DEAR EDITOR,
Genome-wide association studies (GWAS) have identified multiple single nucleotide polymorphisms (SNPs) or small indels robustly associated with schizophrenia; however, the functional risk variations remain largely unknown. We investigated the 10q24.32 locus and discovered a 339 bp Alu insertion polymorphism (rs71389983) in complete linkage disequilibrium (LD) with the schizophrenia GWAS risk variant rs7914558. The presence of the Alu insertion at rs71389983 strongly repressed transcriptional activities in in vitro luciferase assays. This polymorphism may be a target for future mechanistic research. Our study also underlines the importance and necessity of considering previously underestimated Alu polymorphisms in future genetic studies of schizophrenia.
Schizophrenia is a severe chronic psychiatric disorder with high heritability (Sullivan et al., 2003), and depicting the genetic architecture of schizophrenia is essential for understanding its pathophysiology. So far, GWAS have identified numerous risk loci (Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), and several studies have attempted to identify causative risk variations and underlying biological mechanisms from the massive tagged single nucleotide polymorphisms (SNPs) (Duan et al., 2014; Huo et al., 2019; Wu et al., 2017, 2019 Yang et al., 2018). However, one potential limitation of current GWAS platforms is that they have primarily focused on SNPs and small indels, ignoring other sequence variations that have also been implicated in the genetic risk of human disorders including schizophrenia (Payer et al., 2017; Song et al., 2018; Yang et al., 2019) and in non-human primates (Liu et al., 2018). For instance, Song et al. (2018) previously identified a functional human-specific tandem repeat in the CACNA1C gene as a potential causative variation for schizophrenia and bipolar disorder.
The chromosomal 10q24.32 region is a critical locus showing genome-wide significant associations with schizophrenia. For example, rs7914558 is reported to be the most significant SNP in the 10q24.32 region in the PGC1 GWAS of European populations (P=1.82×10–9, n=51 695) (Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011), and its association with schizophrenia has been further confirmed in subsequent GWAS with increased sample size (P=3.49×10–15, n=79 845) (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Intriguingly, according to data from a recent GWAS of East Asian populations, rs7914558 is also significantly associated with schizophrenia genome-wide (P=3.50×10–8, n=58 140) (Lam et al., 2019). In the present study, through population genetic analyses, in vitro luciferase assays, and expression quantitative trait loci (eQTL) data, we identified a functional 339 bp Alu insertion polymorphism (rs71389983) within the 9th intron of the AS3MT gene in complete LD with rs7914558.
The study protocol was approved by the Institutional Review Board of the Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS). Informed consent was obtained before any study-related procedures were carried out. Genotyping of rs71389983 and rs7914558 was conducted using polymerase chain reaction (PCR) on 38 European and 39 Han Chinese subjects, with amplicons analyzed using agarose gel and Sanger sequencing to determine differences in alleles. The PCR primers were: 5'- ATGTAACTGGTATATCCATCGCCT-3' (forward) and 5'- AGAAGACTCAAACAGATGAACGGA-3' (reverse) for rs71389983; and 5'-CTCTACTTGCCCCCTTACAGC-3' (forward) and 5'-GAACCGTATCAGTAATCCAACAGA-3' (reverse) for rs7914558.
The HEK293T (human embryonic kidney 293T) and U87MG (human glioblastoma astrocytoma) cell lines used were originally obtained from the Kunming Cell Bank, KIZ, and the Cell Bank of Type Culture Collection of the CAS, respectively. Both cell lines were checked regularly for mycoplasma infection using PCR and microscopy. No cells were found to be contaminated during the study. The HEK293T cells were cultured in a humidified 5% CO2 incubator at 37 °C in DMEM basic (Dulbecco's Modified Eagle's Medium) (Gibco, USA) supplemented with 10% fetal bovine serum, 1% non-essential amino acids, 1% sodium pyruvate, and 1% penicillin-streptomycin. The U87MG cells were cultured in a humidified 5% CO2 incubator at 37 °C in MEM (Minimum Essential Medium) supplemented with 10% fetal bovine serum, 1% sodium pyruvate, 2.2 g/L NaHCO3, and 1% penicillin-streptomycin.
For the reporter gene assays, DNA fragments encompassing rs71389983 with either allele were amplified from human genomic DNA using primers 5'-GGCTGCCAGGTTCAAGTAAT- 3' (forward) and 5'-CACACTGGAATACTATTCAGACTT-3' (reverse). The sequences were then cloned into the pGL3-promoter vector (Promega, USA) upstream of the SV40 promoter. The recombinant clones were verified through Sanger sequencing to ensure they only differed at the rs71389983 locus. The pGL3-promoter reporters were transiently co-transfected into cells together with the pRL-TK plasmid (Promega, USA) using Lipofectamine 3000 (Thermo Fisher Scientific, USA). All plasmids were accurately quantified and equal amounts were used for transfection. All transfection procedures lasted 36–48 h, and the cells were then collected to measure luciferase activity using the Dual-Luciferase Reporter Assay System (Promega, USA). The activity of firefly luciferase was normalized to that of Renilla luciferase to control for variations in transfection efficiency. All assays were performed with at least three biological replicates in independent experiments, and statistical analyses were performed by two-tailed t-tests.
We also examined the impacts of risk SNPs on gene mRNA expression using two public RNA-seq brain eQTL datasets, i. e., BrainSeq Phase 2 (http://eqtl.brainseq.org/phase2/eqtl/) and GTEx (https://www.gtexportal.org/) (Collado-Torres et al., 2019; GTEx Consortium et al., 2017). Briefly, from the BrainSeq dataset, we obtained eQTL data of the dorsolateral prefrontal cortex (DLPFC) from 397 individuals, which were calculated using linear regression by covarying diagnosis, gender, genotyping principal components, and expression principal components. From the GTEx dataset, we retrieved the eQTL association results from the frontal cortex (BA9) of 175 subjects, which were calculated using linear regression by covarying genotyping principal components, gender, genotyping platforms, and additional covariates.
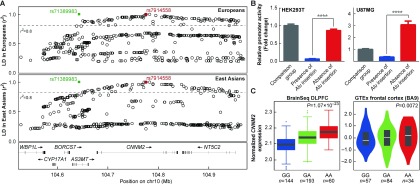
Recent study has shown that a subset of Alu insertion polymorphisms exhibit moderate to strong LD (r2>0.7) with GWAS risk SNPs of complex illnesses (Payer et al., 2017). We therefore examined whether there were Alu insertion polymorphisms within the 10q24.32 region. Using public genomic variation databases (i.e., UCSC, http://genome.ucsc. edu/) followed by Sanger sequencing of target regions, we identified an Alu insertion polymorphism (339 bp) rs71389983 in intron 9 of AS3MT, which was in complete LD with rs7914558 in the Han Chinese and European populations (both r2=1.00, Figure 1A). The presence of the Alu insertion at rs71389983 was linked with the schizophrenia risk G-allele at rs7914558, and therefore may be associated with increased risk of schizophrenia. We note that the frequency of rs71389983 (and rs7914558) showed divergence between the two populations (frequency of Alu insertion at rs71389983: 0.423 in Han Chinese vs. 0.605 in Europeans). We also compared the LD structures of the 10q24.32 region between Europeans and East Asians using genotype data from the 1000 Genomes Project (Genomes Project Consortium et al., 2015), and found that the LD structures were relatively similar across distinct populations, despite showing tiny differences (Figure 1A), in agreement with the significant associations of this genomic area in both populations.
1. Linkage disequilibrium (LD) analysis of rs7914558 and nearby variations (including rs71389983) in European and East Asian populations (A); Reporter gene assay testing regulatory activity of rs71389983 in HEK293T and U87MG cells (B); Expression quantitative trait loci (eQTL) analyses of rs7914558 with CNNM2 mRNA in BrainSeq and GTEx datasets (C) .
Rs7914558 is located in intron of CNNM2, and Alu polymorphism rs71389983 is located in intron 9 of AS3MT. Both variations are in complete LD in both Europeans and East Asians. Effects of rs71389983 allele variation on pGL3 promoter activity in HEK293T and U87MG cells are shown. “Comparison group”in figure represents empty pGL3 promoter. Values represent fold change in luciferase activity relative to control pGL3 vector. Means and standard deviations of at least three independent experiments are shown. ****: P<0.000 01.
The DNA sequence covering rs7914558 is conserved across humans and non-human primates, whereas the Alu polymorphism rs71389983 appears to be human-unique. We thus performed bioinformatics functional prediction of rs7914558 using the HaploReg v4.1 dataset ( https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) (Ward & Kellis, 2012). However, we found that it was unlikely located at any DNA segments showing open-chromatin peaks or directly binding to transcription factors or histone markers (e.g., H3K4me1, H3K4me3, H3K9ac, and H3K27ac). On the other hand, Alu insertions have been found to affect both transcription and post-transcriptional processes (Häsler & Strub, 2006). Considering that rs71389983 was found in intron 9 of AS3MT, we hypothesized that it may be within the enhancer/repressor region of the genome. To test this, we amplified the DNA fragments spanning rs71389983 from individuals carrying different homozygotes (PCR product length: presence of Alu insertion: 589 bp; absence of Alu insertion: 250 bp), and then sub-cloned them into the pGL3 promoter vector. These plasmids were then transfected into the human HEK293T and U87MG cell lines, and reporter gene assays were carried out to examine their regulatory effects. In the HEK293T cells, the transcriptional activity of the pGL3 promoter containing the Alu insertion at rs71389983 was significantly lower than that of the promoter without the allele (P<0.000 01,Figure 1B) and that of the empty vector (P<0.000 01). In the U87MG cells, this trend was reproduced and the presence of theAlu insertion at rs71389983 corresponded to significantly lower activity of the pGL3 promoter compared with that of the pGL3 promoter without the allele (P<0.000 01,Figure 1B) and the empty vector (P<0.000 01). Therefore, rs71389983 is likely a functional variation and theAlu insertion at this locus likely exerts repressive effects on transcription. In addition to the consistent trend of the effect of the rs71389983 Alu insertion on both cell lines, a slight difference between the HEK293T and U87MG cell results was observed, as the pGL3 promoter carrying the "absence of Alu insertion" at rs71389983 showed higher transcriptional activity than the empty vector in the U87MG cells, but lower activity than the empty vector in the HEK293T cells. This inconsistency could be explained by the different genetic and physiological backgrounds between the different cell lines.
To further confirm the regulatory effects of the Alu polymorphism (rs71389983) on gene expression, we examined two public RNA-seq eQTL datasets (i.e., BrainSeq and GTEx-brain) in human brains (Collado-Torres et al., 2019; GTEx Consortium et al., 2017). As rs71389983 is not genotyped in those eQTL databases, we used rs7914558 as an index SNP. In the BrainSeq dataset, which included DLPFC tissues from 397 individuals, the schizophrenia risk G-allele at rs7914558 was significantly associated with increased gene expression of BORCS7 (P=9.28×10–27), as well as decreased mRNA expression of CNNM2 (P=1.07×10–23, Figure 1C) and CYP17A1-AS1 (P=7.98×10–4). In the 175 frontal cortex (BA9) tissues of the GTEx dataset, the G-allele at rs7914558 was also strongly associated with increased gene expression of BORCS7 (P=1.00×10–9) and decreased mRNA expression of CNNM2 (P=0.007 2, Figure 1C), but not with the expression of CYP17A1-AS1 (P=0.94).
Translating the GWAS risk associations of complex disorders into biological mechanisms remains an urgent task (Barešic et al., 2019; Birnbaum & Weinberger, 2017; Edwards et al., 2013; Forrest et al., 2018; Gandal et al., 2016). However, most genetic risk loci are located in noncoding regions, which may affect transcription factor binding affinities, gene expression, or even cellular physiological processes (Duan et al., 2014; Forrest et al., 2017; Li et al., 2011; Roussos et al., 2014). We identified a 339 bp Alu insertion polymorphism (rs71389983) in the 10q24.32 locus, and reporter gene assays showed that different alleles of rs71389983 exhibited significantly different regulatory activities. The promoter carrying the "absence of Alu insertion" at rs71389983 exhibited more than 10-fold higher transcriptional activity than the promoter carrying the "presence of Alu insertion", suggesting that the Alu insertion sequence likely confers function as a gene silencer. Although this effect is usually caused by certain epigenetic modifications such as DNA methylation or noncoding RNA, the current genome-wide sequencing technologies do not provide ideal tools for answering this question. For example, the ENCODE datasets are mostly based on short DNA reads (≤250 bp) (Encode Project Consortium, 2012), and such methods are not able to precisely map retrotransposons, like Alu regions, as Alu elements contain multiple highly similar sequences (>300 bp) across the genome. Thus, it is difficult to identify the epigenetic or regulatory markers at rs71389983 (as reflected in the UCSC browser, which shows no ChIP-seq data at the rs71389983 locus). To resolve this problem, long-read sequencing technologies should be applied.
We found that in both the BrainSeq and GTEx-brain tissues, the schizophrenia risk allele at rs71389983 (i.e., its complete linked SNP rs7914558) predicted lower expression of CNNM2, consistent with the results of our in vitro luciferase assays. Therefore, CNNM2 is likely a schizophrenia risk gene, in agreement with previous study (Thyme et al., 2019). However, the present results do not necessarily mean that rs71389983 directly regulates CNNM2 expression, unless further functional studies (e.g., CRISPR/Cas9 genome editing) are carried out. The significant association of risk SNPs (e.g., rs7914558) at 10q24.32 with BORCS7 expression is also consistent with earlier research (Duarte et al., 2016; Li et al., 2016a).
Previous studies have demonstrated that Alu insertion polymorphisms are significantly associated with multiple complex human disorders and traits, including multiple sclerosis, obesity, height, Alzheimer's disease, breast cancer, and blood pressure (Payer et al., 2017). Our recent study also identified a functional Alu polymorphism at 3p21.1 affecting DNA regulatory activity, which was significantly associated with increased risk of psychiatric disorders (e.g., schizophrenia, bipolar disorder, and major depressive disorder) and cognitive disfunctions (Yang et al., 2019). Combined with the present data, these studies suggest that such types of sequence variations may play essential roles in shaping phenotypes during primate or human evolution (Deininger, 2011; Häsler & Strub, 2006). However, our previous study showed that the Alu insertion sequence at 3p21.1 increased regulatory activity (Yang et al., 2019), and herein the Alu insertion sequence at 10q24.32 reduced transcriptional activities. Although the majority of the Alu sequences across the genome show high similarity, their functional regulatory effects may be distinct.
In summary, we discovered a human-unique Alu insertion in strong LD with the schizophrenia GWAS risk SNP at 10q24.32. Schizophrenia is hypothesized to be specific to or dominant in humans, and its evolutionary mechanism may be related to unique human variations. For example, we previously identified a human-specific allele rs13107325 atSLC39A8 undergoing Darwinian natural selection, which enabled humans to adapt to cold environments in Europe, but simultaneously also increased the risk of schizophrenia (Li et al., 2016b). The schizophrenia risk allele at SLC39A8 is also significantly associated with cognitive function and brain structures in human populations (Davies et al., 2018; Elliott et al., 2018; Luo et al., 2019; Savage et al., 2018). Assuming that the human-unique Alu insertions play pivotal roles in shaping humanity, such as development of the dorsolateral prefrontal cortex and higher order human features (e.g., higher cognitive processing) (Wang & Arnsten, 2015), they may also deliver some susceptible or deleterious effects to human health, such as predisposition to schizophrenia. Investigations of such human-unique variations in non-human primates or other species (such as tree shrews) that are evolutionarily close to humans or in human-induced pluripotent stem cells (hiPSC) or reprogrammed cells via genome editing, may provide novel insights into the pathophysiology of schizophrenia and other human-dominant disorders (Falk et al., 2016; Hoffman et al., 2019; Luo et al., 2016; Xiao et al., 2017; Xu et al., 2013; Yao, 2017).
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS' CONTRIBUTIONS
Z.H.Y., Y.L., and M.L. designed the study and interpreted the results. Z.H.Y., N.Q., and X.C. performed DNA extraction, Alu genotyping, population genetic analysis, and primary experiments. L.J.Z., B.L.Z., S.F.Z., J.C., B.X., H.Y.J., D.Y.Z., W.L., H.C., and X.X. contributed to design and helped in the experiments. Z.H.Y., X.X., and M.L. drafted the manuscript.
Funding Statement
This work was supported by grants from Yunnan Applied Basic Research Projects (2018FB051 to X.X. and 2018FB136 to H. C.); Hubei Province Health and Family Planning Scientific Research Project (WJ2015Q033 to N. Q.); Population and Family Planning Commission of Wuhan (WX14B34 to N. Q.); Open Program of Henan Key Laboratory of Biological Psychiatry (ZDSYS2018001 to H. C.); and Program for Scientific Research of Yunnan Health and Family Planning Commission (2016NS025 to H.Y.J.)
References
- 1.Barešić A, Nash AJ, Dahoun T, Howes O, Lenhard B Understanding the genetics of neuropsychiatric disorders: the potential role of genomic regulatory blocks. Molecular Psychiatry. 2020;25:6–18. doi: 10.1038/s41380-019-0518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnbaum R, Weinberger DR Genetic insights into the neurodevelopmental origins of schizophrenia. Nature Reviews Neuroscience. 2017;18(12):727–740. doi: 10.1038/nrn.2017.125. [DOI] [PubMed] [Google Scholar]
- 3.Collado-Torres L, Burke EE, Peterson A, Shin J, Straub RE, Rajpurohit A, Semick SA, Ulrich WS, BrainSeq C, Price AJ, Valencia C, Tao R, Deep-Soboslay A, Hyde TM, Kleinman JE, Weinberger DR, Jaffe AE Regional heterogeneity in gene expression, regulation, and coherence in the frontal cortex and hippocampus across development and schizophrenia. Neuron. 2019;103(2):203–216. doi: 10.1016/j.neuron.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, Hagenaars SP, Ritchie SJ, Marioni RE, Fawns-Ritchie C, Liewald DCM, Okely JA, Ahola-Olli AV, Barnes CLK, Bertram L, Bis JC, Burdick KE, Christoforou A, DeRosse P, Djurovic S, Espeseth T, Giakoumaki S, Giddaluru S, Gustavson DE, Hayward C, Hofer E, Ikram MA, Karlsson R, Knowles E, Lahti J, Leber M, Li S, Mather KA, Melle I, Morris D, Oldmeadow C, Palviainen T, Payton A, Pazoki R, Petrovic K, Reynolds CA, Sargurupremraj M, Scholz M, Smith JA, Smith AV, Terzikhan N, Thalamuthu A, Trompet S, van der Lee SJ, Ware EB, Windham BG, Wright MJ, Yang J, Yu J, Ames D, Amin N, Amouyel P, Andreassen OA, Armstrong NJ, Assareh AA, Attia JR, Attix D, Avramopoulos D, Bennett DA, Böhmer AC, Boyle PA, Brodaty H, Campbell H, Cannon TD, Cirulli ET, Congdon E, Conley ED, Corley J, Cox SR, Dale AM, Dehghan A, Dick D, Dickinson D, Eriksson JG, Evangelou E, Faul JD, Ford I, Freimer NA, Gao H, Giegling I, Gillespie NA, Gordon SD, Gottesman RF, Griswold ME, Gudnason V, Harris TB, Hartmann AM, Hatzimanolis A, Heiss G, Holliday EG, Joshi PK, Kähönen M, Kardia SLR, Karlsson I, Kleineidam L, Knopman DS, Kochan NA, Konte B, Kwok JB, Le Hellard S, Lee T, Lehtimäki T, Li SC, Liu T, Koini M, London E, Longstreth WT Lopez, Loukola A, Luck T, Lundervold AJ, Lundquist A, Lyytikäinen LP, Martin NG, Montgomery GW, Murray AD, Need AC, Noordam R, Nyberg L, Ollier W, Papenberg G, Pattie A, Polasek O, Poldrack RA, Psaty BM, Reppermund S, Riedel-Heller SG, Rose RJ, Rotter JI, Roussos P, Rovio SP, Saba Y, Sabb FW, Sachdev PS, Satizabal CL, Schmid M, Scott RJ, Scult MA, Simino J, Slagboom PE, Smyrnis N, Soumaré A, Stefanis NC, Stott DJ, Straub RE, Sundet K, Taylor AM, Taylor KD, Tzoulaki I, Tzourio C, Uitterlinden A, Vitart V, Voineskos AN, Kaprio J, Wagner M, Wagner H, Weinhold L, Wen KH, Widen E, Yang Q, Zhao W, Adams HHH, Arking DE, Bilder RM, Bitsios P, Boerwinkle E, Chiba-Falek O, Corvin A, De Jager PL, Debette S, Donohoe G, Elliott P, Fitzpatrick AL, Gill M, Glahn DC, Hägg S, Hansell NK, Hariri AR, Ikram MK, Jukema JW, Vuoksimaa E, Keller MC, Kremen WS, Launer L, Lindenberger U, Palotie A, Pedersen NL, Pendleton N, Porteous DJ, Räikkönen K, Raitakari OT, Ramirez A, Reinvang I, Rudan I, Dan R, Schmidt R, Schmidt H, Schofield PW, Schofield PR, Starr JM, Steen VM, Trollor JN, Turner ST, Van Duijn CM, Villringer A, Weinberger DR, Weir DR, Wilson JF, Malhotra A, McIntosh AM, Gale CR, Seshadri S, Mosley TH Bressler, Lencz T, Deary IJ Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nature Communications. 2018;9(1):2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deininger P Alu elements: know the SINEs. Genome Biology. 2011;12(12):236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan J, Shi J, Fiorentino A, Leites C, Chen X, Moy W, Chen J, Alexandrov BS, Usheva A, He D, Freda J, O'Brien NL, Molecular Genetics of Schizophrenia Collaboration, Genomic Psychiatric Cohort Consortium, McQuillin A, Sanders AR, Gershon ES, DeLisi LE, Bishop AR, Gurling HM, Pato MT, Levinson DF, Kendler KS, Pato CN, Gejman PV A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. American Journal of Human Genetics. 2014;95(6):744–753. doi: 10.1016/j.ajhg.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte RRR, Troakes C, Nolan M, Srivastava DP, Murray RM, Bray NJ Genome-wide significant schizophrenia risk variation on chromosome 10q24 is associated with altered cis-regulation of BORCS7, AS3MT, and NT5C2 in the human brain. American Journal of Medical Genetics Part B Neuropsychiatric Genetics. 2016;171(6):806–814. doi: 10.1002/ajmg.b.32445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards SL, Beesley J, French JD, Dunning AM Beyond GWASs: illuminating the dark road from association to function. American Journal of Human Genetics. 2013;93(5):779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, Marchini J, Smith SM Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562(7726):210–216. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falk A, Heine VM, Harwood AJ, Sullivan PF, Peitz M, Brüstle O, Shen S, Sun YM, Glover JC, Posthuma D, Djurovic S Modeling psychiatric disorders: from genomic findings to cellular phenotypes. Molecular Psychiatry. 2016;21(9):1167–1179. doi: 10.1038/mp.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrest MP, Parnell E, Penzes P Dendritic structural plasticity and neuropsychiatric disease. Nature Reviews Neuroscience. 2018;19(4):215–234. doi: 10.1038/nrn.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrest MP, Zhang H, Moy W, McGowan H, Leites C, Dionisio LE, Xu Z, Shi J, Sanders AR, Greenleaf WJ, Cowan CA, Pang ZP, Gejman PV, Penzes P, Duan J Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell. 2017;21(3):305–318. e8. doi: 10.1016/j.stem.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH The road to precision psychiatry: translating genetics into disease mechanisms. Nature Neuroscience. 2016;19(11):1397–1407. doi: 10.1038/nn.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GTEx Consortium, Laboratory Data Analysis, Coordinating Center-Analysis Working Group, Statistical Methods Groups-Analysis Working Group, Enhancing GTEx Groups, NIH Common Fund, Nih/Nci, Nih/Nhgri, Nih/Nimh, Nih/Nida, Biospecimen Collection Source Site Ndri, Biospecimen Collection Source Site Rpci, Biospecimen Core Resource Vari, Brain Bank Repository-University of Miami Brain Endowment Bank, Leidos Biomedical-Project Management, Elsi Study, Genome Browser Data Integration, Visualization EBI, Genome Browser Data Integration, Visualization-Ucsc Genomics Institute, University of California Santa Cruz, Lead analysts, Laboratory Data Analysis & Coordinating Center, NIH program management, Biospecimen collection, Pathology: eQTL manuscript working group, Battle A, Brown CD, Engelhardt BE, Montgomery SB Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Häsler J, Strub K Alu elements as regulators of gene expression. Nucleic Acids Research. 2006;34(19):5491–1597. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman GE, Schrode N, Flaherty E, Brennand KJ New considerations for hiPSC-based models of neuropsychiatric disorders. Molecular Psychiatry. 2019;24(1):49–66. doi: 10.1038/s41380-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huo Y, Li S, Liu J, Li X, Luo XJ Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nature Communications. 2019;10(1):670. doi: 10.1038/s41467-019-08666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X, Gaspar H, Ikeda M, Benyamin B, Brown BC, Liu R, Zhou W, Guan L, Kamatani Y, Kim SW, Kubo M, Kusumawardhani A, Liu CM, Ma H, Periyasamy S, Takahashi A, Xu Z, Yu H, Zhu F, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Indonesia Schizophrenia Consortium, Genetic REsearch on schizophreniA neTwork-China and the Netherlands (GREAT-CN), Chen WJ, Faraone S, Glatt SJ, He L, Hyman SE, Hwu HG, McCarroll SA, Neale BM, Sklar P, Wildenauer DB, Yu X, Zhang D, Mowry BJ, Lee J, Holmans P, Xu S, Sullivan PF, Ripke S, O'Donovan MC, Daly MJ, Qin S, Sham P, Iwata N, Hong KS, Schwab SG, Yue W, Tsuang M, Liu J, Ma X, Kahn RS, Shi Y, Huang H Comparative genetic architectures of schizophrenia in East Asian and European populations. Nature Genetics. 2019;51:1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y, Chen Q, Li C, Jia Y, Ohi K, Maher BJ, Brandon NJ, Cross A, Chenoweth JG, Hoeppner DJ, Wei H, Hyde TM, McKay R, Kleinman JE, Weinberger DR A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nature Medicine. 2016a;22(6):649–656. doi: 10.1038/nm.4096. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Luo XJ, Xiao X, Shi L, Liu XY, Yin LD, Diao HB, Su B Allelic differences between Han Chinese and Europeans for functional variants in ZNF804A and their association with schizophrenia. The American Journal of Psychiatry. 2011;168(12):1318–1325. doi: 10.1176/appi.ajp.2011.11030381. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Wu DD, Yao YG, Huo YX, Liu JW, Su B, Chasman DI, Chu AY, Huang T, Qi L, Zheng Y, CHARGE Nutrition Working Group, DietGen Consortium, Luo XJ Recent positive selection drives the expansion of a schizophrenia risk nonsynonymous variant at SLC39A8 in Europeans. Schizophrenia Bulletin. 2016b;42(1):178–190. doi: 10.1093/schbul/sbv070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu SX, Hou W, Zhang XY, Peng CJ, Yue BS, Fan ZX, Li J Identification and characterization of short tandem repeats in the Tibetan macaque genome based on resequencing data. Zoological Research. 2018;39(4):291–300. doi: 10.24272/j.issn.2095-8137.2018.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Q, Chen Q, Wang W, Desrivieres S, Quinlan EB, Jia T, Macare C, Robert GH, Cui J, Guedj M, Palaniyappan L, Kherif F, Banaschewski T, Bokde ALW, Buchel C, Flor H, Frouin V, Garavan H, Gowland P, Heinz A, Ittermann B, Martinot JL, Artiges E, Paillere-Martinot ML, Nees F, Orfanos DP, Poustka L, Frohner JH, Smolka MN, Walter H, Whelan R, Callicott JH, Mattay VS, Pausova Z, Dartigues JF, Tzourio C, Crivello F, Berman KF, Li F, Paus T, Weinberger DR, Murray RM, Schumann G, Feng J Association of a schizophrenia-risk nonsynonymous variant with putamen volume in adolescents: a voxelwise and genome-wide association study. JAMA Psychiatry. 2019;76(4):435–445. doi: 10.1001/jamapsychiatry.2018.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo X, Li M, Su B Application of the genome editing tool CRISPR/Cas9 in non-human primates. Zoological Research. 2016;37(4):214–219. doi: 10.13918/j.issn.2095-8137.2016.4.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payer LM, Steranka JP, Yang WR, Kryatova M, Medabalimi S, Ardeljan D, Liu C, Boeke JD, Avramopoulos D, Burns KH Structural variants caused by Alu insertions are associated with risks for many human diseases. Proceedings of the Natlional Academy of Sciences of the United States of America. 2017;114(20):E3984–E3992. doi: 10.1073/pnas.1704117114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, Stahl EA, Georgakopoulos A, Ruderfer DM, Charney A, Okada Y, Siminovitch KA, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Plenge RM, Raychaudhuri S, Fromer M, Purcell SM, Brennand KJ, Robakis NK, Schadt EE, Akbarian S, Sklar P A role for noncoding variation in schizophrenia. Cell Reports. 2014;9(4):1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, Nagel M, Awasthi S, Barr PB, Coleman JRI, Grasby KL, Hammerschlag AR, Kaminski JA, Karlsson R, Krapohl E, Lam M, Nygaard M, Reynolds CA, Trampush JW, Young H, Zabaneh D, Hägg S, Hansell NK, Karlsson IK, Linnarsson S, Montgomery GW, Muñoz-Manchado AB, Quinlan EB, Schumann G, Skene NG, Webb BT, White T, Arking DE, Avramopoulos D, Bilder RM, Bitsios P, Burdick KE, Cannon TD, Chiba-Falek O, Christoforou A, Cirulli ET, Congdon E, Corvin A, Davies G, Deary IJ, DeRosse P, Dickinson D, Djurovic S, Donohoe G, Conley ED, Eriksson JG, Espeseth T, Freimer NA, Giakoumaki S, Giegling I, Gill M, Glahn DC, Hariri AR, Hatzimanolis A, Keller MC, Knowles E, Koltai D, Konte B, Lahti J, Le Hellard S, Lencz T, Liewald DC, London E, Lundervold AJ, Malhotra AK, Melle I, Morris D, Need AC, Ollier W, Palotie A, Payton A, Pendleton N, Poldrack RA, Räikkönen K, Reinvang I, Roussos P, Rujescu D, Sabb FW, Scult MA, Smeland OB, Smyrnis N, Starr JM, Steen VM, Stefanis NC, Straub RE, Sundet K, Tiemeier H, Voineskos AN, Weinberger DR, Widen E, Yu J, Abecasis G, Andreassen OA, Breen G, Christiansen L, Debrabant B, Dick DM, Heinz A, Hjerling-Leffler J, Ikram MA, Kendler KS, Martin NG, Medland SE, Pedersen NL, Plomin R, Polderman TJC, Ripke S, van der Sluis S, Sullivan PF, Vrieze SI, Wright MJ, Posthuma D Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nature Genetics. 2018;50(7):912–919. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schizophrenia Psychiatric Genome-Wide Association Study Consortium Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43(10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song JHT, Lowe CB, Kingsley DM Characterization of a human-specific tandem repeat associated with Bipolar Disorder and Schizophrenia. American Journal of Human Genetics. 2018;103(3):421–430. doi: 10.1016/j.ajhg.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan PF, Kendler KS, Neale MC Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of General Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 34.Thyme SB, Pieper LM, Li EH, Pandey S, Wang Y, Morris NS, Sha C, Choi JW, Herrera KJ, Soucy ER, Zimmerman S, Randlett O, Greenwood J, McCarroll SA, Schier A. F Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions. Cell. 2019;177(2):478–491. e20. doi: 10.1016/j.cell.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, Arnsten AF Physiological approaches to understanding molecular actions on dorsolateral prefrontal cortical neurons underlying higher cognitive processing. Zoological Research. 2015;36(6):314–319. doi: 10.13918/j.issn.2095-8137.2015.6.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward LD, Kellis M HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Bi R, Zeng C, Ma C, Sun C, Li J, Xiao X, Li M, Zhang DF, Zheng P, Sheng N, Luo XJ, Yao YG Identification of the primate-specific gene BTN3A2 as an additional schizophrenia risk gene in the MHC loci. EbioMedicine. 2019;44:530–541. doi: 10.1016/j.ebiom.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.W u, Y, Yao YG, Luo XJ SZDB: A database for schizophrenia genetic research. Schizophrenia Bulletin. 2017;43(2):459–471. doi: 10.1093/schbul/sbw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao J, Liu R, Chen CS Tree shrew (Tupaia belangeri) as a novel laboratory disease animal model . Zoological Research. 2017;38(3):127–137. doi: 10.24272/j.issn.2095-8137.2017.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L, Zhang Y, Liang B, Lu LB, Chen CS, Chen YB, Zhou JM, Yao YG Tree shrews under the spot light: emerging model of human diseases. Zoological Research. 2013;34(2):59–69. doi: 10.3724/SP.J.1141.2013.02059. [DOI] [PubMed] [Google Scholar]
- 41.Yang CP, Li X, Wu Y, Shen Q, Zeng Y, Xiong Q, Wei M, Chen C, Liu J, Huo Y, Li K, Xue G, Yao YG, Zhang C, Li M, Chen Y, Luo XJ Comprehensive integrative analyses identify GLT8D1 and CSNK2B as schizophrenia risk genes. Nature Communications. 2018;9(1):838. doi: 10.1038/s41467-018-03247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Zhou D, Li H, Cai X, Liu W, Wang L, Chang H, Li M, Xiao X The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function, and mushroom dendritic spine. Molecular Psychiatry. 2020;25:48–66. doi: 10.1038/s41380-019-0592-0. [DOI] [PubMed] [Google Scholar]
- 43.Yao YG Creating animal models, why not use the Chinese tree shrew (Tupaia belangeri chinensis)? . Zoological Research. 2017;38(3):118–126. doi: 10.24272/j.issn.2095-8137.2017.032. [DOI] [PMC free article] [PubMed] [Google Scholar]