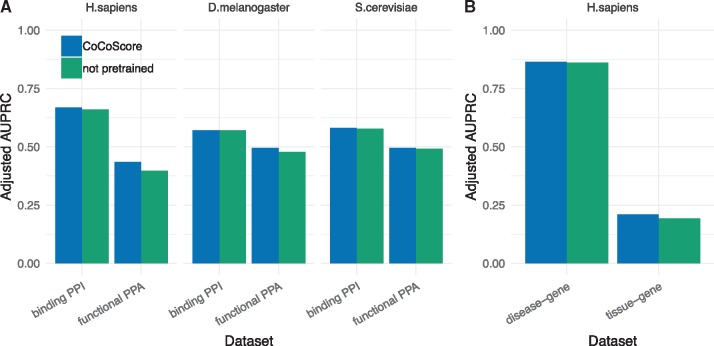
Fig. 3.
Performance using pretrained and not pretrained word embeddings. Not pretrained embeddings are learned at training time. The performance of CoCoScore with (blue) and without (green) pretrained word embeddings is shown for functional PPA and binding PPI datasets (A) as well as disease–gene and tissue–gene associations (B). Performance is depicted as adjusted area under the precision-recall curve (AUPRC). All performance standard deviations after bootstrap resampling were <0.006. The difference between models using pretrained and learned embeddings was significant for all datasets except disease–gene associations and binding PPIs in D.melanogaster at a significance level of 0.001 (Color version of this figure is available at Bioinformatics online.)