Abstract
Programmable RNA editing enables reversible recoding of RNA information for research and disease treatment. Previously, we developed a programmable A to I RNA editing approach by fusing catalytically inactivated RNA-targeting CRISPR-Cas13 (dCas13) with the adenine deaminase domain of ADAR2. Here, we report a C to U RNA editor, referred to as RNA Editing for Specific C to U Exchange (RESCUE), by directly evolving ADAR2 into a cytidine deaminase. RESCUE doubles the number of pathogenic mutations targetable by RNA editing and enables modulation of phospho-signaling-relevant residues. We apply RESCUE to drive β-catenin activation and cellular growth. Furthermore, RESCUE retains A to I editing activity, enabling multiplexed C to U and A to I editing through the use of tailored guide RNAs.
One Sentence Summary:
Programmable cytidine to uridine RNA editing with an evolved ADAR2 fused to CRISPR-Cas13 expands the RNA editing toolbox.
We previously developed a RNA base editing technology called REPAIR (RNA editing for programmable A to I (G) replacement), which uses the RNA targeting CRISPR effector Cas13 (1–6) to direct the catalytic domain of ADAR2 to specific RNA transcripts to achieve adenine to inosine conversion with single-base precision (7). However, REPAIR, along with a number of other RNA editing technologies(8–15), only allow for A-to-I conversions. Technologies for precise RNA editing of cytidine to uridine would greatly expand the range of addressable disease mutations and protein modifications (fig. S1A).
Although natural enzymes capable of catalyzing C to U conversion have been harnessed for DNA base editing (16, 17), they only operate on single stranded substrates (18), exhibit off-targets (19–21), and deaminate multiple bases within a window. Therefore, we took a synthetic approach to evolve the adenine deaminase domain of ADAR2 (ADAR2dd), which naturally acts on double-stranded RNA substrates and preferentially deaminates a target adenine mispaired with a cytidine, into a cytidine deaminase. We fused this evolved cytidine deaminase to dCas13 to develop programmable RNA Editing for Specific C to U Exchange (RESCUE) in mammalian cells (fig. S1B), which we used to activate β-catenin and modulate cell growth. Lastly, we improved the specificity of RESCUE more than 10-fold via rational mutagenesis, generating a highly specific and precise C to U RNA editing tool.
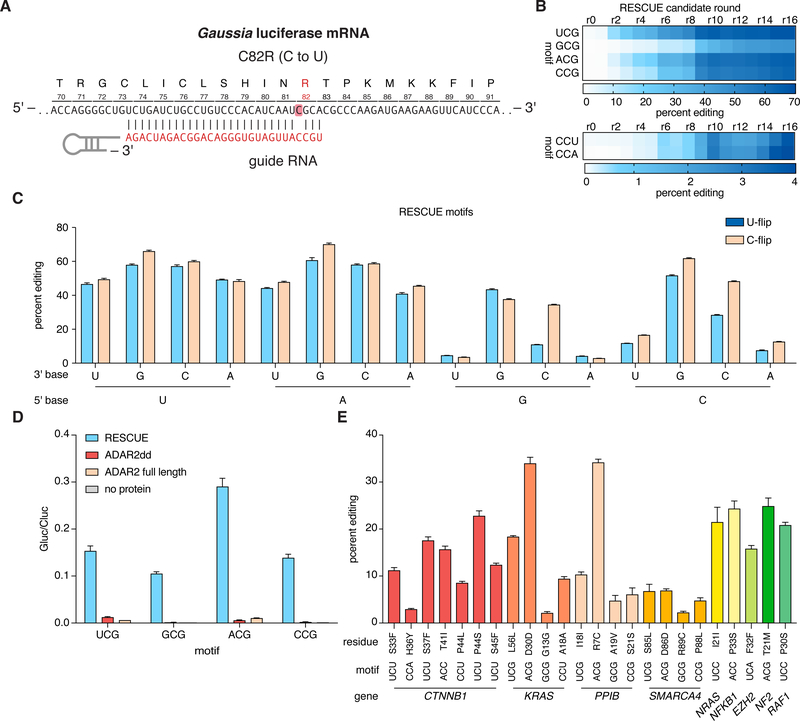
Comparison of the E. coli cytidine deaminase and the human ADAR2dd showed remarkable structural homology between their catalytic cores (22), suggesting the possibility of evolving ADAR2dd into a cytidine deaminase (fig. S1B). We selected residues of ADAR2dd contacting the RNA substrate (23) for three rounds of rational mutagenesis on an ADAR2dd fused to the catalytically inactive Cas13b ortholog from Riemerella anatipestifer (dRanCas13b), yielding RESCUE round 3 (RESCUEr3), with 15% editing activity (Fig. 1A–B, fig. S2–3). We then began directed evolution across ADAR2dd to identify additional candidate mutations that increase the activity of RESCUE in yeast (see Supplementary Methods and Table S1). Sixteen rounds of evolution, culminating with the final construct RESCUEr16 (hereafter referred to as just RESCUE), resulted in increased cytidine deamination activity across all target combinations of neighboring 5ánd 3´ bases (Fig. 1C, fig. S4–S7). We additionally characterized guide features necessary for robust activity, finding that RESCUE is optimally active with C or U base-flips across the target base using a 30-nt guide (Fig. 1C and fig. S8–9). Moreover, as dRanCas13b and the catalytically inactive Cas13b ortholog from Prevotella sp. P5–125 (dPspCas13b) were equivalent, the final RESCUE construct used dRanCas13b (fig. S10).
Figure 1: Evolution of an ADAR2 deaminase domain for cytidine deamination.
A. Schematic of RNA targeting of the catalytic residue mutant (C82R) of Gaussia luciferase reporter transcript.
B. Heatmap depicting the percent editing levels of RESCUEr0-r16 on cytidines flanked by varying bases on the Gluc transcript. More favorable editing motifs are shown at the top, while less favorable motifs (5Ć) are shown at the bottom.
C. Editing activity of RESCUE on all possible 16 cytidine flanking bases motifs on the Gluc transcript with U-flip or C-flip guides.
D. Activity comparison between RESCUE, ADAR2dd without Cas13, full-length ADAR2 without Cas13, or no protein.
E. Editing efficiency of RESCUE on a panel of endogenous genes covering multiple motifs. The best guide for each site is shown with the entire panel of guides displayed in fig. S19.
The 16 mutations in RESCUE are distributed throughout the structure of ADAR2dd (fig. S11A), indicating both direct interactions of the evolved residues with the RNA target within the catalytic pocket as well as indirect effects (Fig. S11B). These mutations enable fitting of either adenosine or cytidine, as RESCUE is capable of both adenosine and cytidine deamination (fig. S12). We evaluated the role of each mutant by individually adding them to REPAIR or removing them from RESCUE. (fig. S13). We found that mutations in the catalytic core (V351G, K350I) and contacting the RNA target (S486A, S495N) were integral to RESCUE activity, while others had minor effects. Biochemical characterization of RESCUE mutations on purified ADAR2dd showed no activity on dsDNA, ssDNA, or DNA-RNA heteroduplexes, with the evolved mutations improving the kinetics of C to U editing on dsRNA substrates in vitro (fig. S14). We also found that ADAR2 or alternative RNA editing platforms without Cas13 (8, 9, 11, 13, 24) with introduced RESCUE mutations had markedly reduced editing compared to Cas13b-based RESCUE (Fig. 1D, fig S15–18).
We next evaluated the efficiency of RESCUE on endogenous transcripts in HEK293FT cells via bulk sequencing of cell populations. We tested a variety of guide designs across 24 different sites across nine genes as well as on 24 synthetic disease-relevant mutation targets from ClinVar and found editing rates up to 42% (Fig. 1E, fig. S19–22; see Table S2). Across the guides tested (See Tables S3–5), we found multiple guide design rules, most notably related to features of the motif (5´ U or A preferred) and guide mismatch position (See Supplementary Materials and Methods).
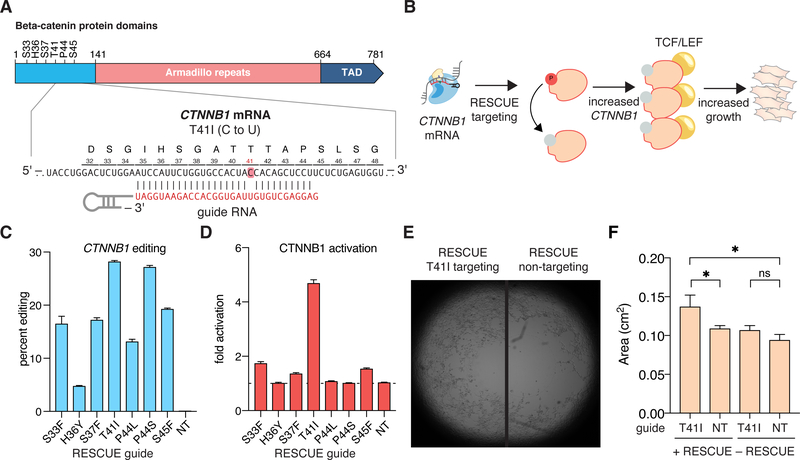
We next applied RESCUE to alter activation of the STAT and Wnt/β-catenin pathways via modulation of key phosphorylation residues, which inhibits ubiquitination and degradation (25) (Fig. 2A–B, fig. S23). We tested a panel of guides targeting the β-catenin transcript (CTNNB1) at known phosphorylation residues and observed editing levels between 5% and 28% (Fig. 2C), resulting in up to 5-fold activation of Wnt/β-catenin signaling (Fig. 2D) and increased cell growth in HEK293FT (Fig. 2E–F) and human umbilical vein endothelial cells (HUVECs) (fig. S24). As therapeutic applications with RESCUE will require shorter constructs for viral delivery, we also evaluated RESCUE activity with C-terminal truncations of dRanCas13b and found either similar or improved deaminase activity (fig. S25).
Figure 2: Phenotypic outcomes of RESCUE on cell growth and signaling.
A. Schematic of β-catenin domains and RESCUE targeting guide.
B. Schematic of β-catenin activation and cell growth via RESCUE editing.
C. Percent editing by RESCUE at relevant positions in the CTNNB1 transcript.
D. Activation of Wnt/β-catenin signaling by RNA editing as measured by β-catenin-driven (TCF/LEF) luciferase expression.
E. Representative microscopy images of RESCUE CTNNB1 targeting and non-targeting guides in HEK293FT cells.
F. Quantitation of cellular growth due to activation of CTNNB1 signaling by RNA editing in HEK293FT cells.
Since RESCUE retains adenosine deaminase activity (fig. S12), the native pre-crRNA processing activity of Cas13b (4) enables multiplexed adenine and cytosine deamination. By delivering RESCUE along with a pre-crRNA targeting an adenine and a cytosine in the CTNNB1 transcript (Fig. 3A), we found that RESCUE could edit both targeted residues S33F and T41A at rates of ~15% and 5%, respectively (Fig. 3B). However, in these experiments, as well as single-plex assays, we found A to I off-targets near the targeted cytosine (fig. S26–27). To eliminate these off-targets, we introduced disfavorable guanine mismatches in the guide across from off-target adenosines (Fig. 3C), significantly reducing off-target editing while minimally disrupting the on-target editing (Fig. 3D).
Figure 3: RESCUE and REPAIR multiplexing and specificity enhancement via guide engineering.
A. Schematic of multiplexed C to U and A to I editing with pre-crRNA guide arrays
B. Simultaneous C to U and A to I editing on CTNNB1 transcripts
C. Schematic of rational engineering with guanine base flips to prevent off-target activity at neighboring adenosine sites.
D. Percent editing at on-target C and off-target A sites for Gaussia luciferase (left) and KRAS (right) using rational introduction of disfavored base flips.
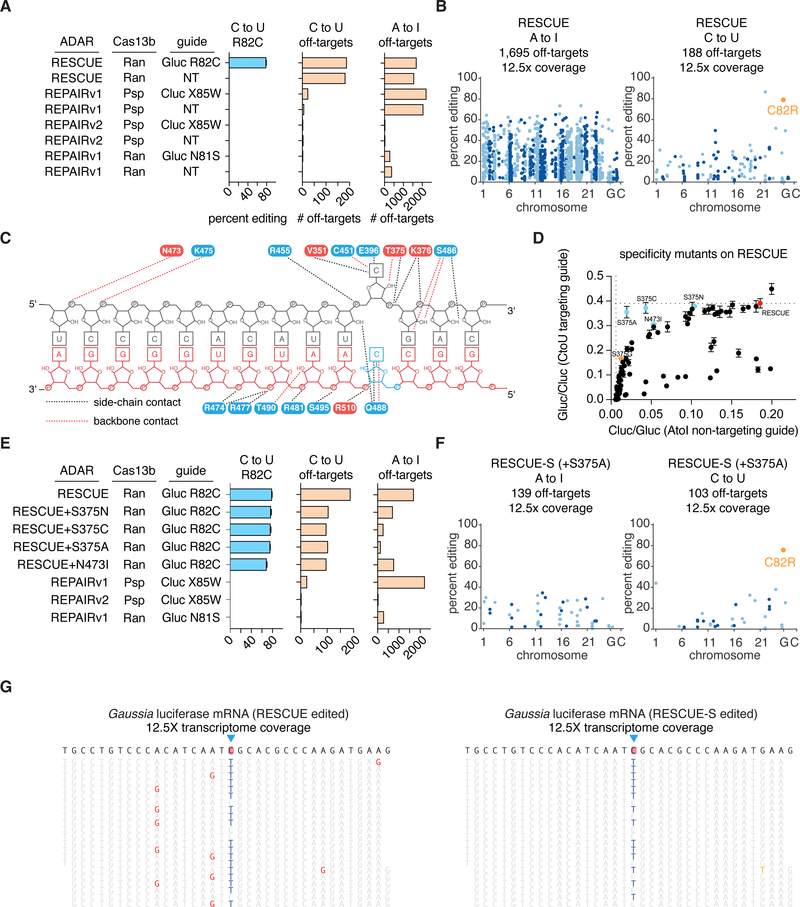
Because of off-targets observed near the target site, we profiled off-targets with whole-transcriptome RNA-sequencing, finding that while RESCUE had ~80% C to U editing on the Gluc transcript (Fig. 4A), it had 188 C to U off-targets and 1,695 A to I off-targets, comparable to A to I off-targeting with REPAIRv1 (7)(Fig. 4A,B). To improve the specificity of RESCUE we performed rational mutagenesis of ADAR2dd at residues interacting with the RNA target (Fig. 4C), resulting in improved specificity RESCUE mutants (Fig. 4D–G). The top specificity mutant, S375A on RESCUE (hereafter referred to as RESCUE-S), maintained ~76% on-target C to U editing (Fig. 4E), but only had 103 C to U off-targets and 139 A to I off-targets (Fig. 4E–G), with reduced missense mutations and differentially-regulated transcripts (fig. S28–S31). We also found that RESCUE-S retained similar efficiency as RESCUE at endogenous sites with higher specificity (fig. S32–34).
Figure 4: Transcriptome-wide specificity of RESCUE.
A. On-target C to U editing and summary of C to U and A to I transcriptome-wide off-targets for RESCUE compared to REPAIR.
B. Manhattan plots of RESCUE A to I (left) and C to U (right) off-targets. The on-target C to U edit is highlighted in orange.
C. Schematic of ADAR2dd interactions with RNA. Residues mutated for improving specificity are highlighted in red.
D. Luciferase values for C to U activity with a targeting guide (y-axis) and A to I activity with a non-targeting guide (x-axis) shown for RESCUE and 95 RESCUE mutants. RESCUE is highlighted in red and mutants with better specificity in blue. The T375G mutant (REPAIRv2) is shown in orange.
E. On-target C to U editing and summary of C to U and A to I transcriptome-wide off targets of RESCUE, REPAIR, and top specificity mutants.
F. Manhattan plot of RESCUE-S (+S375A) A to I (left) and C to U (right) off-targets. The on-target C to U edit is highlighted in orange.
G. Representative RNA sequencing reads surrounding the on-target Gluc editing site (blue triangle) for RESCUE (left) and RESCUE-S (right). A to I edits are highlighted in red; C to U (T) edits are highlighted in blue; sequencing errors are highlighted in yellow.
RESCUE is a programmable base editing tool capable of precise cytidine to uridine conversion in RNA. Using directed evolution, we demonstrate that adenosine deaminases can be relaxed to accept other bases, resulting in a novel cytidine deamination mechanism that can edit dsRNA. The larger targetable amino acid codon space of RESCUE enables modulation of more post-translational modifications, such as phosphorylation, glycosylation, and methylation, as well as expanded targeting of common catalytic residues, disease mutations, and protective alleles, such as ApoE2 (fig. S1, S35). Overall, RESCUE extends the RNA targeting toolkit with new base editing functionality, allowing for expanded modeling and potential treatment of genetic diseases.
Supplementary Material
Acknowledgments:
We would like to thank R. Munshi, A. Baumann, A. Philippakis, and E. Banks for computational guidance; J. Strecker, N. Gaudelli, S. Kannan, M. Alimova, R. Wu, A. Lee, R. Wu, I. Slaymaker for helpful discussions; P. Rogers, C. Otis, S. Saldi, and N. Pirete for flow cytometry assistance; A. Farina for help with yeast supplies; R. Macrae, R. Belliveau, and the entire Zhang lab for discussions and support.
Funding: O.O.A. is supported by a NIH F30 NRSA 1F30-CA210382. F.Z. is a New York Stem Cell Foundation-Robertson Investigator. F.Z. is supported by NIH grants (1R01-HG009761, 1R01- MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell and G. Harold and Leila Mathers Foundations; the Poitras Center for Affective Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; J. and P. Poitras; The Phillips Family; R. Metcalfe; and D. Cheng.
Footnotes
Competing interests: J.S.G., O.O.A, and F.Z. are co-inventors on patent applications filed by the Broad Institute relating to work in this manuscript. J.S.G., O.O.A., and F.Z. are co-founders of Sherlock Biosciences. F.Z. is a co-founder and advisor of Beam Therapeutics, Editas Medicine, Arbor Biotechnologies, and Pairwise Plants. O.O.A. and J.S.G. are advisors for Beam Therapeutics.
Data and materials availability: Sequencing data are available at Sequence Read Archive under BioProject accession number PRJNA525232. Expression plasmids are available from Addgene under UBMTA; support forums and computational tools are available via the Zhang lab website (https://zhanglab.bio/).
References and Notes:
- 1.Abudayyeh OO et al. , C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassidy-Amstutz C et al. , Identification of a Minimal Peptide Tag for in Vivo and in Vitro Loading of Encapsulin. Biochemistry 55, 3461–3468 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Shmakov S et al. , Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shmakov S et al. , Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 15, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.East-Seletsky A et al. , Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abudayyeh OO et al. , RNA targeting with CRISPR-Cas13. Nature 550, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox DBT et al. , RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merkle T et al. , Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat Biotechnol 37, 133–138 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Vogel P et al. , Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat Methods 15, 535–538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda M et al. , Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. Sci Rep 7, 41478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wettengel J, Reautschnig P, Geisler S, Kahle PJ, Stafforst T, Harnessing human ADAR2 for RNA repair - Recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res 45, 2797–2808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montiel-Gonzalez MF, Vallecillo-Viejo IC, Rosenthal JJ, An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Res 44, e157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel P, Schneider MF, Wettengel J, Stafforst T, Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew Chem Int Ed Engl 53, 6267–6271 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJ, Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci U S A 110, 18285–18290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees HA, Liu DR, Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19, 770–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishida K et al. , Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, (2016). [DOI] [PubMed] [Google Scholar]
- 18.Salter JD, Bennett RP, Smith HC, The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci 41, 578–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin S et al. , Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science, (2019). [DOI] [PubMed] [Google Scholar]
- 20.Zuo E et al. , Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grünewald J et al. , Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macbeth MR et al. , Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309, 1534–1539 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews MM et al. , Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat StructMol Biol 23, 426–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katrekar D et al. , In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat Methods 16, 239–242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald BT, Tamai K, He X, Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17, 9–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chee MK, Haase SB, New and Redesigned pRS Plasmid Shuttle Vectors for Genetic Manipulation of Saccharomycescerevisiae. G3 (Bethesda) 2, 515–526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laughery MF et al. , New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32, 711–720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macbeth MR, Bass BL, Large-scale overexpression and purification of ADARs from Saccharomyces cerevisiae for biophysical and biochemical studies. Methods Enzymol 424, 319331 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng H, Dean N, Dramatic Improvement of CRISPR/Cas9 Editing in Candida albicans by Increased Single Guide RNA Expression. mSphere 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heim R, Prasher DC, Tsien RY, Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A 91, 12501–12504 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Beal PA, Probing RNA recognition by human ADAR2 using a high-throughput mutagenesis method. Nucleic Acids Res 44, 9872–9880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz RD, Schiestl RH, Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. NatProtoc 2, 38–41 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT, Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 13, 680–685 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.