-pot reaction can detect synthetic DNA 1 with a sensitivity of 20 aM (Figure 5d).
Acknowledgements
We would like to thank W. Blake and C. Brown for helpful feedback, and R. Macrae, R. Belliveau, E. Blackwell, and the entire Zhang lab for discussions and support.
Funding: O.O.A. is supported by a NIH F30 NRSA 1F30-CA210382. F.Z. is a New York Stem Cell Foundation–Robertson Investigator. F.Z. is supported by NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell and G. Harold and Leila Mathers Foundations; the Poitras Center for Affective Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and J. and P. Poitras, R. Metcalfe, and D. Cheng.
Footnotes
Competing interests: M.J.K, J.S.G., O.O.A, and F.Z. are co-inventors on patent applications filed by the Broad Institute relating to work in this manuscript. J.S.G., O.O.A., and F.Z. are co-founders of Sherlock Biosciences. J.S.G. is a campus advisor of Benchling, Inc.
Data and materials availability: Reagents from the Zhang lab are widely available to the academic community through Addgene and additional information and resources can be found on the Zhang lab website (www.genome-engineering.org) and GitHub (github.com/fengzhanglab).
References
- 1.Piepenburg O, Williams CH, Stemple DL & Armes NA DNA detection using recombination proteins. PLoS Biol 4, e204, doi: 10.1371/journal.pbio.0040204 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharyya RP, Thakku SG & Hung DT Harnessing CRISPR Effectors for Infectious Disease Diagnostics. ACS Infect Dis 4, 1278–1282, doi: 10.1021/acsinfecdis.8b00170 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Batista AC & Pacheco LGC Detecting pathogens with Zinc-Finger, TALE and CRISPR- based programmable nucleic acid binding proteins. J Microbiol Methods 152, 98–104, doi: 10.1016/j.mimet.2018.07.024 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Li S, Wang J & Liu G CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol, doi: 10.1016/j.tibtech.2018.12.005 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Shmakov S et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 60, 385–397, doi: 10.1016/j.molcel.2015.10.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abudayyeh OO et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573, doi: 10.1126/science.aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771, doi: 10.1016/j.cell.2015.09.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SY et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res 28, 491–493, doi: 10.1038/s41422-018-0022-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JS et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, doi: 10.1126/science.aar6245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.East-Seletsky A et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273, doi: 10.1038/nature19802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smargon AA et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell 65, 618–630 e617, doi: 10.1016/j.molcel.2016.12.023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konermann S et al. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell, doi: 10.1016/j.cell.2018.02.033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan WX et al. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol Cell, doi: 10.1016/j.molcel.2018.02.028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gootenberg JS et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442, doi: 10.1126/science.aam9321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasper Spoelstra W et al. CRISPR-based DNA and RNA detection with liquid phase separation. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gootenberg JS et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science, doi: 10.1126/science.aaq0179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomita N, Mori Y, Kanda H & Notomi T Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3, 877–882, doi: 10.1038/nprot.2008.57 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Vincent M, Xu Y & Kong H Helicase-dependent isothermal DNA amplification. EMBO Rep 5, 795–800, doi: 10.1038/sj.embor.7400200 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuzil P, Zhang C, Pipper J, Oh S & Zhuo L Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res 34, e77, doi: 10.1093/nar/gkl416 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrar JS & Wittwer CT Extreme PCR: efficient and specific DNA amplification in 15–60 seconds. Clin Chem 61, 145–153, doi: 10.1373/clinchem.2014.228304 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Ameur A, Kloosterman WP & Hestand MS Single-Molecule Sequencing: Towards Clinical Applications. Trends Biotechnol 37, 72–85, doi: 10.1016/j.tibtech.2018.07.013 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Li SY et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov 4, 20, doi: 10.1038/s41421-018-0028-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Li S & Wang J CRISPR-Cas12b-assisted nucleic acid detection platform. bioRxiv, 362889, doi: 10.1101/362889 (2018). [DOI] [Google Scholar]
- 24.Hajian R et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nature Biomedical Engineering, doi: 10.1038/s41551-019-0371-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abudayyeh OO et al. RNA targeting with CRISPR-Cas13. Nature 550, 280–284, doi: 10.1038/nature24049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34, 863–868, doi: 10.1038/nbt.3609 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Kleinstiver BP et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 34, 869–874, doi: 10.1038/nbt.3620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myhrvold C et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448, doi: 10.1126/science.aas8836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemz A, Ferguson TM & Boyle DS Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol 29, 240–250, doi: 10.1016/j.tibtech.2011.01.007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye J et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134, doi: 10.1186/1471-2105-13-134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.East-Seletsky A, O’Connell MR, Burstein D, Knott GJ & Doudna JA RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol Cell 66, 373–383 e373, doi: 10.1016/j.molcel.2017.04.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]