Abstract
The somatic DNA strand-specific imprinting to effect gene regulation and selective chromatid segregation model was previously proposed to produce developmentally nonequivalent sister cells in mitosis. Such a mechanism might explain generation of stem-cell pattern of cell division in eukaryotes. The developmentally controlled process involves a pair of homologous chromosomes at a specific cell division to establish embryonic left-right body axis asymmetry. As a result, visceral organs in the two sides of vertebrate’s body develop asymmetrically. The model was specifically proposed to explain the well-known axis randomization phenotype of the left-right dynein mutant mice where one-half of animals develop with standard visceral organ’s positioning and the balance develops with the inverted arrangement. The model postulated that the specific dynein, a microtubule-based molecular motor protein, promotes the selective chromatid segregation process in mitosis. Thus, random segregation involving sister chromatids of a pair of specific chromosomes leads to axis randomization of the mutant. Moreover, the model uniquely predicts that 50 percent mutant embryos should produce symmetrical cell divisions because of random segregation; consequently, their either visceral side would develop as mirror image of the other side resulting in embryonic lethality. In view of this prediction, validity of prominent body axis-determination models is scrutinized here. Results supporting the cell-type regulated chromosome 6 and chromosome 7 selective chromatids segregation phenomenon existing in mouse cells are reviewed. Published results with the mutant mice are consistent with the chromosome segregation model for axis determination.
Keywords: Biased DNA-strand segregation phenomenon, asymmetric cell division, epigenetic model for development, left/right axis determination, developmental disorders
INTRODUCTION
While the anterior-posterior (AP) and dorso-ventral (DV) body axes in vertebrates can be established in embryogenesis by exogenous cues of sperm entry point or gravity, but how the left-right (LR) axis is initially established remains unknown. Because the LR axis determination is an offshoot of AP and DV axes, defining the gene function of mutations that disrupt axes association to one another should help us define the mechanism of LR axis determination. Vertebrates exhibit a strikingly conserved LR asymmetry of internal organs, such as heart, stomach, lungs and liver. Xenopus, zebra fish, chick, and mouse studies have produced a mechanism of LR determination based on differential gene expression on the two sides of the embryo. Proteins such as Nodal, Lefty1, Lefty2 – all encoding signaling molecules – and the downstream transcription factor Pitx2, are expressed only on the left side of an early embryo, while Activin-βB, Snail, and Fibroblast growth factor-8 are expressed only on the right side [25,42].
Two prominent mechanisms have been proposed to explain the initiating event that results in side-specific asymmetric gene expression through a cascade of genetic interactions. The most prominent “nodal flow hypothesis” postulates that anti-clockwise rotation of monocilia on cells of the midline embryonic structure, the node, directs the flow of extra-cellular fluid carrying putative “morphogen” molecules to one side of the node in the embryo [10]. Alternatively, an equivalent model of self-enhancement and lateral inhibition system for gene regulation might establish laterality [30]. Second, fluid flow might result in directional bending of mechanosensor immotile cilia in the node that triggers a left-sided asymmetric Ca2+ flux [27]. Such distribution is thought to create an asymmetric pattern of gene expression over a larger region in the embryo in the side-specific fashion. These mechanisms are based on a crucial role for nodal ciliary function. Brown and Wolpert [5] first proposed this class of “morphogen gradient” model, a theoretical concept for the generation of LR asymmetry. The model theorizes that an initial break in embryonic symmetry is due to a hypothetical diffusible “handed” molecule that is distributed in a graded fashion over the embryo from which cells in the embryo derive the asymmetric information for their LR specification. The model was invoked to explain the fascinating phenotype of the famous spontaneous iv−/iv− (situs inversus viscerum) mouse mutation discovered nearly a one-half century ago [11]. Random distribution of the LR asymmetry occurs in the mutant such that 50 percent mice develop with normal (situs solitus) and 50 percent develop with mirror-imaged placement of internal organs (situs inversus). The situs phenotype is usually determined by noting the location of the milk-filled stomach seen through the translucent abdominal wall of newly born pups; stomach normally lies on the left side of the body [24].
Molecular characterization of the iv gene was much anticipated to help provide clues for the LR axis determination mechanism. The iv gene encodes a molecular motor protein, the axonemal Left-Right Dynein (LRD), and the gene is therefore named lrd1 [40]. This motor moves its cargo along microtubules. The cilia developed on nodal cells normally exhibit anti-clockwise rotation and that rotation is proposed to asymmetrically distribute the morphogen to two sides of the developing embryo in mice. To explain the visceral randomization phenotype of the lrd1 mutant by the prominent models quoted above, the orientation of the morphogen gradient is hypothesized somehow to be distributed to the left- versus right-side in 50:50 ratio in embryos owing to the immotile cilia [10,30]. However, how random distribution is precisely accomplished, what is the nature of the morphogen(s), what is the basis for its precise graded distribution in the embryo, and how would cells vary their response to different concentrations of the morphogen, how does the morphogen regulate differential gene expression in cells of different cell types are just a few of the many questions that remain unanswered. In addition, many of the genes implicated in LR specification, including lrd1, are also expressed in non-ciliated cells where they perform other functions. Consequently, although popular, this hypothesis remains controversial [25,42]. Most investigators point out with urgency that the mechanism of symmetry breaking initially in the embryo remains a key “Holy Grail” unanswered question and it raises interesting theoretical issues in developmental biology [10,25,30,42]. The question addressed here is how initially (“the first event”) the LR asymmetry in embryogenesis is established. Here tests of an alternative model that were not considered in previous studies are discussed.
RESULTS AND DISCUSSION
The somatic strand-specific imprinting and selective chromatid segregation (SSIS) model
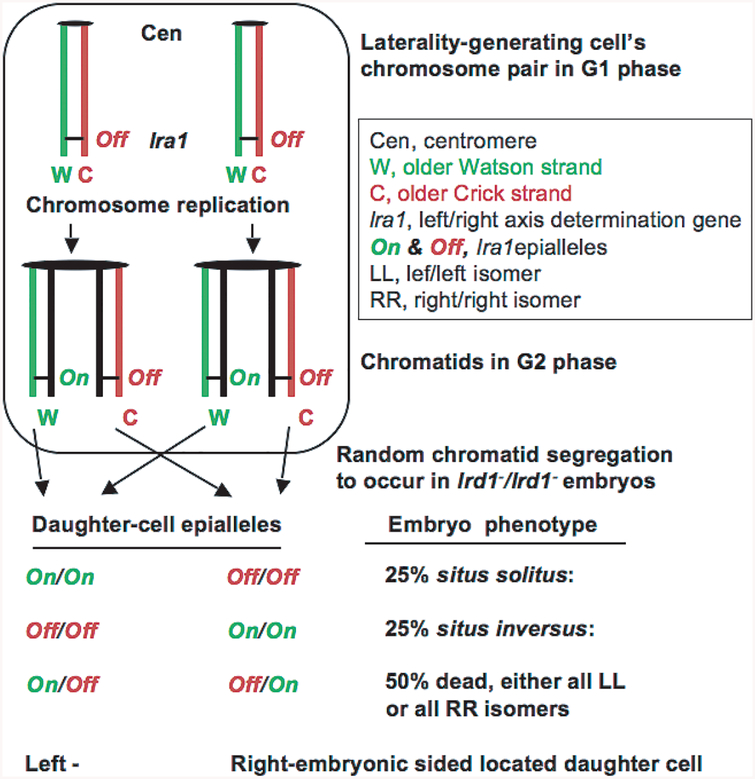
An alternative chromosome-based model (Fig. 1) was proposed [17] as a mechanism for initially breaking the embryonic symmetry, specifically to explain the random laterality phenotype of the lrd1−/lrd1− mutant. The SSIS model proposed a cell-lineage based developmental mechanism whereby a single laterality-generating progenitor cell in the embryo produces one left- and another right-side-generating daughter cell through an asymmetric cell division. LRD, possibly acting through the centromere [2], is proposed to selectively sort sister chromatids that had become nonequivalent during replication by epigenetic means at the left/right axis-determining (lra1) gene. Accordingly, random situs in the mutant mice would result from random segregation of chromatids owing to the LRD defect [3]. However, experimental verification of the model is problematic, as many questions remain unanswered, including: which cell division in embryogenesis is relevant for axis determination considering that inner cell mass cells are totipotent; which chromosome is epigenetically modified; what is the mechanism of strand-specific imprinting for differentially regulating the lra1 gene in sister chromatids; how could biased chromatid segregation of a specific chromosome in mitosis be accomplished and its existence tested, etc. As stated above, because the popular nodal flow models remain controversial [25,42],we recently demonstrated the existence of the selective strand/chromatid segregation phenomenon in mouse cells [2,3], and because several specific phenotypes of the LRD mutant (discussed below) have not been explained by prominent models, these considerations provide motivation to scrutinize the validity of the axis-determining models.
Fig. 1.
Predictions of the Somatic Strand-specific Imprinting and selective chromatid Segregation (SSIS) model concerns lrd1−/lrd1− embryos’ phenotype. According to the model (Klar 1994), the laterality-generating cell contains a hypothetical left/right axis-determining (lra1) gene in both homologs of an unknown chromosome in the epigenetically silenced (Off) state (top). It suggests generation of nonequivalent sister chromatids of the chromosomes pair whereby DNA replication activates (On) the lra1 gene in the “older Watson” (W) strand-containing chromatid but it remains Off in the “older Crick” (C) strand-containing chromatid. Subsequently, the lrd1 gene-encoded LRD motor protein, by acting directly or indirectly through the centromere, promotes selective chromatid segregation. Consequently, both older W-containing chromatids are segregated to the daughter cell placed on the left side of the embryo that thereby becomes left-side-organs generating progenitor cell. As a result, both older C-containing chromatids are delivered to the other right-side-program generating progenitor daughter cell placed on the right side of the embryo. With reference to older strands and to simplify presentation, this is designated as the “WW:CC” segregation pattern [2]. Consequently, an asymmetric cell division results from asymmetric expression of the lra1 gene in sister cells when initially the embryonic asymmetry is determined. This causes all wild-type embryos to develop with normal situs. However, random chromatid segregation is predicted to occur in the lrd1−/lrd1− mutant; consequently, the predicted number and variety of epigenetic state combinations in embryos and their resulting visceral phenotypes are indicated at the bottom of the drawing (see text for detailed description of the phenotypes). DNA strands that normally exist in helix are represented as straight lines for clarity and are arbitrarily designated as W and C based on their 5’−3’ orientation. The older W strand in each chromatid is indicated in green colour, older C in red, and all “younger” strands synthesized in the most recent DNA replication cycle are coloured in black. The black arrows represent distribution of a specific strand/chromatid, along with the indicated lra1-epiallele, to specific daughter cell.
Genetic tests of the SSIS model
A primary developmental event should depend on a mechanism that is independent of the gene regulatory cascade. The SSIS model proposes that the double helix structure of DNA constitutes the primary event because the two strands are inherently different from one another. The lrd1 mutation does not eliminate LR axis development; it randomizes its distribution to specific sides of the embryo. Thus, LRD somehow controls non-random distribution of a binary choice in the side-specific fashion that likely occurs early in embryogenesis. The SSIS model makes four predictions concerning results of the lrd1−/lrd1− mutant, and the last three of them are unique to the model (Fig. 1). First, the number of embryos per pregnant mother should not be altered in the mutant. Second, three kinds of embryos will be generated in the ratio of 25 percent situs solitus: 25 percent situs inversus: 50 percent “isomers” where either side develops as the mirror image of the other side. Third, isomers resulting from the On/Off lra1 epiallelic constitution inherited by both sister cells are predicted to die. Fourth, such moribund embryos should be all left/left (LL) isomers, if the lra1-On epiallelic state promotes specifically the left-sided organs identity development, or should be all right/right (RR) isomers, if it promotes only the right-sided development. That is, the isomers should be either all LL or all RR, but not a mixture of both types. The observation of 50 percent solitus and 50 percent inversus mice found in the LRD mutant is prominently highlighted in all the studies concerning mice laterality development. Should that indeed be true, such a result formally rules out the SSIS model. This is because while considering all embryos, the model predicts that one-half of them should die specifically because of isomerism, and only those that survive should be randomized (Fig. 1). Might it be that the existence of dead embryos not fully appreciated in previous studies? For example, they might have escaped notice should the isomeric embryos die too early during embryogenesis.
Keeping these predictions in mind, we reviewed literature and found an earlier and most extensive study [24] that highlighted embryonic lethality result for which no satisfactory explanation has been advanced in a very large number of studies published thus far. This study satisfied above described predictions of the SSIS model as follows: First, the number of embryos per pregnant female in homozygous mutant crosses were similar to that found in control crosses. Second, after counting several hundred pups and “by comparing the average number of mutant embryos per litter counted in utero with the average number per litter weaned”, Layton noted that “the mortality rate calculated in this way was about 50%” [!] Third, the dead embryos, or the newborn mice that were moribund due to heart malformations, often exhibited some form of the so-called heterotaxia. Heterotaxia indicates when placement of some visceral organs, such as lung, spleen, liver, or gut, is not coordinated with the specific side of the embryo. For example, the mouse normally has one lobe in the left-side lung and four lobes in the right-side lung. Therefore, in addition to stomach positioning, lung anatomy is commonly used as another convenient indicator of visceral LR identity. Among 61 newborns with congenital heart malformations analyzed, 26 cases of one lobe in each lung (LL isomers), 5 cases with three lobes in each lung, but no case of four lobes in each lung was found. Importantly, in the most frequent heterotaxic cases only the LL lobation pattern was observed. Overall, these results are consistent with predictions of the SSIS model and explain development of a specific kind of visceral isomers and of 50% embryonic lethality (Fig. 1).
The observation of minority and novel (five) cases with three lobes in each lung remains unexplained by any model. It is possible that, because of a gene expression threshold consideration, the novel On/Off “heterozygous” epigenetic state of the lra1 gene causes development of a minority of cases with an intermediate phenotype that is neither fully LL nor fully RR isomer. Alternative possibility is that arrested development due to LRD mutation in a minority of embryos may cause them not to reach a developmental stage where specification of fully developed left identity occurs (see below). Said another way, such anomalous cases might not be the direct consequence of the LRD deficiency; rather they may be due to a secondary consequence of earlier developmental anomalies. Moreover, factors in the genetic background may influence the frequency of these unusual cases. Despite this caveat, because the LL type comprised a predominant class (26 among 31), the simplest interpretation is that the On/Off gene configuration promotes the left identity developmental program. Therefore, we suggest that the lra1-on state causes left-sided organs development, and the off state causes the default mode of development that activates the right-side organs identity program.
The SSIS model proposed that a single cell division in embryogenesis primarily establishes the LR axis, specifically concerning the stomach placement. Although the result of 50 percent dead embryos is supportive of the model, the model does not readily explain another finding. That is, 28.1 to 40.8 percent of the viable pups, the number varying with genetic background, exhibit heterotaxia of the large veins of the thorax or of abdomen [24]. We can imagine two possibilities to explain this phenotype. One possibility is that situs is primarily determined by a single asymmetric cell division in the endoderm layer of cells, especially concerning stomach placement, and that other LRD-dependent asymmetric cell division(s) may likewise control the laterality development of mesoderm tissues from which other organs, such as heart, develop. Consistent with this interpretation, these malformations occur with equal frequency in mice with situs solitus and those with situs inversus defined by the criteria of stomach placement. So, the LRD function may be also required later on at the organogenesis stage. Supporting this possibility, we find that the lrd1 gene is expressed in cell lineages that exhibit selective segregation of chromosome 7 chromatids and it is not expressed in others that exhibit random segregation [3]. Second, as suggested [43], secondary mechanisms may have evolved to improve the performance of the major laterality establishing process. That is, some other system might be activated to repair the situs anomalous organs, including deviations due to developmental noise or mixed genetic background that cause heterotaxia. For comparing our interpretation with that of prevailing models, we note that the morphogen gradient model also fails to explain not only heterotaxia, but also the 50 percent embryonic lethality and the predominantly LL isomer heterotaxia phenotype of the LRD mutant embryos. Interestingly, an analogous visceral randomization due to DNAH11 dynein gene mutations that result in the “Kartagener’s syndrome” in humans does not involve heterotaxia of individual organs [1]. Whether a 50% embryonic lethality and visceral isomerization also occurs in humans remains to be determined.
The axis randomization phenotype is exhibited by all three iv alleles known to date. The original mutation discussed above is a missense, the legless mutation is a larger deletion covering several genes and the third one contains a targeted deletion of the ATP binding domain (ivΔP1) [40,41]. Therefore, the reversed-organs phenotype in 50 percent of the mice cannot be easily attributed to leakiness in any of the three mutations. An apparent contradiction to our explanation might originate from results with the ivΔP1 allele where an absence of embryonic lethality was noted [41]. However, their study only analyzed 85 progeny of matings between iv−/+ heterozygote mice. Because the mutation is recessive, therefore analysis of only one-quarter (85/4 = 21) homozygous mutant embryos impacts the lethality issue. If one-half of the 21 homozygous embryos should die according to the SSIS model, then 10 or 11 homozygous viable progeny is expected. Therefore, the finding of 16, as opposed to predicted 10 or 11 in the study, is not necessarily contradictory to the SSIS model on statistical grounds. Clearly, analysis of a larger number of progeny, preferably from homozygous ivΔP1 mutant matings, needs to be performed. Unfortunately, further mutant analysis is not possible because the ivΔP1 stock was not saved (M. Brueckner, personal communication).
One argument advanced to support the cilia motility model is that the LR axis orientation can be changed by artificially altering the orientation of the extra-embryonic fluid flow [10]. It should be noted that the monocilia originate from the basal body and that is associated with centrosome, a structure found at the pole of the mitotic spindle. As the LRD is found in the basal body, both ciliary motility and some mitotic spindle function might be affected by the LRD deficiency. Therefore, it is formally possible that the experimentally induced liquid flow might mechanically stimulate immotile cilia to influence some event of the spindle, such as chromatid segregation, thereby axis orientation might be experimentally altered. Moreover, because cytoplasmic dynein is required for mitotic spindle orientation, chromosome orientation and it binds to mitotic kinetochores, such activities might facilitate the selective chromatid segregation process [8]. In this context, here discussion of the origin of the cilia motility hypothesis should be helpful. Prior studies of the human Kartagener’s syndrome had motivated investigators subsequently to consider ciliary motility as a mechanism for LR axis determination in mice. This human syndrome likewise causes 50% incidence of situs inversus, and the syndrome is associated with sperm immotility and impaired ciliary motility on epithelial cells lining the respiratory airways. Their dynein defect was discovered through elctronmicroscopic studies designed primarily to understand the cause of sperm immotility [1]. In contrast, sperms of the iv−/iv− mutant mouse are motile. It was therefore initially thought that mouse must have an axis determination mechanism different from that of humans. However, when iv was discovered to encode dynein, and because workers were influenced by the human sperm motility defect observation, motility defects in other kinds of cilla of the mutant mouse were subsequently investigated to gain clues to the axis determination mechanism. Indeed, as normally motile monocilia found on nodal cells are immotile in the mutant [10], such a comparison between the two systems motivated researchers to focus on defining the role of nodal ciliary motility in axis determination of mice. It remains to be seen whether this flow of logic is helpful or whether it has led workers to stray away from the field.
How to explain the situs inversus phenotype found in nearly all mutant animals of another gene, inv (inversion of embryonic turning) [45]? It may be suggested by the morphogen gradient model that the gradient’s orientation is inverted compared to that of wild-type mice through an unknown mechanism, but this notion remains to be experimentally verified [42]. Notably the ciliary rotation of the LRD mutant is not reversed, as might be predicted by the fluid flow model. Instead, inversion of embryonic turning during gastrulation observed in the mutant provides a simpler explanation for the Situs inversus phenotype. Another conceptually related possibility is that the DV axis may be inverted in the inv mutant with respect to the LR axis to result in the observed phenotype. Accordingly, as lrd mutations uncouple DV axis from the LR axis, homozygous double mutants for iv and inv genes should be randomized in situs. Our analysis here is however limited only for explaining the LRD mutants’ phenotypes.
LR axis specification occurs very early during embryogenesis in model organisms
Recent studies suggest that cell fate is determined by cleavage pattern and positions of blastomeres in the embryo in vertebrates, although early embryos are subject to regulative development. Interestingly, both neuronal and visceral asymmetry in Caenorhabditis elegans is linked to the AP axis through the invariant mitotic spindle orientation in specific blastomeres, as early or earlier than the six-cell stage of the embryo [31]. Notably, the axis asymmetry is initially established as early as at the two-cell stage of the embryo in Xenopus and newts [25,42] but it is reprogrammed by “de-differentiation” if the two blastomeres are completely separated from each other where two whole embryos develop. However, paradoxically, if one of the two cleavage cells in the frog embryo is destroyed, by poking it with a red-hot needle, the surviving cell does not develop into an embryo. Why not? Interestingly, the presence of cell membrane of the damaged blastomere blocks development of the undamaged blastomere [36]. Thus, cell-cell contact is essential to maintain identity of left- versus right-sided developmental program of respective blastomeres. Perhaps the SSIS model for explaining cellular asymmetry in early balstomeres of other species also should be contemplated. Considering these cases, it is possible that the LR asymmetry in mammals is also established primarily at an as yet unknown single cell division. Indeed, asymmetric cell divisions do occur prior to 32-cell stage of the mouse embryo [4]. However, other mechanisms are also thought to be involved in determining the laterality of visceral organs in other vertebrates such as chick and Xenopus. These include ion flux and gap-junction communication that function in the LR pathway long before the appearance of cilia or a mature node in embryos of those species [10,25,30,42]. We hypothesize that ion flux or gap-junction communication mechanisms may promote asymmetric cell division by establishing planar cellular polarity because the SSIS model requires it both to accomplish biased chromatid segregation and to non-randomly place the resulting left- and right-sided-generating progenitor cells in the context of the DV and AP axes (Fig. 1). Therefore, we suggest that, drugs inhibiting H+/K+-ATPase transporters that regulate ion influx [42] might indirectly affect axis laterality by perturbing cellular polarity.
Discovery of the selective chromatid segregation phenomenon
With respect to replication history, sister chromatid copies made by chromosome replication are inherently nonequivalent as one chromatid “WC′”) contains an older W and a newer C′(′indicates younger) strand and the sister chromatid is always W′C. When both WC′ chromatids of a homologous pair of chromosomes are segregated to a specific daughter cell and thereby both W′C chromatids are delivered to the other daughter cell (Fig. 1), the “WW:CC” segregation pattern is designated to have occurred [19]. As a result of strand/chromatid-specific imprinting to modulate gene expression, the WW:CC segregation pattern will produce nonequivalent, while the WC:WC pattern will produce equivalent sister cells. In principle, controlling segregation pattern during mitosis can alter developmental fate of daughter cells. The discovery of the selective chromosome-specific segregation phenomenon required genetic analysis of chromosomal recombinants of mouse cells. Normally, G2 recombination events between non-sister chromatids produce a mixture of homozygous and heterozygous daughter cells concerning a marker located distal to the crossover point. Surprisingly, all 432 Cre/loxP-induced G2 phase embryonic stem (ES) cells Chr. 7 recombinants analyzed were found to be homozygous [26]. To explain this finding, existence of the nonrandom WW:CC strand/chromatid segregation phenomenon was invoked [19]. Endoderm cells also follow the WW:CC pattern, while neuroectoderm cells exhibit the WC:WC pattern or recombination occurs there in G1. However, the pancreatic, mesoderm, and cardiomyocyte cell cultures follow the usually presumed random distribution mode because both homozygous and heterozygous recombinants are obtained [2]. Different sets of chromosomes might be subject to different cell-type-regulated segregation processes because Chr. 11 ES cells produce a mixture of homozygous and heterozygous recombinants [26].
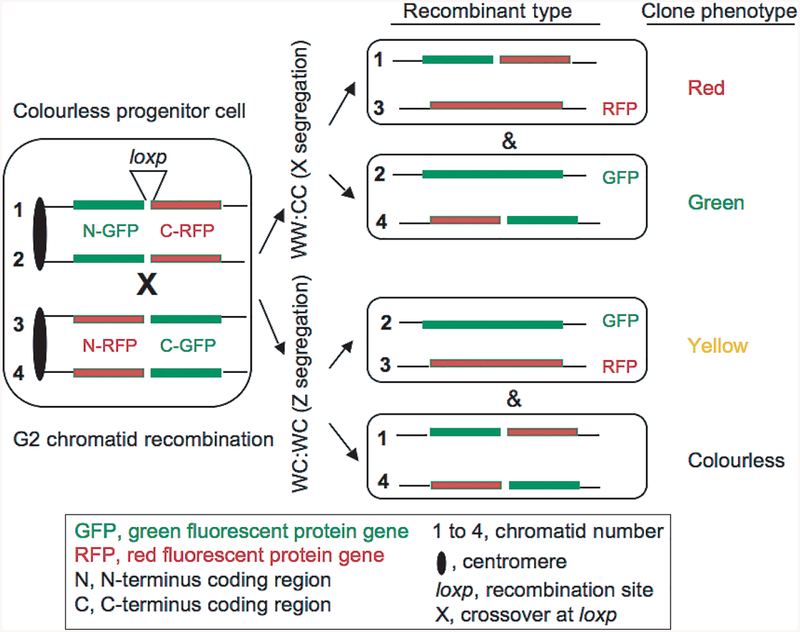
Conceptually, similar analysis of Cre/loxp-induced recombinants of cell clones produced during development can be exploited to determine the segregation pattern of a particular chromosome. In a study designed to simultaneously label and produce gene knockout in clones of somatic cells to help track cell-lineage, two reciprocally chimeric genes, each containing the N-terminus of one marker and C-terminus of the other marker interrupted by a loxp-containing intron sequences, were placed at the mouse Chr. 6 ROSA26 locus (Fig. 2). Both chimeric genes consisted of part of the coding sequences of green fluorescent protein (GFP) and red fluorescent protein (RFP). The chimeric genes before recombination do not produce functional proteins, and therefore, cells in tissues with these trans genes are colorless. Recombination in G2 would restore functional expression cassettes where one recombined chromatid would produce GFP and the other RFP. Following recombined cell’s mitosis, the recombinant chromatids/chromosomes can segregate into different daughter cells (so-called “X segregation”, Fig. 2) or into the same daughter cell (“Z segregation”). The X segregation events will result in red-and green-colored mosaic clones of cells, whereas the Z segregation events will produce double-colored (i.e., yellow) clones. Remarkably, in tissues of liver, heart and kidney predominantly red and green patches of cells were produced. This result requires that recombination predominantly occurs in G2 as G1 recombinants will produce yellow clones, that recombination is restricted between non-sister chromatids number 2 and #3 (and/or #1 and #4; with respect to replication history of DNA strands, both possibilities are equivalent with one another), followed by X segregation (Fig. 2). As homozygosis of a marker located distal to the crossover point occurs in all green and red clones expected from X segregation [46], we interpret these results to mean that predominantly WW:CC segregation occurs in these tissues. Thus, biased chromatid segregation phenomenon is proposed here as a mechanism to produce recombinants that only follow X segregation. In contrast, predominantly yellow-coloured ones (26 among 26) were produced in granule cell clones. This result can be explained by one or a combination of three possibilities; Z segregation (WC:WC, Fig. 2), recombination limited between chromatids #1 and #3 and/or #2 and #4 followed by WW:CC segregation, or recombination occurs in G1. Likely, this result indicates selection of which pair of chromatids is able to recombine followed by operation of the selective chromatid segregation process. In contrast, all three types of colored patches were found indicating unbiased segregation process operating in cortical cap tissues. Thus, a chromosome-specific, cell-type-regulated, selective versus random strand-segregation mechanism exists as an ordinary cellular process in mouse cells. Future studies should be directed to define the mechanism of biased chromatid segregation, to determine generality, and to decipher the function of such a phenomenon in biology at large.
Fig. 2.
The mouse mitotic recombination genetic model used to label cell lineages in vivo [46]. The indicted chimeric gene recombination cassettes are placed at the ROSA26 locus in the mouse homologous Chr. 6 pair. The Cre-induced mitotic recombination occurs in rare cells, because of inefficient Cre gene expression, between recombination cassettes at the loxp site in cells of live animals. This produces red, green or yellow clones of cells after they have inherited functional GFP- and RFP-encoding recombined genes as diagrammed. Here this model is exploited to determine the chromatid segregation pattern occurring specifically at the centromere of chromosome 6 (this study). To simplify presentation, the loxp element is omitted in daughter cells. Monitoring the distribution of coloured cell clones on colourless tissue background helps define the segregation pattern during development in particular tissues.
Comparing the Cairns’ immortal strand inheritance with the SSIS model
The SSIS model differs in several respects, but it is conceptually similar in one aspect to the genome-wide “immotal” strand inheritance feature of the Cairns hypothesis [6]. Cairns postulated biased delivery of the “oldest” DNA strands to asymmetrically dividing “stem” cells, such as those in the gut, blood and skin tissues. According to this model, which is gaining considerable support in some studies [7,14,28,32–34,37,39] but not in all studies [21], stem cells would thus avoid inheriting potentially cancer-causing DNA replication errors. A few studies concerning non-stem cell divisions have investigated segregation of base-labeled strands of the entire genome and found random [12,13] and nonrandom [23,35] distribution patterns. We note that the segregation pattern of a specific chromosome cannot be determined by base-labeling DNA of the entire genome, the procedure used in studies to test validity of the Cairns model. Such a procedure would provide interpretable information only if older versus younger DNA strands were to nonrandomly segregate in all the chromosomes and in all the cells of the tissues under examination. Instead of the proposal of avoiding replication errors by the Cairns model, the chromosome(s)-specific SSIS process is proposed strictly as a mechanism for cellular differentiation that exploits the W versus C nature of DNA strands, strands replication history and epigenetics differences between sister chromatids. This has been proposed to specify visceral laterality in mice [17] and to specify brain hemispheric asymmetry in humans where analyses of chromosome 11 translocations have provided support to the model [18,19,38]. As a criticism to the interpretation of results partially supporting Cairns immortal stand hypothesis, Lansdorp [22] recently proposed a biased chromosome- and chromatid-specific segregation model for explaining cellular differentiation, a model that is identical to the SSIS model proposed previously.
The SSIS process constitutes a new mitogenetic paradigm for development
The conventional use of term imprinting, monoallelic gene expression depending on its parent-of-origin, indicates an epigenetic process that is installed in the gamete and that is propagated as such in the progeny. Instead, this term in the SSIS model reflects somatically installed epigenetic events that control expression of developmentally important genes in chomatid-specific fashion in somatic lineages. Thus, the SSIS model comprises a new mitotic genetics (“mitogenetic”) paradigm to explain cellular differentiation in development in somatic cells. It should be distinguished from the Mendelian genetics paradigm that concerns meiotic inheritance of gene factors and that does not directly address studies of development. It should be appreciated that asymmetric cell division based on inheritance of DNA strands of particular chromosome is only tested and demonstrated in fission yeast cells. As asymmetric cell division is likely to be the central tenet of eukaryotic development, by following the yeast example plus by evolving mechanisms for biased chromaitd segregation in diploid organisms, we suggest that such chromosome-based mechanisms might explain development in general [15,16,20]. Structural foundations of the SSIS model comprise the DNA double-helix structure [44], its semi-conservative mode of replication [29], and segregation of a copy of each chromosome to daughter cells. The model is based on cell lineage in development and it is designed to exploit both the inherent base sequence differences in the W and C strands and of the strand-specific epigenetic events presumably installed at the time of chromosome replication [15]. Furthermore, somatically installed heterochromatin-based epigenetic states of gene expression are inherited as conventional Mendelian/chromosomal markers in multiple rounds of mitoses in fission yeast [9]. As a consequence of these processes, sister cells would have been modified to differentially regulate the pattern of their gene expression. Such a mechanism to regulate expression of developmentally important genes can evolve independently and over and over again in organisms in which DNA constitutes the genetic material. Thus, just as the mechanism of Mendelian heredity employs chromosomes for passing on genetic characteristics from one generation to the next through meiosis, chromosomes might also carry decisions for accomplishing cellular differentiation in mitotic cell lineages. We speculate that the SSIS process may involve different sets of chromosomes with specific epigenetic states to establish multiple arrays of cell types in complex organisms.
CONCLUSIONS
Overall, results of LRD mutant embryos [24] support several genetic predictions of the SSIS model and support the idea that the LRD motor protein plays a cytoplasmic role in LR patterning through the selective chromatid segregation process that does not involve the extra-cellular ciliary motion proposed by other models. Also, we suggest that the lra1 gene in the On epiallelic state activates the left-sided developmental program, while the Off state promotes the default mode to activate the right-sided program. It is easy to imagine that even occasional disruption of the SSIS process, such as by spontaneous unwanted somatic recombination both between sister and non-sister chromatids, it can create imbalance in epigenotype of progeny cells. For example, such loss of heterozygosity of imprints could result in cancer development without any gene being mutated. Thus, investigating the SSIS mechanism that was initially designed to explain cellular differentiation and development has implications for understanding origin of disease and developmental disorders such as psychoses. We point out that the SSIS model provides a mechanism only for initially breaking symmetry in the embryo, and in addition, we have implicated LRD in the process of selective chromatid segregation [3]. However, the model does not necessarily concern subsequent development of organs where the morphogen gradient/nodal liquid flow, SSIS or some other model such as the two-cilia model [27,42] might be functional. This perspective should provide the impetus for further research to scrutinize validity of prevailing models for axis determination. Clearly, more work is needed before the mechanism of axis determination is understood.
ACKNOWLEDGEMENTS
We thank Dr. Peter Donovan for discussions about mouse embryology, and Dan Burke for critically reading the manuscript. Because of limited scope of the perspective, the author apologizes to scientists for not discussing a vast amount of literature published on axis determination. Several review articles quoted here should be consulted for in-depth treatise. The Intramural Research Program of the National Institute of Health, National Cancer Institute at Frederick, USA, has supported this research.
REFERENCES
- [1].Afzelius BA, Cilia-related diseases, J Pathol 204(4) (2004), 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Armakolas A and Klar AJS, Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis, Science 311 (2006), 1146–1149. [DOI] [PubMed] [Google Scholar]
- [3].Armakolas A and Klar AJS, Left-right dynein motor implicated in selective chromatid segregation in mouse cells, Science 315 (2007), 100–101. [DOI] [PubMed] [Google Scholar]
- [4].Bischoff M, Parfitt DE and Zernicka-Goetz M, Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions, Development 135(5) (2008), 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brown NA and Wolpert L, The development of handedness in left/right asymmetry, Development 109 (1990), 1–9. [DOI] [PubMed] [Google Scholar]
- [6].Cairns J, Mutation selection and the natural history of cancer, Nature 255 (1975), 197–200. [DOI] [PubMed] [Google Scholar]
- [7].Conboy MJ, Karasov AO and Rando TA, High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny, PLoS Biol 5(5) (2007), e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Faulkner NE, Vig B, Echeverri CJ, Wordeman L and Vallee RB, Localization of motor-related proteins and associated complexes to active, but not inactive, centromeres, Hum Mol Genet 7(4) (1998), 671–677. [DOI] [PubMed] [Google Scholar]
- [9].Grewal SI and Klar AJS, Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis, Cell 86(1) (1996), 95–101. [DOI] [PubMed] [Google Scholar]
- [10].Hirokawa N, Tanaka Y, Okada Y and Takeda S, Nodal flow and the generation of left-right asymmetry, Cell 125(1) (2006), 33–45. [DOI] [PubMed] [Google Scholar]
- [11].Hummel KP and Chapman DB, Visceral inversion and associated anomalies in the mouse, J Hered 50 (1959), 10–13. [Google Scholar]
- [12].Ito K and McGhee JD, Parental DNA strands segregate randomly during embryonic development of Caenorhabditis elegans, Cell 49(3) (1987), 329–336. [DOI] [PubMed] [Google Scholar]
- [13].Ito K, McGhee JD and Schultz GA, Paternal DNA strands segregate to both trophectoderm and inner cell mass of the developing mouse embryo, Genes Dev 2(8) (1988), 929–936. [DOI] [PubMed] [Google Scholar]
- [14].Karpowicz P, Morshead C, Kam A, Jervis E, Ramuns J et al. , Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro, J Cell Biol 170 (2005), 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Klar AJS, Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast, Nature 326(6112) (1987), 466–470. [DOI] [PubMed] [Google Scholar]
- [16].Klar AJS, The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands, EMBO J 9(5) (1990), 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Klar AJS, A model for specification of the left-right axis in vertebrates, Trends Genet 10(11) (1994), 392–396. [DOI] [PubMed] [Google Scholar]
- [18].S Klar AJ, The chromosome 1;11 translocation provides the best evidence supporting genetic etiology for schizophrenia and bipolar affective disorders, Genetics 160(4) (2002), 1745–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Klar AJS, A genetic mechanism implicates chromosome 11 in schizophrenia and bipolar diseases, Genetics 167(4) (2004), 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Klar AJS, Lessons learned from studies of fission yeast mating-type switching and silencing, Annual Review of Genet 41 (2007), 213–236. [DOI] [PubMed] [Google Scholar]
- [21].Kuroki T and Murakami Y, Random segregation of DNA strands in epidermal basal cells, Jpn J Cancer Res 80(7) (1989), 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lansdorp PM, Immortal strands? Give me a break, Cell 129(7) (2007), 1244–1247. [DOI] [PubMed] [Google Scholar]
- [23].Lark KG, Consigli RA and Minocha HC, Segregation of sister chromatids in mammalian cells, Science 154(753) (1966), 1202–1205. [DOI] [PubMed] [Google Scholar]
- [24].Layton WM Jr., Heart malformations in mice homozygous for a gene causing situs inversus, Birth Defects Orig Artic Ser 14(7) (1978), 277–293. [PubMed] [Google Scholar]
- [25].Levin M, Left-right asymmetry in embryonic development: a comprehensive review, Mech Dev 122(1) (2005), 3–25. [DOI] [PubMed] [Google Scholar]
- [26].Liu P, Jenkins NA and Copeland NG, Efficient Cre-loxP-induced mitotic recombination in mouse embryonic stem cells, Nat Genet 30(1) (2002), 66–72. [DOI] [PubMed] [Google Scholar]
- [27].McGrath J, Somlo S, Makova S, Tian X and Brueckner M, Two populations of node monocilia initiate left-right asymmetry in the mouse, Cell 114(1) (2003), 61–73. [DOI] [PubMed] [Google Scholar]
- [28].Merok JR, Lansita JA, Tunstead JR and Sherley JL, Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics, Cancer Res 62(23) (2002), 6791–6795. [PubMed] [Google Scholar]
- [29].Messelson M and Stahl FW, The replication of DNA in Escherichia coli, Proc Natl Acad Sci U S A 44 (1958), 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nakamura T, Mine N, Nakaguchi E, Mochizuki A, Yamamoto M et al. , Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system, Dev Cell 11(4) (2006), 495–504. [DOI] [PubMed] [Google Scholar]
- [31].Poole RJ and Hobert O, Early embryonic programming of neuronal left/right asymmetry in C elegans, Curr Biol 16(23) (2006), 2279–2292. [DOI] [PubMed] [Google Scholar]
- [32].Potten CS, Keratinocyte stem cells, label-retaining cells and possible genome protection mechanisms, J Investig Dermatol Symp Proc 9(3) (2004), 183–195. [DOI] [PubMed] [Google Scholar]
- [33].Potten CS, Hume WJ, Reid P and Cairns J, The segregation of DNA in epithelial stem cells, Cell 15(3) (1978), 899–906. [DOI] [PubMed] [Google Scholar]
- [34].Rambhatla L, Ram-Mohan S, Cheng JJ and Sherley JL, Immortal DNA strand cosegregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells, Cancer Res 65(8) (2005), 3155–3161. [DOI] [PubMed] [Google Scholar]
- [35].Rosenberger RF and Kessel M, Nonrandom sister chromatid segregation and nuclear migration in hyphae of Aspergillus nidulans, J Bacteriol 96 (1968), 1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schmidt GA, Schnurungs- und Durchschneidungsversuche am Amphibien-keim, Roux Arch 129 (1933), 1–44. [DOI] [PubMed] [Google Scholar]
- [37].Shinin V, Gayraud-Morel B, Gomes D and Tajbakhsh S, Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells, Nat Cell Biol 8(7) (2006), 677–682. [DOI] [PubMed] [Google Scholar]
- [38].Singh G and Klar AJ, A hypothesis for how chromosome 11 translocations cause psychiatric disorders, Genetics 177(2) (2007), 1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Smith GH, Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands, Development 132(4) (2005), 681–687. [DOI] [PubMed] [Google Scholar]
- [40].Supp DM, Witte DP, Potter SS and Brueckner M, Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice, Nature 389(6654) (1997), 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Supp DM, Brueckner M, Kuehn MR, Witte DP, Lowe LA et al. , Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries, Development 126(23) (1999), 5495–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tabin C, Do we know anything about how left-right asymmetry is first established in the vertebrate embryo? J Mol Histol 36(5) (2005), 317–323. [DOI] [PubMed] [Google Scholar]
- [43].Tamakoshi T, Itakura T, Chandra A, Uezato T, Yang Z et al. , Roles of the Foxj1 and Inv genes in the left-right determination of internal organs in mice, Biochem Biophys Res Commun 339(3) (2006), 932–938. [DOI] [PubMed] [Google Scholar]
- [44].Watson JD and Crick FH, Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid, Nature 171(4356) (1953), 737–738. [DOI] [PubMed] [Google Scholar]
- [45].Yokoyama T, Copeland NG, Jenkins NA, Montgomery CA, Elder FF et al. , Reversal of left-right asymmetry: a situs inversus mutation, Science 260(5108) (1993), 679–682. [DOI] [PubMed] [Google Scholar]
- [46].Zong H, Espinosa JS, Su HH, Muzumdar MD and Luo L, Mosaic analysis with double markers in mice, Cell 121(3) (2005), 479–492. [DOI] [PubMed] [Google Scholar]