Abstract
There is a growing appreciation for the specific health benefits conferred by commensal microbiota on their hosts. Clinical microbiota analysis and animal studies in germ-free or antibiotic-treated mice have been crucial for improving our understanding of the role of the microbiome on the host mucosal surface; however, studies on the mechanisms involved in microbiome-host interactions remain limited to small animal models. Here, we demonstrated that rhesus monkeys under short-term broad-spectrum antibiotic treatment could be used as a model to study the gut mucosal host-microbiome niche and immune balance with steady health status. Results showed that the diversity and community structure of the gut commensal bacteria in rhesus monkeys were both disrupted after antibiotic treatment. Furthermore, the 16S rDNA amplicon sequencing results indicated that Escherichia-Shigella were predominant in stool samples 9 d of treatment, and the abundances of bacterial functional genes and predicted KEGG pathways were significantly changed. In addition to inducing aberrant morphology of small intestinal villi, the depletion of gut commensal bacteria led to increased proportions of CD3+ T, CD4+ T, and CD16+ NK cells in peripheral blood mononuclear cells (PBMCs), but decreased numbers of Treg and CD20+ B cells. The transcriptome of PBMCs from antibiotic-treated monkeys showed that the immune balance was affected by modulation of the expression of many functional genes, including IL-13, VCAM1, and LGR4.
Keywords: Gut microbiome, Rhesus macaque, Antibiotic treatment, Immune response, Pathological changes
INTRODUCTION
There is growing appreciation for the importance of commensal microbiota in shaping host development and physiology (Hooper & Gordon, 2001; Schmidt et al., 2018). Critically, the commensal microbiome is an important regulator of anti-infection immunity and colonizes the host for its lifetime (Abt et al., 2012; Postler & Ghosh, 2017). Several reports and clinical cases have indicated an imbalance in immune cell subsets and abnormal up-regulation of some cytokines following gut microbiome dysbiosis, which is defined as an altered state of the microbial community (Ivanov & Honda, 2012; Langhorst et al., 2009; Soderborg & Friedman, 2018; Sprouse et al., 2019; Yang et al., 2015). These data are also supported by intestinal histopathological and immunological characteristics from experiments using mice treated with antibiotics (Kernbauer et al., 2014; Thackray et al., 2018), suggesting that the interaction between the microbiome and host shaped by commensal colonization could lead to a unique immune response. Studies on germ-free and antibiotic-treated mice have greatly improved our understanding of the specific health benefits conferred by commensal microbiota on the host immune system, digestive system, metabolic system, inflammation, and brain function (Belkaid & Hand, 2014; Desbonnet et al., 2015; Ferrer et al., 2014; Shapiro et al., 2014). However, in-depth studies on this interaction in mice have not provided sufficient data to help clarify the pathogenesis of immune disorders related to changes in the mucosal niche in the gut. Mechanisms that facilitate the establishment and stability of the gut microbiota remain poorly described. Moreover, rodent models have shortcomings such as high mortality, instability, autoimmune defects, and non-transformability, which limit the application of these research results (Vandamme, 2015). Thus, there is an urgent need for a suitable non-human primate (NHP) animal model to study the interactions among commensal microbiota and hosts.
Previous experimental results have indicated an essential role of the gut microbiome and probiotics in the intestinal mucosal barrier (Allaire et al., 2018; Yousefi et al., 2019). Furthermore, gene transcription profiling has been used to understand the systemic immune response that correlates with the function of microbiota in regulating the immune system (Gury-BenAri et al., 2016). Both innate immune and inflammatory responses after bacterial dysbiosis, which are responsible for regulating the variable integrated functions of the immune system, should be emphasized (Belkaid & Hand, 2014). In this work, the process of gut bacteria dysbiosis was verified using 12-month-old rhesus macaques (Macaca mulatta). Three rhesus monkeys were treated with a combination of antibiotics for 21 d to observe dynamic immunology and pathology process characteristics. Results showed that both the diversity and structure of the commensal bacteria were disrupted, and intestinal commensal bacterial species decreased by 95.6%–98.7% after antibiotic treatment. This depletion of gut commensal bacteria induced aberrant small intestinal morphology. In addition, we systemically analyzed and correlated the immune response and modulation of gene expression in peripheral blood mononuclear cells (PBMCs) from these monkeys. Gene transcripts in PBMCs that were up- and down-regulated after antibiotic treatment were identified, including those categorized under the GO terms of immune system process, cell communication, and cell activation. The results of this study may provide support for evaluating how changes in commensal bacteria affect immune status, as reflected by PBMCs. Moreover, our study indicates that long-term oral antibiotic treatment could result in neurological symptoms. Therefore, the relationship between gut commensal bacteria and host immunity could be studied using this NHP model.
MATERIALS AND METHODS
Animals
Four one-year-old male rhesus monkeys (ID No.: 1, 2, 3, 4) were reared separately in a large cage (BSL-2 conditions) with sufficient fresh air and natural light, allowing visual, olfactory, and auditory interactions with other monkeys. All rhesus monkeys were healthy and weighed 2±0.5 kg. A temperature control valve was installed in each room to guarantee a room temperature of ~25 °C, with food, water, and fruit readily available. All animal experiments were performed with approval of the Yunnan Provincial Experimental Animal Management Association (Approval No.: SYXK (Dian) K2015-0006) and the Experimental Animal Administration and Ethics Committee of the Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College (Approval No.: DWSP201803006) and in accordance with the principles of the “Guide for the Care and Use of Laboratory Animals” and “Guidance to Experimental Animal Welfare and Ethical Treatment”. All animals were fully under the care of veterinarians at the Institute of Medical Biology, Chinese Academy of Medicine Science.
Antibiotic treatment
Rhesus monkeys (ID No.: 1, 2, 3) were treated with a cocktail of antibiotics (1 g of ampicillin, 1 g of kanamycin, 1 g of metronidazole, 1 g of neomycin, and 1 g of vancomycin (Sigma-Aldrich, USA)) in 15 mL of 10% sucrose solution per day for three weeks, while ensuring health. The antibiotics were chosen based on previous research (Kernbauer et al., 2014) and taken orally, with care taken to avoid any confounding effects resulting from chronic stress caused by oral administration. The fourth rhesus monkey was treated with 10% sucrose solution per day for two weeks.
Sample collection and DNA extraction
Conventional animal samples were collected from other healthy rhesus monkeys of the same age. Fecal samples were collected every morning and frozen at –80 °C within 1 h of sampling. Blood samples were collected from the femoral or saphenous veins of monkeys without anesthesia every few days. Blood was divided into EDTA-K2 tubes for cell detection and into separate serum tubes for the detection of cytokines. One of the antibiotic-treated monkeys (ID No. 2) was anesthetized with isoflurane, with blood then collected, followed by sacrifice via exsanguination. All tissues and contents were flash frozen in liquid nitrogen and stored at –80 °C until use, with a portion of the tissues fixed in formalin for histomorphometric analyses. Genomic DNA was extracted from stool using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) following standard protocols. The concentration of genomic DNA was measured using a NanoDrop 2000 (Thermo Fisher Scientific, USA).
Hematology
A complete blood count (CBC) analysis was performed on whole blood immediately after sampling on day 14 after commencement of antibiotic treatment using an automatic hematological analyzer (XT-2000IV, Sysmex Corporation, Japan). Hematological values included white blood cells (WBCs), red blood cells (RBCs), neutrophils, lymphocytes, monocytes, eosinophils, basophils, mean corpuscular hemoglobin concentration (MCHC), and platelets.
16S rDNA amplicon sequencing and analyses
Polymerase chain reaction (PCR) amplification was performed to generate amplicons using bar-coded primers targeting the V3-V4 region of the bacterial 16S rRNA gene. The primers included 338F 5'-ACTCCTACGGGAGGCAGCA-3' and 806R 5'-GGACTACHVGGGTWTCTAAT-3'. Amplicons were extracted from 2% agarose gels, purified by the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, USA), and quantified using QuantiFluor™-ST (Promega Biosciences LLC, USA) according to standard protocols. The purified amplicons were then pooled and analyzed using a TruSeqTM DNA Sample Prep Kit and paired-end sequenced (2×300) on an Illumina MiSeq platform (Illumina Inc., USA) according to the manufacturer’s instructions. Pairs of reads obtained by MiSeq sequencing were merged based on the overlapping relationship of paired-end reads. The quality of reads and the effect of merging were filtered by quality control. Effective sequences for each sample were obtained as operational taxonomic units (OTUs), which were clustered with a cut-off of 97% similarity using Usearch (v7.0, http://drive5.com/uparse), with single sequences removed. RDP Classifier (v2.2, http://sourceforge.net/projects/rdp-classifier/) was used to analyze the taxonomy of each representative OTU sequence against the SILVA (Release 128) 16S rRNA database with a confidence threshold of 0.7 (Amato et al., 2013; Quast et al., 2013). The community composition of each sample was counted at each taxonomic level. Alpha (α) diversity was analyzed by Mothur (Schloss et al., 2009), and statistical significance between two groups was calculated using Student’s t-tests. Beta (β) diversity was estimated by computing the weighted UniFrac distance metric (Lozupone et al., 2006). Principal component analysis (PCA) was conducted according to Euclidean distance. Functional prediction analysis was performed to normalize the OTU abundance table by PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, which stores Cluster of Ortholog Genes (COG) and KEGG Ortholog (KO) information corresponding to Greengene ID), removing the influence of the 16S marker gene on the number of copies in the genome, then obtaining COG family and KO information corresponding to each OTU and calculating the abundance.
Real-time PCR (RT-PCR) validation
Changes in gut bacterial community were validated by RT-PCR using a SYBR Premix Ex Taq II kit (TaKaRa, Japan). We used DNA extracted from fecal samples as the template. Primers were designed to amplify specific regions in the hypervariable region V3 of the bacterial 16S rRNA gene (5'-ATTACCGCGGCTGCTGG-3' (F) and 5'-CTACGGAGGCAGCAG-3' (R)), Enterobacteriaceae (5'-CATTGACGTTACCCGCAGAAGAAGC-3' (F) and 5'-CTCTACGAGACTCAAGCTTGC-3' (R)), Bacteroides-Prevotella (5'-GAAGGTCCCCCACATTG-3' (F) and 5'-CAATCGGAGTTCTTCGTG-3' (R)), Bifidobacterium (5'-GGGTGGTAATGCCGGATG-3' (F) and 5'-TAAGCCATGGACTTTCACACC-3' (R)), Lactobacillus (5'-AGCAGTAGGGAATCTTCCA-3' (F) and 5'-ATTYCACCGCTACACATG-3' (R)), Enterococcus-Vagococcus (5'-AACCTACCCATCAGAGGG-3' (F) and 5'-GACGTTCAGTTACTAACG-3' (R)), Clostridium phoceensis (5'-GATGGCCTCGCGTCCGATTAG-3' (F) and 5'-CCGAAGACCTTCTTCCTCC-3' (R)), Clostridium cluster I (5'-TACCHRAGGAGGAAGCCAC-3' (F) and 5'-GTTCTTCCTAATCTCTACGCAT-3' (R)), Clostridium cluster XI (5'-ACGCTACTTGAGGAGGA-3' (F) and 5'-GAGCCGTAGCCTTTCACT-3' (R)), and Clostridium cluster XIV (5'-GAWGAAGTATYTCGGTATGT-3' (F) and 5'-CTACGCWCCCTTTACAC-3' (R)). We used the 2-ΔCt method to calculate the richness of the gut bacteria.
Flow cytometry and LiquiChip
Peripheral blood (100 μL) was incubated with antibodies for 30 min at room temperature in the dark and then incubated with red blood cell lysis buffer and washed with phosphate-buffered saline (PBS). The cells were analyzed using a CytoFLEX flow cytometer (Beckman Coulter, USA) according to the manufacturer’s instructions. The following antibodies (clones) were used for staining: CD3 (SP34-2), CD4 (L200), CD20 (2H7), CD8 (RPA-T8), and CD25 (M-A251), all from BD Bioscience (USA). All antibodies were titered in advance and used at optimal concentrations for flow cytometry. FlowJo v.10 was used to analyze the data. Measurement of cytokines and chemokines in the serum was performed using a MILLIPLEX® MAP NHP cytokine magnetic bead panel kit (Millipore Corporation, USA) and detected by a Bio-Plex 200 System (Bio-Rad Laboratories, USA).
Agilent genome microarray
The PBMCs were isolated by density gradient centrifugation with Lymphoprep medium (Ficoll-Paque PREMIUM; GE Healthcare, USA). Total RNA was extracted using TRIzol Reagent (Cat#15596-018, Life Technologies, USA), following the manufacturer’s instructions, and checked for RNA integrity numbers (RIN) to determine integrity using an Agilent Bioanalyzer 2100 (Agilent Technologies, USA). Microarray analysis was performed using the Agilent Rhesus Macaque Genome Microarray (USA, 4×44K). The arrays were hybridized, washed, and scanned according to the standard protocols. Gene chip tests were performed by the Shanghai Biochip Company (China). Data were extracted with Feature Extraction software v10.7 (Agilent Technologies, USA). Raw data were normalized by the quantile algorithm limma packages in R. The log-transformed expression values were adjusted, and fold-change statistical method was used to select differentially expressed genes (DEGs). Gene Ontology (GO), pathway enrichment, and network analysis of significant DEGs were systematically conducted.
Morphology
The small intestine and brain were fixed in formalin and embedded in paraffin according to standard histological protocols. Paraffin-embedded sections were deparaffinized and stained with hematoxylin-eosin-safran (H&E). The slides were scanned by a Pannoramic MIDI scanner (3DHISTECH, Hungary), and all measurements were made using CaseViewer software v2.2.
Data analysis
Significant differences between the values of two groups were calculated using Student’s t-tests. IBM SPSS statistics v23 was used for data evaluation. Statistical significance was considered at P<0.05.
RESULTS
Health status of rhesus monkeys during oral antibiotic treatment
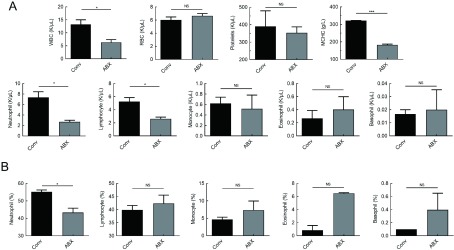
The three rhesus monkeys exhibited normal food and water intake, activities, and mental state during antibiotic treatment. Diarrhea symptoms appeared on day 2 after antibiotic treatment, which recovered after one week (Table 1). Hematological levels remained normal after 14 d of antibiotic (ABX) treatment (Figure 1). Compared with conventional (Conv) rhesus monkeys, the mean values for white blood cells, neutrophils, lymphocytes, and mean corpuscular hemoglobin concentration (MCHC) were significantly lower in rhesus monkeys treated with antibiotics (Figure 1A, B). Therefore, the body status tended to be stable after 14 d of antibiotic treatment.
1. Antibiotic treatment induces diarrhea symptoms in rhesus monkeys.
| Days post-ABX treatment | Severity of diarrhea | ||
| 1 | 2 | 3 | |
| 1, 2, 3 are IDs of rhesus monkeys. Severity of diarrhea was determined by number of defecations, characteristics of feces, and degree of dehydration. +: Mild diarrhea; ++: Diarrhea; +++: Severe diarrhea; –: Normal stool. | |||
| 0 | – | – | – |
| 1 | ++ | ++ | ++ |
| 2 | +++ | +++ | +++ |
| 3 | +++ | +++ | +++ |
| 4 | +++ | +++ | +++ |
| 5 | +++ | +++ | +++ |
| 6 | ++ | ++ | ++ |
| 7 | + | + | + |
| 8 | – | + | – |
| 9 | – | – | – |
| 10 | – | – | – |
1. Hematological changes in rhesus monkeys after 14 d of antibiotic treatment.
A: Hematological values of white blood cells (WBC), red blood cells (RBC), platelets, mean corpuscular hemoglobin concentration (MCHC), neutrophils, lymphocytes, monocytes, eosinophils, and basophils. B: Proportion of neutrophils, lymphocytes, monocytes, eosinophils, and basophils in blood. Conv: Conventional or untreated rhesus monkeys. Graphs show means±standard error of mean (SEM). n=3 rhesus monkeys per group. Significant differences between values were calculated using an unpaired two-tailed t-test. NS: Not significant, *: P<0.05, **: P<0.01, ***: P<0.001.
Antibiotic-treated rhesus monkeys display a dramatic shift in bacterial community structure in the intestine
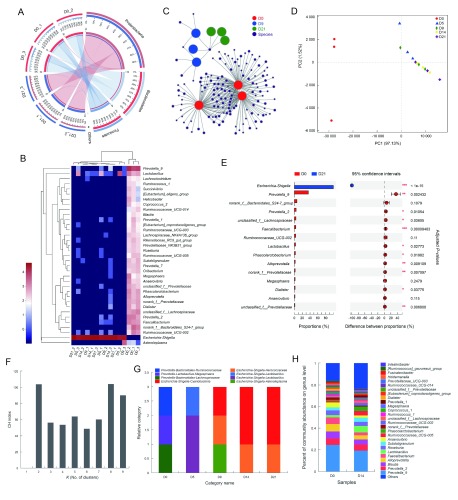
Antibiotic treatment resulted in a significant reduction in the abundance of commensal bacteria and reorganization of bacterial composition. According to the results of 16S rDNA amplicon sequencing, 538 722 effective sequences were obtained from samples, with an average length of 448 nt. Taxonomic analysis of OTUs resulted in 1, 1, 12, 20, 37, 61, 143, 218, and 324 different categories at the domain, kingdom, phylum, class, order, family, genus, species, and OTU levels, respectively. Based on α diversity analysis, the abundance and diversity of each sample were sufficient to fully describe the composition of the bacteria, and the sequencing depth was higher than 0.99 (Table 2). After antibiotic treatment, the Shannon index decreased significantly, and the number of OTUs decreased by 96.0%–98.6% after 21 d (Table 2). At the phylum level, the intestinal commensal bacteria in untreated rhesus monkeys mainly consisted of Bacteroidetes and Firmicutes (Figure 2A), and dominant bacteria included Prevotella, Lactobacillus, Bacteroides, Lachnospira, Phascolarctobacterium, Faecalibacterium, Ruminococcus, Megasphaera, and Subdoligranulum (Figure 2B). The diversity and abundance of the commensal bacteria decreased significantly after 21 d; only a few Proteobacteria were detected, and most were antibiotic-resistant Escherichia and Shigella (Figure 2B, C). Based on statistical analysis and PCA, we identified a significant difference in the structure of the commensal bacterial community between untreated and antibiotic-treated rhesus monkeys, and the reorganized bacterial structure maintained stability (Figure 2D, E). We next investigated the enterotype according to the clustering of dominant bacterial communities (Arumugam et al., 2011). Results showed that the bacterial communities were most naturally categorized into eight clusters during treatment (Figure 2F), and antibiotic treatment in rhesus monkeys resulted in a change from Prevotella enterotype to Escherichia-Shigella enterotype (Figure 2G). In addition, the diversity and community structure of the intestinal bacteria in the monkey (ID No. 4) treated only with sucrose remained unchanged (Figure 2H).
2. Alpha diversity estimators of 16S rDNA amplicon sequencing.
| ID_days post ABX treatment | Abundance index | Diversity index | Coverage | ||||
| Sobs | Ace | Chao | Shannon | Simpson | |||
| Data were divided as individuals described in the text. Number of alpha diversity indices are shown. | |||||||
| 1_D0 | 284 | 286.31 | 286.55 | 4.175 399 | 0.045 147 | 0.999 685 | |
| 1_D5 | 28 | 86.14 | 41 | 0.101 25 | 0.966 94 | 0.999 573 | |
| 1_D14 | 13 | 18.79 | 16.33 | 0.011 107 | 0.997 727 | 0.999 874 | |
| 1_D21 | 4 | 5.59 | 4 | 0.003 219 | 0.999 342 | 0.999 97 | |
| 2_D0 | 227 | 247.43 | 254 | 3.398 891 | 0.112 415 | 0.999 196 | |
| 2_D5 | 8 | 43.56 | 11 | 0.477 874 | 0.702 516 | 0.999 877 | |
| 2_D14 | 7 | 15.84 | 8.5 | 0.005 4 | 0.998 908 | 0.999 918 | |
| 2_D21 | 6 | 6.83 | 6 | 0.009 683 | 0.997 853 | 0.999 977 | |
| 3_D0 | 249 | 256.00 | 260.33 | 3.843 893 | 0.060 07 | 0.999 246 | |
| 3_D5 | 28 | 117.74 | 49 | 0.190 655 | 0.926 19 | 0.999 52 | |
| 3_D14 | 32 | 149.41 | 95.33 | 0.027 369 | 0.994 332 | 0.999 415 | |
| 3_D21 | 10 | 91.57 | 17.5 | 0.013 941 | 0.996 685 | 0.999 844 | |
2. Changes in commensal bacteria spectrum in stool after antibiotic treatment.
A: Relationship between samples and bacterial community at phylum level. Data were rendered by Circos (Krzywinski et al., 2009). Left half-circle represents composition of species in sample, color of outer ribbon represents which group it comes from, and colors of inner ribbon represent phyla. Right half-circle represents distribution proportion of each phylum in samples from different days, outer ribbons represent phyla, inner ribbon color represents different groups. Length of bars from each phylum indicates relative abundance of that phylum in corresponding sample. B: Abundances of top 35 genera in each sample were selected and compared with abundances of these genera in other samples by heatmap. C: Network analysis elucidating distribution of samples and species (abundance>50) at OTU level, highlighting similarities and differences between samples. D: PCA plot displaying variation in community structure during treatment at OTU level. Each point represents an individual. E: Bar plot showing significantly different phylotypes between pre- and post-ABX-treated rhesus monkeys at genus level. Statistical analysis was performed by Student’s t-test and P-values were adjusted by FDR. n=3, in each group. *: P<0.05, **: P<0.01, ***: P<0.001. F, G: Enterotype analysis. Clustering analysis using Jensen-Shannon distance (JSD) and partitioning around medoids (PAM) method. Calinski-Harabasz (CH) index was then used to calculate optimal clustering K value (K=8), and bar plot was used for visualization of each sample’s enterotype. H: Bar plot displaying variation in community structure of monkey receiving sucrose-treatment only at genus level.
To verify the 16S rDNA amplicon sequencing results, we used quantitative RT-PCR to detect the composition and abundance of intestinal bacteria in the three rhesus monkeys. Results showed that the dominant commensal bacteria in untreated rhesus monkeys consisted of Bacteroides, Prevotella, Lactobacillus, and Clostridium (Supplementary Figure S1A–C). We also tested antibiotic resistance by selective agar medium, and found no antibiotic-resistant strain in the normal commensal bacteria. After 3 d of antibiotic treatment, the copies of commensal bacteria per milligram of feces decreased by 10 000 to 100 000 times (Supplementary Figure S2). After 14 d of antibiotic treatment, the abundance of the 16S rRNA gene decreased significantly in the feces, and predominant bacteria were depleted (Supplementary Figure S1D–F). Furthermore, antibiotics induced the rapid rise of resistant Escherichia coli and Shigella, which did not exist before (Supplementary Figure S1G–I).
Predictive functional profiling changes driven by microbial shifts
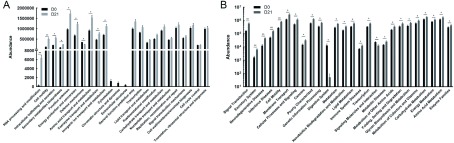
After standardization of OTU abundances, the 16S functional predictions were obtained from COG family information according to the corresponding Greengene ID of each OTU. Functional abundances were acquired by analyzing the descriptive and functional information of COG classifications in the eggNOG database. According to the analysis of bacterial functional genomics, we found that the abundance of functional genes in RNA processing and modification, cell motility, and intracellular trafficking were the most significantly increased (P<0.001) after antibiotic treatment (Figure 3A).
3. Microbial functions altered by restructured bacteria.
COG function (A) and KEGG pathway (B) prediction analyses of bacterial microbiome. n=3 per group. *: P<0.05, **: P<0.01, ***: P<0.001.
To characterize the functional alterations of commensal bacteria in antibiotic-treated rhesus monkeys, we predicted the functional composition profiles from the 16S rDNA sequencing data under PICRUSt pre- and post-antibiotic treatment. We found that multiple KEGG (level 2) categories were disturbed. The changes in pathways enriched in signal transduction, excretory system, neurodegenerative diseases, infectious diseases, and cell motility showed the most significant differences (P<0.001) between untreated and antibiotic-treated rhesus monkeys (Figure 3B). Strikingly, abundances in neurodegenerative disease, infectious disease, cancer, and metabolism pathways were significantly increased after commensal bacteria depletion. In addition, abundances in the digestive system pathway decreased along with signaling molecules and interaction pathways.
Impact of commensal bacteria alteration on host immune profile in PBMCs
As the immune system is modulated by the gut microbiota, we examined whether oral antibiotic treatment affected lymphocyte populations in peripheral blood. We observed increased numbers of CD3+ T cells and CD16+ NK cells after antibiotic treatment, as well as greater CD4+ and CD8+ cells (Figure 4B). In contrast to CD3+ T cells, we observed decreased numbers of Treg cells and CD20+ B cells after antibiotic treatment, suggesting a potential defect in the humoral immune response (Figure 4B). In the cytokine expression profile, we found that CD40L maintained a stable level in serum, indicating a pivotal role in co-stimulation and regulation of the adaptive immune response. The concentration of inflammatory cytokines also varied with the development of intestinal bacterial depletion (Figure 4C).
4. Host immune responses evoked by alterations in microbiome.
A: Gating strategies for detection of lymphocyte subsets by flow cytometry. B: Changes in frequency of CD3+, CD3+CD4+, CD3+CD8+, CD20+, CD16+, and CD4+CD25+ T cells among lymphocytes in PBMCs after antibiotic treatment. Red line represents mean value at each point. C: Heatmap displaying change in different cytokines in serum during antibiotic treatment. Concentrations (pg/mL) were detected by multiplex immunoassay and values were log2 transformed.
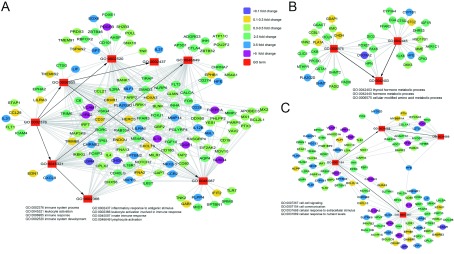
In addition, we performed an Agilent genome microarray with PBMC samples to acquire gene expression profiling of immune cells. In biological process analysis, we focused on the metabolic process, immune system process, cell communication, and cell activation GO terms. The GO terms of leukocyte activation, immune response, cell-cell signaling, and thyroid hormone metabolic process were significantly different pre- and post-antibiotic treatment (Figure 5). In immune system response and cell-cell signaling, the expression level of LGR4 decreased more than 10-fold. The protein encoded by this gene is a G-protein coulped receptor that binds R-spondins, activates the Wnt signaling pathway, and is associated with osteoporosis (Styrkarsdottir et al., 2013). The IFNA2, CXCL10, and TLR7 genes were also down-regulated, which may affect host susceptibility to viral infection (Karst, 2016; Spurrell et al., 2005; Wu et al., 2013). At the same time, the levels of VCAM1 and IL-13 were upregulated more than 5-fold (Figure 5A, C). It has been reported that IL-13 induces several changes in the gut that can lead to detachment of organisms from the gut wall (Seyfizadeh et al., 2015), and overexpression of IL-13 may contribute to some features of allergic lung diseases such as airway hyperresponsiveness, goblet cell metaplasia, and mucus hypersecretion (Wills-Karp et al., 1998). Several effector factors of immune checkpoint proteins, tissue remodeling, and cell adhesion, such as the MMP14, ABL1, and LILRA6 genes, were markedly elevated. Additionally, several cell communication genes, including GRIA2, BCAN, CACNG3, SLC12A5, SOX17, TDGF1, and VCAM1, were up-regulated (Figure 5C). These results suggest that the absence of commensal bacteria may alter the immune system and disease status of the host.
5. Network plots showing host-gene responses significantly regulated after antibiotic treatment.
Co-occurrence analysis of significantly different GO terms and gene expression profiling, including immune system process (A), metabolic process (B), and cell communication (C). Square nodes represent GO terms, elliptical nodes represent functional genes, and different colors show different fold-change values.
Depletion of commensal bacteria impairs development of small intestinal morphology
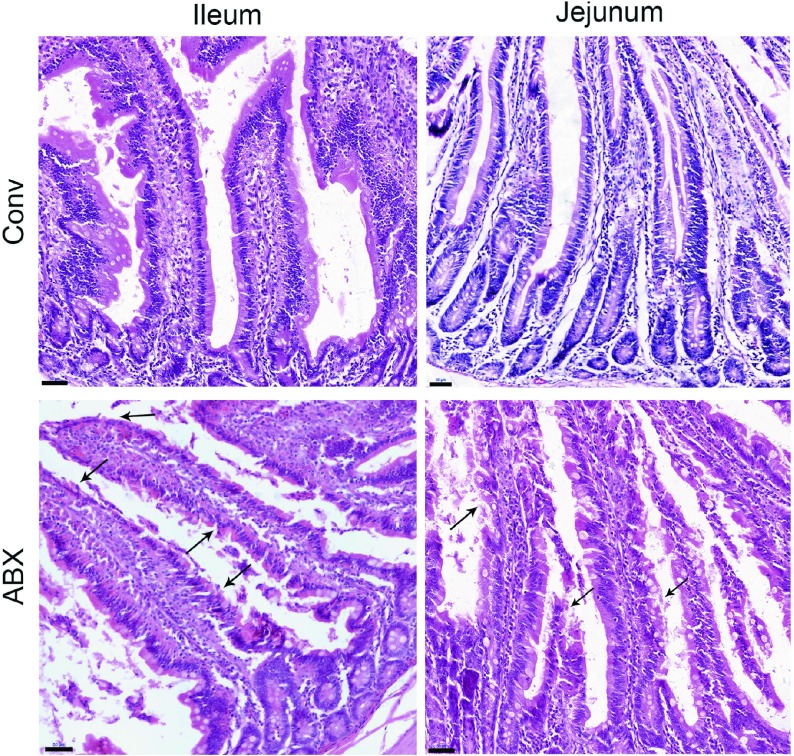
Compared with the conventional rhesus macaques, the antibiotic-treated monkeys showed impaired morphology of the small intestine. The top of the villi of the intestinal mucosa were diminished, and the epithelial cells of the intestinal mucosa were denatured, necrotic, and shedding, forming extensive superficial erosion (Figure 6). Thus, our results demonstrated that dysbiosis of commensal bacteria could impair the small intestine, as reported in previous animal studies (Kernbauer et al., 2014; Yeruva et al., 2016).
6. Histopathological changes in small intestine in antibiotic-treated rhesus monkeys.
Comparison of ileum and jejunum between conventional (Conv) and antibiotic-treated (ABX) rhesus monkeys. n=1 per group. Tissue sections stained with H&E. Images were taken at 20× magnification; Scale bars: 50 μm. Black arrows show aberrant intestinal morphology, including diminished top of villi and denatured epithelial cells of intestinal mucosa.
Long-term use of antibiotics leads to severe adverse reactions in a rhesus monkey
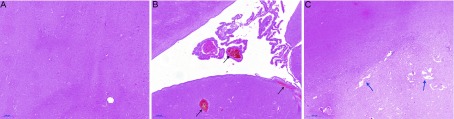
Although drugs are a known cause of neurological symptoms, antibiotics have not been taken seriously and the frequency of severe central nervous system (CNS) events associated with antibiotics is reported to be less than 1% (Bhattacharyya et al., 2016; Owens & Ambrose, 2005). However, the results of a recent retrospective study suggest that bacteria-associated neurogenic disease may be underestimated (Sandler et al., 2000). In our study, one rhesus monkey (ID No. 2) exhibited sudden myoclonus after 20 d of oral antibiotic treatment, which was improved by the reduction of the antibiotic to one third of the designed dose, although convulsive seizure occurred as long as antibiotic treatment continued. Finally, we found abnormal brain morphology in this rhesus monkey. The thalamus appeared congested (Figure 7B), and the area between the inner and outer bundles was sparse, similar to softening foci (Figure 7C).
7. Histopathological changes in brain after long-term antibiotic treatment.
A: Thalamus of conventional rhesus monkey. B, C: Congestion (black arrows) and lacunar state (blue arrows) pathological changes in thalamus of antibiotic-treated rhesus monkey. Images were taken at 20× magnification; Scale bars: 200 μm.
DISCUSSION
Under conventional conditions, gut commensal bacteria inhabit the surface of the intestinal epithelium, forming a stable symbiotic relationship with the host and shaping a physical barrier on the surface (Baumgart & Dignass, 2002; Davenport et al., 2017). The presence of intestinal commensal bacteria reduces the colonization, translocation, and growth of conditional pathogens (Kamada et al., 2013) and is therefore referred to as “colonization resistance”. Dysbiosis or lack of commensal bacteria can cause extensive proliferation of pathogenic bacteria or other pathogens in the intestine, even causing diseases in the host (Thackray et al., 2018; Van den Bergh et al., 2016; Wienhold et al., 2018). With the development of genome sequencing technology, we are aware of the relationship between commensal bacteria and the host. However, the mechanism of interactions among dominant bacterial species, pathogens, and hosts remains unclear.
NHPs exhibit high similarity with humans in anatomical structure, physiological metabolism, and immune system. However, few studies have systematically compared gut commensal microbiomes between human and NHPs (Davenport et al., 2017). Rhesus macaques can still be used in basic studies that cannot be completed or simulated with rodent models. However, due to limitations of body size, feeding conditions, and growth conditions, no studies have reported on commensal microbiota using the rhesus monkey as an animal model. In addition to germ-free animal models, antibiotic-treated animal models are also commonly used to study intestinal bacteria (Karst, 2016). In this study, four broad-spectrum antibiotics, including aminoglycoside, penicillin, glycopeptide, and nitroimidazole, were co-administered, and were found to deplete commensal bacteria in the intestines of rhesus monkeys. This study has some limitations, including that we were unable to completely deplete the intestinal bacterial community. The relative abundance of the 16S rRNA gene decreased by more than 80% on day 9 of treatment according to RT-PCR analysis, and the subsequent bacterial structure remained relatively stable. Based on the 16S rDNA amplicon sequencing results, the OTU level of intestinal symbionts decreased by more than 90% on day 9 after antibiotic treatment. After 14 d, the dominant bacterial group changed to Escherichia-Shigella, and both the richness and diversity of the intestinal bacterial community were depleted stably and continuously. Overall, the aim of eliminating enteric commensal bacteria in rhesus monkeys can be achieved 9 d after high-dose oral antibiotic treatment.
The pathological findings of the small intestine indicated that the absence of commensal bacteria can lead to aberrant intestinal morphology, including that of intestinal epithelial cells and villi. Flow cytometry results also showed alterations in lymphocyte populations, with increased numbers of T lymphocytes in peripheral blood. At the same time, the abundance of infectious diseases in KEGG prediction analysis of the microbiome significantly increased after antibiotic treatment. Thus, the lack of commensal bacteria could disrupt colonization resistance and affect the host immune system. Microarray analysis of the transcriptome performed on PBMCs isolated from antibiotic-treated monkeys also showed that the immune balance was affected by modulation in the expression of many different genes. The innate immunity-associated genes demonstrated cross-talk in response to bacterial dysbiosis with a high level of variation. These genes, including IL-13, CD28, CCR2, IL-12β, IL-1RL2, VCAM1, and FCER1A, exhibited obvious up-regulation with activation of the MHC system, T cell activity, and inflammatory responses. In addition, genes encoding cytokines and chemokines, such as IFNα2, CXCL10, and IL-4, were down-regulated. The alteration in the expression of these genes likely contributes to the effective functional activation of the innate immune response (Cross et al., 2004). These results suggest that bacterial dysbiosis caused by antibiotic treatment may affect the outcome of intestinal infectious diseases, including both bacterial and viral infections.
Furthermore, frequent oral antibiotic treatment could lead to the production of highly resistant E. coli (Van den Bergh et al., 2016). This study found that drug-resistant E. coli colonies appeared 4 d after antibiotic treatment, whereas no Escherichia-Shigella strains were present in the gut before treatment. This result should serve as an alarm suggesting that extensive clinical trials and optimization of antibiotic treatment should be conducted to minimize the possibility of strain resistance. In this study, the use of antibiotics caused adverse reactions such as diarrhea and vomiting in the short term. In addition, long-term antibiotic use resulted in serious symptoms such as myoclonus. Predictive functional profiling of bacteria also showed that abundance of the KEGG pathway of neurodegenerative diseases increased significantly, suggesting that the diversity and stability of intestinal commensal bacteria are important for the maintenance of the nervous system.
In conclusion, our data suggest that early childhood represent a critical period during which perturbations to the gut microbiota and dysregulation of microbiota-gut-immunity axis communication may significantly impact the host immune system in adulthood. Although the precise mechanisms by which gut microbiota mediate changes in the host have not yet been elucidated, our findings indicate that rhesus monkeys with short-term high-dose antibiotic treatment represent a useful model for assessing the importance of gut microbiota to the host during distinct stages of early life, without disruption to host health. This is an unavailable feature of the germ-free animal model. Further studies focusing on the effects of bacterial depletion on the host are required to identify its potential relevance to neurodevelopmental and infectious diseases and functional mechanisms.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
L.D.L. and J.J.W. designed the study. Z.L.H. and Y.Z. performed the animal study. H.Z.L., N.L., X.H., H.L., H.W.Z., Z.N.Y., H.T.F., M.M.C., J.X.Y., and Q.W.W. collected samples. H.Z.L analyzed the data and drafted the manuscript. L.D.L. and L.G. revised the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Animal Laboratory Center personnel Feng-Mei Yang and Bin Xie for vivarium help with the monkeys. We thank Majorbio Biological Technology Co., Ltd., which contributed to the 16S rDNA amplicon sequencing project.
Funding Statement
This work was supported by the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2016-I2M-1-014)
References
- 1.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA The intestinal epithelium: Central coordinator of mucosal immunity. Trends in Immunology. 2018;39(9):677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. The ISME Journal. 2013;7(7):1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Meta HITC, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgart DC, Dignass AU Intestinal barrier function. Current Opinion in Clinical Nutrition and Metabolic Care. 2002;5(6):685–694. doi: 10.1097/00075197-200211000-00012. [DOI] [PubMed] [Google Scholar]
- 6.Belkaid Y, Hand TW Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya S, Darby RR, Raibagkar P, Gonzalez Castro LN, Berkowitz AL Antibiotic-associated encephalopathy. Neurology. 2016;86(10):963–971. doi: 10.1212/WNL.0000000000002455. [DOI] [PubMed] [Google Scholar]
- 8.Cross ML, Ganner A, Teilab D, Fray LM Patterns of cytokine induction by gram-positive and gram-negative probiotic bacteria. FEMS Immunology & Medical Microbiology. 2004;42(2):173–180. doi: 10.1016/j.femsim.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Davenport ER, Sanders JG, Song SJ, Amato KR, Clark AG, Knight R The human microbiome in evolution. BMC Biology. 2017;15(1):127. doi: 10.1186/s12915-017-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desbonnet L, Clarke G, Traplin A, O'Sullivan O, Crispie F, Moloney RD, Cotter PD, Dinan TG, Cryan JF Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain, Behavior, and Immunity. 2015;48:165–173. doi: 10.1016/j.bbi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer M, Martins dos Santos VA, Ott SJ, Moya A Gut microbiota disturbance during antibiotic therapy: A multi-omic approach. Gut Microbes. 2014;5(1):64–70. doi: 10.4161/gmic.27128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, Shapiro H, Dori-Bachash M, Pevsner-Fischer M, Lorenzo-Vivas E, Keren-Shaul H, Paul F, Harmelin A, Eberl G, Itzkovitz S, Tanay A, Di Santo JP, Elinav E, Amit I The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166(5):1231–1246. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 13.Hooper LV, Gordon JI Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 14.Ivanov II, Honda K Intestinal commensal microbes as immune modulators. Cell Host and Microbe. 2012;12(4):496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamada N, Chen GY, Inohara N, Núñez G Control of pathogens and pathobionts by the gut microbiota. Nature Immunology. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karst SM The influence of commensal bacteria on infection with enteric viruses. Nature Reviews Microbiology. 2016;14(4):197–204. doi: 10.1038/nrmicro.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kernbauer E, Ding Y, Cadwell K An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516(7529):94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA Circos: An information aesthetic for comparative genomics. Genome Research. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langhorst J, Junge A, Rueffer A, Wehkamp J, Foell D, Michalsen A, Musial F, Dobos GJ Elevated human beta-defensin-2 levels indicate an activation of the innate immune system in patients with irritable bowel syndrome. The American Journal of Gastroenterology. 2009;104(2):404–410. doi: 10.1038/ajg.2008.86. [DOI] [PubMed] [Google Scholar]
- 20.Lozupone C, Hamady M, Knight R Unifrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7(1):371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens RC, Jr., Ambrose PG Antimicrobial safety: Focus on fluoroquinolones. Clinical Infectious Diseases. 2005;41(Suppl 2):S144–157. doi: 10.1086/428055. [DOI] [PubMed] [Google Scholar]
- 22.Postler TS, Ghosh S Understanding the holobiont: How microbial metabolites affect human health and shape the immune system. Cell Metabolism. 2017;26(1):110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research. 2013;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandler RH, Finegold SM, Bolte ER, Buchanan CP, Maxwell AP, Väisänen ML, Nelson MN, Wexler HM Short-term benefit from oral vancomycin treatment of regressive-onset autism. Journal of Child Neurology. 2000;15(7):429–435. doi: 10.1177/088307380001500701. [DOI] [PubMed] [Google Scholar]
- 25.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt TSB, Raes J, Bork P The human gut microbiome: From association to modulation. Cell. 2018;172(6):1198–1215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 27.Seyfizadeh N, Seyfizadeh N, Gharibi T, Babaloo Z Interleukin-13 as an important cytokine: A review on its roles in some human diseases. Acta Microbiologica et Immunologica Hungarica. 2015;62(4):341–378. doi: 10.1556/030.62.2015.4.2. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro H, Thaiss CA, Levy M, Elinav E The cross talk between microbiota and the immune system: Metabolites take center stage. Current Opinion in Immunology. 2014;30:54–62. doi: 10.1016/j.coi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Soderborg TK, Friedman JE Imbalance in gut microbes from babies born to obese mothers increases gut permeability and myeloid cell adaptations that provoke obesity and NAFLD. Microbial Cell. 2018;6(1):102–104. doi: 10.15698/mic2019.01.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprouse ML, Bates NA, Felix KM, Wu HJ Impact of gut microbiota on gut-distal autoimmunity: A focus on t cells. Immunology. 2019;156(4):305–318. doi: 10.1111/imm.13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spurrell JC, Wiehler S, Zaheer RS, Sanders SP, Proud D Human airway epithelial cells produce ip-10(CXCL10) in vitro and in vivo upon rhinovirus infection. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2005;289(1):L85–L95. doi: 10.1152/ajplung.00397.2004. [DOI] [PubMed] [Google Scholar]
- 32.Styrkarsdottir U, Thorleifsson G, Sulem P, Gudbjartsson DF, Sigurdsson A, Jonasdottir A, Jonasdottir A, Oddsson A, Helgason A, Magnusson OT, Walters GB, Frigge ML, Helgadottir HT, Johannsdottir H, Bergsteinsdottir K, Ogmundsdottir MH, Center JR, Nguyen TV, Eisman JA, Christiansen C, Steingrimsson E, Jonasson JG, Tryggvadottir L, Eyjolfsson GI, Theodors A, Jonsson T, Ingvarsson T, Olafsson I, Rafnar T, Kong A, Sigurdsson G, Masson G, Thorsteinsdottir U, Stefansson K Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497(7450):517–520. doi: 10.1038/nature12124. [DOI] [PubMed] [Google Scholar]
- 33.Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, Briseño CG, Theisen DJ, Tan Q, Hykes BL, Jr., Lin H, Lucas TM, Desai C, Gordon JI, Murphy KM, Virgin HW, Diamond MS Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Reports. 2018;22(13):3440–3453.e6. doi: 10.1016/j.celrep.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van den Bergh B, Michiels JE, Wenseleers T, Windels EM, Boer PV, Kestemont D, De Meester L, Verstrepen KJ, Verstraeten N, Fauvart M, Michiels J Frequency of antibiotic application drives rapid evolutionary adaptation of escherichia coli persistence. Nature Microbiology. 2016;1(9):16020. doi: 10.1038/nmicrobiol.2016.20. [DOI] [PubMed] [Google Scholar]
- 35.Vandamme TF Rodent models for human diseases. European Journal of Pharmacology. 2015;759:84–89. doi: 10.1016/j.ejphar.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 36.Wienhold SM, Macrì M, Nouailles G, Dietert K, Gurtner C, Gruber AD, Heimesaat MM, Lienau J, Schumacher F, Kleuser B, Opitz B, Suttorp N, Witzenrath M, Müller-Redetzky HC Ventilator-induced lung injury is aggravated by antibiotic mediated microbiota depletion in mice. Critical Care. 2018;22(1):282. doi: 10.1186/s13054-018-2213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD Interleukin-13: Central mediator of allergic asthma. Science. 1998;282(5397):2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 38.Wu S, Jiang ZY, Sun YF, Yu B, Chen J, Dai CQ, Wu XL, Tang XL, Chen XY Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza a virus infection. Current Microbiology. 2013;67(4):414–422. doi: 10.1007/s00284-013-0380-z. [DOI] [PubMed] [Google Scholar]
- 39.Yang LY, Nossa CW, Pei ZH. 2015. Microbial dysbiosis and esophageal diseases. <italic>In</italic>: Highlander SK, Rodriguez-Valera F, White BA (eds). Encyclopedia of Metagenomics. Boston, MA: Springer, 379-384, <a href="https://doi.org/10.1007/978-1-4899-7475-4_65">https://doi.org/10.1007/978-1-4899-7475-4_65</a>.
- 40.Yeruva L, Spencer NE, Saraf MK, Hennings L, Bowlin AK, Cleves MA, Mercer K, Chintapalli SV, Shankar K, Rank RG, Badger TM, Ronis MJ Formula diet alters small intestine morphology, microbial abundance and reduces VE-cadherin and IL-10 expression in neonatal porcine model. BMC Gastroenterology. 2016;16:40. doi: 10.1186/s12876-016-0456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yousefi B, Eslami M, Ghasemian A, Kokhaei P, Farrokhi AS, Darabi N Probiotics importance and their immunomodulatory properties. Journal of Cellular Physiology. 2019;234(6):8008–8018. doi: 10.1002/jcp.27559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.