Abstract
Motivation
Automatic biomedical named entity recognition (BioNER) is a key task in biomedical information extraction. For some time, state-of-the-art BioNER has been dominated by machine learning methods, particularly conditional random fields (CRFs), with a recent focus on deep learning. However, recent work has suggested that the high performance of CRFs for BioNER may not generalize to corpora other than the one it was trained on. In our analysis, we find that a popular deep learning-based approach to BioNER, known as bidirectional long short-term memory network-conditional random field (BiLSTM-CRF), is correspondingly poor at generalizing. To address this, we evaluate three modifications of BiLSTM-CRF for BioNER to improve generalization: improved regularization via variational dropout, transfer learning and multi-task learning.
Results
We measure the effect that each strategy has when training/testing on the same corpus (‘in-corpus’ performance) and when training on one corpus and evaluating on another (‘out-of-corpus’ performance), our measure of the model’s ability to generalize. We found that variational dropout improves out-of-corpus performance by an average of 4.62%, transfer learning by 6.48% and multi-task learning by 8.42%. The maximal increase we identified combines multi-task learning and variational dropout, which boosts out-of-corpus performance by 10.75%. Furthermore, we make available a new open-source tool, called Saber that implements our best BioNER models.
Availability and implementation
Source code for our biomedical IE tool is available at https://github.com/BaderLab/saber. Corpora and other resources used in this study are available at https://github.com/BaderLab/Towards-reliable-BioNER.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
PubMed contains over 30 million publications and is growing rapidly. Accurate, automated text mining tools are needed to maximize discovery and unlock structured information from this massive volume of text (Cohen and Hunter, 2008; Rzhetsky et al., 2009). Biomedical named entity recognition (BioNER) is the task of identifying biomedical named entities—such as genes and gene products, diseases, chemicals and species—in raw text. Biomedical named entities have several characteristics that make their recognition in text challenging (Zhou et al., 2004), including the use of descriptive entity names (e.g. ‘normal thymic epithelial cells’) leading to ambiguous term boundaries, and several spelling forms for the same entity (e.g. ‘N-acetylcysteine’, ‘N-acetyl-cysteine’ and ‘NAcetylCysteine’). Many solutions for reliable BioNER have been proposed (e.g. Delėger et al., 2016; Kim et al., 2004) and current state-of-the-art approaches employ a domain-independent approach, based on deep learning and statistical word embeddings, called bidirectional long short-term memory network-conditional random field (BiLSTM-CRF; Habibi et al., 2017; Lample et al., 2016; Ma and Hovy, 2016) or modifications to this approach [e.g. transfer learning (Giorgi and Bader, 2018) and multi-task learning (Wang et al., 2018)].
While BiLSTM-CRF paired with pre-trained word embeddings as inputs is a powerful approach to sequence labeling (Huang et al., 2015), it has many trainable parameters which could lead to a reduction in generalizability. Here, we are concerned with the ability of a model to generalize from training on one corpus to testing on another for the same biomedical entity. This simulates a real-world scenario where the model is used to annotate text which is outside the corpus it was trained on, but still within the biomedical domain. Generalization across biomedical corpora appears to be a problem even for less-parameterized machine learning methods. For example, Gimli, an open-source tool for BioNER based on a CRF classifier, achieved an F1 score of 87.17% when trained and tested on the GENETAG corpus (Campos et al., 2013a), but only a 45–55% F1 score when trained on the GENETAG corpus and tested on the CRAFT corpus for genes and proteins (Campos et al., 2013b). Galea et al. (2018) explore this further by demonstrating that performance of a CRF model for BioNER trained on individual corpora decreases substantially for recognition of the same biomedical entity in independent corpora. They conclude that bias in the available BioNER evaluation corpora is partly to blame. In a simple orthographic feature analysis (i.e. what does a word look like?), they find that features which significantly predict biomedical named entities in one corpus (e.g. number of digits or capital letters, text span length) are not necessarily useful to predict those same entities in a different corpus. We need to address this problem if BiLSTM-CRF models are to be useful in real-world scenarios involving the large-scale annotation of diverse articles.
To address this challenge, we evaluate successful ideas from recent work on BiLSTM-CRF models for BioNER (Giorgi and Bader, 2018; Habibi et al., 2017; Wang et al., 2018) and sequence labeling with BiLSTM-CRFs in general (Reimers and Gurevych, 2017) and propose several model-based strategies to improve generalization, namely: additional regularization via variational dropout, transfer learning and multi-task learning. We assessed the performance of the model on the same corpus it was trained on (‘in-corpus’ performance) and when trained on one corpus and tested on another corpus annotated for the same entity class (‘out-of-corpus’ performance). All proposed strategies achieved an improvement in out-of-corpus performance without degrading the average in-corpus performance. The best improvement resulted from a combination of multi-task learning and variational dropout, which boosts out-of-corpus performance by 10.75%. We make available to the community a user-friendly, open-source tool for BioNLP (‘Saber’) which incorporates these successful model features (https://github.com/BaderLab/saber).
2 Materials and methods
We evaluated three modifications to a BiLSTM-CRF model’s architecture and training strategy aimed at improving generalization, described briefly below (details in Supplementary Material). We use the BiLSTM-CRF neural network architecture introduced by Lample et al. (2016) as our baseline (BL) model (Supplementary Fig. S1).
2.1 Variational dropout
As neural networks have many parameters, model regularization is critical for generalization performance. Traditionally, the ‘dropout’ technique, which randomly drops units during training, is used for this purpose (Srivastava et al., 2014). Previous applications of BiLSTM-CRF models for BioNER (Giorgi and Bader, 2018; Habibi et al., 2017) have only applied dropout to the character-enhanced word embeddings, the final inputs to the word-level BiLSTM layers, as originally proposed by Lample et al. (2016). However, no regularization technique is applied to the recurrent layers of the model, which contain the majority of the model’s trainable parameters. Part of the reason for this is likely that the standard dropout implementation is ineffective for regularizing recurrent connections, as it disrupts the recurrent neural networks ability to retain long-term dependencies (Bayer et al., 2013; Pachitariu and Sahani, 2013; Zaremba et al., 2014). Variational dropout, where the same units are dropped across multiple time steps (in our case, tokens of an input sentence), has been proposed to overcome this (Gal and Ghahramani, 2016) (Supplementary Fig. S2). We hypothesize that regularizing the recurrent layers of the BiLSTM-CRF model via variational dropout will improve out-of-corpus generalization. To test this, we compare the performance of two BiLSTM-CRF models for the task of BioNER: one with the standard dropout strategy (Giorgi and Bader, 2018; Habibi et al., 2017; Lample et al., 2016), and a second where variational dropout is additionally applied to the input, recurrent and output connections of all recurrent layers.
2.2 Transfer learning
Transfer learning is a machine learning research problem which aims to perform a task on a ‘target’ dataset using knowledge learned from a ‘source’ dataset (Li, 2012; Pan and Yang, 2010; Weiss et al., 2016). Ideally, transfer learning reduces training time on the target dataset and the amount of labeled data needed to obtain high performance. It can be used to improve model generalization by training on very large, but usually lower quality silver standard corpus (SSC), and then using the learned parameters to initialize training on a smaller, manually generated and more reliable gold-standard corpus (GSC). We recently showed that transfer learning for BioNER (Giorgi and Bader, 2018) reduces the amount of labeled data needed to achieve high performance from a BiLSTM-CRF model, but we did not assess its impact on generalizability. As in Giorgi and Bader (2018), here we apply transfer learning by first training on a large, semi-automatically generated and lower quality SSC and then transfer to continued training on a GSC. Like Giorgi and Bader (2018), we use CALBC-SSC-III (Collaborative Annotation of a Large Biomedical Corpus) as our SSC (Kafkas et al., 2012; Rebholz-Schuhmann et al., 2010).
2.3 Multi-task learning
Multi-task learning (Caruana, 1993) is a machine learning method in which multiple learning tasks are solved at the same time. The idea is that by sharing representations between tasks, we can exploit commonalities, leading to improved learning efficiency, prediction accuracy and generalizability for the task-specific models, when compared to training the models separately (Baxter et al., 2000; Caruana, 1997; Thrun, 1996). Recently, Crichton et al. (2017) demonstrated that a neural network multi-task convolutional neural network (CNN) model outperforms a comparable single-task model, on average, for the task of BioNER. Similarly, Wang et al. (2018) found that multi-task learning outperforms single-task learning for BioNER with a BiLSTM-CRF. Neither study explored the effect of multi-task learning on the model’s ability to generalize across corpora. We hypothesize that multi-task learning will improve across-corpora generalization, potentially as a result of exposing the model to more training data. To test this, we compare the performance of a single-task and multi-task BiLSTM-CRF model for the task of BioNER.
Our multi-task model (MTM) builds off this BiLSTM-CRF architecture. The MTM is a global model comprised of distinct, task-specific input and output layers, while the hidden layers (and their parameters) are shared across all tasks (Supplementary Fig. S3). We follow Wang et al. (2018) who found it best to share all hidden layers of the BiLSTM-CRF model for BioNER. During training, all corpora are used to update the parameters of the hidden layers of the model but the output ‘task’ layers are trained using only their corresponding corpus. In our multi-task experiments, two corpora of the same entity type are used to train the model. The ‘train’ corpus is used to train the output layer that is then evaluated on the ‘test’ corpus. The ‘partner’ corpus contributes to hidden layer training and is used to train its output layer, but the corresponding output is not used for performance evaluation. During training, the model optimizes the log-probability of the correct tag sequence for each corpus. In practice, the model is trained in an end-to-end fashion.
3 Results
3.1 Establishing a baseline
To establish a baseline for each corpus used in this study, we performed 5-fold cross-validation using the hyperparameters presented in Supplementary Material, Section 3.5. We use the dropout strategy proposed by Lample et al. (2016) and employed by Habibi et al. (2017) and Giorgi and Bader (2018), which applies a single dropout layer (with a dropout rate of 0.3) to the character-enhanced word embeddings, which is the final input to the word-level BiLSTM layers. In Supplementary Table S3, we present our baseline F1 scores along with the best reported F1 scores from Habibi et al. (2017), Wang et al. (2018) and Giorgi and Bader (2018), all of whom use nearly identical BiLSTM-CRF models and the same word embeddings as used in this study, and Crichton et al. (2017), who used a CNN-based model for BioNER. Our baseline significantly outperforms previous results obtained with BiLSTM-CRF models for 7 out of the 12 corpora evaluated and was comparable to the best method in the remaining 5 cases. Since our architecture is nearly identical and paired with the same word embeddings, this is likely due to our hyperparameter choice as presented by Reimers and Gurevych (2017). For the remainder of the study, we compare all performance scores to our baseline.
3.2 Can BiLSTM-CRF generalize for BioNER?
To test the out-of-corpus generalizability of a BiLSTM-CRF model for BioNER, we trained the model on various corpora and evaluated its performance on independent corpora annotated for the same entity type. Even when corpora are annotated for the same entity type, they may have different biases, given that they are typically independently developed with different annotation guidelines. To control for the expected drop in performance due to differing annotation guidelines, we used a relaxed matching criterion in our evaluation (described in Supplementary Material, Section 3.7). We found that, even with a relaxed matching criterion, performance of the model as measured by F1 score falls by an average of 31.16% when the model is evaluated on a corpus other than the one it was trained on (Supplementary Table S4). Out-of-corpus performance was worst for chemicals, falling by an average of 33.45%, followed by genes/proteins (29.27%), species (28.47%) and disease (26.09%). These results demonstrate the dramatically poor out-of-corpus generalizability of BiLSTM-CRF for BioNER, even though the model obtains state-of-the-art results when trained and tested on the same corpus (Supplementary Table S3).
3.3 Improved regularization
We next explored the effect that additional regularization of the BiLSTM-CRF model via variational dropout (see Section 2.1) has on both in-corpus and out-of-corpus performance. In Table 1, we compare the in-corpus performance of the baseline (BL) BiLSTM-CRF model employing a simple dropout strategy—proposed by Lample et al. (2016) and used in previous applications of BiLSTM-CRF to BioNER (Giorgi and Bader, 2018; Habibi et al., 2017)—compared to a model in which variational dropout has been additionally applied to the recurrent layers. We use dropout ratios of 0.3, 0.3 and 0.1 for the input, output and recurrent connections, respectively. In Table 2, we measure the effect that variational dropout has on the generalizability of the model, by comparing out-of-corpus performance to the baseline.
Table 1.
In-corpus (IC) performance, measured by F1-score, of the baseline (BL) BiLSTM-CRF compared to a BiLSTM-CRF with variational dropout (VD)
| BL |
VD |
||||
|---|---|---|---|---|---|
| Entity | Corpus | Average | σ | Average | σ |
| Chemicals | BC4CHEMD | 88.46 | 0.61 | 88.71 | 0.76 |
| BC5CDR | 92.82 | 0.80 | 93.08 | 0.82 | |
| CRAFT | 84.98 | 1.98 | 85.22 | 1.37 | |
| Disease | BC5CDR | 84.49 | 0.33 | 85.10 | 0.56 |
| NCBI-disease | 87.01 | 1.17 | 87.60 | 1.50 | |
| Variome | 85.75 | 2.83 | 85.69 | 3.81 | |
| Species | CRAFT | 96.28 | 2.21 | 96.38 | 2.26 |
| Linnaeus | 89.44 | 3.91 | 89.66 | 7.47 | |
| S800 | 72.75 | 2.42 | 77.39 | 4.17 | |
| Genes/proteins | BC2GM | 81.48 | 0.48 | 83.10 ** | 0.50 |
| CRAFT | 84.46 | 6.08 | 86.09 | 5.19 | |
| JNLPBA | 80.92 | 2.50 | 81.95 | 2.62 | |
Note: In the BL model, dropout is applied only to the character-enhanced word embeddings. In the VD model, dropout is additionally applied to the input, recurrent and output connections of all LSTM layers. IC performance is derived from 5-fold cross-validation, using exact matching criteria. Statistical significance is measured through a two-tailed t-test. Bold, best scores, σ, standard deviation.
Significantly different than the BL (P ≤ 0.01).
Table 2.
Out-of-corpus (OOC) performance, measured by F1 score, of the baseline (BL) BiLSTM-CRF compared to a BiLSTM-CRF with variational dropout (VD)
| Entity | Train | Test | BL | VD | |
|---|---|---|---|---|---|
| Chemicals | BC4CHEMD | BC5CDR | 90.90 | 90.61 | −0.29 |
| CRAFT | 47.44 | 47.67 | 0.23 | ||
| BC5CDR | BC4CHEMD | 71.81 | 72.41 | 0.60 | |
| CRAFT | 39.55 | 41.30 ** | 1.74 | ||
| CRAFT | BC4CHEMD | 40.50 | 42.65 | 2.14 | |
| BC5CDR | 41.59 | 56.64 ** | 15.05 | ||
| Diseases | BC5CDR | NCBI-disease | 76.67 | 80.86 * | 4.19 |
| Variome | 74.03 | 74.83 | 0.81 | ||
| NCBI-disease | BC5CDR | 69.62 | 74.96 ** | 5.33 | |
| Variome | 74.98 | 75.69 | 0.72 | ||
| Variome | BC5CDR | 22.45 | 30.38 * | 7.93 | |
| NCBI-disease | 40.17 | 45.16 ** | 4.99 | ||
| Species | CRAFT | Linnaeus | 45.32 | 53.25 * | 7.93 |
| S800 | 36.88 | 46.10 ** | 9.21 | ||
| Linnaeus | CRAFT | 82.49 | 82.85 | 0.36 | |
| S800 | 62.90 | 66.93 * | 4.02 | ||
| S800 | CRAFT | 57.09 | 76.44 ** | 19.34 | |
| Linnaeus | 61.43 | 67.05 * | 5.62 | ||
| Genes/proteins | BC2GM | CRAFT | 56.04 | 58.17 | 2.12 |
| JNLPBA | 69.77 | 70.79 ** | 1.02 | ||
| CRAFT | BC2GM | 44.11 | 49.12 * | 5.01 | |
| JNLPBA | 52.88 | 56.30 | 3.42 | ||
| JNLPBA | BC2GM | 51.03 | 55.61 | 4.57 | |
| CRAFT | 44.29 | 49.08 | 4.79 |
Note: In the BL model, dropout is applied only to the character-enhanced word embeddings. In the VD model, dropout is additionally applied to the input, recurrent and output connections of all LSTM layers. OOC performance is derived by training on one corpus (train) and testing on another annotated for the same entity type (test) using a relaxed, right-boundary matching criteria. Bold, best scores.
Significantly different than the BL (P ≤ 0.05).
Significantly different than the BL (P ≤ 0.01).
In general, variational dropout has a small positive impact on in-corpus performance. For at least two corpora, S800 and BC2GM, variational dropout leads to a large improvement in performance, but only the latter case is statistically significant. For out-of-corpus performance, variational dropout improves performance for nearly every train/test corpus pair we evaluated, with an average F1 score improvement of 4.62%. In some cases, variational dropout leads to sizable improvements in out-of-corpus performance, such as when the model was trained on S800 and tested on CRAFT (19.34%), or trained on CRAFT and tested on BC5CDR (15.05%). In one case—when trained to recognize chemicals on BC4CHEMD and tested on BC5CDR—variational dropout reduced out-of-corpus performance, although the performance difference was minimal (<0.5%). Thus, variational dropout improves the out-of-corpus performance of the model, without degrading in-corpus performance. Regularization of the recurrent layers of a BiLSTM-CRF model via variational dropout, therefore, can improve model generalizability for BioNER.
3.4 Transfer learning
In this experiment, we measure the effect that a transfer learning strategy for BiLSTM-CRF has on both in-corpus and out-of-corpus performance. In Table 3, we compare the in-corpus performance of the baseline BiLSTM-CRF model (BL) to that of the model trained with transfer learning and in Table 4 we similarly compare out-of-corpus performance of the two models. In the transfer learning setting, the model was pre-trained on the CALBC-SSC-III corpus, which is annotated for chemicals, diseases, species and genes/proteins, before being trained on one of the 12 GSCs, each annotated for a single entity class. The state of the optimizer is reset during this transfer, but model weights for all layers beside the final CRF layer are retained (see Section 2.2 and Supplementary Material, Section 3.5).
Table 3.
In-corpus (IC) performance, measured by F1-score, of the baseline (BL) BiLSTM-CRF compared to a BiLSTM-CRF trained with transfer learning (TL)
| BL |
TL |
||||
|---|---|---|---|---|---|
| Entity | Corpus | Average | σ | Average | σ |
| Chemicals | BC4CHEMD | 88.46 | 0.61 | 88.98 | 0.63 |
| BC5CDR | 92.82 | 0.80 | 92.20 | 0.86 | |
| CRAFT | 84.98 | 1.98 | 85.80 | 1.74 | |
| Disease | BC5CDR | 84.49 | 0.33 | 84.41 | 0.24 |
| NCBI-disease | 87.01 | 1.17 | 87.66 | 0.86 | |
| Variome | 85.75 | 2.83 | 86.69 | 3.03 | |
| Species | CRAFT | 96.28 | 2.21 | 96.55 | 1.72 |
| Linnaeus | 89.44 | 3.91 | 90.72 | 4.90 | |
| S800 | 72.75 | 2.42 | 74.93 | 3.27 | |
| Genes/proteins | BC2GM | 81.48 | 0.48 | 80.65* | 0.57 |
| CRAFT | 84.46 | 6.08 | 85.50 | 4.59 | |
| JNLPBA | 80.92 | 2.50 | 81.56 | 2.73 | |
Note: The TL model was pre-trained on the CALBC-Small-III corpus. IC performance is derived from 5-fold cross-validation, using exact matching criteria. Statistical significance is measured through a two-tailed t-test. Bold, best scores, σ, standard deviation.
Significantly different than the BL (P ≤ 0.05).
Table 4.
Out-of-corpus (OOC) performance, measured by F1 score, of the baseline (BL) BiLSTM-CRF compared to a BiLSTM-CRF trained with transfer learning (TL)
| Entity | Train | Test | BL | TL | |
|---|---|---|---|---|---|
| Chemicals | BC4CHEMD | BC5CDR | 90.90 | 90.73 | −0.17 |
| CRAFT | 47.44 | 47.02 | −0.42 | ||
| BC5CDR | BC4CHEMD | 71.81 | 74.27 * | 2.46 | |
| CRAFT | 39.55 | 41.20 * | 1.64 | ||
| CRAFT | BC4CHEMD | 40.50 | 46.15 * | 5.64 | |
| BC5CDR | 41.59 | 58.57 ** | 16.98 | ||
| Diseases | BC5CDR | NCBI-disease | 76.67 | 78.51 * | 1.83 |
| Variome | 74.03 | 77.18 | 3.16 | ||
| NCBI-disease | BC5CDR | 69.62 | 73.19 | 3.56 | |
| Variome | 74.98 | 76.95 * | 1.97 | ||
| Variome | BC5CDR | 22.45 | 50.28 ** | 27.83 | |
| NCBI-disease | 40.17 | 58.64 ** | 18.47 | ||
| Species | CRAFT | Linnaeus | 45.32 | 53.37 | 8.04 |
| S800 | 36.88 | 46.46 ** | 9.57 | ||
| Linnaeus | CRAFT | 82.49 | 83.07 | 0.57 | |
| S800 | 62.90 | 67.64 * | 4.73 | ||
| S800 | CRAFT | 57.09 | 69.56 ** | 12.47 | |
| Linnaeus | 61.43 | 67.21 * | 5.78 | ||
| Genes/proteins | BC2GM | CRAFT | 56.04 | 56.84 | 0.79 |
| JNLPBA | 69.77 | 70.27 | 0.50 | ||
| CRAFT | BC2GM | 44.11 | 49.69 * | 5.58 | |
| JNLPBA | 52.88 | 57.91 * | 5.03 | ||
| JNLPBA | BC2GM | 51.03 | 57.81 * | 6.78 | |
| CRAFT | 44.29 | 56.90 ** | 12.61 |
Note: The TL model was pre-trained on the CALBC-Small-III corpus. OOC performance is derived by training on one corpus (train) and testing on another annotated for the same entity type (test) using a relaxed, right-boundary matching criteria. Statistical significance is measured through a two-tailed t-test. Bold, best scores.
Significantly different than the BL (P ≤ 0.05).
Significantly different than the BL (P ≤ 0.01).
In general, transfer learning had a small positive effect on in-corpus performance, boosting the average F1 score by ∼1%. In contrast, transfer learning had a large positive effect on out-of-corpus performance, improving performance for nearly every train/test pair we evaluated for an average improvement of 6.48%. In a handful of cases, such as when the model was trained on the CRAFT corpus and tested on the BC5CDR corpus, performance improved by over 10%. In a single case (i.e. when the model was trained on Variome and tested on BC5CDR) transfer learning doubled out-of-corpus performance over the baseline. Thus, the use of transfer learning improves the generalizability of BiLSTM-CRF models for BioNER, in some cases dramatically, while preserving in-corpus performance.
3.5 Multi-task learning
To assess the effect that a multi-task learning strategy for BiLSTM-CRF has on both in-corpus and out-of-corpus performance, we evaluate a model trained on all corpus pairs within an entity class. Each model is simultaneously trained on a ‘train’ and ‘partner’ corpus, each defined as a separate task, and a ‘test’ corpus is used to evaluate performance on the ‘train’ task (see Section 2.3 and Supplementary Material, Section 3.5). In Table 5 we compare the in-corpus performance of the single-task, baseline BiLSTM-CRF model, to that of the MTM. In Table 6 we similarly compare out-of-corpus performance of the BL and MTM.
Table 5.
In-corpus (IC) performance, measured by F1-score, of the baseline (BL) BiLSTM-CRF compared to the multi-task model (MTM)
| BL |
MTM |
|||||
|---|---|---|---|---|---|---|
| Entity | Train | Partner | Average | σ | Average | σ |
| Chemicals | BC4CH. | BC5CDR | 88.46 | 0.61 | 88.81 | 0.60 |
| CRAFT | — | — | 88.67 | 0.50 | ||
| BC5CDR | BC4CH. | 92.82 | 0.80 | 93.00 | 0.55 | |
| CRAFT | — | — | 91.52* | 0.68 | ||
| CRAFT | BC4CH. | 84.98 | 1.98 | 85.06 | 1.49 | |
| BC5CDR | — | — | 84.74 | 1.33 | ||
| Diseases | BC5CDR | NCBI-disease | 84.49 | 0.33 | 83.85 | 0.64 |
| Variome | — | — | 83.29* | 0.80 | ||
| NCBI-disease | BC5CDR | 87.01 | 1.17 | 86.89 | 1.74 | |
| Variome | — | — | 86.27 | 1.44 | ||
| Variome | BC5CDR | 85.75 | 2.83 | 86.13 | 2.49 | |
| NCBI-disease | — | — | 85.73 | 2.46 | ||
| Species | CRAFT | Linnaeus | 96.28 | 2.21 | 96.82 | 1.51 |
| S800 | — | — | 96.90 | 1.31 | ||
| Linnaeus | CRAFT | 89.44 | 3.91 | 89.72 | 4.51 | |
| S800 | — | — | 92.18 | 3.42 | ||
| S800 | CRAFT | 72.75 | 2.42 | 74.80 | 2.98 | |
| Linnaeus | — | — | 74.43 | 1.90 | ||
| Genes/proteins | BC2GM | CRAFT | 81.48 | 0.48 | 79.41** | 0.14 |
| JNLPBA | — | — | 79.60** | 0.53 | ||
| CRAFT | BC2GM | 84.46 | 6.08 | 87.76 | 2.65 | |
| JNLPBA | — | — | 85.36 | 4.74 | ||
| JNLPBA | BC2GM | 80.92 | 2.50 | 81.61 | 2.53 | |
| CRAFT | — | — | 81.15 | 2.04 | ||
Note: The MTM is trained on pairs of corpora (train, partner), where each corpus is used during training to update the parameters of all hidden layers. IC performance is derived from 5-fold cross-validation, using exact matching criteria. Statistical significance is measured through a two-tailed t-test. Bold, best scores, σ, standard deviation.
Significantly different than the BL (P ≤ 0.05).
Significantly different than the BL (P ≤ 0.01).
Table 6.
Out-of-corpus (OOC) performance, measured by F1 score, of the baseline (BL) BiLSTM-CRF compared to the multi-task model (MTM)
| Entity | Train | Partner | Test | BL | MTM | |
|---|---|---|---|---|---|---|
| Chemicals | BC4CHEMD | BC5CDR | CRAFT | 47.44 | 47.74 | 0.29 |
| CRAFT | BC5CDR | 90.90 | 90.97 | 0.07 | ||
| BC5CDR | BC4CHEMD | CRAFT | 39.55 | 44.79 ** | 5.24 | |
| CRAFT | BC4CHEMD | 71.81 | 72.54 | 0.73 | ||
| CRAFT | BC4CHEMD | BC5CDR | 41.59 | 71.68 ** | 30.09 | |
| BC5CDR | BC4CHEMD | 40.50 | 49.80 ** | 9.30 | ||
| Diseases | BC5CDR | NCBI-disease | Variome | 74.03 | 76.84 * | 2.81 |
| Variome | NCBI-disease | 76.67 | 77.33 | 0.66 | ||
| NCBI-disease | BC5CDR | Variome | 74.98 | 76.32 * | 1.34 | |
| Variome | BC5CDR | 69.62 | 70.72 | 1.10 | ||
| Variome | BC5CDR | NCBI-disease | 40.17 | 69.35 ** | 29.18 | |
| NCBI-disease | BC5CDR | 22.45 | 60.06 ** | 37.61 | ||
| Species | CRAFT | Linnaeus | S800 | 36.88 | 50.26 | 13.38 |
| S800 | Linnaeus | 45.32 | 57.80 ** | 12.48 | ||
| Linnaeus | CRAFT | S800 | 62.90 | 67.90 | 4.99 | |
| S800 | CRAFT | 82.49 | 82.69 | 0.20 | ||
| S800 | CRAFT | Linnaeus | 61.43 | 67.90 ** | 6.46 | |
| Linnaeus | CRAFT | 57.09 | 80.04 ** | 22.94 | ||
| Genes/proteins | BC2GM | CRAFT | JNLPBA | 69.77 | 70.26 | 0.49 |
| JNLPBA | CRAFT | 56.04 | 57.17 | 1.12 | ||
| CRAFT | BC2GM | JNLPBA | 52.88 | 58.78 * | 5.89 | |
| JNLPBA | BC2GM | 44.11 | 45.12 | 1.01 | ||
| JNLPBA | BC2GM | CRAFT | 44.29 | 52.78 * | 8.49 | |
| CRAFT | BC2GM | 51.03 | 57.35 ** | 6.32 |
Note: The MTM is trained on pairs of corpora (train, partner), where each corpus is used during training to update the parameters of all hidden layers, but only the train corpus task is used for evaluation on another corpus annotated for the same entity type (test) using a relaxed, right-boundary matching criteria. Bold, best scores.
Significantly different than the BL (P ≤ 0.05).
Significantly different than the BL (P ≤ 0.01).
Multi-task learning, as applied here, appears to have little impact on in-corpus performance. Average performance of the BL and the MTM were nearly identical, at 85.74 and 85.99% respectively, though in a few cases, such as when the model was trained on BC2GM alongside CRAFT or JNLPBA, the MTM significantly underperforms the baseline. On the other hand, multi-task learning improved out-of-corpus performance for every train/partner/test corpus set we evaluated, with an average improvement of 8.42%. In some cases, this improvement was substantial, such as when the model was trained on the Variome and NCBI-Disease corpora and tested on the BC5CDR corpus (37.61%). However, we do observe significant variability overall in the degree of improvement, suggesting that multi-task learning is sensitive to the choice of train/partner pairs. Thus, a multi-task learning strategy of simultaneously training on multiple corpora can substantially boost out-of-corpus performance of BiLSTM-CRF models for BioNER.
3.6 Combining modifications
We next evaluate if combinations of the above modifications improve BiLSTM-CRFs model performance above individual modifications (Fig. 1). In general, all combinations of the proposed modifications improve average out-of-corpus performance without degrading in-corpus performance. However, not all combinations are additive. For example, multi-task learning improves out-of-corpus performance by 8.42%, transfer learning by 6.48% but together only by 5.61%. The biggest boost to out-of-corpus performance is achieved by the MTM paired with additional regularization of the recurrent layers via variational dropout, improving average performance by 10.75%. Therefore, we recommend using this combination to produce a model with the highest expected out-of-corpus performance.
Fig. 1.
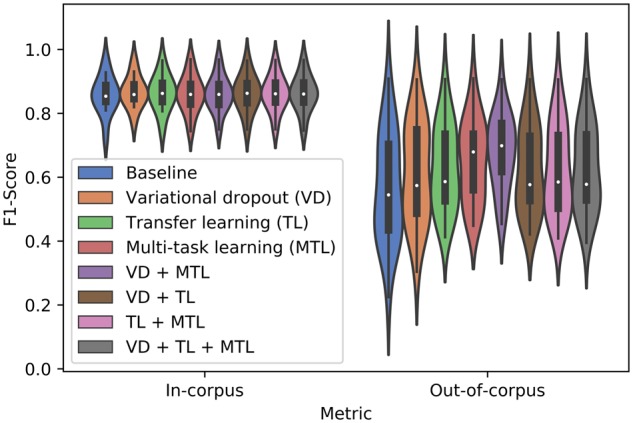
Violin plot of the average in-corpus (IC) and out-of-corpus (OOC) performance, measured by F1 score, of the BiLSTM-CRF model. IC performance is derived from 5-fold cross-validation, using exact matching criteria. OOC performance is derived by training on one corpus (train) and testing on another corpus annotated for the same entity type (test) using a relaxed, right-boundary matching criterion. The average performance of a model employing one of each of the proposed modifications: variational dropout (VD), transfer learning (TL) and multi-task learning (MTL) independently as well as models which employ all combinations of these methods are shown
4 Discussion
We demonstrate that BiLSTM-CRF, a popular deep learning-based approach to BioNER and sequence labeling in general, does a poor job generalizing to corpora other than the one it was trained on. While some drop in performance is expected whenever a model is evaluated on data outside the train set, the magnitude of decrease we observed was substantial, falling by an average of over 30% F1 score. This was true even though we used a liberal scoring criterion and any two corpora in our experiments were annotated for the same entity type (and sometimes even used similar annotator guidelines, such as the case with BC5CDR and NCBI-Disease).
There are two possible explanations for the poor out-of-corpus generalization we observed—either the corpora are biased or insufficient or the model is prone to training in a corpora specific manner. Likely both reasons contribute to the problem. We elected to explore improving the model because corpus creation is extremely laborious and modification of existing corpora would make it difficult to compare our methods against existing solutions. The three modifications we evaluated—variational dropout, transfer learning and multi-task learning—are straightforward to implement and improve across-corpora generalizability without degrading average in-corpus performance. On average, variational dropout improves out-of-corpus performance by 4.62%, transfer learning by 6.48% and multi-task learning by 8.42%, and the best combination of these (multi-task learning and variational dropout) improves average out-of-corpus performance by 10.75%. We provide our model to the community as an easy-to-use tool for BioNLP under a permissive MIT license (https://github.com/BaderLab/saber).
We note three limitations of our work. First, the most promising modification to BiLSTM-CRF, multi-task learning, appears to be sensitive to the choice of train/partner corpus pairs. A user should, therefore, evaluate multiple train/partner corpora to determine the most beneficial combination for their use case. Second, while transfer learning is not part of our recommended combination, we previously found it to significantly improve performance on smaller corpora (<6000 labels) (Giorgi and Bader, 2018). While we did not evaluate the effect of corpus size, presumably, transfer learning would still be useful and should be considered for small corpora. Third, in our multi-task experiments, we restricted ourselves to training only on two corpora, which were annotated for the same entity type. These restrictions were made only to keep the number and the training time of the multi-task experiments within a reasonable range and not because of inherent limitations in the model’s architecture. Previous work has suggested that multi-task learning with deep neural networks for the task of BioNER may increase performance even when trained on corpora that do not annotate the same entity class. Crichton et al. (2017) report, for example, that the best partner for Linnaeus (Species) out of 15 corpora was NCBI-Disease (Disease), not another Species corpus. Additionally, previous work (Wang et al., 2018) has suggested that training a BiLSTM-CRF on many corpora (i.e. >2) in a multi-task setting leads to sizable improvements in BioNER. Thus, a further direction for our work could be to explore performance improvements as increasing numbers of corpora annotated for different entity types are used for training. We suspect that this could significantly boost out-of-corpus performance. Further, these results suggest that a single model trained on many corpora (and even additionally pre-trained on an extremely large SSC) may produce a robust and reliable tagger suitable for deployment on massive literature databases (such as PubMed).
Our strategy for transfer learning involves pre-training a model on a large SSC and transferring the learned weights to initialize training on a smaller, but typically much higher quality, GSC. As we were writing this paper, a novel transfer learning strategy for NLP demonstrated state-of-the-art performance on many benchmark corpora (https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf; Devlin et al., 2018; Howard and Ruder, 2018). This transfer learning strategy involves first training a language model on a massive corpus of unlabeled text. The task of a language model is to predict the next most probable word given a sequence of words (or more recently, to predict randomly masked words). By learning this task, the model is required to capture both syntax and semantics and is also required to encode something akin to common sense. This is followed by the addition of task-specific layers, which take the output of the language model as input and are trained on labeled data in order for the model to learn a specific classification task such as NER. This transfer learning strategy has already been applied to BioNER with some success (Lee et al., 2019; Sachan et al., 2018). In the future, we plan to explore this transfer learning strategy for BioNER, and also for other tasks in the biomedical text mining pipeline, such as relation and event extraction.
5 Conclusion
While BioNER has recently made substantial advances in performance with the application of deep learning, current applications suffer from poor generalizability in real-world scenarios. We show that out-of-corpus performance of a BiLSTM-CRF model for BioNER (training on one corpus and testing on another annotated for the same entity type) suffers when using a current state-of-the-art model. Straightforward model modifications (variational dropout, transfer learning, and multi-task learning and their combinations) substantially improve across-corpora generalization performance. We propose that our model will significantly outperform previous applications of BiLSTM-CRF models to BioNER when deployed for the large-scale annotation of widely diverse articles, such as the articles found in databases like PubMed. We make our model accessible as an easy-to-use BioNLP tool, Saber (https://github.com/BaderLab/saber).
Supplementary Material
Acknowledgements
This research was enabled in part by support provided by Compute Ontario (https://computeontario.ca/) and Compute Canada (www.computecanada.ca). We also gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research.
Funding
This research was funded by the US National Institutes of Health [grant number 5U41 HG006623-02].
Conflict of Interest: none declared.
References
- Baxter J. et al. (2000) A model of inductive bias learning. J. Artif. Intell. Res., 12, 3. [Google Scholar]
- Bayer J. et al. (2013) On fast dropout and its applicability to recurrent networks. arXiv preprint arXiv: 1311.0701.
- Campos D. et al. (2013a) Gimli: open source and high-performance biomedical name recognition. BMC Bioinformatics, 14, 54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos D. et al. (2013b) A modular framework for biomedical concept recognition. BMC Bioinformatics, 14, 281.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana R. (1993) Multitask learning: a knowledge-based source of inductive bias. In: Proceedings of the Tenth International Conference on Machine Learning, pp. 41–48. Morgan Kaufmann, Citeseer.
- Caruana R. (1997) Multitask learning Mach. Learn., 28, 41–75. [Google Scholar]
- Cohen K.B., Hunter L. (2008) Getting started in text mining. PLoS Comput. Biol., 4, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton G. et al. (2017) A neural network multi-task learning approach to biomedical named entity recognition. BMC Bioinformatics, 18, 368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delėger L. et al. (2016) Overview of the bacteria biotope task at BioNLP shared task 2016. In: Proceedings of the 4th BioNLP Shared Task Workshop, pp. 12–22.
- Devlin J. et al. (2018) BERT: pre-training of deep bidirectional transformers for language understanding. CoRR, abs/1810.04805.
- Gal Y., Ghahramani Z. (2016) A theoretically grounded application of dropout in recurrent neural networks In: Lee D.D.et al. (eds.), Advances in Neural Information Processing Systems, Vol. 29. Curran Associates, Inc, pp. 1019–1027. [Google Scholar]
- Galea D. et al. (2018) Exploiting and assessing multi-source data for supervised biomedical named entity recognition. Bioinformatics, 34, 2474–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi J.M., Bader G.D. (2018) Transfer learning for biomedical named entity recognition with neural networks. Bioinformatics, 34, 4087–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi M. et al. (2017) Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics, 33, i37–i48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J., Ruder S. (2018) Universal language model fine-tuning for text classification. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, Melbourne, Australia, pp. 328–339.
- Huang Z. et al. (2015) Bidirectional LSTM-CRF models for sequence tagging. CoRR, abs/1508.01991.
- Kafkas S. et al. (2012) CALBC: releasing the final corpora. In: Proceedings of the Eighth International Conference on Language Resources and Evaluation (LREC-2012) European Languages Resources Association (ELRA), Istanbul, Turkey, pp. 2923–2926.
- Kim J.-D. et al. (2004) Introduction to the bio-entity recognition task at JNLPBA. In: Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications, pp. 70–75. Association for Computational Linguistics.
- Lample G. et al. (2016) Neural architectures for named entity recognition. arXiv preprint arXiv:1603.01360.
- Lee J. et al. (2019) BioBERT: pre-trained biomedical language representation model for biomedical text mining. arXiv preprint arXiv: 1901.08746. [DOI] [PMC free article] [PubMed]
- Li Q. (2012) Literature Survey: Domain Adaptation Algorithms for Natural Language Processing. Department of Computer Science the Graduate Center, The City University of New York, pp. 8–10.
- Ma X., Hovy E. (2016) End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, Berlin, Germany, pp. 1064–1074.
- Pachitariu M., Sahani M. (2013) Regularization and nonlinearities for neural language models: when are they needed? arXiv preprint arXiv: 1301.5650.
- Pan S.J., Yang Q. (2010) A survey on transfer learning. IEEE Trans. Knowl. Data Eng., 22, 1345–1359. [Google Scholar]
- Rebholz-Schuhmann D. et al. (2010) CALBC silver standard corpus. J. Bioinform. Comput. Biol., 8, 163–179. [DOI] [PubMed] [Google Scholar]
- Reimers N., Gurevych I. (2017) Optimal hyperparameters for deep LSTM-networks for sequence labeling tasks. CoRR, abs/1707.06799.
- Rzhetsky A. et al. (2009) Getting started in text mining: part two. PLoS Comput. Biol., 5, e1000411.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachan D.S. et al. (2018) Effective use of bidirectional language modeling for transfer learning in biomedical named entity recognition. In: Doshi-Velez,F. et al (eds.). Proceedings of the 3rd Machine Learning for Healthcare Conference PMLR, Palo Alto, California pp. 383–402.
- Srivastava N. et al. (2014) Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res., 15, 1929–1958. [Google Scholar]
- Thrun S. (1996) Is learning the n-th thing any easier than learning the first? In: Advances in Neural Information Processing Systems, The MIT Press, pp. 640–646.
- Wang X. et al. (2018) Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics, 35, 1745–1752. [DOI] [PubMed] [Google Scholar]
- Weiss K. et al. (2016) A survey of transfer learning. J. Big Data, 3. [Google Scholar]
- Zaremba W. et al. (2014) Recurrent neural network regularization. CoRR, abs/1409.2329.
- Zhou G. et al. (2004) Recognizing names in biomedical texts: a machine learning approach. Bioinformatics, 20, 1178–1190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


