Abstract
Muscle wasting, or muscle atrophy, can occur with age, injury, and disease; it affects the quality of life and complicates treatment. Insulin-like growth factor 1 (IGF1) is a key positive regulator of muscle mass. The IGF1/Igf1 gene encodes multiple protein isoforms that differ in tissue expression, potency, and function, particularly in cellular proliferation and differentiation, as well as in systemic versus localized signaling. Genome engineering is a novel strategy for increasing gene expression and has the potential to recapitulate the diverse biology seen in IGF1 signaling through the overexpression of multiple IGF1 isoforms. Using a CRISPR-Cas9 gene activation approach, we showed that the expression of multiple IGF1 or Igf1 mRNA variants can be increased in human and mouse skeletal muscle myoblast cell lines by using a single guide RNA (sgRNA). We found increased IGF1 protein levels in the cell culture media and increased cellular phosphorylation of AKT1, the main effector of IGF1 signaling. We also showed that the expression of Class 1 or Class 2 mRNA variants can be selectively increased by changing the sgRNA target location. The expression of multiple IGF1 or Igf1 mRNA transcript variants in human and mouse skeletal muscle myoblasts promoted myotube differentiation and prevented dexamethasone-induced atrophy in myotubes in vitro. Our findings suggest that this novel approach for enhancing IGF1 signaling has potential therapeutic applications for treating skeletal muscle atrophy.
Keywords: insulin-like growth factor 1, muscle atrophy, gene regulation
INTRODUCTION:
Muscle wasting, or muscle atrophy, is defined as a progressive decrease in muscle mass that nutrition alone cannot correct. In addition to being a hallmark of aging, muscle atrophy also occurs after an injury, such as in severe burn patients and in patients with serious and often chronic diseases, such as diabetes, cancer, or kidney or heart failure (1–3). Muscle atrophy impairs quality of life, and it complicates treatment and recovery (4, 5).
Insulin-like growth factor 1 (IGF1) is a pleiotropic protein that regulates a diverse set of biological processes during development, adulthood, and old age, such as growth, differentiation, apoptosis, and regeneration (6, 7). IGF1 increases muscle mass by increasing satellite cell proliferation, enhancing myoblast differentiation, and inducing skeletal muscle cell hypertrophy, making it a well-suited target for treating muscle atrophy (8, 9). Furthermore, IGF1 prevents the loss of muscle mass by reducing atrophy-related signaling in muscle cells (10), and it promotes blood vessel growth, inhibits apoptosis, and increases neurite outgrowth (11–15).
IGF1 has multiple isoforms that vary in potency and tissue localization (14, 16, 17). The IGF1 gene, which is highly conserved between humans (IGF1) and mice (Igf1), has six exons. Alternative splicing of these exons generates the mRNA transcript variants that encode the different pre-pro IGF1 isoforms. Pre-pro IGF1 isoforms contain a leader sequence, the core mature IGF1 sequence, and a carboxy-terminal E-peptide. Exon 1 and exon 2 encode two mutually exclusive leader sequences; mRNA transcript variants that contain exon 1 or exon 2 are classified as Class1 or Class 2 mRNA transcripts, respectively. Exon 3 and exon 4 encode the core peptide sequence, which is the same for all IGF1 isoforms. Rodent IGF1 has two different E-peptides (Ea and Eb), whereas human IGF1 has three (Ea, Eb, and Ec). Leader sequence cleavage forms pro IGF1, which cells can store or process further either by removing the E-peptide to generate mature IGF1 or by glycosylating the asparagines in Ea domain. Cells can secrete cleaved E-peptides and potentially three different IGF1 protein isoforms: mature IGF1, glycosylated or unglycosylated IGF1Ea, IGF1Eb or in humans only IGF1Ec (9). Skeletal muscle cells predominantly express Class 1 IGF1Ea protein, although they also express isoforms that contain the Eb peptide (9, 16).
Experimental evidence suggests that these trimmed signals regulate the functional balance between local (autocrine or paracrine) versus systemic (endocrine) IGF1 signaling, as well as the potency of IGF1 signaling (17). In mice, both the glycosylated IGF1Ea and unglycosylated IGF1Eb isoforms of pro IGF1 bind to the extracellular matrix, thereby localizing the signal, whereas mature IGF1 does not bind to the extracellular matrix (17). When secreted, the pro IGF1 isoforms have increased binding affinity to the IGF1 receptor (IGF1R), which, in conjunction with the retention or pro IGF1 isoforms in the extracellular matrix, may locally enhance IGF1 potency. Pro IGF1 isoforms that contain the Eb peptide or the Ea peptide improve myoblast proliferation. However, IGF1Ea may promote myogenic fusion and differentiation better than IGF1Eb, which is thought to be more potent at stimulating proliferation (16). In rodents, free Eb peptides may act as growth factors, and free E-peptides may enhance IGF1R activation by IGF1 (16, 18).
The various IGF1 protein isoforms have been proposed to have different functions during regeneration and recovery from disease and injury (14). During injury and exercise, IGF1Eb expression increases rapidly and then decreases, whereas IGF1Ea expression increases after a delay. Increased expression of IGF1Ea correlates with a decline in IGF1Eb expression, suggesting that IGF1Eb has a role in the early stages of regeneration. The role of IGF1Ea is more prominent during the later stages of regeneration (14). With aging, the transient response of IGF1Eb decreases, and circulating levels of IGF1 generally decline. Thus, the general decline in IGF1 expression and the reduction in the IGF1Eb response to injury may contribute to the decline in the regenerative capacity of skeletal muscle and the increased propensity for muscle atrophy that occurs with age (19).
Because experimental evidence suggests that each IGF1 protein isoform and E-peptide plays a slightly different but important role in IGF1 signaling, we hypothesized that the overexpression of multiple IGF1 isoforms would better recapitulate the rich and complex signaling that underlies IGF1 activity than would overexpression of a single isoform. One way to achieve this goal of overexpressing multiple IGF1 protein isoforms is to use a CRISPR-Cas9–based genome engineering approach to activate or enhance endogenous IGF1 expression. The CRISPR-Cas9 system is divided into two major classes and multiple types (20). The type 2 CRISPR-Cas9 system has been widely studied and is popular in genome engineering. In this system, transactivating CRISPR RNA (tracrRNA) binds to and aids in the maturation of the CRISPR RNA (crRNA) sequence. crRNA sequences contain the guide sequence that targets Cas proteins to foreign DNA (21). Genome engineering typically involves the use of single-guide RNAs (sgRNAs), which are a fusion of tracrRNA and crRNA. Another CRISPR-Cas9 genome engineering application, gene activation, involves the use of a deactivated Cas9 (dCas9) protein fused to an activation domain. This fusion protein is then targeted to the promoter of a gene of interest by using sequence-specific sgRNAs. An example of this approach is the synergistic activation mediator (SAM) CRISPR-Cas9 gene activation system, which can achieve robust gene activation by using only one sgRNA (21). This system is composed of a nuclear localization signal containing dCas9 with VP64 fused at the N-terminus, a sgRNA modified by inserting an MS2 bacteriophage coat protein–binding aptamer into its tetraloop and stem-loop 2, and a cofactor fusion protein consisting of the MS2 bacteriophage coat protein and the activation domains of p65 and human heat-shock factor 1 (HSF1) (22).
In this study, we implemented a strategy using the SAM-mediated CRISPR-CAS9 gene activation system to induce or enhance the expression of multiple IGF1 or Igf1 mRNA variants in human and mouse skeletal myoblasts and skeletal myotubes. We describe the effectiveness of using this strategy both in vitro to prevent dexamethasone (DEX)-induced atrophy in human and mouse skeletal myotubes.
MATERIALS & METHODS:
Cell Lines.
HEK 293T cells (ATCC CRL-3216), C2C12 myoblasts (ATCC CRL-1772), and NIH/3T3 cells (ATCC CRL-1658) were purchased from ATCC. HEK 293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM with 4.5 g/L D-glucose, 4 mM glutamine, 25 mM HEPES buffer, 1 mM sodium pyruvate, 1X MEM non-essential amino acids, 100 U/mL penicillin-streptomycin, and 10% fetal bovine serum [FBS]). C2C12 myoblasts and C2C2-derived cell lines were maintained under growth conditions by culturing cells in DMEM with 4.5 g/L D-glucose, 4 mM glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin-streptomycin, and 10% FBS. NIH/3T3 cells were cultured in DMEM with 4.5 g/L D-glucose, 4 mM glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin-streptomycin, and 10% newborn calf serum. HSMMs (catalog #CC-2580) were purchased from Lonza (Lonza Viral-Based Therapeutics). HSMMs and HSMM-derived cell lines were maintained under growth conditions by culturing cells in SkGM-2 BulletKit Medium (catalog #CC-3245) from Lonza. All cell lines were passaged according to the manufacturer’s guidelines by using trypsin. Myoblast cell lines were maintained at 50% to 70% or less confluency before differentiation.
Viral Vectors, Viral Packaging, and Cell Line Creation.
The lentiviral vectors that compose the SAM gene activation system (a gift from Feng Zhang, Broad Institute) included lenti dCAS-VP64_Blast (Addgene plasmid # 61425), lenti MS2-P65-HSF1_Hygro (Addgene plasmid # 61426), and lenti sgRNA(MS2)_zeo backbone (Addgene plasmid # 61427). Potential sgRNAs were identified and cloned into the lenti sgRNA(MS2)_zeo backbone as previously described (22). The empty vector used in all experiments was the lenti sgRNA(MS2)_zeo backbone plasmid without a sgRNA target sequence.
Briefly, lentiviral vectors were packaged by transfecting 10 μg of transfer plasmid (lenti dCAS-VP64_Blast, lenti MS2-P65-HSF1_Hygro, or lenti sgRNA[MS2]_zeo), 5 μg of packaging plasmid (psPAX2, Addgene plasmid #12260), and 5 μg of envelope plasmid (pMD2.G, Addgene plasmid #12259) into confluent HEK 293T cells in a 100-mm plate by using FugeneHD (Promega Corporation). Media was changed after 24 hours. Conditioned media was subsequently harvested, and fresh media was added every 24 hours for the next 72 hours. Conditioned media was centrifuged (1,000 x g) for 5 minutes and then passed through a 0.45-micron filter. Harvested media from each transfected cell line was pooled and stored at 4°C before viral particles were concentrated by precipitation using the Lenti-X Concentrator solution (Clontech catalog #631231) according to the manufacturer’s guidelines. The resulting supernatant was removed, and the remaining virus-containing pellets were suspended in phosphate-buffered saline (PBS). The virus was stored in single-use aliquots at −80°C.
Cell lines (HEK 293T cells, C2C12 myoblasts, and NIH/3T3 cells) expressing components of the SAM gene activation system were generated by performing sequential viral particle infection. After each infection, viral integration was confirmed by using the following drug selection protocol: 10 μg/mL blasticidin after infection with lenti dCAS-VP64_Blast viral particles, 500 μg/mL hygromycin after infection with lenti MS2-P65-HSF1_Hygro, and either 500 μg/mL zeocin for HEK 293T cell lines or 1,000 μg/mL zeocin for C2C12 myoblast and NIH/3T3 cell lines after infection with lenti sgRNA(MS2)_zeo viral particles. Periodically, cell lines underwent drug selection to confirm the maintenance of the SAM gene activation system components. Each round of viral particle infection was performed after we observed the elimination of all cells on control uninfected plates that underwent drug selection. HSMM cell lines expressing all three components of the SAM gene activation system were generated by performing a sequential viral infection of each component without drug selection. For the generation of the HSMM cell lines, viral particles were incubated with the cells for 24 hours, the media was changed, and the next round of viral particle infection was performed. All infections were performed in the presence of 8 μg/mL polybrene.
HSMM or C2C12 Cell Line Differentiation.
HSMM cell lines were plated and grown to approximately 70% confluency before replacing the growth media with differentiation media (DMEM with 4.5 g/L D-glucose, 4 mM glutamine, 100 U/mL penicillin-streptomycin, and 0.05% bovine serum albumin [BSA]). HSMM cell lines were then cultured under differentiation conditions for 8 days with periodic media changes every 3 days. C2C12 cell lines were plated on 0.1% gelatin-coated dishes and grown to 70% confluency before the media was replaced with differentiation media (DMEM with 4.5 g/L D-glucose, 4 mM glutamine, 100 U/mL penicillin-streptomycin, and 0.05% BSA). Differentiating C2C12 cell lines were then maintained under these media conditions for the indicated amount of time (typically 3 to 4 days).
Isolation of RNA, cDNA Synthesis, semi-quantitative PCR, and Analysis.
To harvest RNA from cells, cell lines were washed with PBS, and cells were suspended in TRI-Reagent (Zymo Research, catalog #R2050–1). Samples were immediately processed or stored at –80°C. RNA from the samples was extracted by using either Direct-zol RNA Miniprep or Direct-zol RNA Microprep kits (Zymo Research, catalog #R2050 or #R2052) according to the manufacturer’s guidelines. The RNA was quantified by using a Nanodrop spectrophotometer (ThermoFisher Scientific). cDNA was then synthesized with the qScript cDNA SuperMix reagent (Quanta Biosciences, catalog #95048) according to the manufacturer’s guidelines. For all samples associated with a given experiment, the same amount of total RNA was used for cDNA synthesis reactions. After cDNA synthesis, samples were diluted in water and used for semi-quantitative PCR. Semi-quantitative PCR was performed by using the Power SYBR Green PCR Master Mix reagent (ThermoFisher Scientific, catalog #4367659) and either an Applied Biosystems 7900HT Real-Time PCR System or a QuantStudio 6 Flex Real-Time PCR System (ThermoFisher Scientific). All PCR primers are shown in Tables-1 and 2. Samples were analyzed using the comparative Ct method (23). The relative Ct value of each mRNA transcript was first calculated by subtracting the Ct value of a housekeeping gene from the Ct value of the mRNA transcript of interest. These relative Ct values were used to determine the relative quantity of the mRNA transcript compared to a control, which was the empty vector cell line in most cases. For each experiment, the relative Ct value used to determine the fold-change is indicated. For samples derived from human cell lines (i.e. HEK 293T cells and HSMM cells), the GAPDH mRNA transcript Ct values were used as the housekeeping gene. For mouse cell lines (i.e., NIH/3T3 cells and C2C12 cells), Gapdh mRNA transcript Ct values or Csnk2a2 mRNA transcript Ct values, respectively, were used as the housekeeping gene. Statistical significance was determined by performing unpaired t-tests. Results were considered statistically significant for p-values less than or equal to 0.05.
Table-1.
Previously described semi-quantitative PCR primer pairs for human IGF1 and mouse Igf1 transcript variants.
| Gene Isoform | Primer Sequence (5’ to 3’) | RefSeq Number | Ref |
|---|---|---|---|
| Human Primer Pairs | |||
| IGF1Ea | TCGTGGATGAGTGCTGCTTCCG | NM_000618 | (1) |
| TCAAATGTACTTCCTTCTGGGTCTTG | (1) | ||
| IGF1Eb | ATCTACCAACAAGAACACG | NM_001111285 | (1) |
| TACTTCCAATCTCCCTCC | (1) | ||
| IGF1Ec | ACCAACAAGAACACGAAGTC | NM_001111283 | (1) |
| CATGTCACTCTTCACTCCTC | (1) | ||
| Class 1 IGF1 | CAGCAGTCTTCCAACCCA | NM_000618 | (1) |
| CACAGCGCCAGGTAGAAGAGATGC | (1) | ||
| GAPDH | TGTTGCCATCAATGACCCCTT | NM_002046 | (2) |
| CTCCACGACGTACTCAGCG | (2) | ||
| Mouse Primer Pairs | |||
| Igf1Ea | GCCCAAGACTCAGAAGGAAGTACATTTG1 | NM_01111275 | (3) |
| TGTGGCATTTTCTGCTCCGTGG | (3) | ||
| Igf1Eb | AGCTGCAAAGGAGAAGGAAAGGAAG | NM_010512 | (3) |
| GGTGATGTGGCATTTCCTGCT | (3) | ||
| Gapdh | AGGTCGGTGTGAACGGATTTG | NM_008084 | (4) |
| TGTAGACCATGTAGTTGAGGTCA | (4) | ||
Table-2.
IGF1 and Igf1 semi-quantitative PCR primers commercially available from Integrated DNA Technologies.
| Gene | IDT Assay Name | RefSeq Number |
|---|---|---|
| Human IGF1 (total) | Hs.PT.58.21022358 | NM_000618 |
| Human Class 2 IGF1 | Hs.PT.58.45473990 | Nm_001111284 |
| Mouse Igf1 (total) | Mm.PT.5832726889 | NM_001111275 |
| Mouse Class 1 Igf1 | Mm.PT.58.13167185 | NM_010512 |
| Mouse Class 2 Igf1 | Mm.PT.58.30779893 | NM_001111276 |
| Mouse Csnk2a2 | Mm.PT.58.10226157 | NM_009974 |
Dexamethasone (DEX) Treatment.
Before treating HSMM cell lines with DEX (Sigma, catalog #D4902), media was changed after 3 days of differentiation, and cells were cultured for an additional 3 days to ensure the presence of IGF1 protein in the media. The media was not changed for C2C12 cell lines because they were differentiated for only 2 days before being treated with DEX. After differentiation for the indicated length of time (6 days for HSMM cell lines and 2 days for C2C12 cell lines), HSMM cell lines were treated with 50 μM DEX for 48 hours and C2C12 cell lines were treated with 50 μM DEX for 24 hours. Ethanol was used as a carrier control.
Immunofluorescence Staining.
After the cells were differentiated and treated with DEX, they were washed with PBS and fixed with 4% formaldehyde in PBS for 10 minutes. After fixation, cells were washed with PBS and permeabilized with 0.1% TritonX-100 in PBS for 15 minutes, followed by blocking in PBS with 10% normal goat serum for one hour. Cells were then incubated with MF20 antibody (Developmental Studies Hybridoma Bank) in PBS with 1% BSA at 4°C overnight. Cells were then rinsed with PBS before incubating with goat anti-mouse IgG (H+L) Alexa Fluor 555 (ThermoFisher Scientific) for 1 hour at room temperature. After washing and incubation with 5 μ/mL 4′,6-diamidino-2-phenylindole (DAPI), cells were mounted with Prolong Antifade Diamond (ThermoFisher Scientific). Coverslips were coated with 0.1% gelatin before cell plating.
In vitro Myotube Diameter and Nuclei Quantification.
For both HSMM and C2C12 cell lines, cells were imaged under high magnification after staining. Ten images were randomly captured for each biological replicate. Myotube diameter was measured by using ImageJ software, and the number of nuclei per myotube was counted. In the studies using the C2C12 cell line the following number of cells were looked at for each test condition: 194 (Empty vector plus Dex), 252 (Empty vector plus ethanol), 362 (sgRNA plus ethanol), and 423 (sgRNA plus Dex). The number for the HSMM cells examined are as follows: 59 (Empty vector plus Dex), 64 (Empty vector plus ethanol), 64 (sgRNA plus ethanol), and 67 (sgRNA plus Dex). Statistical significance was determined by performing a two-way analysis of variance (ANOVA) and Tukey’s multiple comparison t-tests. Results were considered statistically significant for p-values less than or equal to 0.05.
IGF1 ELISA.
Media was removed from C2C12 cell lines that were differentiated for 3 days without a media change or from proliferating C2C12 myoblasts with conditioned media. Media was centrifuged (500 x g for 3 min) and snap-frozen in single-use aliquots with liquid nitrogen. Samples were thawed on ice and analyzed by using a Mouse/Rat IGF1 Quantikine ELISA Kit according to the manufacturer’s guidelines (R&D Systems, catalog #MG100). The amount of IGF1 measured in the media was normalized to the total number of cells and the volume of conditioned media. Statistical significance was determined by performing two-way ANOVA and Tukey’s multiple comparison t-tests. Results were considered statistically significant for p-values less than or equal to 0.05.
Western Blotting.
C2C12 cell lines were differentiated for 3 days, washed with PBS, and harvested with radioimmunoprecipitation assay buffer (RIPA) buffer (ThermoFisher Scientific, catalog #89900) supplemented with protease inhibitors (ThermoFisher Scientific catalog # 88669) before shearing chromatin with a Bioruptor UCD-300 (Diagenode). Lysed and sheared cells were centrifuged at maximum speed (~21,130 x g) for 10 minutes at 4°C. The total protein content of the sample lysate was determined by using a bicinchoninic acid assay kit (BCA) (ThermoFisher Scientific, catalog # 23225). For each sample, 5 micrograms of total protein were loaded and separated on a Nupage 4–12% Bis-Tris Protein gel (ThermoFisher Scientific, catalog # NP0322BOX) before being transferred to a PVDF membrane. Membranes were washed with tris-buffered saline (TBS) containing 0.1% Tween-20 (TBST) before blocking in TBST with 5% BSA. Membranes were incubated with the appropriate antibodies overnight at 4°C in TBST with 5% BSA. Membranes were washed with TBST before incubating with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 hour. After a final wash with TBST, the signal from the HRP-conjugated secondary antibodies was detected with SignalFire Enhanced Chemiluminescence (ECL) Reagent (Cell Signaling, catalog #6883S) according to the manufacturer’s guidelines. Membranes were washed with TBST and stripped with Restore PLUS Western Blot Stripping Buffer (ThermoFisher Scientific, catalog #46430). Membranes were blocked again with TBST containing 5% BSA before being probed with additional primary and secondary antibodies. Primary antibodies rabbit polyclonal IgG anti-(pan) AKT (catalog #4691) and rabbit polyclonal IgG anti-phospho-AKT(Ser473) (catalog #4060) were purchased from Cell Signaling. Secondary antibody goat anti-mouse IgG-HRP (catalog #sc-2031) was purchased from Santa Cruz Biotechnology.
RNA-Seq and Data Analysis.
C2C12 cell lines were differentiated for 3 days and treated with 50 μM DEX for 24 hours. RNA was then isolated from C2C12 cell lines with an RNeasy Plus Micro Kit (Qiagen, catalog #74034). RNA concentrations were determined by using a Qubit Fluorometer (ThermoFisher Scientific). A total of 1 μg of RNA was used from each sample to generate RNA-seq libraries by using a ScriptSeq Complete Kit (Human/Mouse/Rat) (Illumina) according to the manufacturer’s guidelines for low-input starting materials. RNA-seq libraries were multiplexed and pair-end sequenced at the University of Houston’s Seq-N-Edit Core with an Illumina NextSeq 500.
Quality control and sequence read trimming were performed with FastQC (24) and cutadapt (25), respectively. Sequence reads were mapped by using HISAT2 with the m. musculus GRCm38 index (26). Next, mRNA transcripts were assembled and quantified by using StringTie (27), and gene counts were determined by using featureCounts (28). Differential expression analysis and gene ontology (GO) enrichment analysis were performed by using edgeR (29, 30).
RESULTS:
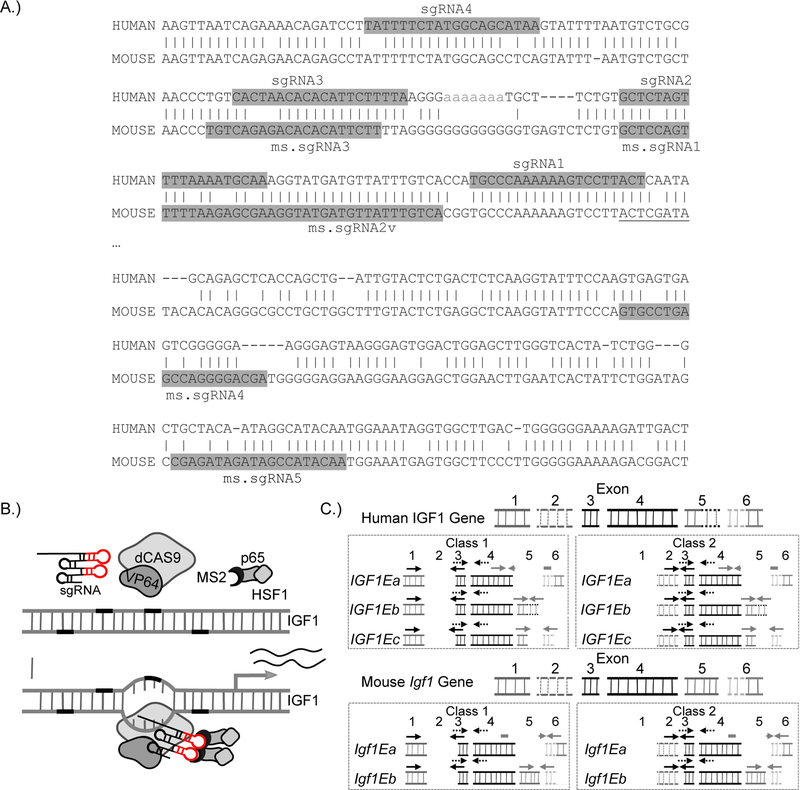
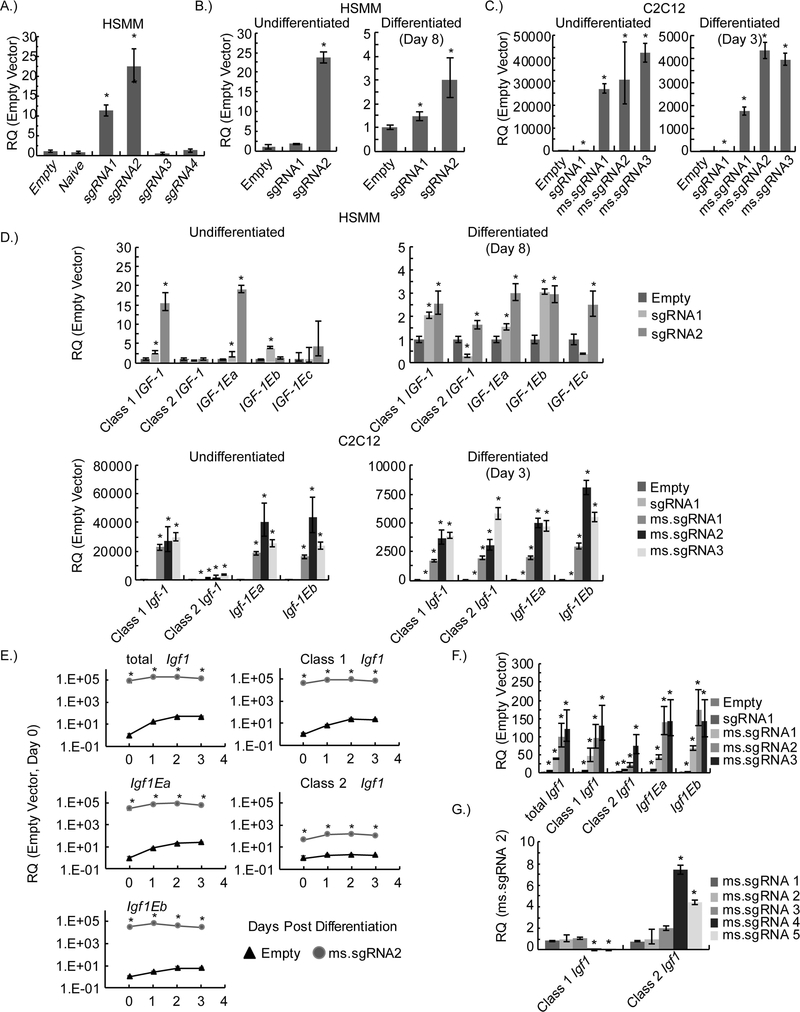
We designed a set of human and mouse sgRNAs (sgRNA1–4 and ms.sgRNA1–5) that target the proximal promoter region of IGF1/Igf1 from the 5’ end to the first exon and first transcriptional start site (TSS) of IGF1/Igf1 (Figure-1A and Table-3). A lentiviral vector expressing one of the sgRNAs and lentiviral vectors expressing the two components of the SAM gene expression system (i.e., the chimeric cofactor MS2-P65-HSF1 and dCAS9 fused with VP64) were transfected into different cells to generate stable cell lines (Figure-1B). To determine which IGF1 isoforms were affected by the SAM gene activation system, we designed semi-quantitative PCR primers specific for the different IGF1 or Igf1 mRNA transcript variants (Figure-1C). Total expression levels of IGF1 mRNA were determined by semi-quantitative PCR, and we found that the sgRNAs that targeted IGF1 closest to the start of the first exon resulted in higher expression of total IGF1 mRNA (Figure-2A). Each of the two best-performing human sgRNAs (sgRNA1 and sgRNA2) and the two other components of the SAM gene activation system were introduced into human skeletal myoblasts (HSMMs) by lentiviral transduction in order to confirm their enhancement of total IGF1 mRNA expression in a skeletal muscle cell line. Total IGF1 mRNA expression was significantly increased in both undifferentiated HSMMs and differentiated myotubes expressing sgRNA2 when compared to those expressing the empty vector control. In addition, total IGF1 mRNA expression was significantly increased in differentiated HSMM-derived myotubes expressing sgRNA1when compared to those expressing the empty vector control (Figure-2B).
Figure-1.
Use of CRISPR-Cas9–mediated gene activation to increase IGF1 or Igf1 expression in human or mouse myoblasts and myotubes. (A) Schematic representation of the alignment of human and mouse genomic DNA showing the conservation of human sgRNA-targeting sites in mice. The sgRNA-targeting sequences are highlighted in gray, and the start of the first exon is underlined. (B) A schematic illustrating the transcriptional activation of IGF1 (or Igf1) by the CRISPR-Cas9–based three-component SAM gene activation system. (C) A schematic of the semi-quantitative PCR strategy for determining the relative quantities of total human IGF1 or mouse Igf1 mRNA transcripts and the relative quantities of their respective mRNA transcript variants. Arrows indicate the binding sites for the semi-quantitative PCR primers used in expression profiling.
Table-3.
Human and Mouse sgRNA sequences targeting the IGF1 or Igf1 promoter.
| ID | sgRNA | Base pairs from the 5’ of the End of Exon 1 |
|---|---|---|
| Human sgRNA1 | ||
| sgRNA1 | AGTAAGGACTTTTTTGGGCA | −51 |
| sgRNA2 | GCTCTAGTTTTAAAATGCAA | −94 |
| sgRNA3 | CACTAACACACATTCTTTTA | −134 |
| sgRNA4 | TTATGCTGCCATAGAAAATA | −179 |
| Mouse sgRNA2 | ||
| ms.sgRNA1 | GCTCCAGTTTTTAAGAGCGA | −60 |
| ms.sgRNA2 | AGGTATGATGTTATTTGTCA | −40 |
| ms.sgRNA3 | TGTCAGAGACACACATTCTT | −107 |
| ms.sgRNA4 | GTGCCTGAGCCAGGGGACGA | +1974 |
| ms.sgRNA5 | CGAGATAGATAGCCATACAA | +2043 |
Figure-2.
Expression of human IGF1 or mouse Igf1 mRNA and their respective transcript variants due to CRISPR-Cas9-mediated gene activation. (A) The relative quantity (RQ) of total IGF1 mRNA in HEK 293T cell lines measured by semi-quantitative PCR. Data are shown for different sgRNAs. Naïve cells are uninfected HEK 293T cells and empty cells are HEK 293T cell lines that have the gene activation system but express a sgRNA without a targeting sequence. (B) The RQ of total IGF1 mRNA in undifferentiated and differentiated (day-8) human skeletal muscle myoblast (HSMM) cell lines measured by semi-quantitative PCR. (C) The RQ of total Igf1 mRNA in undifferentiated and differentiated (day-3) C2C12 cell lines measured by semi-quantitative PCR. (D) RQ of IGF1 mRNA or Igf1 mRNA transcript variants in undifferentiated and differentiated human skeletal muscle myoblast (HSMM) and C2C12 cell lines measured by semi-quantitative PCR. (E) Time course of the RQ of Igf1 mRNA or Igf1 mRNA transcript variants determined by semi-quantitative PCR. (F) Analysis of semi-quantitative PCR data showing total Igf1 mRNA or Igf1 mRNA variant expression in NIH/3T3 cell lines expressing the SAM-mediated gene activation system and various Igf1 sgRNAs. (G) Analysis of semi-quantitative PCR data from undifferentiated C2C12 cell lines expressing different sgRNAs. The RQ of samples was normalized to that of Igf1 mRNA transcript variants (Class 1 or Class 2) in C2C12 cells expressing ms.sgRNA2. Samples in A-D and F were normalized to the corresponding empty vector sample to determine the RQ of total IGF1 mRNA transcript expression. Samples in E were normalized to empty vector C2C12 cells on day-0, and samples in G are normalized to that of Igf1 mRNA transcript variants (Class 1 or Class 2) in C2C12 cells expressing ms.sgRNA2. Three biological replicates were performed in all experiments. In A-D and F, * indicates a statistically significant difference when compared with the corresponding empty vector sample (P<0.05). In E, * indicates a significant difference (P<0.05) between the RQ of mRNA in cells expressing ms.sgRNA2 and the RQ of mRNA in cells expressing empty vector at the same time point. In G, * indicates a statistical significance between the mRNA transcript and the ms.sgRNA2 sample. Error bars represent the standard deviation.
When we aligned human and mouse IGF1 and Igf1 sequences containing the genomic region near the first exon, we found that the target sequence for human sgRNA1 is conserved in mice, except that the mouse sequence lacks a PAM sequence (Figure-1A). Due to this conservation, we tested sgRNA1 in the mouse cell lines. Each of the mouse-specific sgRNAs or human sgRNA1, along with the two other components of the SAM gene activation system were then introduced into mouse C2C12 myoblasts by lentiviral transduction. Total Igf1 mRNA expression was examined in undifferentiated C2C12 myoblasts and differentiated C2C12-derived myotubes. All tested mouse-specific sgRNAs (ms.sgRNA1–3) significantly increased Igf1 mRNA expression in both undifferentiated and differentiated cells. In addition, human sgRNA1, which lacked a PAM sequence, increased Igf1 mRNA expression the least but still significantly above the background (Figure-2C).
In undifferentiated HSMMs expressing the two components of the SAM gene activation system, sgRNA1 or sgRNA2 upregulated the expression of Class 1 mRNA transcript variants (which include the different E protein variants) and the IGF1Ea mRNA variant (Figure-2D). Only sgRNA1 upregulated the IGF1Eb mRNA variant in undifferentiated HSMMs. Neither sgRNA1 nor sgRNA2 upregulated IGF1Ec or Class 2 mRNA transcript variants. In differentiated HSMMs, sgRNA1 and sgRNA2 preferentially upregulated expression of Class 1 mRNA transcript variants, whereas sgRNA1 downregulated and sgRNA2 upregulated expression of Class 2 mRNA variants (Figure-2D). Both sgRNA1 and sgRNA2 upregulated expression of IGF1Ea and IGF1Eb mRNA variants, whereas the only sgRNA2 upregulated the IGF1Ec mRNA variant (Figure-2D). In C2C12 myoblasts and differentiated C2C12 myotubes expressing the SAM gene activation system, the sgRNAs tested (sgRNA1, ms.sgRNA1, ms.sgRNA2, and ms.sgRNA3) significantly upregulated expression of all Igf1 mRNA transcript variants (Class 1, Class 2, Igf1Ea, and Igf1Eb). However, in undifferentiated C2C12 myoblasts, these sgRNAs preferentially upregulated expression of Class 1 mRNA transcript variants over Class 2 mRNA variants (Figure-2D). In differentiated C2C12-derived myotubes, all tested sgRNAs increased expression of Class 1 and Class 2 mRNA variants to a similar extent. The equivalent induction of Class 1 and Class 2 mRNA levels is related to the fact that Class 1 mRNA transcript variant expression increases during differentiation, while Class 2 mRNA transcript variant expression remains relatively constant. This has the effect of reducing the relative expression level of Class 1 mRNA variants while the relative expression level of Class 2 mRNA variants remains constant (Figure-2E). A general trend in the data shows a reduced response (i.e. elevation in Igf1 mRNA expression) in myotubes compared to myoblasts. This is because of the SAM-mediated gene activation of Igf1. As C2C12 cells differentiate, Igf1 mRNA variant and total mRNA levels increase, which in turn reduces the overall relative increase in Igf1 expression because the SAM gene activation system has potentially saturated Igf1 mRNA expression. As the cells that express the SAM gene activation system differentiate, the expression level of Igf1 remains constant. The proposed saturation in expression due to gene activation is supported by the unchanged expression of total and variant Igf1 mRNA observed during differentiation (Figure-2E).
Notably, the capacity of the SAM gene activation system to increase the expression of total Igf1 mRNA and all transcript variants of Igf1 mRNA is not restricted to myoblasts and myotubes. We also observed this phenomenon in NIH/3T3 cells. Similar to our observations in HSMM and undifferentiated C2C12 cell lines, Class 1 mRNA transcript variant expression was preferentially upregulated over Class 2 mRNA transcript variant expression in NIH/3T3 cells (Figure-2F). This preference for the expression of Class 1 over Class 2 mRNA transcript variants could be shifted by using sgRNAs that target downstream of exon 1 and closer to exon 2 (ms.sgRNA4 and 5), which substantiates the tunability of the system (Figure-2F).
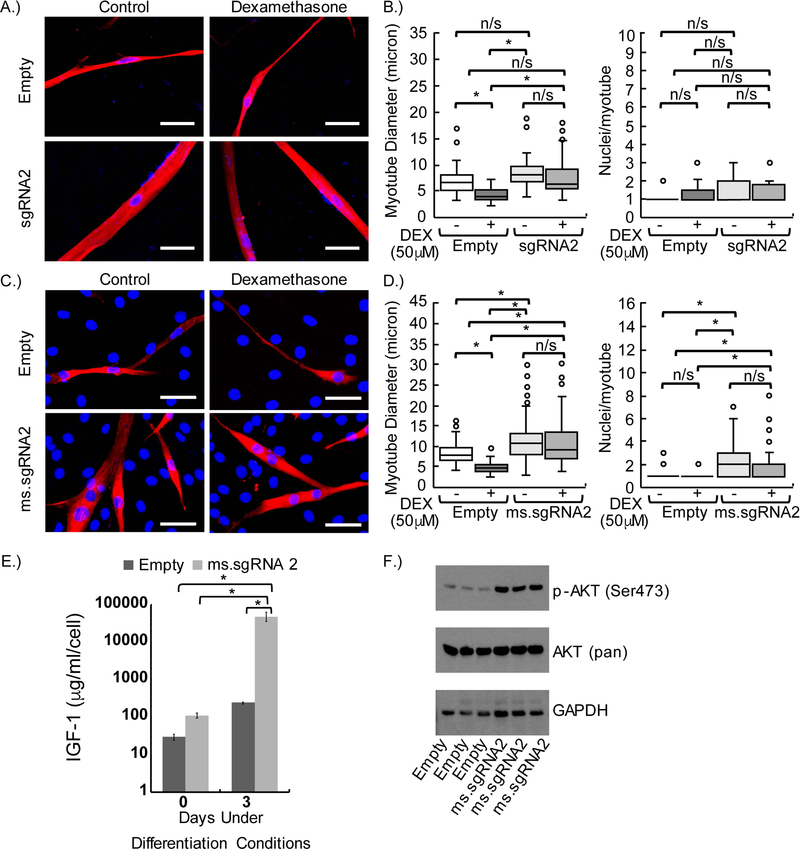
Next, we used an in vitro DEX-induced model of atrophy to examine whether the SAM gene activation system with our sgRNAs mediated the production of functional IGF1 protein. HSMMs expressing the SAM gene activation system with sgRNA2 or empty vector were first differentiated and then treated with or without DEX (Figure-3A). The fusion index was similar between HSMM-derived myotubes expressing empty vector and those expressing sgRNA2. Although a trend toward thicker myotubes was observed for HSMMs expressing sgRNA2 compared to empty vector controls, the difference was not statistically significant (Figure-3B). After DEX treatment, myotube diameter was significantly smaller in HSMM-derived myotubes expressing empty vector than in untreated HSMM-derived myotubes expressing empty vector. Furthermore, the mean myotube diameter was significantly larger in both untreated and DEX-treated myotubes expressing sgRNA2 than in DEX-treated HSMM myotubes expressing empty vector. No significant difference was observed in the mean myotube diameter of untreated versus DEX-treated HSMM-derived myotubes expressing sgRNA2 (Figure-3B).
Figure-3.
Inhibition of dexamethasone (DEX)-induced atrophy in human and mouse myotubes by CRISPR-Cas9–mediated gene activation to increase the expression of IGF1 or Igf1 mRNA variants. (A) Anti-myosin (MF20) staining of human skeletal muscle myoblast (HSMM)-derived myotubes expressing empty vector or human sgRNA2 after 6 days of differentiation and 2 days of treatment or no treatment (control) with 50 μM DEX. (B) Quantification of myotube diameter and fusion index (nuclei per myotube) of MF20-positive HSMM-derived myotubes after DEX treatment. (C) MF20 staining of C2C12-derived myotubes expressing empty vector or mouse ms.sgRNA2 after 2 days of differentiation and 1 day of treatment or no treatment with 50 μM DEX. (D) Quantification of myotube diameter and fusion index of MF20-positive C2C12-derived myotubes after DEX treatment. (E) Enzyme-linked immunosorbent assay (ELISA) results showing the concentration of IGF1 in the conditioned media of C2C12 cells before and after differentiation for 3 days. (F) Western blot analysis showing the levels of phosphorylated AKT (p-AKT) in C2C12 cells expressing empty vector or mouse ms.sgRNA2 after 3 days of differentiation. Scale bars, 50 μm. DAPI-positive nuclei are blue and MF20-positive myotubes are red. In all DEX-treated cells, ethanol was used as a carrier control. * P<0.01; n/s, no statistically significant difference between samples; braces show the statistical comparisons performed. Experiments were performed in biological triplicates in panels (A), (B), (E), and (F), and four biological replicates were performed in panels (C) and (D). Error bars in panel (E) represent 95% confidence intervals.
We also examined the effects of DEX on myotube diameter in a C2C12 cell line expressing the SAM gene activation system and ms.sgRNA2 (Figure-3C). Unlike what we observed in the HSMM cell line, both DEX-treated and untreated C2C12-derived myotubes expressing ms.sgRNA2 had a significantly larger myotube diameter than did DEX-treated or untreated C2C12-derived myotubes expressing empty vector (Figure-3D). Similar to what we observed in the HSMM cell line, the mean diameter between DEX-treated and untreated C2C12-derived myotubes expressing ms.sgRNA2 was not significantly different. DEX-treated or untreated C2C12-derived myotubes expressing ms.sgRNA2 had a significantly higher fusion index than did DEX-treated or untreated C2C12-derived myotubes expressing empty vector, but no difference in fusion index was observed between DEX-treated and untreated groups (Figure-3D).
To further characterize Igf1 gene activation mediated by the SAM gene activation system and ms.sgRNA2, we examined protein expression and pathways associated with IGF1 signaling. Using an enzyme-linked immunosorbent assay (ELISA) to detect IGF1 in the conditioned media of C2C12 cells, we observed a significant increase in IGF1 protein levels in C2C12-derived myotubes but not C2C12 myoblasts expressing the SAM gene activation system and ms.sgRNA2 (Figure-3E). In addition, western blot analysis showed phosphorylation of AKT at Ser473 in C2C12-derived myotubes expressing the SAM gene activation system and ms.sgRNA2 (Figure-3F).
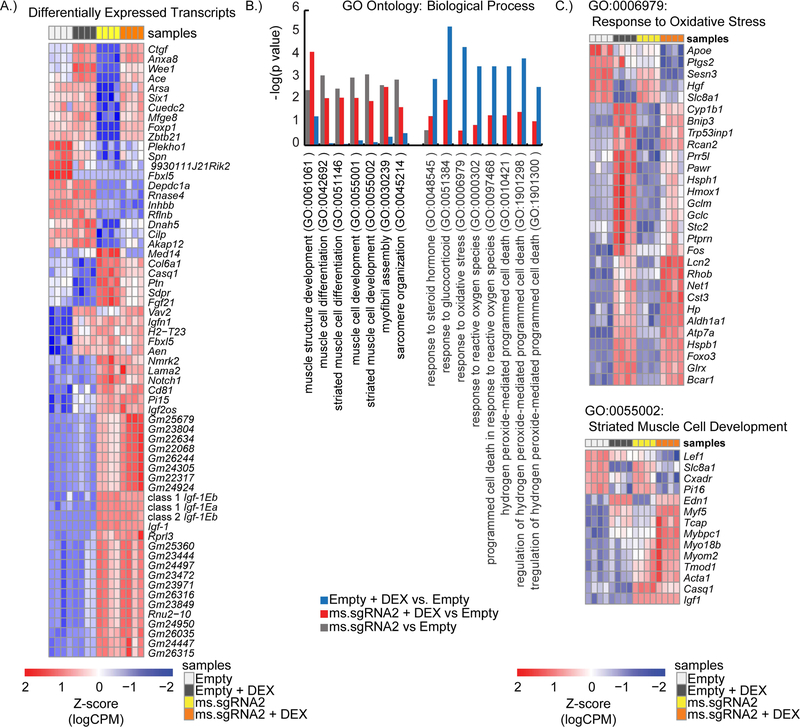
To better assess IGF1 signaling and the response to DEX treatment, we performed next-generation sequencing (RNA-seq) on RNA harvested from differentiated C2C12 cell lines expressing the SAM gene activation system and either ms.sgRNA2 or the empty vector control that were treated with DEX or left untreated. After filtering to remove mRNA transcripts with low counts (<1 count-per-million) and mRNA transcripts that did not appear in at least four of the sample libraries, we mapped 28,660 different mRNA transcripts to the samples. These 28,660 mRNA transcript corresponded to 12,773 genes. Upon our initial evaluation of the RNA-seq data, we identified all mRNA transcripts that had at least a 1.5-fold change in expression between untreated C2C12 cells expressing ms.sgRNA2 and untreated C2C12 cells expressing the empty vector control. Of the 63 mRNA transcripts that met this criterion, 36 were upregulated in the cells expressing ms.sgRNA2. Among these 36 upregulated genes, three were specific for different Igf1 mRNA transcript variants (Class 1 Igf1Ea, Class 1 Igf1Eb, and Class 2 Igf1Eb), which confirmed that the SAM gene activation system with ms.sgRNA2 induced the expression of multiple Igf1 transcripts (Figure-4A). When we performed go analysis (GO) analysis on genes upregulated by at least 2-fold in cells expressing ms.sgRNA2 compared to untreated cells expressing the empty vector control, we observed enrichment in biological processes involved in muscle development and differentiation (Figure-4B). We observed a similar trend in GO analysis when the same comparison was performed on DEX-treated cell lines (i.e. genes that were 2-fold upregulated in DEX-treated cells expressing empty vector compared to DEX-treated cells expressing ms.sgRNA2). Specifically, cells expressing ms.sgRNA2 were enriched for terms related to muscle development regardless of DEX exposure. In addition, GO analysis showed that cells expressing ms.sgRNA2 did not have as strong of enrichment for biological processes related to cell death and glucocorticoid stimulation cells (Figure-4B). Heatmaps of the genes represented by GO terms related to glucocorticoid stimulation and muscle development are shown in Figure-4C.
Figure-4.
Enhanced muscle cell differentiation and reduced response to glucocorticoids in C2C12 myotubes expressing multiple Igf1 mRNA transcript variants. (A) Heatmap of the Z-scores for mRNA transcripts that showed a significant change in expression (at least a 1.5-fold change) between untreated C2C12 myotubes expressing ms.sgRNA2 and untreated C2C12 myotubes expressing empty vector. mRNA variant data were determined by RNA-seq. (B) From the RNA-seq data, P-values are shown for representative gene ontology (GO) terms associated with muscle development, the response to dexamethasone (DEX), and cell death. (C) Representative heatmaps of genes that are differentially expressed in untreated C2C12-derived myotubes expressing ms.sgRNA2, DEX-treated C2C12-derived myotubes, and DEX-treated C2C12-derived myotubes expressing empty vector when compared to untreated C2C12-derived myotubes expressing empty vector. Genes shown are associated with the GO terms Response to Oxidative Stress and Striated Muscle Cell Development. Four biological replicates were performed per condition.
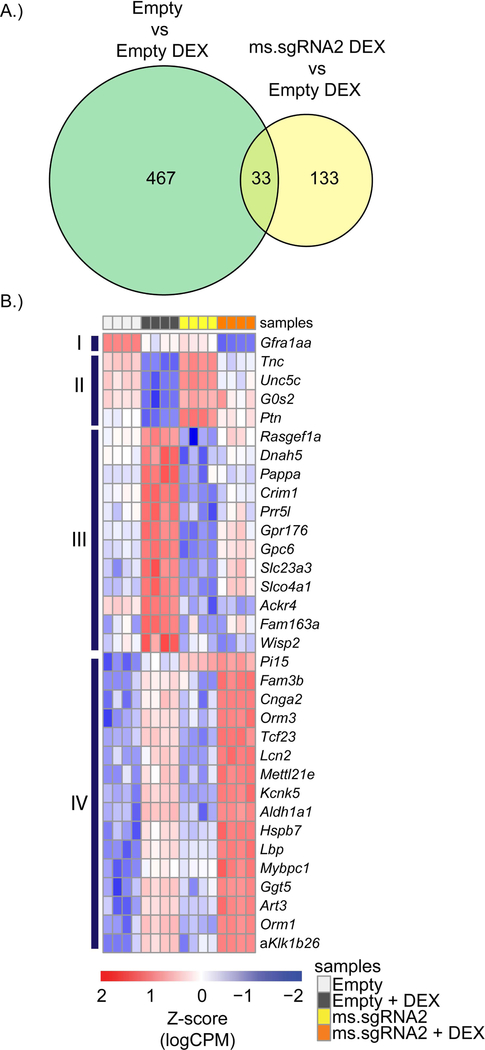
Using the RNA-seq data, we sought to identify genes related to DEX-induced atrophy that were differentially regulated by the activation of Igf1 by the SAM gene activation system with ms.sgRNA2. We identified 500 differentially regulated genes that had at least a 2-fold change in expression in DEX-treated myotubes expressing empty vector compared to untreated myotubes expressing empty vector. These genes represent the signature for DEX activity in C2C12 myotubes. Next, we identified 166 differentially regulated genes that had at least a 2-fold change in expression in DEX-treated myotubes expressing empty vector compared to DEX-treated myotubes expressing ms.sgRNA2. This comparison, made against a background of DEX treatment, was used to identify DEX-associated genes that are regulated by IGF1 signaling. The intersection of these data sets revealed 33 genes that are regulated by both DEX and IGF1 signaling (Figure-5A). Clustering of these genes revealed 4 genes in cluster II that are upregulated and 12 genes in cluster III that are downregulated in DEX-treated C2C12 myotubes expressing ms.sgRNA2 relative to DEX-treated C2C12 myotubes (Figure-5B).
Figure-5.
Differential regulation of a subset of genes by IGF1 signaling in dexamethasone (DEX)-treated C2C12 cells. (A) A Venn diagram summarizing data from the RNA-seq analysis showing the number of differentially regulated genes that were in common between the comparison of untreated C2C12 cells expressing empty vector versus DEX-treated C2C12 cells expressing empty vector (Empty vs Empty DEX) and the comparison of DEX-treated C2C12 cells expressing empty vector versus DEX-treated C2C12 cells expressing ms.sgRNA2 (ms.sgRNA2 DEX vs Empty DEX). (B) A heatmap of the Z-scores for the 33 genes that were identified in (A). Four biological replicates were performed.
DISCUSSION:
In this study, we showed in human and mouse skeletal muscle myoblast cell lines that the SAM gene activation strategy can be used to effectively increase the expression of multiple IGF1 or Igf1 mRNA transcript variants. In fact, our data suggest that this system saturates mRNA production from the IGF1 promoter of both mouse and human (Figure-2E). Furthermore, we showed that this strategy can be used to select between Class 1 and Class 2 IGF1 or Igf1 mRNA transcript variants because of the structural organization of the IGF1 or Igf1 gene.
The promoters of the human and mouse (IGF1 and Igf1) genes are unique in that they have multiple TSSs within the first two exons (16, 17). Konermann and colleagues (22) previously showed in their characterization of the SAM gene activation system that there is a small window proximal to a TSS in which the system could effectively enhance gene expression. This property makes the system ideally suited for selection between Class 1 and Class 2 IGF1 or Igf1 mRNA transcript variants. In the case of the IGF1 or Igf1 gene, simply shifting where a sgRNA targets in relationship to exons 1 and 2 can result in a switch between the expression of Class 1 and Class 2 IGF1 or Igf1 mRNA transcript variants (Figure-2F). However, with the current system, data suggest that the different E-peptides are nonspecifically upregulated.
IGF1 overexpression studies usually rely only on the expression of the IGF1Ea isoform, thereby excluding any benefits that may arise from the transient upregulation of IGF1Eb, which is a more potent inducer of proliferation than is IGF1Ea (15, 31, 32). In transgenic mice that overexpress only IGF1Ea, Chakravarty and colleagues (33) showed that a loss of enhanced satellite cell replication was observed with age, underscoring the importance of multiple isoform expression. The more potent increase in differentiation versus proliferation induced by IGF1Ea may have prematurely depleted the satellite cell population. The addition of IGF1Eb, which is a more potent mitogen than IGF1Ea, may have recovered the observed loss of satellite cell proliferation and subsequent decrease in hypertrophy; at the least, it may have extended the duration of increased proliferation and hypertrophy before the apparent stem cell exhaustion occurred (33). Similarly, the expression of IGF1Eb alone may not be effective; in aged mice, Barton and colleagues (34) showed that only IGF1Ea and not IGF1Eb could induce hypertrophy. The importance of E-peptides in IGF1 signaling is further supported by other experiments by Barton and colleagues (35) in young developing mice showing that the local overexpression of mature IGF1 did not induce hypertrophy, but the overexpression of IGF1Ea and IGF1Eb did. Therefore, with the importance of multiple isoform expression in mind, we propose that overexpressing multiple IGF1 isoforms with the SAM gene activation strategy will have greater therapeutic value than the overexpression of a single IGF1 isoform.
To initially assess the feasibility of using the SAM gene activation system to enhance IGF1 signaling through the increased expression of multiple IGF1 or Igf1 mRNA transcript variants, we established four criteria that must be met. First, this strategy must treat or at least reduce the disease phenotype of an in vitro muscle atrophy model. Second, this strategy must increase IGF1 protein secretion. Third, this strategy must activate a downstream component of the IGF1 signaling pathway. In this study, we showed that increasing the expression of multiple transcript variants of IGF1 or Igf1 by using the SAM gene activation system reduces the effects of DEX-induced atrophy, even in the absence of hypertrophy (Figure 3A-D). We also showed that this strategy increased the secretion of IGF1 into the media (Figure-3E). Moreover, we showed that this strategy induced AKT phosphorylation. A central component of the IGF1 signaling pathway is the phosphorylation and subsequent activation of AKT1. Phosphorylated AKT1 signals through multiple pathways to promote regeneration and reduce muscle atrophy (33, 36). For instance, activated AKT1 phosphorylates Forkhead box protein O1 (FOXO1) transcription factors, leading to the exclusion of FOXO1 protein from the nucleus and its degradation. FOXO1 transcription factors upregulate a core set of muscle atrophy genes that are conserved in several different muscle atrophy models (2). Phosphorylated AKT1 also indirectly activates mTOR, which promotes skeletal muscle hypertrophy (37). Furthermore, AKT1 phosphorylation inhibits numerous proapoptotic proteins, such as caspase 9 (38). Therefore, our findings suggest that the SAM gene activation system can be used to increase functional IGF1 signaling and has the potential to be a novel approach for treating skeletal muscle atrophy.
How the combination of IGF1 and DEX affects gene expression in C2C12 myotubes or whether IGF1 signaling and DEX competitively regulate these genes are not well understood. Latres and colleagues (39) reported that IGF1 signaling negatively regulates atrophy, whereas DEX positively regulates atrophy through the PI3K/AKT1/mTOR pathway. Latres and colleagues (39) identified genes in C2C12 myotubes that are differentially regulated in the presence of IGF1 or DEX alone, but they did not examine the effect of IGF1 and DEX together. We found that the expression of the atrophy-associated genes identified by Latres and colleagues (e.g. MT-1, MT-2, MAFbx, and MuRF1) was significantly increased in C2C12 myotubes treated with DEX compared to untreated myotubes expressing empty vector (data not shown). In addition, atrophy-associated gene expression was not significantly different between untreated C2C12 myotubes expressing ms.sgRNA2 and untreated C2C12 myotubes expressing empty vector (data not shown), nor was it significantly different between DEX-treated C2C12 myotubes expressing ms.sgRNA2 and DEX-treated myotubes expressing empty vector. These findings suggest that IGF1 signaling and DEX treatment do not competitively regulate these atrophy-associated genes.
Our data suggest that IGF1 signaling in the presence of DEX regulates a different set of genes than the atrophy-associated genes identified by Latres and colleagues (39). To identify genes that are competitively regulated by IGF1 signaling in the presence of DEX, we examined which differentially expressed genes were in common between untreated myotubes expressing empty vector versus DEX-treated myotubes expressing empty vector and DEX-treated myotubes expressing empty vector versus myotubes expressing ms.sgRNA2 (Figure-5). The genes identified represent those that are differentially regulated by DEX and that are targeted by IGF1 signaling. When we performed this analysis, we identified 33 possible genes, of which 4 were upregulated and 12 were downregulated in DEX-treated myotubes expressing ms.sgRNA2 versus DEX-treated myotubes expressing empty vector.
This set of 16 genes included Ptn and Prr5l, which were increased and decreased, respectively. This observation suggests that IGF1 signaling directly attenuates the effects of DEX by promoting survival signaling. PTN is a growth factor that reduces apoptosis by promoting AKT1 phosphorylation (40). In an in vivo context, an increase in PTN promotes neuronal survival and potentially muscle reinnervation. A decrease in PRR5L also promotes survival by reducing apoptosis or by increasing the stability of ZFP36-associated mRNAs (41).
In summary, we have shown that genome engineering and specifically the SAM gene activation system can increase myotube diameter in an in vitro model. Our approach has the potential for therapeutic applications because we demonstrated that it increases IGF1 signaling and reduced steroid-induced atrophy in vitro. Our data indicate that this technique enhanced the expression of all IGF1 or Igf1 isoforms. Our results indicate that when there are multiple mRNA splice variants with different TSSs in different exons, the sgRNA targeting sequence can be shifted further or closer to the TSS of interest to bias the expression of a specific mRNA transcript variant. This approach can be applied universally to any gene with multiple TSS-containing exons. Finally, because of the modular nature of the SAM gene activation system, different regulation domains can be selected that may in turn bias mRNA transcript splicing. Future studies will focus on identifying domains or splicing proteins that can shift E-peptide splicing, explore the translation of this strategy to in vivo models of skeletal muscle atrophy, and focus on optimizing the delivery of each component of the activation system to the single muscle fiber.
SIGNIFICANCE STATEMENT:
Therapies involving insulin-like growth factor 1 (IGF1) usually deliver a single IGF1 isoform. For the first time to our knowledge, we showed that a CRISPR-Cas9 gene activation system can be used to upregulate the expression of multiple IGF1 or Igf1 mRNA variants. This strategy prevented dexamethasone-induced atrophy in both human and mouse skeletal muscle myotubes in vitro, demonstrating its therapeutic capabilities as a regulator of muscle differentiation. For cases in which multiple transcriptional start sites regulate mRNA transcript variant expression, we showed that CRISPR-Cas9 gene activation can selectively express a specific mRNA variant on the sole basis of its proximity to a transcriptional start site. This novel approach for enhancing IGF1 signaling has potential therapeutic applications in vivo.
ACKNOWLEDGMENTS:
We would like to thank Nicole Prodan and Ahad Azimuddin for their technical support. Nicole Stancel, PhD, ELS of the Section of Scientific Publications at Texas Heart Institute provided editorial support.
FUNDING:
The research reported in this manuscript was supported in part by a grant from the Center for the Advancement of Science in Space (R.J.S). Research was also supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) under Award Number R15 HL141963 to (B.K.M.), R15 HL124458 to (B.K.M.), American Heart Association (AHA) under Award Number 18AIREA 33960175 (B.K.M.), and a grant from Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (B.K.M.). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
ABBREVIATIONS:
- ANOVA
Analysis of Variance
- BCA
Bicinchoninic Acid Assay
- dCas9
Cas9
- CRISPR
Clustered Regularly-Interspaced Short Palindromic Repeats
- crRNA
CRISPR RNA
- GO
Gene Ontology
- DEX
Dexamethasone
- DAPI
4′,6-Diamidino-2-Phenylindole
- ECL
Enhanced Chemiluminescence
- ELISA
Enzyme-Linked Immunosorbent Assay
- FOXO1
Forkhead Box Protein O1
- HSF1
Heat-Shock Factor 1
- HRP
Horseradish Peroxidase
- HSMMs
Human skeletal myoblasts
- IGF1
Insulin-like Growth Factor 1
- PBS
Phosphate-Buffered Saline
- RIPA
Radioimmunoprecipitation Assay Buffer
- RQ
Relative Quantity
- RNA-Seq
RNA Sequencing
- sgRNA
single-guide RNA
- SAM
Synergistic Activation Mediator
- tracrRNA
transactivating CRISPR RNA
- TSS
Transcriptional Start Site
- TBS
Tris Buffered Saline
Footnotes
COMPETING INTERESTS:
Authors declare no conflicts of interests.
REFERENCES:
- 1.Jackman RW, and Kandarian SC (2004) The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol 287, C834–843 [DOI] [PubMed] [Google Scholar]
- 2.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, and Goldberg AL (2004) Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18, 39–51 [DOI] [PubMed] [Google Scholar]
- 3.Fanzani A, Conraads VM, Penna F, and Martinet W. (2012) Molecular and cellular mechanisms of skeletal muscle atrophy: an update. J Cachexia Sarcopenia Muscle 3, 163–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egerman MA, and Glass DJ (2014) Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol 49, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lok C. (2015) Cachexia: The last illness. Nature 528, 182–183 [DOI] [PubMed] [Google Scholar]
- 6.Lee WS, and Kim J. (2017) Insulin-like growth factor-1 signaling in cardiac aging. Biochim Biophys Acta [DOI] [PubMed] [Google Scholar]
- 7.Demonbreun AR, and McNally EM (2017) Muscle cell communication in development and repair. Curr Opin Pharmacol 34, 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman ME, DeMayo F., Yin KC, Lee HM, Geske R., Montgomery C., and Schwartz RJ (1995) Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J Biol Chem 270, 12109–12116 [DOI] [PubMed] [Google Scholar]
- 9.Philippou A., and Barton ER (2014) Optimizing IGF-I for skeletal muscle therapeutics. Growth Horm IGF Res 24, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stitt TN, Drujan D., Clarke BA, Panaro F., Timofeyva Y., Kline WO, Gonzalez M., Yancopoulos GD, and Glass DJ (2004) The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14, 395–403 [DOI] [PubMed] [Google Scholar]
- 11.Cen Y., Liu J., Qin Y., Liu R., Wang H., Zhou Y., Wang S., and Hu Z. (2016) Denervation in Femoral Artery-Ligated Hindlimbs Diminishes Ischemic Recovery Primarily via Impaired Arteriogenesis. PLoS One 11, e0154941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrucki LW, Tsutsumi Y., Kalinowski L., Dean J., Gavin M., Sen S., Mendizabal M., Sinusas AJ, and Aikawa R. (2010) Analysis of angiogenesis induced by local IGF-1 expression after myocardial infarction using microSPECT-CT imaging. J Mol Cell Cardiol 48, 1071–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apel PJ, Ma J., Callahan M., Northam CN, Alton TB, Sonntag WE, and Li Z. (2010) Effect of locally delivered IGF-1 on nerve regeneration during aging: an experimental study in rats. Muscle Nerve 41, 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matheny RW Jr., Nindl BC, and Adamo ML (2010) Minireview: Mechano-growth factor: a putative product of IGF-I gene expression involved in tissue repair and regeneration. Endocrinology 151, 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabinovsky ED, Gelir E., Gelir S., Lui H., Kattash M., DeMayo FJ, Shenaq SM, and Schwartz RJ (2003) Targeted expression of IGF-1 transgene to skeletal muscle accelerates muscle and motor neuron regeneration. FASEB J 17, 53–55 [DOI] [PubMed] [Google Scholar]
- 16.Shavlakadze T., Winn N., Rosenthal N., and Grounds MD (2005) Reconciling data from transgenic mice that overexpress IGF-I specifically in skeletal muscle. Growth Horm IGF Res 15, 4–18 [DOI] [PubMed] [Google Scholar]
- 17.Oberbauer AM (2013) The Regulation of IGF-1 Gene Transcription and Splicing during Development and Aging. Front Endocrinol (Lausanne) 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brisson BK, and Barton ER (2012) Insulin-like growth factor-I E-peptide activity is dependent on the IGF-I receptor. PLoS One 7, e45588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldspink G. (2006) Impairment of IGF-I gene splicing and MGF expression associated with muscle wasting. Int J Biochem Cell Biol 38, 481–489 [DOI] [PubMed] [Google Scholar]
- 20.Wang H., La Russa M., and Qi LS (2016) CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 85, 227–264 [DOI] [PubMed] [Google Scholar]
- 21.Dominguez AA, Lim WA, and Qi LS (2016) Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17, 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konermann S., Brigham MD, Trevino AE, Joung J., Abudayyeh OO, Barcena C., Hsu PD, Habib N., Gootenberg JS, Nishimasu H., Nureki O., and Zhang F. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmittgen TD, and Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 24.Bioinformatics B. (2011) FastQC: a quality control tool for high throughput sequence data. Cambridge, UK: Babraham Institute [Google Scholar]
- 25.Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal 17, pp. 10–12 [Google Scholar]
- 26.Kim D., Langmead B., and Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pertea M., Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Y., Smyth GK, and Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 [DOI] [PubMed] [Google Scholar]
- 29.McCarthy DJ, Chen Y., and Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, and Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallego-Colon E., Sampson RD, Sattler S., Schneider MD, Rosenthal N., and Tonkin J. (2015) Cardiac-Restricted IGF-1Ea Overexpression Reduces the Early Accumulation of Inflammatory Myeloid Cells and Mediates Expression of Extracellular Matrix Remodelling Genes after Myocardial Infarction. Mediators Inflamm 2015, 484357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rae FK, Suhaimi N., Li J., Nastasi T., Slonimsky E., Rosenthal N., and Little MH (2012) Proximal tubule overexpression of a locally acting IGF isoform, Igf-1Ea, increases inflammation after ischemic injury. Growth Horm IGF Res 22, 6–16 [DOI] [PubMed] [Google Scholar]
- 33.Chakravarthy MV, Fiorotto ML, Schwartz RJ, and Booth FW (2001) Long-term insulin-like growth factor-I expression in skeletal muscles attenuates the enhanced in vitro proliferation ability of the resident satellite cells in transgenic mice. Mech Ageing Dev 122, 1303–1320 [DOI] [PubMed] [Google Scholar]
- 34.Barton ER (2006) Viral expression of insulin-like growth factor-I isoforms promotes different responses in skeletal muscle. J Appl Physiol (1985) 100, 1778–1784 [DOI] [PubMed] [Google Scholar]
- 35.Barton ER, DeMeo J., and Lei H. (2010) The insulin-like growth factor (IGF)-I E-peptides are required for isoform-specific gene expression and muscle hypertrophy after local IGF-I production. J Appl Physiol (1985) 108, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravarthy MV, Abraha TW, Schwartz RJ, Fiorotto ML, and Booth FW (2000) Insulin-like growth factor-I extends in vitro replicative life span of skeletal muscle satellite cells by enhancing G1/S cell cycle progression via the activation of phosphatidylinositol 3’-kinase/Akt signaling pathway. J Biol Chem 275, 35942–35952 [DOI] [PubMed] [Google Scholar]
- 37.Rommel C., Bodine SC, Clarke BA, Rossman R., Nunez L., Stitt TN, Yancopoulos GD, and Glass DJ (2001) Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3, 1009–1013 [DOI] [PubMed] [Google Scholar]
- 38.Kim AH, Khursigara G., Sun X., Franke TF, and Chao MV (2001) Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol Cell Biol 21, 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latres E., Amini AR, Amini AA, Griffiths J., Martin FJ, Wei Y., Lin HC, Yancopoulos GD, and Glass DJ (2005) Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem 280, 2737–2744 [DOI] [PubMed] [Google Scholar]
- 40.Li J., Wei H., Chesley A., Moon C., Krawczyk M., Volkova M., Ziman B., Margulies KB, Talan M., Crow MT, and Boheler KR (2007) The pro-angiogenic cytokine pleiotrophin potentiates cardiomyocyte apoptosis through inhibition of endogenous AKT/PKB activity. J Biol Chem 282, 34984–34993 [DOI] [PubMed] [Google Scholar]
- 41.Holmes B., Artinian N., Anderson L., Martin J., Masri J., Cloninger C., Bernath A., Bashir T., Benavides-Serrato A., and Gera J. (2012) Protor-2 interacts with tristetraprolin to regulate mRNA stability during stress. Cell Signal 24, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]