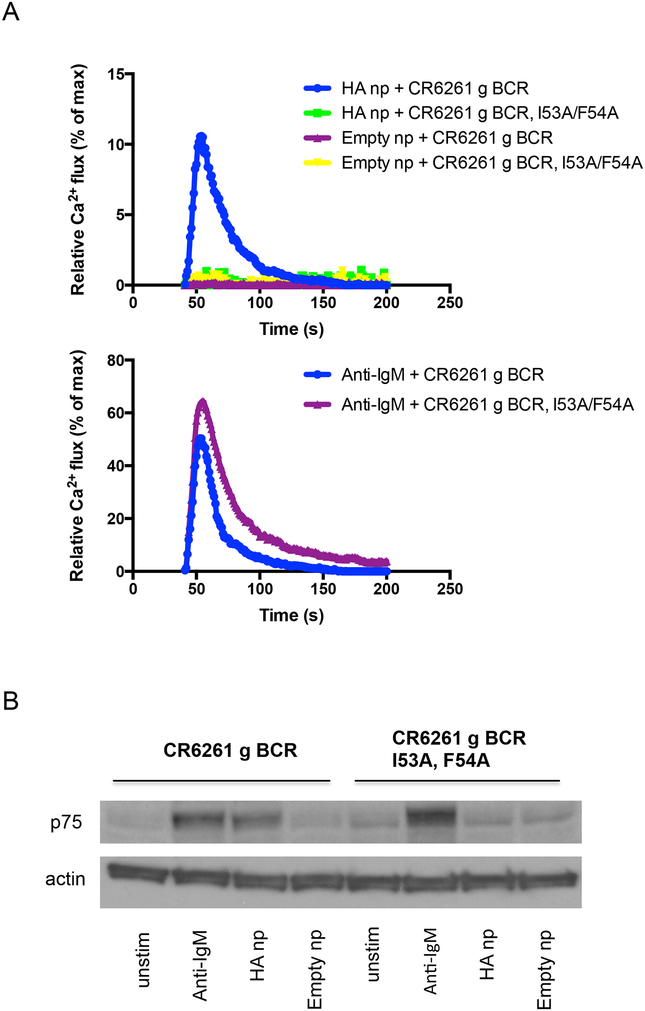
Abstract
Predicting immune responses prior to vaccination is challenging due the complexity of the governing parameters. Nevertheless, recent work has shown that B cell receptor (BCR) antigen engagement in vitro can prove a powerful means of informing the design of antibody-based vaccines. We have developed this principle into a two-phased immunogen evaluation pipeline to rank-order vaccine candidates. In Phase 1, recombinant antigens are screened for reactivity to the germline precursors that produce the antibody responses of interest. To both mimic the architecture of initial antigen engagement and facilitate rapid immunogen screening, these antibodies are expressed as membrane anchored IgMs in 293F indicator cells. In Phase 2, the binding hits are multimerized by nanoparticle or proteoliposome display and evaluated for BCR triggering in an engineered B cell line displaying the IgM sequences of interest. Key developments that complement existing methodology in this area include: 1) introduction of a high throughput screening step prior to evaluation of more time intensive BCR triggering analyses; 2) generalizable multivalent antigen display platforms needed for BCR activation; and 3) engineered use of a human B cell line that does not display endogenous antibody, only ectopically expressed BCR sequences of interest. Through this pipeline, the capacity to initiate favorable antibody responses is evaluated. The entire protocol can be completed within 2.5 months.
Editorial Summary:
This protocol describes an immunogen evaluation pipeline containing two main components that enable vaccine candidates to be rank-ordered.
INTRODUCTION
The mechanistic basis for differences in vaccine immunogenicity is poorly understood and represents a significant hurdle to elicit effective antibody responses against pathogenic agents. While it is clear that sustained antibody responses are a function of interactions between dendritic cells, B cells and T cells during germinal center reactions1–3, there is no systematically defined correlate for predicting vaccine efficacy prior to use2,4. This problem is exacerbated by the fact that certain pathogens display a structure that is both variable and immunogenic, effectively manipulating immunogenicity to ensure that antibody responses are unable to neutralize their targets4,5. We, and others, have recently developed a new approach to evaluate vaccine candidates6–12. It centers on the premise that antibody responses can be, in part, predicted through a single parameter: reconstituted interactions between the antigen and the germline B cell receptor. Our work has shown that the human antibody VH gene, IGHV1–69, encodes for germline BCRs that naturally engage and signal in response to the influenza spike protein hemagglutinin (HA)6. Antigen engagement is specific to the HA stem region, a functionally conserved epitope for broadly neutralizing antibodies (bnAbs) against influenza virus13–17. Affinity to the HA stem is provided by a hydrophobic CDRH2 that is not reconfigured during antibody recombination, predicting that humans are genetically hardwired to generate broad immunity to this virus6,18,19. Indeed this recognition is then correlated with biased usage of IGHV1–69 for influenza bnAb development13–19 suggesting that reconstituted IGHV1–69 BCR-antigen interactions can also be used to inform the design of HA-based subunit vaccines.
Comparison with alternative methods
Since the first demonstration of a relationship between reconstituted BCR signaling and influenza antibody response patterns6, the biochemical reconstruction of germline BCR stimulation has been applied to screen immunogen candidates for other pathogens, namely HIV7–12,20, and is consequently emerging as an important tool to inform vaccine design4,16,17,21–23. In all cases, the methodology relies on 1) establishing a B cell reporter system in which selected BCR sequences can be ectopically expressed and systematically evaluated for signaling, and 2) generating multivalent displays of antigens to trigger BCR activation. However, there are some key differences in this protocol that are outlined here.
Firstly, the B cell reporter lines used in other studies including K46 or WEHI-231 mouse B cells7,8, A20 mouse B cells11 or the DG-75 human B cell line7,9–12 are not used in our protocol as they express endogenous BCR, which can complicate monospecific germline antibody display. For example, DG-75 offers clear advantages over mouse analogs as a human reporter line, but it presents its own IgM BCR, meaning that germline BCRs of interest are expressed and enriched by FACS in an IgG format7,9–12. Germline activation naturally takes place through IgM BCR, and recent studies have demonstrated marked differences in the kinetics and mechanistic basis of signaling through IgM versus IgG BCR24,25. To generate a human B cell reporter line that facilitates expression of only IgM BCR sequences of interest, this protocol describes generating a clone of the Ramos Burkitt’s lymphoma B cell line that presents no endogenous surface antbody6,20.
Triggering in response to a multimerized antigen is measured kinetically by calcium flux and by tyrosine phosphorylation of downstream effectors of BCR signaling, as is the case with all the methodology in this area6–12,20. However, in contrast to other studies, this protocol employs a rapid pre-selection step wherein candidate immunogens are first evaluated for reactivity to membrane anchored IgM (mIgM) which is displayed by 293F cells6,19. This step facilitates expedient rank-ordering of candidate immunogens during which time the B cell reporter system and multivalent antigen display needed for BCR triggering can be established. For antigen multivalency, HIV signaling studies have largely relied on the trivalency of gp140 structure7,9–12, however we have found that trivalent antigen can be in some cases suboptimal for initiating BCR signaling, particularly since germline affinity for antigen is often low6. Consequently, this protocol also presents procedures for further arraying antigen through established nanoparticle26 and proteoliposomal6,8 platforms.
Experimental Design
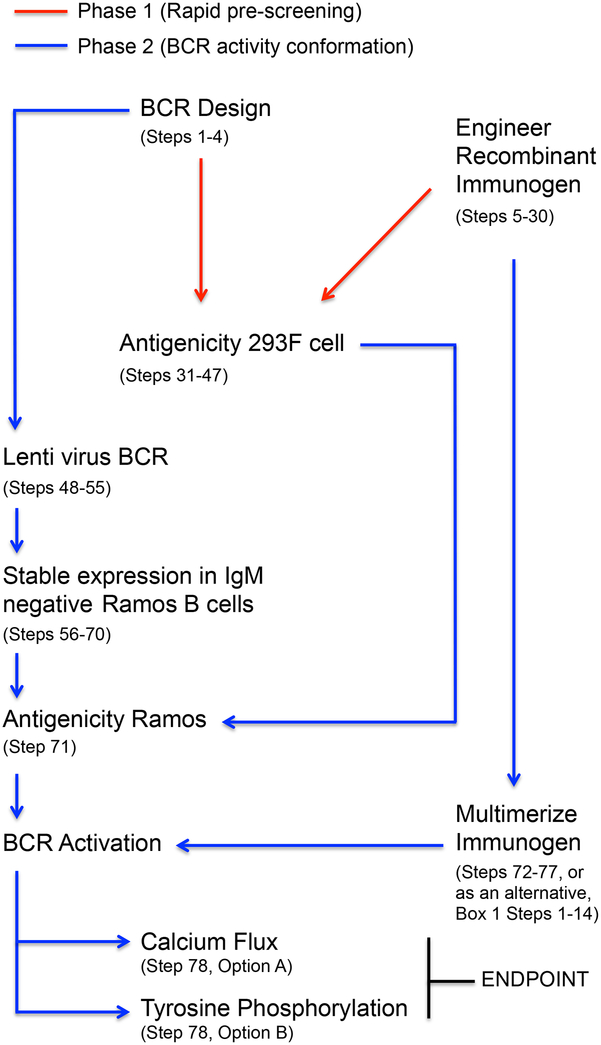
This protocol provides instructions for performing our immunogen rank-ordering procedure that was originally developed in the influenza context6,18, and has since been adapted to other pathogens20 (Figure 1). There are two key phases in this pipeline: 1) Rapid pre-screening of an antigen’s capacity to bind transiently transfected germline membrane anchored IgM (mIgM) of interest; and 2) a more time intensive confirmation of activation via stable expression of BCR-formatted IgM and analysis of triggering in response to multimerized configurations of that same antigen. Dividing the pipeline into two steps relates to the efficiency of the screening process. To activate a BCR independently from the antigen presentation machinery, the candidate immunogen must ultimately be multimeric27. This requires a structure-based design step that, while specifically addressed in this protocol, is time consuming as it must often be tailored to one particular antigen structure7,26,28. To more rapidly rank-order BCR engagement prior to this step, a mIgM recognition assay that only requires the use of fluorescently labeled recombinant antigen is presented. Membrane IgM is expressed in 293F cells and binding is rapidly assessed by flow cytometry. This quick screening step is unique to this approach, however, the principle of membrane display was first identified as important for recognition of substrate at low affinity in the context of directed evolution29. We have shown that it is also a design-feature that enhances antigen specificity prior to affinity maturation, and can be critical for identifying germline antibody binding partners since these molecular interactions are weak and are often not measurable in solution6. In our experience, all cases of mIgM recognition transduce into BCR triggering in response to multimerized versions of that antigen6,20.
Figure 1.
BCR antigen recognition pipeline. This protocol is used to assess whether candidate immunogens can engage and stimulate BCRs of interest, namely germline antibody sequences that give rise to effective humoral responses following infection with pathogen. The protocol is divided into two phases: 1) a rapid screening step wherein antigen interactions with low-affinity germline mIgM are evaluated at the 293F cell surface; and 2) a subsequent analysis of the capacity to stimulate BCR activity through the engagement specificity identified in Phase 1.
For BCR triggering, receptor signaling is evaluated through a stably expressed IgM BCR in a human B cell line, a clone of Ramos Burkitt’s lymphoma that expresses no endogenous surface IgM. This contrasts other BCR surface expression systems where the reporter B cell lines are either not human7,8,11 or unavoidably display endogenous/irrelevant BCR at the plasma membrane7–12. Engagement of fluorescently labeled antigen is first confirmed for the BCR of interest, and then triggering is assessed by calcium flux and tyrosine phosphorylation of downstream BCR signaling effectors6–12,20. In this protocol, calcium flux in response to BCR stimulation is evaluated by flow cytometry as the ratio of the Ca2+ bound/unbound states of the membrane permeable dye Fura Red30. Indo-1 is another ratiometric dye that can be used, however, it requires UV laser settings that are not standard to many FACS analyzers. Ratiometric dyes are the most quantitative as they correct for variations in fluorescence due to inconsistences in dye loading, changes in the focus of the equipment, and fluorescent bleaching effects31. Emission/excitation maxima for Fura Red are: 472/657 (low Ca2+) and 436/637 (high Ca2+). Non-ratiometric dyes commonly used for flux experiments include Fluo-3, Fluo-4 and their derivatives, all of which exhibit large fluorescence intensity increases on binding Ca2+ 31. Unlike Fura Red and Indo-1, there is no accompanying spectral shift. In all cases, baseline emission is recorded and then calcium flux is measured in real time following addition of antigen multimer.
To generate multivalent antigen presentations required to initiate signaling, this protocol also provides instructions for generating nanoparticle and proteoliposome configurations of HA, the latter requiring no tailored antigen configuration or structure-design effort (Box 1). For nanoparticle design, we employ the recently developed HA ferritin nanoparticle platform26 as the BCR stimulating ligand. In this platform, HA monomers are fused with ferritin, an iron sequestering protein that self-assembles into nanoparticles composed of 24 identical polypeptides26. Monomeric HA is inserted at the interface of ferritin subunits such that eight trimeric HA spikes are formed during nanoparticle assembly26. By contrast, for protoliposome display, pre-purified his-tagged HA trimer is arrayed on the surface of preformed 100nm liposomes containing Ni-linked phospholipids. Nanoparticles and proteoliposomes can substitute for one another as BCR stimulating ligands in this protocol. Some HIV-BCR signaling studies have relied on the multivalency provided by recombinant gp140 envelope trimer8–10,12, while we and others have employed higher order multimers of monomeric gp1207,20. For influenza, we have found that HA trimer alone has a very limited capacity to induce germline BCR signaling and as a general rule would recommend the use of multimerized antigen formats when the valency and affinity of engagement is low or not yet defined. This is clearly not a requirement for gp1408–10,12. Nevertheless, increased antigenic valency has long been correlated with heightened immunogenicity for antibody-based vaccines and should therefore be considered during immunogen design32.
BOX 1. Proteoliposome Array of His-tagged Antigen.
This procedure presents the steps to array antigen (produced during Steps 6–30) on liposomal surfaces. It can act as a substitute for nanoparticle or virus-like particle presentations of antigen that often require more structural design effort.
-
79
Under a gentle stream of nitrogen, evaporate one gram of DOPC/DGS-NTA(Ni ) that has been pre-mixed in a 1:1 molar ratio. This can be done in a 2ml Eppendorf microtube.
-
80
For 30 min, rehydrate the dry lipid film in 1000 μl of HBS, all the time being heated above the melting temperature (Tm) of the lipid mixture. The melting temperature of DOPC is low, but to insure even distribution with DGS-NTA(Ni), maintain the mixture at least 50°C. Intermittent vortexing will also promote lipid-lipid miscibility.
PAUSEPOINT. A 30 minute incubation is sufficient, however longer periods (e.g. 2 h) may be employed without adverse effects to the experiment.
-
81
Subject resulting suspension to ten freeze-thaw cycles using liquid nitrogen
and a heating block.
-
82
Assemble mini-extruder (100-nm pore polycarbonate membrane should be sandwiched between two filter supports on either side that have been pre-soaked in H2O).
-
83
Place extruder on heating block and allow it to warm to 50°C.
-
84
Equilibrate the extruder in 1 ml of HBS and then remove from the system.
-
85
Extrude the lipid mixture twenty-one times through a 100-nm pore polycarbonate membrane the mini-extruder at 50°C.
CRITICAL STEP. The extrusion step should be performed at 50°C or above to insure an even liposome composition.
-
86
Equilibrate the 100 nm liposomes to RT and mix with His-tagged antigen at a protein/lipid molar ratio of 1/900 for 1 hr.
CRITICAL STEP. Although liposome volume is lost in the void during extrusion, the lipid concentration is maintained and thus an accurate protein to lipid ratio can be calculated.
-
87
Adjust the liposome/protein sample to 12.5% iodixanol (in 1.25 ml HBS) and carefully overlay with 1.75 ml, 0.5 ml and 0.5 ml of 10%, 2.5% and 0% iodixanol in HBS, respectively. This can be done in a 11 × 60 mm ultracentrifuge tube.
CRITICAL STEP. Layer gradient gently to prevent disruption of the gradient.
-
88
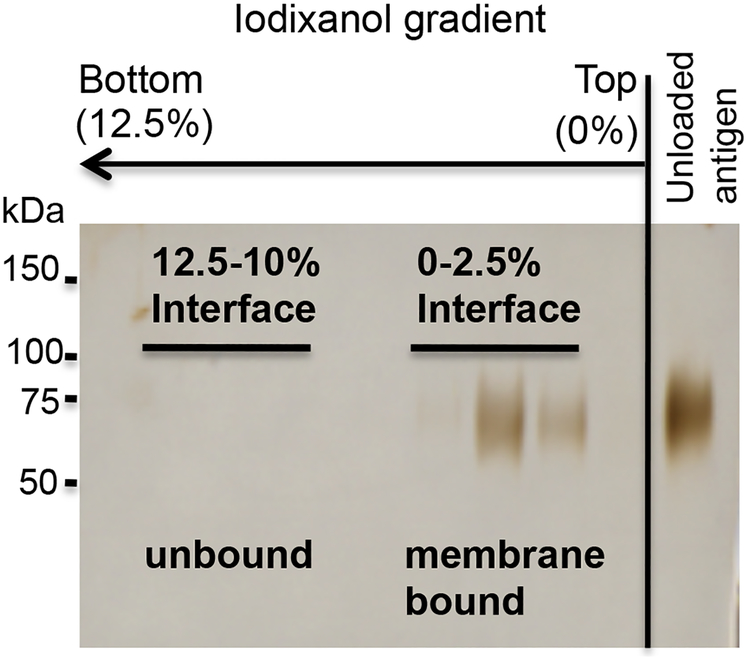
Centrifuge samples for 200,000 × g in a TH660 rotor for 2 hr. The proteoliposome fraction concentrates at the 2.5%−0% iodixanol interface, unbound protein is found in the fractions below. Floating lipid can be visualized by illuminating the gradient with a flashlight (Figure 8).
CRITICAL STEP. Ensure that the centrifuge tubes are completely filled otherwise they will collapse during centrifugation.
-
89
Collect proteoliposome fraction and dialyze for at least 3 h using a 10 kDa cutoff slide-A-Lyzer Dialysis Cassette to remove density gradient material.
-
90
Following dialysis, pellet the (200,000 × g, 2 h, TH660 rotor).
-
91
Resuspend in HBS and measure using a BCA protein assay.
CRITICAL STEP. Take care not to dislodge pellet when aspirating tube
PAUSEPOINT. Proteoliposomes should be aliquoted, snap frozen in liquid nitrogen and then stored for several months at −80°C.
-
92
Apply proteoliposomes in place of nanoparticles to trigger BCR signaling during Step 78 [Option A (iv) for calcium flux; Option B (i) for tyrosine phosphorylation].
TIMING Step 1, 30 min; Step 2, 30 min; Step 3, 20 min; Steps 4–7, 30 min; Step 8 1 h; Step 9, 20–30 min; Step 10, 2 h 30 min; Step 11, 3 h; Step 12, 2 h; Step 13, 1h; Step 14, 2h (Option A), 1–2 days (Option B).
Troubleshooting
Step 10: If the lipids are not floating in the gradient this may be due to the protein being overloaded. This problem can be addressed by decreasing the protein/lipid ratio.
Step 13: If the protein yields are low this could be because the protein has been underloaded. This problem can be addressed by increasing the protein/lipid ratio
Phase 1 (Steps 1–47)
In the first phase of this protocol (Figure 1), the germline precursor antibody is initially defined through publically available computer software and then experimentally configured as a membrane anchored IgM. Next, the germline mIgM is expressed on the surface of 293F cells and is assessed for reactivity to fluorescently labeled antigen. Phase 1 necessitates the use of the following gene constructs:
The variable domains of germline-reverted heavy and light chains from an antibody of interest. Publically available algorithms that revert antibody sequences to their germline precursors define these sequences.
The constant domain for membrane anchored IgM, obtained from Genbank.
For the germline antibody of interest and for an irrelevant control antibody, either separate plasmids containing mIgM heavy chain and light chain6,20 or a single plasmid containing the mIgM heavy chain heavy and light chain combined. When cloned together in the same plasmid, sequences should be separated by a furin cleavage site and/or a F2A peptide sequence8,9.
A mammalian expression vector encoding recombinant antigen with an avi-tag for site-specific biotinylation and fluorescent labeling.
Experimental Controls for Phase 1.
The use of mature antibody is not obligate, but it can prove a useful positive control. If structural information is available, we recommend that antibody-binding mutants be designed as negative controls. Mutations can be introduced into the recombinant antigen and/or the surface expressed IgM; both control forms are presented in this protocol. To attenuate recognition of the HA-stem at the level of the antigen, we introduce I45R/T49R mutations6 or alternatively an N-linked glycosylation site (I45N/G47T)26 within the stem region. To attenuate HA-stem recognition at the level of the antibody (IGHV1–69 mIgM), we mutate the HA stem contacting CDRH2 (I53A/I54A)6. If structural information is not available, specific engagement to the germline mIgM of interest can be defined relative to reactivity to irrelevant mIgM. In all cases, surface IgM expression is co-monitored by fluorescent anti-Ig.
Phase 2 (Steps 48–78)
In the second phase of this protocol (Figure 1) the mIgM constructs are expressed in a reporter B cell line so that the corresponding IgM BCR is produced at the cell surface6,20. Other approaches have employed ectopic expression of membrane anchored IgG or IgM in B cell lines that contain endogenous surface immunoglobulin7–12. For germline stimulation, we favor use of IgM expressed in Ramos B cells, both because IgM is a natural germline BCR configuration, and in this cell line, clones negative for surface IgM can be easily derived, making mono-specific BCR display possible6,20. To stability integrate mIgM sequences of interest, we use the FEEKW lentiviral expression system that was originally designed to deliver exogenous antibody sequences to hematopoietic B cell progenitors33. B cells expressing BCRs of interest are sorted by FACS and then cultured. Following a minimum of two additional cycles of sorting and enrichment, signaling is then assessed in response to multimerized versions of the antigen used in Phase 1.
Phase 2 requires the use of following gene constructs:
A lentiviral system for transducing B cell lines with BCRs of interest8,20,33. Transient transfection/electroporation with mammalian expression vectors encoding BCRs of interest can also be used6,8,9. We have found that establishing stable cell lines helps reduce the workload per BCR activation experiment, particularly when multiple BCRs are being tested.
To confirm the antigenicity seen in Phase 1, the same mammalian expression vector encoding recombinant antigen with an avi-tag for site-specific biotinylation and fluorescent labeling.
A mammalian expression vector encoding recombinant antigen displayed in multivalent format. The HA ferritin nanoparticle platform26 is described in this protocol. Alternatively, the monomeric/low valency form can be multimerized by a proteoliposome display platform described in Box 1. In all cases, multimeric antigen is used to trigger BCRs of interest.
Experimental Controls for Phase 2.
The binding mutations used in Phase 1 are also recommended as negative controls for Phase 2; either as mutant antigen and/or mutations in the BCR itself. If structural information is unavailable, triggering of a BCR of interest can still be defined in comparison to the response observed through irrelevant control BCR. In this protocol, we use both CDRH2 (I53A/I54A) BCR that is unable to engage the HA-stem6 and empty ferritin nanoparticles26 as negative controls. To confirm that both control and experimental BCRs have similar surface expression, total receptor activity is monitored by anti-IgM crosslinking.
Limitations of the protocol
Reconstituted interactions between antigen and germline BCRs of interest are predicted to reflect the recognition steps from which favorable antibody responses derive. To evaluate the capability to initiate an antibody response of interest, our protocol tests whether candidate immunogens can engage and stimulate the corresponding germline-reverted B cell receptor. This does not guarantee the elicitation of said responses in the vaccine setting. There are a number of other immunological factors regulating antibody-vaccine efficacy (e.g. adjuvant effects, lymph node delivery and antigen presentation to B cells34–36) that this protocol is unable to address. Rather we present methodology that rank-orders the capacity to stimulate a specific BCR signature and should only be applied as an immunogen design principle in that context.
MATERIALS
Reagents
293F Transfection:
-
FreeStyle™ 293-F Cells (Life Technologies, cat. no. R790–07)
CAUTION: SL1 level biohazard; handle only in tissue culture facility.
CAUTION: The cell lines should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Gibco® FreeStyle™ 293 Expression Medium (Life Technologies, cat. no. 12338–026)
−293fectin™ Reagent (Invitrogen, cat. no. 12347019)
-
Opti-MEM (Life Technologies, cat. no. 51985–091).
CRITICAL: This protocol is optimized for 293F protein expression, other mammalian expression systems are not covered.
Mammalian gene expression vector encoding genes of interest (e.g. VRC 840019).
Plasmid Preparation
Maxiprep kit (e.g. PowerPrep HP Plasmid Maxiprep System, Origene, cat. no. NP100009
QuikChange mutagenesis kit (Agilent Technologies, cat. no. 200523)
Antigen Purification:
Ni Sepharose Excel Affinity Media (GE Healthcare, cat. no. 17-3712-02)
Imidazole (Sigma, cat. no. I5513)
Erythrina cristagalli Gel-ECA-Immobilized Lectin (EY Laboratories, cat. no. A-5901-2)
D-lactose monohydrate (Sigma, cat. no. 61345)
Imidazole (Sigma Aldrich, cat. no. I202)
Potassium phosphate monobasic (KH2PO4; Sigma Aldrich, cat. no. P5655)
Sodium phosphate dibasic heptahydrate (Na2HPO4 ·7H2O; Sigma Aldrich, cat. no. 431478)
-
Liquid nitrogen
CAUTION: Tissue damage or burns can result from exposure. Personal protective equipment includes a full-face shield over safety goggles; loose fitting thermal insulated gloves; and a chemical resistant lab coat.
Antigen Probe Biotinylation and fluorescent conjugation
-
Biotin Protein Ligase Kit (Avidity, cat. no. Bulk BirA).
CRITICIAL: This particular enzyme kit is required for site-specific biotinylation of the Avi-tagged antigen.
Streptavidin APC conjugate (Life Technologies, cat. no. S-32362)
Streptavidin PE conjugate (Life Technologies, cat. no. S-21388)
293F displayed mIgM analysis
Anti-Human Ig kappa Light Chain PE (eBioscience, cat. no. 12-9970-42)
-
Anti-human Light Chain, λ PE (BD Biosciences, cat. no. 555797)
CRITICAL: In our protocol, surface trafficking of mIgM is confirmed using these PE-conjugated anti-light chain antibodies.
BCR Expression:
ProFection Transfection System (Promega, cat. no. E1200)
DMEM (Sigma Aldrich, cat. no. D5671)
RPMI (Sigma Aldrich, cat. no. R0883)
FBS (Sigma Aldrich, cat. no. F-4135)
−1× L-Glutamine (Life Technologies, cat. no. 25030)
−10000 U/mL Penicillin / 10000μg/ml Streptomycin (Life Technologies, cat. no. 15140)
-
Lentivirus BCR expression plasmids [e.g. FEEKW heavy/light chain system expression33; VSVG for envelope pseudotyping37; and a packaging vector, e.g. pΔR8.238]
CAUTION: Potential biohazard, lentivirus systems are engineered for biosafety, but are infectious. The virus produced in this protocol is only capable of a single round of infection. Moreover, the deletion of Envelop protein in pΔR8.2 increases biosafety38. Handle using BSL2 procedures.
Hexadimethrine bromide/polybrene (Sigma Aldrich, cat. no. 107689)
-
−293T cells (ATCC® CRL-3216™)
CAUTION: Will be transduced with lentivirus, handle using BSL2 procedures.
CAUTION: The cell line should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
-
IgM− Ramos Cells (derived from ATCC® CRL-1596™).
CAUTION: Will be transduced with lentivirus, handle using BSL2 procedures.
CAUTION: The cell line should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
LIVE/DEAD® Fixable Violet Dead Cell Stain Kit (ViViD) (Life Technologies, cat. no. L34955)
APC anti-human IgM Antibody (Biolegend, cat. no. 314510)
Anti-Human Ig kappa Light Chain PE (eBioscience, cat. no. 12-9970-42)
Anti-human Light Chain, λ PE (BD Biosciences, cat. no. 555797)
MycoAlert™ mycoplasma detection kit (Lonza, cat no. LT07–118)
-
−2-Propanol (Fisher Scientific, cat. no. A416500)
CAUTION: 2-Propanol is flammable. Add to cell freezing chamber in fumehood, wear safety goggles, chemical resistant lab coat and nitrile gloves to prevent exposure. Store in dedicated steel flammable cabinet.
Proteoliposome Assembly
HEPES (Fisher, cat. no. BP-310)
NaCl (Fisher, PI-28314)
OptiPrep [60% (wt/vol) iodixanol; (Progen Biotechnik, cat. no. 1030061)]
Hydrochloric acid (HCl; Sigma, cat. no. H1758)
Sodium hydroxide (NaOH; Sigma, S8045)
−1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids Inc.)
−1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid) succinyl (DGS-NTA(Ni)) (Avanti Polar Lipids, cat. no. 850375)
Pierce BCR protein assay kit (Thermo Scientific, cat. no. 23227)
-
Chloroform (Alfa Aesar, cat. no. 43685-K2)
CAUTION: This is an organic solvent (halogen class) and suspected human carcinogen. Use in fumehood, wear safety goggles, chemical resistant lab coat and nitrile gloves (min 8mil). Store away from direct sunlight and keep in a dry well-ventilated area. Do not store with oxidizing compounds strong bases or in containers made of aluminum.
-
Methanol (Fisher Scientific, cat. no. BP1105)
CAUTION: Methanol is extremely flammable. Use in fumehood, wear safety goggles, chemical resistant lab coat and nitrile gloves to prevent exposure. Store in dedicated steel flammable cabinet.
-
Liquid nitrogen.
CAUTION: Extensive tissue damage or burns can result from exposure. Personal protective equipment includes: a full face shield over safety goggles; loose fitting thermal insulated gloves; and a chemical resistant lab coat.
BCR Signaling
Fura Red (Life Technologies, cat. no. F-3021)
Mouse anti-human IgM F(ab’)2 (Southern Biotech, cat. no. 9023–01)
Ionomycin (Life Technologies, cat. no. I24222)
Complete Protease Inhibitor Cocktail (PIC) (Roche Applied Science, cat. no. 13352700)
Pierce BCA protein assay kit (Thermo Scientific, cat. no. 23225)
Antibody G410 pY (Millipore, cat. no. 05–0321)
HEPES (Fisher, cat. no. BP-310)
NaCl (Fisher, PI-28314)
−10% Triton X-100 (TX-100) (Fisher, PI-28314)
Monoclonal anti-β-actin (Sigma Aldrich, cat. no. A5316)
Equipment
Instrumentation
BD LSR II (BD Biosciences)
FACS Aria II (BD Biosciences)
Humidity Controlled, Shaking Incubator (Infors Multitron)
Cell culture Incubator (Eppendorf, Galaxy 170)
Tangential Flow Filtration System (Pall)
FPLC system (AKTA Explorer)
Nanodrop 8000 (Thermo Scientific)
Thermomixer (Eppendorf)
Luminometer (e.g. SpectraMax L, Molecular devices)
Tube Rotator (Bioexpress H-5651–1)
Microcentrifuge (Eppendorf 5424)
Benchtop Centrifuge (Sorvall Legend XTR)
Recombinant Antigen Work
−500 ml Conical Centrifuge Tubes (Corning, cat. no. 431123)
VacuCap 0.8/0.2 μm filters (Pall, cat. no. 4628)
−1 L Square Pyrex bottles (Corning, cat. no. 1396–1L)
Polypropylene conical tubes, 50 and 15 ml with graduation (Corning)
Büchner/vacuum flask (Fisher Scientific, cat. no. FB-300–2000)
Glass Econo-Column, 35 ml, 1.5×20 cm (Biorad, cat. no. 7374152)
Econo-Column Funnel (Biorad, cat. no. 7310003)
Amicon Ultra concentrators, 30 kDa and 100 kDa cutoffs (Millipore)
Centrifuge Tubes, 1.5 and 2 ml (Eppendorf)
Superdex 200 10/300 Column (GE Healthcare, cat. no. 17517501)
Superose 6 10/300 Column (GE Healthcare, cat. no. 17517201)
HiLoad 16/10 Q Sepharose HP column (GE Healthcare, cat. no. 17115301)
Low Speed Orbital Shaker (Bioexpress S-3200-LS)
−1 mL Plastic Luer-Lock Syringes (BD, cat. no.309628)
−10 mL Plastic Luer-Lock Syringes (BD, cat. no. 309604)
Slide-A-Lyzer Dialysis Cassette, 10,000 MWCO (Thermo Scientific, cat. no. 66380)
Liquid nitrogen dewar (Finemech, cat. no. KGW-10215)
Opaque Microtubes (Bioexpress, cat. no. C3396–2)
293F mIgM expression and analysis
Steri-Flip Filters (Millipore, cat. no. SCGP00525)
−125 ml vented, baffled flask (Fisher Scientific, cat. no. 10–529)
−250 ml vented, baffled flask (Fisher Scientific, cat. no. 10–530)
−500 ml vented, baffled flask (Fisher Scientific, cat. no. 10–531)
−1000 ml vented, baffled flask (Fisher Scientific, cat. no. 10–532)
−2000 ml vented, baffled flask (Fisher Scientific, cat. no. 10–533)
−96 well v-bottom plates (Corning, cat. no. 3894)
FACS tubes (Corning, cat. no 353054)
Proteoliposome Assembly
Nitrogen cylinder with pressure gauge and regulator
Manifold for distributing nitrogen flow for evaporation
Liquid nitrogen dewar (Finemech, cat. no. KGW-10215)
−2.0 ml microtubes (Bioexpress, cat. no. C-3261–1)
Extruder apparatus with 1000 μl Hamilton syringes (Avanti Polar Lipids, cat. no 610000)
Extruder filter supports (Whatman, cat. no. 230300)
−0.1 μm extruder filter (Whatman, cat. no. 800309)
Extruder heating block (Bioexpress, cat. no. D-2000–1B)
Beckman Optima MAX ultracentrifuge
Thinwall, Ultra-Clear™, 11 × 60 mm ultra centrifuge tubes (Beckman, cat. no. 335649)
TH660 rotor (Thermo Scientific, cat. no. 10502)
Lipid glassware (e.g. Wheaton® Scintillation Vials with polypropylene cap)
−25-gauge (25-G) Needles (BD Medical, cat. no. 305127)
−5-ml Plastic syringes (BD Medical, cat. no. 301027)
Vortexer (Scientific Industries, cat. no. SI-0236, or equivalent)
Slide-A-Lyzer Dialysis Cassette, 10,000 MWCO, (Thermo Scientific, cat. no. 66380)
BCR Expression and Analysis
−0.45 μM syringe filters (Millipore, cat. no. SLHV033RS)
−10 cm culture dishes (Corning, cat. no. 430167)
−96 well culture dishes (Corning, cat. no. 353072)
−48 well culture dishes (Corning, cat. no. 3548)
−24 well culture dishes (Corning, cat. no. 3524)
−12 well culture dishes (Corning, cat. no. 3512)
−6 well plates (Corning, cat. no. 3506)
T25 culture flask (Corning, cat. no. 3056)
T75 culture flask (Corning, cat. no. 430641U)
−0.22 μM filter (Corning, cat. no. 430758)
FACS tubes (Corning, cat. no 353054)
−96 well plates for luminescence (PerkinElmer, cat. no. 6005030)
Cell Freezing container (Thermo Scientific, cat. no. 5100–0001)
Cell culture cryotubes (Thermo Scientific, cat. no. 374081)
REAGENT SETUP
FBS.
Filter at 0.22 μm, snap freeze in liquid nitrogen and distribute into 25 ml aliquots. Store at −80°C for several months.
100× L-Glutamine.
Filter at 0.22 μm, snap freeze in liquid nitrogen and distribute into 5 ml aliquots. Store at −80°C for several months.
100× Penicillin / Streptomycin.
Filter at 0.22 μm, snap freeze in liquid nitrogen and distribute into 5 ml aliquots. Store at −80°C for several months.
Complete RPMI (cRPMI)
(15% FBS + 1× glutamine + 1× of penicillin/ streptomycin) Remove 85 ml RPMI from 500 ml RPMI stock and add 75 ml FBS + 5ml 100× glutamine + 5 ml of 100× penicillin/streptomycin. Filter at 0.22 μm. Store at 4°C for several weeks.
cRPMI Conditioned Media
Grow Ramos cells to a minimum of 1×106 cells/ml in a T75 flask and culture for 24 h in a cell culture incubator set to 37°C and 5% CO2 saturation. Collect culture supernatant and filter at 0.22 μm. Store at 4°C for several weeks.
DMEM
(10% FBS + 1× glutamine + 1× of penicillin/streptomycin) Remove 60 ml from 500 ml DMEM, then add 50 ml FBS, 5ml 100× glutamine + 5 ml of 100× penicillin/streptomycin. Filter at 0.22 μm. Store at 4°C for several weeks.
ViViD staining solution
Dissolve 200 μg stock in 200 μl DMSO, then dilute 1 in 40 in PBS (e.g. 3 μl ViViD in DMSO +117 μl H2O). The diluted solution is applied as described in the procedure and the DMSO stock should be stored at −20 to −80°C. The diluted staining solution should be made fresh for each experiment.
PBS
(155 mM NaCl, 1.5 mM KH2PO4, 2.7 mM Na2HPO4, pH 7.4) Dissolve 0.21 g KH2PO4, 0.73 g Na2HPO4 ·7H2O and 9 g NaCl in H2O, pH to 7.4 and bring to 1 L). Store at room temperature (RT = 23°C); can be used for several weeks.
Imidazole Solutions.
Dissolve 340.39 g of imidazole into 600 ml of H2O (=5 M stock) and pH to 7.4. To make 500 mM imidazole in PBS, add 100ml of 5 M imidazole stock to the PBS recipe, pH to 7.4 and then bring volume to 1L. To make 60 mM imidazole in PBS, add 12 ml of 5 M imidazole stock to the PBS recipe, pH to 7.4 and then bring volume to 1L. The 5 M imidazole stock can be stored at 4°C and used for several months. The 500 mM and 60 mM imidazole solutions can be stored at RT and used for several weeks.
Biotinylation Buffer
(10 mM Tris, pH 8.0) Dissolve 1.21 g Tris in H2O, pH to 8.0 and bring to 1 L. Store at RT and used for several weeks.
Lipid Stock Solutions
Dissolve DOPC and DGS-NTA(Ni) into each of two 25 mg/ml stock solutions in 2:1 chloroform:methanol (v:v). Ensure that lipid glassware is used. These stock solutions should be kept at −20oC and can be used for several months.
5X HEPES Buffered Saline (HBS)
(250 mM HEPES, 750 mM NaCl, pH 7.4)
Dissolve 70.8 g HEPES and 43.83 g NaCl in H2O pH to 7.4 and bring to 1 L. This can be stored at RT and used for several weeks.
10% Iodixanol in 1×HBS
Add 2ml of 5×HBS to 1.67 ml of 60% iodixanol stock and bring to a final volume of 10 ml. This should be made fresh for each experiment.
2.5% Iodixanol in 1×HBS
Add 2ml of 5×HBS to 0.42 ml of 60% iodixanol stock and bring to a final volume of 10 ml. This should be made fresh for each experiment.
Fura Red staining solution
Dissolve 50 μg stock in 50 μl DMSO, then dilute 1/200 in cRPMI (final Fura Red concentration = 3 μM). This should be made fresh for each experiment.
RPMI media for B cell signaling
Remove 10ml from 500ml RPMI, then add 5ml 100× glutamine + 5 ml of 100× penicillin/streptomycin. Do not add FBS to this media as it may influence signaling results. For example, serum can bind positive control anti-IgM and reduce BCR triggering. Filter RPMI at 0.22 μm. Store at 4°C for several weeks.
Cell lysis Buffer
(50 mM HEPES, 150 mM NaCl, 1% Ttiton X-100, 2 mM EDTA, 1× PIC, pH 7.4) (100× PIC = 1 tablet dissolved in 500μl H2O) Dissolve 119 mg HEPES, 88 mg NaCl, and 7.4 mg EDTA in H2O, pH to 7.4, then add 100 μl 10% Triton X-100 and 1× PIC (final volume = 10 ml). 100× PIC can be aliquoted stored at −20°C, but lysis buffer should be made fresh for each experiment.
Low Salt Tris buffer
(20 mM Tris, 50 mM NaCl, pH 8.0) Dissolve 2.42 g Tris and 2.92 g NaCl in H2O, pH to 7.4 and bring to 1 L. Store at RT for several weeks.
High Salt Tris Buffer
(20 mM Tris, 500 mM NaCl, pH 8.0) Dissolve 2.42 g Tris and 58.44 g NaCl in H2O, pH to 7.4 and bring to 1 L. Stored at RT for several weeks.
PROCEDURE
PHASE 1 ANTIGEN SCREENING through 293F surface expression and binding: Engineer membrane anchored germline IgM
TIMING: One day for germline IgM design; three weeks for germline variable region synthesis; transgene cloning can be completed in 3 days.
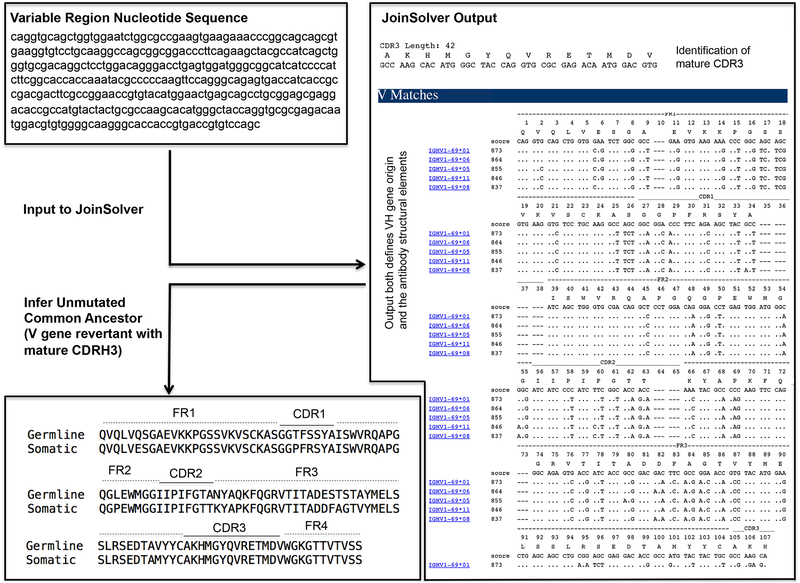
CRITICAL. Antibody heavy and light chains are each encoded by a separate multigene family whose structural elements, namely the framework regions (FRs) and antigen binding loops [termed complementarity-determining regions (CDRs)], are assembled from a variety of V, D, and J gene segments39. The germline precursors for antibody responses of interest are first inferred and then configured into a membrane IgM format.
-
1
Assemble the antibody sequences of interest; a detailed illustration for designing germline revertants using the heavy chain of the HA-stem directed influenza bnAb CR62616 is provided in Figure 2. Enter mature antibody heavy and light chain sequences (preferably in the original nucleotide format40) into the JoinSolver software server (http://joinsolver.niaid.nih.gov) or the International ImMunoGeneTics Information System® (IMGT) server (http://www.imgt.org/IMGT_vquest/vquest) to predict the original gene segments used for assembly. The sequence will be displayed as FR1, CDR1, FR2, CDR2, FR3, CDR3. JoinSolver offers the choice of either Kabat41 or IMGT nomenclature42 for CDR definitions and numbering. As junctional diversity can make the CDR3 and FR4 sequences difficult to predict39, engineer V gene-only reversions of heavy and light chains (FR1, CDR1, FR2, CDR2, FR3) and retain the mature CDR3 and FR4. Both servers will identify the CDR3 sequences, however, the FR4 sequence is not in the standard output. It consists of the remaining variable region sequence downstream to the CDR3 but preceding the immunoglobulin constant region (the variable and downstream constant regions are demarcated by the amino acid sequence …VSS|X…, Figure 2).
-
2
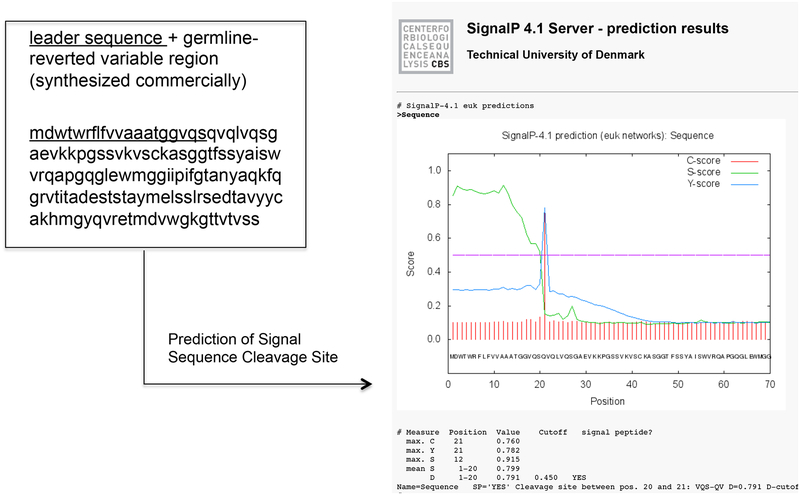
Attach a leader sequence to the V gene-reverted variable region. The variable region (V gene reverted FR1, CDR1, FR2, CDR2, FR3 + mature CDR3, FR4) is synthesized commercially and should be preceded by leader sequence to direct translation into lumen of the endoplasmic reticulum (ER). Leader sequences can be analyzed by the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) to confirm the presence and location of signal peptide cleavage sites (Figure 3). PAUSEPOINT. Synthesized DNA may be stored at −20°C for several months.
-
3
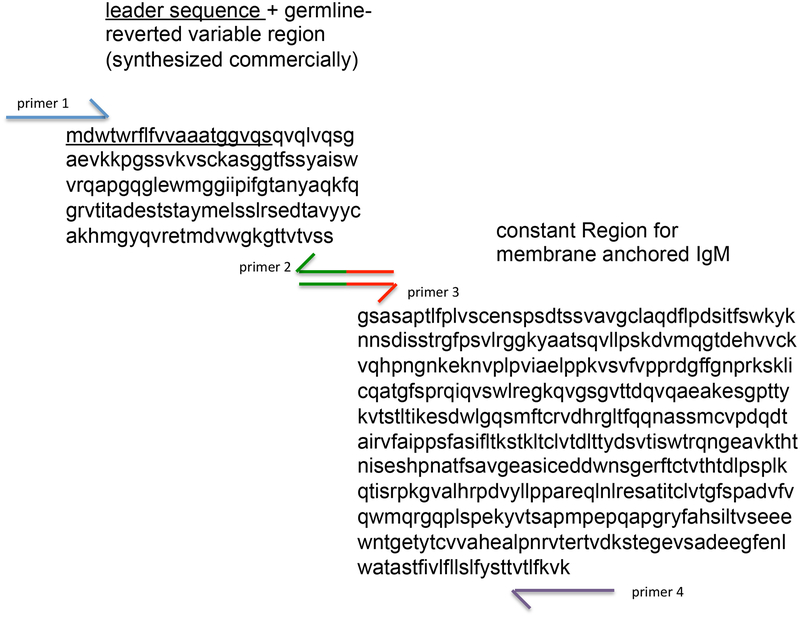
Attach the variable domain of the germline-reverted heavy chain to the constant region encoding membrane-anchored IgM. The sequence for membrane anchored IgM is available in GenBank (Accession X17115) and can also be synthesized commercially. As shown in Figure 2, immunoglobulin variable and constant regions are separated by the amino acid sequence …VSS|X…; consequently conjugating the germline reverted variable region can be performed by overlap PCR between PCR-amplified variable domain fragments and the PCR-amplified membrane IgM constant domain6. This procedure is follows standard PCR methodology and is illustrated using CR6261 as an example in Figure 4.
-
4
Clone germline membrane IgM heavy and light chains into expression vectors for efficient expression in mammalian cells. This involves standard amplification, digestion and ligation of the immunoglobulin gene sequences into the desired expression plasmid. We typically use VRC840043, a plasmid containing the CMV IE Enhancer/Promoter, HTLV-1 R Region and Splice Donor site, and the CMV IE Splice Acceptor site upstream of the open reading frame. Deploy heavy chain and light chains in separate expression vectors. Cloning the antibody sequences into the multiple cloning site of VRC8400 can be conveniently achieved using Xba1 and BamH1 restriction sites. For IGHV1–69-influenza studies, use a QuikChange mutagenesis kit to generate an antigen-binding mutant via introduction of an I53A,I54A substitution6 within the HA stem-contacting CDR2 heavy chain.
PAUSEPOINT. Once the expression vectors have been constructed, they can be stored at −20°C.
Figure 2.
Inferring a germline reverted antibody. Presented is an example using the heavy chain variable region from CR6261, an IGHV1–69-derived bnAb against influenza virus6,48. The nucleotide sequence for the variable region (FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4) was entered into JoinSolver and analyzed using the Kabat nomenclature41 setting. The two important features of the output are: 1) the highest scoring V gene alignment (=V gene origin); and 2) the position and sequence of the mature CDR3. Information on the D and J genes used to generate the CDR3 are also provided in the output, but they are not displayed or used in this protocol as this junctional information it is often not clear39 and is consequently not standard to germline reversion.
Figure 3.
A leader sequence should precede the variable region of germline-reverted antibody. This will insure that the mIgM ectodomain is assembled in the lumen of the ER. For CR6261, the leader sequence “mdwtwrflfvvaaatggvqs” (underlined) is used. Inputting the leader + variable region sequence into the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) confirms the presence and position of the introduced signal peptide cleavage site.
Figure 4.
Overlap PCR to generate membrane anchored IgM configuration of germline reverted antibodies. Two fragments, the variable region (+leader sequence, underlined) and mIgM constant region are first amplified by individual PCR. Primers for these reactions are displayed in color. We recommend a minimum of 20bp overlap for the design of primers 3 and 4. Following amplification and gel purification of the two fragments, they are combined and subject to another round of PCR using only primers 1 and 4. The resultant combined fragment should run at ~1.8 kb. Primers 1 and 4 should be designed with the appropriate restriction enzyme sites to ligate the combined product into the desired expression vector.
PHASE 1 ANTIGEN SCREENING through 293F surface expression and binding: Antigen probe design
TIMING: 2–3 weeks for antigen probe construction
-
5
Clone the antigen probe into a mammalian expression vector (e.g. VRC8400). The antigen probe should contain, at either N- or C- terminus, the biotinylatable avi-tag sequence GLNDIFEAQKIEWHE, followed by a 6xHis-tag for affinity purification. Influenza probes are engineered into VRC8400 as the extracellular domain of HA (A/New Caledonia/20/1999, an H1N1 strain44, GenBank AY289929) fused at the C-terminus to the trimeric foldon domain of T4 fibritin followed by the avi + his tag19. To remove indiscriminant cell surface sialic acid binding activity by HA, incorporate the mutation Y98F into the receptor binding site19. For HA stem binding mutants, introduce I45R,T49R substitutions6 or alternatively an N-linked glycosylation site (I45N,G47T)26 in the stem region. All cloning steps involve standard amplification, digestion and ligation of antigen probe nucleotide sequence into the desired expression vector. Cloning the HA sequence into the multiple cloning site of VRC8400 can be conveniently achieved using Xba1 and BamH1 restriction sites. For mutagenesis, use a QuikChange mutagenesis kit.
PAUSEPOINT. Once the expression vector has been constructed, it can be stored at −20°C.
PHASE 1 ANTIGEN SCREENING through 293F surface expression and binding: Antigen probe expression in 293F
TIMING: 5–6 days
CRITICAL. This section describes our optimized transfection conditions: 500 μg/L of plasmid + 1 mL 293Fectin transfection reagent per liter culture volume with a culture density of 1.2×106 cells/ml. Depending on size of the experiment, cells can be transfected in 250 ml, 500 ml, 1, or 2L baffled flasks with vented lids. To ensure adequate mixing, the culture volume should not exceed 1/3 of the flask volume when all components are present in the culture. We present conditions to transfect recombinant antigen into a 500 ml culture volume, with the transfection taking place in a 2L baffled flask. For HA antigen, 500 ml culture should provide at least 5 mg of protein which should last for several months during the course of this protocol. Generally protein yield can range from <1 to 15 mg/L, so depending on the construct and expression vector used, culture conditions may have to be scaled up (or down).
-
6
Grow 293F cells in FreeStyle media to a culture density of 1.2×106 cells/ml in a humidity controlled shaking incubator set to 37°C, 8% CO2 saturation, 78% humidity and 120 rpm.
CRITICAL STEP. Do not let cell density exceed 3×106 cells/ml as the cells will move out of log growth phase and will not respond well to transfection.
-
7
In a sterile 50ml Falcon tube, add 250 μg of filtered DNA from Step 5 to 12.5 ml of pre-warmed Opti-Mem.
CRITICAL STEP. Filter DNA, Opti-MEM, and Freestyle media to avoid contamination as 293F cannot be grown with antibiotics. There is no antibiotic in the 293F media.
-
8
In another sterile 50ml Falcon tube, add 500 μl of 293Fectin to 12.5 ml of pre-warmed Opti-Mem. Incubate for 5 min at RT.
-
9
Combine the two volumes and incubate for 25 min at RT.
-
10
During this incubation, transfer 500 ml of 293F culture (density = 1.2×106 cells/ml) to a 2 L baffled flask with a vented lid.
-
11
After the 25 min incubation, add the 25 ml transfection solution from Step 9 to the 2L flask and incubate cells in a humidified, shaking incubator set to 37°C, 8% CO2 saturation, 78% humidity and 120 rpm.
-
12
Incubate cells in the cell culture incubator for 5 days.
PHASE 1 ANTIGEN SCREENING through 293F surface expression and binding: Antigen probe purification
TIMING: 1–2 days
CRITICAL. Purification involves affinity- followed by size exclusion chromatography. For the latter, an FPLC set-up is utilized. Different FPLC manufacturers will have different instrument settings, however the steps listed should be universal.
-
13
Following the five days of culture described in Step 12, harvest media by centrifugation at 2,000 × g, and filter through VacuCap 8/0.2 μm filters.
PAUSEPOINT. Supernatant may be stored for several days at 4°C, however long term storage is not advisable as protein aggregation may occur.
-
14
Using a tangential flow filtration (TFF) setup (30 kDa cut off), concentrate the supernatant and exchange the buffer with PBS. For a 500 ml supernatant, concentrate sample down to 100ml and then PBS to bring it back to a 500 ml volume. Repeat and concentrate sample to 100 ml.
CRITICAL STEP. Ensure that the TFF system is thoroughly washed between steps with at least 4–6 L of H2O between different samples. Clean the system with 2L of 0.1N NaOH after use. Insure that sample lines are clamped to insure there is no backflow when the system is not in use.
CRITICAL STEP. If immunogens are eventually going be used in non-human primates, then the system must be cleared of endotoxin via an overnight soak in 1N NaOH prior to use.
-
15
Equilibrate Ni sepharose with 50 ml of H2O. Use 1 ml of resin per 2.5 liter of expression culture. Spin at 500 × g for 5 min and re-equilibrate in 50 ml of PBS.
-
16
Load resin onto a 1.5×20 cm Glass Econo-Column and wash through with another 50 ml of PBS.
-
17
Add imidazole to the protein concentrate from Step 14 to give a final concentration of 20 mM imidazole (from 5M imidazole stock, add 0.4 ml to the 100 ml concentrate), and load on to column prepared in Step 15.
-
18
Wash column with 6 column volumes of PBS containing 60 mM imidazole. Discard washes.
-
19
Elute antigen probe from column with 2–5 column volumes of PBS containing 500 mM imidazole.
-
20
If proceeding to immediate use, concentrate the eluted protein to >1ml using an Amicon Ultra centrifugal concentrator (30 kDa cutoff) prior to proceeding to Step 21.
PAUSEPOINT. For overnight storage, dialyze the eluted protein overnight in 4–5 L of PBS using a Slide-A-Lyzer Dialysis Cassette. The next day, concentrate the sample as described in this step prior to proceeding to size exclusion chromatography (Steps 21–30).
CRITICAL STEP. Some FPLC systems have automated sample infusion via a dedicated sample pump. If using one of these systems, it is not necessary to concentrate samples to a low injection volume, thus this step is not needed.
TROUBLESHOOTING
-
21
In preparation for size exclusion FPLC, degas 500ml of PBS using a Büchner/vacuum flask
CRITICAL STEP. Ensure that degassed PBS is used to avoid bubbles from developing in the column.
-
22
Use double distilled water (DDW) to wash through flow path without column.
-
23
Connect Superdex 200 10/300 column to the flow path FPLC system. Connect during flow to ensure the column remains under pressure.
CRITICAL STEP. Do not exceed maximal flow rate (0.5 ml/min) as it will damage the column.
-
24
Wash through column with 1.5 column volumes PBS.
-
25
Wash 1 ml manual injection loop with 15 ml H2O.
-
26
Inject 1 ml sample from Step 20 into a 1ml sample loop or use the automated sample infusion system of the FPLC system if appropriate.
TROUBLESHOOTING
-
27
Run size exclusion chromatography program. Key program parameters include: emptying the sample loop with 1.1 ml of PBS and automated fraction collection of protein.
-
28
Collect fractions (e.g. trimeric HA will elute between 10–12ml).
-
29
Pool and concentrate the fractions corresponding to the correct antigen size to 1–2 mg/mL protein. Calculate final protein concentration by dividing the absorbance at 280nm by the extinction coefficient of the protein.
PAUSEPOINT. Samples may be aliquoted, snap-frozen in liquid nitrogen and stored at −80°C.
-
30
Equilibrate FPLC pump and then column in PBS containing 20% ethanol or 0.02% sodium azide in preparation for storage.
CRITICAL STEP. Without preservatives, the column will eventually become contaminated.
PHASE 1 ANTIGEN SCREENING through 293F surface expression and binding: Antigen probe biotinylation and fluorescent labeling
TIMING: 1–2 days
CRITICAL. This section describes the procedure to fluorescently label the antigen probe by means of a site-specific avi-tag, such that the antigen probe can be used for flow cytometry.
-
31
Buffer-exchange concentrated protein from Step 29 into 10 mM Tris pH 8.0 using 10 kDa cut off Slide-A-Lyzer Dialysis Cassettes for at least 2h at 4°C.
PAUSEPOINT. Buffer exchange can be extended to overnight at 4°C.
-
32
Using the components of the Protein Biotin Ligase kit, combine 40 μM of antigen probe with 7 parts Biomix-A, 7 parts Biomix-B, and 2.5 μg of BirA biotin ligase per nmol substrate, and incubate at 30°C for one hour.
CRITICAL STEP. BirA has optimal activity in 10 mM Tris pH 8.0. Biotinylation in PBS is possible, however, longer incubation times and higher enzyme levels are needed.
-
33
33. Re-purify the resulting biotinylated antigen probe by size exclusion chromatography in PBS, as described in Steps 21–30.
TROUBLESHOOTING
-
34
Calculate the amount of antigen that needs to be combined with fluorescent label, such that 25 flow cytometry assays can be performed wherein each assay uses 5 μl of antigen that contains 0.4 μg of protein. In this conjugation, add fluorescent streptavidin (e.g. PE or APC) to biotinylated antigen probe in five increments, such that the molar ratio of probe to streptavidin valency (4 protein binding sites) is 1:1. For an example calculation, see Box 2.
-
35
Perform conjugation according to the calculation made in Step 34. In the example provided in Box 2, 10 μl of antigen at 1 μg/μl is added to 103.5 μl PBS, and then fluorescent PE-streptavidin is added in 2.3 μl increments with a total of 5 increments. After each stepwise addition of fluorescent label, incubate the mixture for 20 min and set rotating at 4°C.
PAUSEPOINT. Fluorescently labeled probes can be stored at 4°C in opaque microtubes to prevent light damage but should be used within 1–2 months.
BOX 2: Example calculation fluorescent labeling of antigen.
The following is a calculation for labeling biotinylated antigen with fluorescent streptavidin through iterative complex formation. The biotinylated antigen is labeled in five increments, such that the molar ratio of probe to streptavidin valency (4 protein binding sites) is 1:1.
Parameters
Number of assays = 25
Volume per assay= 5 μl
Amount of protein per assay = 0.4 μg
Molecular weight HA monomer = 65 000 g/mol
Concentration of biotinylated HA = 1 μg/μl
Molecular weight of fluorescent label: APC = 164 000 g/mol; PE = 300 000 g/mol
Concentration of fluorescent label = 1 μg/μl
Ratio of Streptavidin-protein binding sites to fluorescent label = 4
Antigen monomer:label excess ratio = 1
Number of Conjugation Steps = 5
-
xxixCalculation for Antigen
- Total protein required (μg)
- =(number of assays) × (amount protein per assay)
- =25 × 0.4 μg
- =10 μg
- Volume of monomer per test (μl)
- =[(number of assays) × (amount of protein per assay)]/ (concentration biotinylated protein)
- =(25 × 0.4 μg)/ 1 μg/μl
- =10 μl
- Moles monomer per μg protein (mol/μg)
- = 1/(65 000 g/mol) × 1 g/(106 μg)
- = 1.53 × 10−11 mol/μg.
- Total moles protein for assay (mol)
- = (amount protein per assay) × (moles monomer per μg protein) × (number of assays)
- = (0.4 μg) × (1.53 × 10−11 mol/μg) × 25
- =1.53× 10−10 mol
-
xxxCalculation for fluorescent antigen
- Molar concentration of fluorescent label (e.g. PE) (mol/L)
- =(concentration of fluorescent label) / (molecular weight of fluorescent label)
- =1 g/L / (300 000 g/mol)
- =3.33×10−6 mol/L
- Molar concentration of protein binding sites (mol/L)
- = (molar concentration of fluorescent label) × (ratio of streptavidin-protein binding
- sites to fluorescent label)
- = 3.33×10−6 mol/L × 4
- =1.33×10−5 mol/L
- Desired moles of label per monomer (mol)
- =(Total moles of protein for assay) / (antigen monomer:label excess ratio)
- = 1.53× 10−10 mol / 1
- = 1.53× 10−10 mol
- Total label volume (μl)
- = (desired moles of label per monomer) / (molar concentration of concentration of protein binding sites) × 106 μl/L
- = (1.53× 10−10 mol) / (1.33×10−5 mol/L) × 106 μl/L
- =11.5 μl
-
xxxiCalculation for Conjugation
- Label volume per conjugation step (μl)
- =(Total label volume) / (number of conjugation steps)
- =11.5 μl / 5
- =2.3 μl
- Volume of PBS to reach desired final volume(μl)
- = [(number of assays) × (volume per assay)] – (total label volume)- (volume of monomer per test)
- =(25 × 5 μl) - 11.5 μl – 10 μl
- =103.5 μl
- Accordingly,10 μl of biotinylated HA at 1 μg/μl should be first added to 103.5 μl PBS. Then fluorescent PE-streptavidin at 1 μg/μl should then be added in 5 increments, with an incremental volumne of 2.3 μl.
PHASE 1 ANTIGEN SCREENING through 293F surface expression and binding: Expression of membrane IgM in 293F
TIMING: 72 h
CRITICAL. In this section we describe the optimized transfection conditions we have developed to express mIgM in reporter 293F cells. Our optimized transfection conditions use 500 μg/L of plasmid + 1 mL 293Fectin transfection reagent per liter culture volume with a culture density of 1.2×106 cells/ml. In this section, we present conditions to express mIgM in 125ml flasks with 20ml culture volume. We find that this is a convenient volume for flow cytometry assays and should not be confused with the larger 293F volumes needed for protein purification in Step 6.
-
36
Grow 293F cells in FreeStyle media to a culture density of 1.2×106 cells/ml in a humidity controlled shaking incubator set to 37°C, 8% CO2 saturation, 78% humidity and 120 rpm.
CRITICAL STEP. Do not let cell density exceed 3×106 cells/ml as the cells will move out of log growth phase and will not respond well to transfection.
-
37
In a sterile 2ml Eppendorf tube, add 10 μg of filtered DNA (generated in Steps 1–4; mIgM heavy chain +light chain) to 500 μl of pre-warmed Opti-Mem.
CRITICAL STEP. Filter DNA, Opti-MEM, and Freestyle media to avoid contamination as 293F cannot be grown with antibiotics. There is no antibiotic in the 293F media.
-
38
In another sterile 2ml Eppendorf tube, add 20 μl of 293Fectin to 500 μl of pre-warmed Opti-Mem. Incubate for 5 min at RT.
-
39
Combine the volumes from Steps 37 and 38 and incubate for 25 min at RT. During the 25 min incubation, proceed with next step.
-
40
During the incubation in Step 39, transfer 20 ml aliquots of 293F culture (density = 1.2×106 cells/ml) to separate 125mL baffled flasks with vented lids (use one flask for each of the following: each mIgM of interest; each IgM control; and each mutant or irrelevant antibody).
-
41
After the 25 min incubation, add 1 ml transfection solution from Step 39 to each flask.
-
42
Incubate cells for 72h in the humidified, shaking incubator a humidity controlled shaking incubator set to 37°C, 8% CO2 saturation, 78% humidity and 120 rpm.
PHASE 1 ANTIGEN SCREENING through 293F surface expression and binding: 293F Membrane IgM Recognition Assay
TIMING: 3.5 h
CRITICAL. This section assays whether fluorescent antigen (generated from Steps 34 and 35) engages the germline mIgM constructed during Steps 1–4 and expressed during Steps 36–42.
-
43
After the seventy-two hour incubation for transfection (Step 42), spin down cells (1,000 × g, 5 min) and resuspend IgM expressing 293F in 5 ml of PBS. Transfer 95 μl of cells to each well of a 96 well format plate containing V- bottomed wells. Run samples in duplicate. As described in Step 34, the antigens can be labeled with either PE or APC. To simultaneously confirm surface trafficking of mIgM, PE-conjugated anti-light chain antibodies are also applied to the transfected cells (see Step 45). Consequently, when using APC-conjugated antigens, it will require two wells for each antigen where each well will contain the fluorescent antigen and anti-light chain combined together (can test 48 antigens per plate). If the antigen is labeled with PE, then two are needed for the sample, and two additional wells are needed for the light chain antibody (can test 24 per plate). Each fluorescent antigen and fluorescent antibody will be applied in a single fixed concentration, as detailed in Step 45.
-
44
After adding the cells to the wells, place on ice and add 5 μl ViVid staining solution (total volume = 100 μl of cell suspension in each well). Stain on ice for 15 min.
-
45
Wash each well with PBS, then on ice, stain each well for 1 hour with probes of interest 5 μl fluorescent antigen (which should correspond to 0.4 μg protein; prepared in Steps 34 and 35) and a choice of either 5 μl PE-anti-kappa chain, or 20 μl PE-anti-lambda chain. Each well should be made up to a total volume of 100 μl with PBS.
CRITICAL STEP. The anti-Ig volumes we state here are those recommended by the suppliers.
-
46
Wash each well twice and then place PBS containing 0.5% PFA into each well to fix samples. Fixed samples may be analyzed immediately.
PAUSEPOINT. Fixed samples may also be stored overnight at 4°C and then analyzed the following day.
-
47
Quantify antigen binding by flow cytometry. Apply the following gating scheme: 1) side scatter area × forward scatter area to display cells; 2) side scatter area × vivid intensity to identify the living cells that are not ViVid positive; and 3) a histogram plot of PE or APC intensity for the living cells (Figure 5). Ideally this step is performed using a flow cytometry cell analyzer capable of automated sampling and analysis from a 96 well format [e.g. the LSR II system with high throughput sampler (HTS) by BD].
TROUBLESHOOTING
PAUSEPOINT. This marks the end of Phase 1. Antigens that are found to engage mIgM of interest can proceed to analysis in Phase 2.
Figure 5.
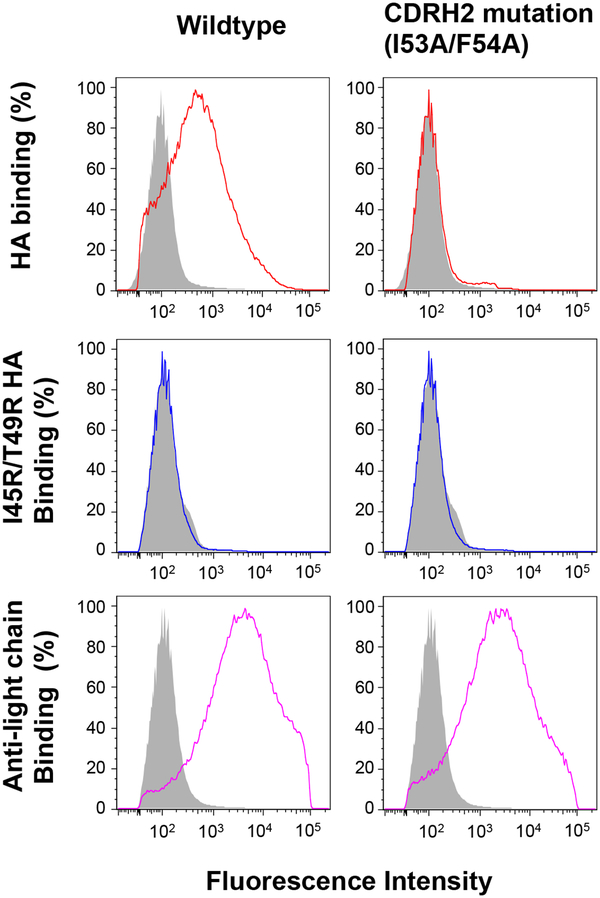
Membrane presented antibody as a means to pre-screen candidate immunogens for the capacity to engage germline BCR sequences of interest. Candidate immunogens are fluorescently labeled and their binding to mIgM antibody expressed on the surface of 239F cells is evaluated by flow cytometry. Presented is CDRH2-dependent recognition of the influenza HA stem region by germline CR6261 (colored lines; grey is binding to 239F expressing VRC01 mIgM, an irrelevant antibody sequence). Surface trafficking of both wildtype and CR6261 germline mIgM containing a I53A/F54A mutation in CDRH26 is confirmed using a fluorescent anti-light chain antibody. Specificity for the HA stem is defined by reactivity to wildtype HA but not HA containing the mutation I45R/T49R in the stem epitope6.
PHASE 2 Assessing BCR activation by candidate antigens: Generate Lentivirus for BCR Delivery
Timing: 4–5 days
CRITICAL. This section describes how to generate a lentivirus encoding the mIgM sequences of interest for stable expression in a B cell line. In this protocol, the FEEKW lentiviral expression system originally designed to deliver exogenous antibody sequences to hematopoietic B cell progenitors33 is employed.
-
48
Clone mIgM heavy chain and light chain sequences generated in Steps 1–3 into a lentiviral expression vector. This involves standard amplification, digestion and ligation of the immunoglobulin gene sequences into the desired expression plasmid. Cloning antibody sequences into the FEEKW plasmid can easily be achieved using Xho1, Xba1, Hpa1, or BamH1 restriction enzyme sites within the multiple cloning sequence this vector. The other components of the FEEKW lentiviral system include a VSVG expressing plasmid for envelope pseudotyping37 and a packaging vector (pΔR8.238) which need not be altered.
-
49
Grow 293T cells to 80% confluence with DMEM in a 10 cm dish in a cell culture incubator set to 37°C and 5% CO2 saturation.
CRITICAL STEP. Insure that 293T are neither fully confluent, nor less than 50% confluent. Both conditions will result in lower viral titer.
-
50
Three hours before transfection, replace medium with fresh DMEM.
-
51
Using the ProFection Transfection System co-transfect: 7.5 μg FEEKW plasmids for heavy and light chains of interest33; 7.5 μg ΔR8.2 (a packaging vector that under CMV expression produces all necessary HIV-1 trans-acting proteins minus Env38); and 1 μg of VSVG (vesicular stomatitis virus glycoprotein for envelope pseudotyping37) as described in manufacturers instructions and incubate cells in the cell culture incubator overnight.
-
52
The next morning after the overnight transfection, replace medium with fresh DMEM.
-
53
Incubate cells for a further forty-eight to seventy-two hours to allow time for cells to produce virus.
-
54
Following the 48–72 h viral production period, harvest virus by spinning cells at 2,000 × g for 5 min and collecting 10 ml supernatant.
-
55
Filter supernatant through a 0.45 μm pore filter.
PAUSEPOINT. Virus may be divided into 1 ml aliquots and snap frozen in liquid nitrogen and stored at −80°C for several months. Alternatively, virus may be stored for up to two weeks at 4°C.
PHASE 2 Assessing BCR activation by candidate antigens: Isolate and expand Ramos B cells that are surface negative for IgM
Timing: 3–5 weeks
CRITICAL. In this section surface negative Ramos B cells are generated that can then be used to express mIgM sequences of interest, generating mono-specific BCR display. In all cases, culturing takes place in a cell incubator set to 37°C and 5% CO2 saturation.
-
56
Culture wildtype Ramos B cells in cRPMI in the cell culture incubator set to 37°C and 5% CO2 saturation.
-
57
Spin down 2 × 106 cells (1,000 × g, 5 min) and resuspend in 100 μl PBS. Add 95 μl cells and 5 μl ViVid staining solution to each well of a 96 V-well plate and incubate on ice for 15 min.
-
58
Wash each well once with PBS then add 5 μl APC-anti-IgM, 20 μl PE-anti-lambda chain and 75 μl PBS (the anti-Ig volumes given here are those recommended by the supplier; these work well for us).
-
59
Wash cells twice in PBS and then sort for light chain/ IgM negative cells by FACS. Apply the following gating scheme: 1) side scatter area × forward scatter area to display cells; 2) side scatter area × vivid intensity to identify the living cells that are not ViVid positive; and 3) on living cells sort for single IgM/lambda negative clones into directly into conditioned cRPMI in 96 well format. This step requires a cell sorter such as FACS Aria II (BD Biosciences), not to be confused with the cell analyzer used in Step 47.
CRITICAL STEP. To culture single Ramos B cells, conditioned media must be used, otherwise cells will die.
-
60
Culture the cells in conditioned media to 50–80% confluency in 100 μl in 96 well format, followed by 300 μl, 500 μl, 1 ml, and 2 ml in 48, 24, 12, and 6 well formats, respectively. This method of scaling up will ensure that the culture density remains high, a feature that we find is important for growth of this cell type. At the 6 well stage, culture medium may be exchanged for non-conditioned cRPMI and cells may be further scaled up in 5–10 ml in a T25 flask and then in 10–20 ml in a T75 flask. Grow up to 1×106 cells/ml into a T75 flask, spin down the cells (1,000 × g, 5 min), resuspend in FBS containing 10% DMSO and transfer to a cryovial and freeze using a cell freezer. Cells will need 3–4 weeks to grow in a T75 flask.
TROUBLESHOOTING
-
61
Perform two additional cycles of sorting and enrichment of light chain/ IgM negative Ramos. In these cycles, sort cells in bulk (e.g. 3×106 cells) and directly culture in 10–20 ml cRMPI in a T75 flask. No conditioned media is needed at this stage.
-
62
Separate a subset of low passage number cells for freezing. Grow in cRPMI to 1×106 cells/ml into a T75 flask, spin down the cells (1,000 × g, 5 min), resuspend in FBS containing 10% DMSO, and transfer to a cryovial and freeze using a cell freezer prefilled with 2-propanol according to the manufacturer’s instructions.
PHASE 2 Assessing BCR activation by candidate antigens: Express mIgM as signaling competent germline BCR
TIMING: 3–4 weeks
CRITICAL. In this section, lentivirus generated in Steps 48–55 is used to generate B cells expressing BCRs of interest from the IgM negative Ramos generated from Steps 56–62. Individual lentivirus preparations may be tittered with surface IgM/anti-light chain co-expression as a readout; the conditions presented give a BCR expression level of less than 1% following initial infection, indicating a multiplicity of infection (MOI) below 1 (i.e. the BCR expressing cells have not been infected with multiple copies of virus). In all cases, culturing takes place in a cell incubator set to 37°C and 5% CO2 saturation.
-
63
Spin down (1,000 × g, 5 min) and resuspend 2×106 IgM surface negative Ramos cells grown up in Step 61 with 2 ml of lentivirus from Step 55 (1 ml of mIgM heavy chain virus + 1 ml light chain virus) containing 8 μg/ml polybrene. The lentivirus can be quantified by p24 ELISA (see Table 1), however this is not initially necessary as we find that the conditions presented consistently generate a BCR expression level with MOI <1.
-
64
After the overnight lentiviral infection, replace medium with cRPMI and culture cells for 3–4 days.
CRITICAL STEP. Insure that media is changed back to cRPMI and cells are allowed to culture to the point where they are again dividing.
TABLE 1.
Troubleshooting
| Step | Problem | Possible reason (s) | Solution |
|---|---|---|---|
| 20 | Protein is not well concentrated. | Expression is low. | Increase culture time to 6 days, vary ratio of DNA to 293Fectin. |
| Protein is stuck at base of concentrator. | Mix concentrate 10 times with a pipette prior to removing. | ||
| Eluted protein is precipitating. | The concentration is too high. | Do not keep the protein unfrozen above 1–2mg/ml for long periods of time. | |
| 26 | Sample is difficult to inject. | There is salt precipitate obstructing the sample flow. | Disconnect sample loop and backflow 0.1N NaOH through the pathway. Wash with H2O. |
| Column has collapsed. | Flow rate was too high. | Invert the column and run at a lower flow rate. | |
| 33 | Is it possible to skip last FPLC step to increase efficiency? | Increase throughput. | Yes, antigen probe may be buffer exchanged back to PBS minus biotinylation components by centrifugal concentration. |
| 47 | Immunogen probe does not bind antibody, even to affinity matured mIgM. | Antigen probe was not properly biotinylated. | Confirm biotinylation by capture with streptavidin-coated plates, and detect by ELISA with an antibody specific to immunogen probe. Adjust biotinylation parameters (e.g. antigen probe concentration, BirA levels, incubation time) to optimize labeling. |
| MIgMs are not trafficking to cell surface. | Plasmid has degraded or 293F cell culture is suboptimal. | Maxiprep fresh plasmid. Ensure that 293F are in log-phase growth. Thaw out low passage number and repeat. | |
| 60 | Sorted IgM− Ramos cells are dying. | Preconditioned media is not being used. | Used preconditioned media and sort a number of 96 well plates. Culture and save low passage numbers for future experiments.. |
| Sorted Ramos are difficult to scale up. | Ramos are difficult to grow at low density. | Keep density high and scale from 96 to 48 to 24 to 12 to 6 well format then to T25 to T75. | |
| Cells die after thawing. | Ramos can be fragile at this stage. | 1) Thaw tube at 37°C and transfer to a 50 ml Falcon tube. | |
| 2) Add DROPWISE 5ml of pre-warmed cRPMI to the cells and incubate at RT for 5 min. (This will ensure slow diffusion of DMSO out of the cells.) | |||
| 4) Add 30–40ml cRPMI and pellet cells. | |||
| 6) Resuspend (6 ml), culture in T25 and scale up. | |||
| 68 | Ramos do not express BCR. | Lentivirus is not produced at high enough titers. | Make fresh helper plasmid stocks. If using pΔR8.2, the level concentration of virus may be titered by p24 levels as determined by the Perkin-Elmer ELISA kit (product# NEK050001KT). |
| Sorted Ramos not growing. | Cells not dense enough. | Culture densely and use conditioned media. | |
| 78 | Calcium flux is weak or not seen. | Inefficient incorporation of calcium sensitive dye. | Increase concentration of dye, increase incubation time, insure that there is a dynamic range in which the positive controls (anti-IgM, ionomycin) work. |
| Cells are dying. | Maintain cells at 37°C in a water bath during run | ||
| 78 | The cell lysate has low protein levels. | The cells are dying. | Insure that cells >95% viable prior to experimentation. |
-
65
Spin down (1,000 × g, 5 min) and resuspend cells in 95 μl PBS, place on ice, add 5 μl of ViVid staining solution and incubate for 15 min.
-
66
Wash cells once with PBS then stain cells with 5 μl APC-anti-IgM + 20 μl PE-anti-lambda chain + 75 μl PBS or 5 μl APC-anti-IgM + 5 μl PE-anti kappa light chain + 90 μl PBS (the anti-Ig volumes are those recommended by the supplier).
-
67
Enrich double BCR positive cells by FACS. Apply the following gating scheme: 1) side scatter area × forward scatter area to display cells; 2) side scatter area × vivid intensity to identify the living cells that are not ViVid positive; and 3) on living cells gate on APC/PE double positives (Figure 6). Double positives will be less than 1% of cells (MOI<1). Sort as many BCR expressing cells as possible into 1 ml of conditioned cRPMI.
-
68
Culture cells to confluency in a 12 well plate in conditioned media and then scale up to 2 ml in a 6 well plate and 5–10 ml in a T25 flask. Conditioned media need not be applied in the T25 flask.
-
69
Perform two or three additional rounds of sorting by FACS to enrich BCR positive B cells to a minimum of 85% positivity for BCR expression (Figure 6). Apply the same gating scheme outlined in Step 67, however, in this step the numbers of positive B cells will be much higher (e.g. 1–3×106 cells) and can be cultured directly in T25 flasks in cRPMI without conditioning.
-
70
Separate a subset of low passage number cells for freezing. Grow in cRPMI to 1×106 cells/ml into a T75 flask, spin down the cells (1,000 × g, 5 min), resuspend in FBS containing 10% DMSO transfer to a cryovial and freeze using a cell freezer prefilled with 2-propanol according to the manufacturer’s instructions.
PAUSEPOINT. Use BCR enriched Ramos in culture for 3 month periods, during which time BCR positivity should be stable.
TROUBLESHOOTING
Figure 6.
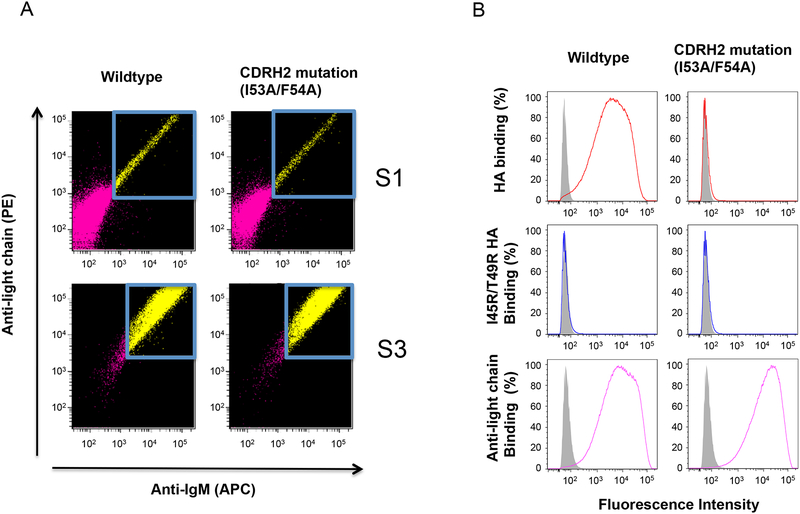
Lentiviral mediated expression of BCR in Ramos. To assess BCR activity in response to antigen, heavy and light chain components are expressed in a clone of Ramos expressing no endogenous mIgM at the cell surface. Following enrichment by FACs (A), BCR antigenicity (B) is assessed by flow cytometry. CDRH2-dependent recognition of the influenza HA stem by germline CR6261 (colored lines) versus irrelevant IgM VRC01 IgM BCR (grey) is assessed by binding of fluorescent HA (but not I45R/T49R HA) to wildtype BCR but not to its mutant form containing a I53A/F54A substitution in CDRH26. S1 refers to the first sort after lentiviral infection and S3 shows the cells after three rounds of BCR sorting/enrichment.
-
71
To confirm that candidate immunogens engage the BCR format of the antibody sequence of interest, repeat Steps 43–47, except instead of using 293F transfected with mIgM, use 2×106 of mIgM expressing Ramos B cells generated during Steps 63–70. Similar antigen binding should be seen by germline antibody sequences of interest displayed in the 293F system (Figure 5) and the Ramos B cell system (Figure 6).
PHASE 2 Assessing BCR activation by candidate antigens: Antigen Multimer Expression in 293F and Purification
TIMING: 5–6 days for protein expression, 2 days for purification
CRITICIAL. In this section, soluble trimeric HA is further multimerized to induce BCR signaling. We use the recently developed HA ferritin nanoparticle platform26 as the BCR stimulating ligand in this section of the procedure. As for the earlier part of the procedure where the monomeric antigen probe is expressed and purified (Steps 6–30), production of HA ferritin involves expression in 293F and purification through affinity chromatography and size exclusion FPLC. Additionally there is an ion-exchange FPLC step that is used to purify control empty ferritin nanoparticles. Different FPLC manufacturers will have different instrument settings, however the steps listed should be universal.
CRITICAL. A high throughput method of arraying the His-tagged antigen probes from Phase 1 (Steps 6–30) on the surface of 100 nm liposomes is also described (Box 1) and can be used as an alternative to Steps 72–77. As nanoparticle systems must be tailored to each individual antigen, a high-throughput method can be helpful. Proteoliposome preparations such as those used in Box 1 can be used in place of nanoparticle scaffolds for BCR triggering analyses6,20.
-
72
Clone HA ferritin nanoparticle26 into a mammalian expression vector (e.g. VRC8400). As with construction of antibody plasmids (Steps 1–4), nucleotide sequences can be synthesized commercially and then incorporated into the desired expression vector using standard PCR-amplification, digestion and ligation methodology. For HA ferritin26, the human CD5 leader sequence (Genbank NM_014207) and a Ser-Gly-Gly spacer is linked to the ectodomain of HA (GenBank AY289929, residues HA1 1-HA2 174) which is then connected to the Helicobacter pylori non-haem iron containing ferritin (Genbank NP_223316, residues 5–167) by means of another Ser-Gly-Gly linker. As described in Step 2, the leader sequence can be analyzed by the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) to determine the presence and location of signal peptide cleavage sites. HA nanoparticles also contain the Y98F mutation to prevent non-selective binding to cell surface sialyl-oligosaccharide19. Empty/control ferritin nanoparticles are encoded by human CD5 leader sequence-Ser-Gly-Gly-ferritin (residues 5–167)26. Cloning the ferritin nanoparticle sequences into the multiple cloning site of VRC8400 can be conveniently achieved using Xba1 and BamH1 restriction sites.
-
73
Grow 293F cells in FreeStyle media to a culture density of 1.2×106 cells/ml in a humidity controlled shaking incubator set to 37°C, 8% CO2 saturation, 78% humidity and 120 rpm.
CRITICAL STEP. Do not let cell density exceed 3×106 cells/ml as the cells will move out of log growth phase and will not respond well to transfection.
-
74
Transfect plasmids encoding nanoparticles or empty ferritin controls generated in Step 72 into 293F cells following the transfection conditions described in Steps 6–12.
CRITICAL STEP. Filter DNA, Opti-MEM, and Freestyle media to avoid contamination as 293F cannot be grown with antibiotics. There is no antibiotic in the 293F media.
-
75
Incubate the cells for 5 days with the transfection reagents and then harvest by centrifugation at 2,000 × g, and filter supernatant containing nanoparticles through VacuCap 8/0.2 μm filters. Cells can be discarded at this stage.
-
76
Concentrate nanoparticles using TFF (as described in Step 14).
-
77
Purify nanoparticles from medium from Step 76 as described in Option A for HA ferritin nanoparticles and Option B for empty control ferritin nanoparticles.
Option A) Purification of HA ferritin nanoparticles
-
i
At 4°C, gently shake 100 ml of TFF-PBS exchanged nanoparticle concentrate with 2 mL of PBS equilibrated Erythrina cristagalli lectin-immobilized resin.
-
ii
Incubate overnight with gentle agitation at 4°C. Lectin affinity is weak, therefore overnight incubation with resin is necessary to increase yields.
-
iii
Following the overnight incubation, load resin onto a 1.5×20 cm Glass Econo-Column and wash through with 5 column volumes of PBS (gravity flow).
-
iv
Elute HA nanoparticles with two column volumes of 0.2 M D-lactose in PBS.
-
v
Concentrate eluted protein in a centrifugal concentrator (100 kDa cut off).
CRITICAL STEP. Some FPLC systems have automated sample infusion via a dedicated sample pump. If using one of these systems, it is not necessary to concentrate samples to a low injection volume, thus this step is not needed.
-
vi
Perform size exclusion FPLC as described in Steps 21–30 except use a Superose 6 10/300 column (HA nanoparticle will elute between 9–10ml) and centrifugal concentrators with 100kDa cut off. As with antigen probes, final protein concentration can be conveniently calculated from its absorbance at 280nm divided by the extinction coefficient of the protein.
CRITICAL STEP. Aliquot and snap freeze the protein in liquid nitrogen after the concentration is measured. Store at −80°C for several months.
Option B) Purification of empty control ferritin nanoparticles
-
vii
Concentrate nanoparticle concentrate from Step 76 to 1 ml using an Amicon Ultra centrifugal concentrator (100 kDa cut off).
CRITICAL STEP. Some FPLC systems have automated sample infusion via a dedicated sample pump. If using one of these systems, it is not necessary to concentrate samples to a low injection volume, thus this step is not needed.
-
viii
Degas 500ml of Low Salt Tris buffer and 500 ml of High Salt Tris Buffer using a Büchner/vacuum flask
CRITICAL STEP. Ensure that degassed buffer is used to avoid bubbles from developing in the column.
-
ix
Equilibrate FPLC pump in degassed Low Salt Tris buffer
-
x
Use double distilled water (DDW) to wash through flow path without column.
-
xi
Connect HiLoad 16/10 Q Sepharose HP column to flowpath. Connect during flow to ensure the column remains under pressure.
CRITICAL STEP. Do not exceed maximal flow rate (1ml/min) as it can damage column.
-
xii
Wash through column with 1.5 column volumes of Low Salt Tris buffer
-
xiii
After washing the 1 ml manual injection loop with 15 ml H2O, inject 1 ml sample from Option B (i) or use the automated sample infusion system of the FPLC system if appropriate.
-
xiv
Run ion exchange chromatography program. Key program parameters include: a flow rate of 0.5 ml/min; emptying the sample loop with 1.1 ml of Low Salt Tris Buffer; sample elution by a linear gradient of 0–50% of High Salt Tris Buffer; and automated fraction collection of protein.
-
xv
Collect empty ferritin nanoparticle peak (will elute at ~250mM NaCl).
-
xvi
Perform size exclusion FPLC as described in Steps 21–30 except use a Superose 6 10/300 column and centrifugal concentrators with 100kDa cut off. As with antigen probes, final protein concentration is calculated from its absorbance at 280nm divided by the extinction coefficient of the protein.
CRITICAL STEP. Aliquot and snap freeze the protein in liquid nitrogen after the measuring its concentration. Store at −80°C for several months.
-
xvii
Equilibrate FPLC pumps and then column in PBS containing 20% ethanol or 0.02% sodium azide as a storage solution.
CRITICAL STEP. Without preservatives, column can and eventually will become contaminated.
PHASE 2 Assessing BCR activation by candidate antigens: BCR activation assays
TIMING: 2h (Option A); 1–2 days (Option B)
-
78
BCR expressing Ramos generated in Step 69 are stimulated by antigen multimerzed in nanoparticle format (Step 77) or by proteoliposomal array (Box 1). BCR triggering can be assessed by measuring calcium flux (Option A) or phosphorylation of downstream BCR effectors (Option B).
Option A) Calcium flux to measure the kinetics of BCR activation
CRITICAL. Calcium flux in response to BCR stimulation is measured by flow cytometry as a ratio between the Ca2+ bound/unbound states of the cell permeable dye Fura Red30.
-
xviii
Spin down (1,000 × g, 5 min) 1×106 BCR-expressing Ramos (generated in Step 69) and resuspend in 100 μl of Fura Red staining solution. Incubate for 30 min at 37°C.
-
xix
Spin down (1,000 × g, 5 min) and resuspend cells in a FACS tube containing a suitable volume of RPMI media for B cell signaling. For example, if using the lowest flow rate setting on a BD instrument, use a 300 μl sample volume as this will allow for 5 min of data acquisition).
CRITICAL STEP. Maintain stained cells in a 37°C water bath during flow cytometry analysis. This will ensure signaling competency over the course of a >2 h experiment.
CRITICAL STEP. Do not put FBS in the media as it may degrade antigen.
-
xx
Acquire a 30 second baseline recording on a flow cytometry cell analyzer. Apply the following gating scheme: 1) side scatter area × forward scatter area to display cells; 2) from the cells, fluorescence emission for Fura Red in Ca2+ unbound state (emission maximum = 657 nm) versus Ca2+ bound state (emission maximum = 637 nm). Most flow cytometers will have the appropriate filters to distinguish between Ca2+ bound/unbound Fura Red.
-
xxi
After 30 seconds, remove tube and spike in either 5 μg/ml anti-human IgM F(ab’)2 as a positive control or multimerized antigen (for germline stimulation, 2.5 μM multimer is a useful starting point). Immediately return FACS tube to instrument (this should take less then 10 seconds).
-
xxii
Record for an additional 3–5 min. Within this time period calcium will flux in response to BCR stimulation and then return to baseline.
-
xxiii
In another tube, perform the same procedure but instead spike in 10 μg/ml of the calcium ionophore ionomycin. The difference between baseline and cells permeablized for Ca2+ represents the total flux capacity. The cells will not return to baseline following addition of the ionophore.
CRITICAL STEP. Insure that sample lines are well washed in-between ionomycin use. Low levels of ionophore carryover have the potential to alter flux measurements in subsequent sample runs.
-
xxiv
Export the intensity readings for high and low calcium emission. These parameters can conveniently be displayed and extracted from FlowJo software (http://www.flowjo.com) and processed in Microsoft Excel. Average ratiometric measures for individual runs (n=2–3) and standardize antigen and anti-IgM flux data to total Ca2+ flux as measured by exposure of cells to ionomycin. If comparing multiple BCRs, then insure that the flux in response to anti-IgM is equivalent (Figure 7).
TROUBLESHOOTING
Figure 7.
BCR activation in response to multimerized candidate immunogen. For a kinetic readout, multimerized antigen is added to cells displaying BCR of interest that have been equilibrated with Fura Red, a ratiometric calcium-reporting fluorescent dye30. Calcium bound versus unbound emission is simultaneously measured following antigen addition. (A) CDRH2-dependent calcium flux through germline CR6261 BCR in response to HA ferritin nanoparticle (np) is presented. Negative controls include empty ferritin nanoparticle and I53A/F54A germline CR6261 BCR that is unable to engage the HA stem. This data is made relative to total membrane calcium flux as measured by incubation with the calcium ionophore ionomycin. When comparing different BCR receptors, it is also important to demonstrate that total receptor calcium flux, as measured via crosslinking by anti-IgM is comparable. Curves represent the average of 2–3 replicates. (B) BCR triggering can also be measured using an end point phosphorylation assay. In Ramos, p75 as measured by the antibody G410 pY serves as a reporter of this receptor output6,45,46. Western blotting for tyrosine phosphorylation following addition of multimerized antigen shows analogous activation of germline CR6261 BCR but not its I53A/F54A stem-binding mutant in response to HA np. As with the kinetic readout, empty ferritin nanoparticle does not induce signaling. Total receptor output is presented as tyrosine phosphorylation in response to anti-IgM, and actin levels are included as a loading control.
Option B) Tyrosine phosphorylation to measure BCR activation
CRITICAL. To complement the kinetic measurements of BCR stimulation, triggering can also be measured via phosphorylation of downstream BCR effectors. BCR expressing B cells are incubated with antigen multimer, lysed and examined for phosphorylation by western blotting.
-
xxv
Spin down (1,000 × g, 5 min) 4×106 of BCR expressing Ramos (generated in Step 69) and resuspend in 100 μl of RPMI media for B cell signaling containing 5 μg/ml anti-human IgM F(ab’)2 or antigen multimer (apply concentration ranges from 0.1–2.5 μM). Perform in 100 μl in 96 well format.
-
xxvi
After 15 minutes of exposure at RT, place cells at 4°C, wash twice with PBS and lyse for 10 min in lysis buffer supplemented with protease inhibitors.
CRITICAL STEP. Insure that protein lysate is kept on ice to minimize proteolysis in the sample.
-
xxvii
Pellet cellular debris at 2000 × g for 5 min and collect and measure protein content of supernatant using a BCA protein assay.
PAUSEPOINT. Samples may be stored at −20°C prior to western blotting.
TROUBLESHOOTING
-
xxviii
Run 50 μg of supernatant on SDS PAGE and western blot for phosphotyrosine MW 75 kDa (p75) using G410 pY6,45,46; anti-human actin can also be used to as a loading control (Figure 7). Quantify total phosphotyrosine intensity by densitometry [Image Processing in Java (Image J) Software (http://imagej.nih.gov/ij/) + curve area density calculation in Microsoft Excel]. p75 intensity can be standardized to the actin signal. For comparing multiple BCRs, the level of phosphorylation following anti-IgM crosslinking must be equivalent.
CRITICAL STEP. Densitometry should reflect the calculation of the total area under the curve, not the peak intensity value.
Anticipated Results.
In our experience, all antigens that engage low affinity germline IgM, expressed either as membrane anchored antibody at the surface of 293F cells or in Ramos BCR format will trigger BCR signaling upon antigen multimerization. For example, using 293F-displayed germline CR6261 IgM, a precursor for developing HA-stem directed bnAbs within the IGHV1–69 lineage6, we observe reactivity to fluorescently labeled HA trimer during Phase 1 (Figure 5). Moreover stem recognition does not occur in the absence of the IGHV1–69 CDRH2 loop (despite equivalent trafficking to the cell surface) or when the I45R/T49R is inserted into the step epitope (Figure 5). Similarly, we do not observe binding to irrelevant mIgM (VRC01 an anti-HIV immunoglobulin) (Figure 5). In Phase 2, this mIgM is then stably expressed as an IGHV1–69 BCR in a clone of Ramos B cells lacking surface Ig (Figure 6). The CR6261 germline mIgM, now in BCR format, shows identical HA-stem binding characteristics as was seen in the 293F system (Figure 6). With this ‘correct’ antigenicity in place, we can also observe BCR signaling in response to HA displayed on a multimeric ferritin nanoparticle (Figure 7). Signaling is measured both by calcium flux and tyrosine phosphorylation of downstream BCR effectors (e.g. p756,45,46). Critically, triggering does not occur in the absence of the stem-contacting IGHV1–69 CDRH2 loop (despite equivalent signaling in response crosslinking by anti-IgM) or in response to empty ferritin nanoparticles (Figure 7). From this data, we can conclude that HA ferritin nanoparticle can engage and induce signaling through the stem-specific germline IGHV1–69 BCR.
While arrayed antigen will ultimately be preferred for vaccine immunogenicity27,47, the ‘equivalency’ between mIgM binding and BCR triggering allows for faster comparison of different antigens, different BCR sequences, and is not reliant on time consuming design of multimeric antigen display. Moreover, once a germline-engaging antigen has been identified, its capacity to induce signaling can then be immediately confirmed in the context of proteoliposome array, during which time other multivalent immunogen display platforms may also be explored.
Figure 8.
Proteoliposome- array of candidate immunogen. To avoid having to design a nanoparticle-array for each antigen of interest, a time consuming structure design effort, his-tagged antigen can be conveniently multimerized on preformed 100 nm liposomes containing DGS-NTA(Ni). The resultant proteoliposomes are isolated by flotation in an iodixanol density gradient. Presented is are the silver stained fractions of that gradient, along with an unloaded control antigen. Membrane bound material is recovered at the 2.5/0% iodixanol interface. Proteoliposomes can then quantified by BCA protein assay and be then used to assess BCR triggering in Phase 2.
Acknowledgements
The authors thank members of the Lingwood lab for critical reading of the manuscript, Steve Perfetto (Vaccine Research Center, NIH) for flow cytometry advice, Jeffery Boyington and James Whittle (Vaccine Research Center, NIH) for key discussions. This work was supported by the following awards to D.L.: a Harvard University CFAR grant (P30 AI060354); a Broad-Ragon ENDHIV Catalytic Grant; the William F. Milton Fund; and the Gilead Sciences Research Scholars Program in HIV.
Footnotes
Competing financial interests The authors declare that they have no competing financial interests.
References
- 1.McHeyzer-Williams LJ & McHeyzer-Williams MG Antigen-specific memory B cell development. Annual review of immunology 23, 487–513 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Slifka MK & Amanna I How advances in immunology provide insight into improving vaccine efficacy. Vaccine 32, 2948–2957 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinuesa CG, Linterman MA, Goodnow CC & Randall KL T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunological reviews 237 72–89 (2010). [DOI] [PubMed] [Google Scholar]
- 4.G Gruell H & Klein F Opening Fronts in HIV Vaccine Development: Tracking the development of broadly neutralizing antibodies. Nature medicine 20, 478–479 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Kirchenbaum GA & Ross TM Eliciting broadly protective antibody responses against influenza. Current opinion in immunology 28C, 71–76 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Lingwood D et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jardine J et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ota T et al. Anti-HIV B Cell lines as candidate vaccine biosensors. Journal of immunology 189, 4816–4824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuire AT et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. The Journal of experimental medicine 210, 655–663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoot S et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS pathogens 9, e1003106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGuire AT, Glenn JA, Lippy A & Stamatatos L Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447–52D. Journal of virology 88, 2645–2657 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGuire AT et al. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science 346, 1380–1383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson CA et al. Pandemic H1N1 Influenza Infection and Vaccination in Humans Induces Cross-Protective Antibodies that Target the Hemagglutinin Stem. Frontiers in immunology 3, 87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti D et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. The Journal of clinical investigation 120, 1663–1673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrammert J et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine 208, 181–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avnir Y et al. Molecular Signatures of Hemagglutinin Stem-Directed Heterosubtypic Human Neutralizing Antibodies against Influenza A Viruses. PLoS pathogens 10, e1004103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pappas L et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Wheatley AK et al. H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. Journal of immunology 195, 602–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whittle JR et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. Journal of virology 88, 4047–4057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou T et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 161,1280–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dosenovic P et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell 161, 1505–1515, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jardine JG et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349,156–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yassine HM et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nature medicine 21, 1065–1070 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Davey AM & Pierce SK Intrinsic differences in the initiation of B cell receptor signaling favor responses of human IgG(+) memory B cells over IgM(+) naive B cells. Journal of immunology 188, 3332–3341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y et al. No receptor stands alone: IgG B-cell receptor intrinsic and extrinsic mechanisms contribute to antibody memory. Cell research 24, 651–664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanekiyo M et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzger H Transmembrane signaling: the joy of aggregation. Journal of immunology 149, 1477–1487 (1992). [PubMed] [Google Scholar]
- 28.Akahata W et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nature medicine 16, 334–338 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cumbers SJ et al. Generation and iterative affinity maturation of antibodies in vitro using hypermutating B-cell lines. Nature biotechnology 20, 1129–1134 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Novak EJ & Rabinovitch PS Improved sensitivity in flow cytometric intracellular ionized calcium measurement using fluo-3/Fura Red fluorescence ratios. Cytometry 17, 135–141 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Wendt ER, Ferry H, Greaves DR & Keshav S Ratiometric analysis of fura red by flow cytometry: a technique for monitoring intracellular calcium flux in primary cell subsets. PloS one 10, e0119532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dintzis HM, Dintzis RZ & Vogelstein B Molecular determinants of immunogenicity: the immunon model of immune response. Proceedings of the National Academy of Sciences of the United States of America 73, 3671–3675 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo XM et al. Engineering human hematopoietic stem/progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Blood 113, 1422–1431 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Bachmann MF & Jennings GT Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nature reviews. Immunology 10, 787–796 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Batista FD & Harwood NE The who, how and where of antigen presentation to B cells. Nature reviews. Immunology 9, 15–27 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Cyster JG B cell follicles and antigen encounters of the third kind. Nature immunology 11, 989–996 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Lois C, Hong EJ, Pease S, Brown EJ & Baltimore D Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Naldini L, Blomer U, Gage FH, Trono D & Verma IM Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proceedings of the National Academy of Sciences of the United States of America 93, 11382–11388 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroeder HW Jr. & Cavacini L Structure and function of immunoglobulins. The Journal of allergy and clinical immunology 125, S41–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabat EA, Wu TT, Perry HM, Gottesman KS & Foeller C Sequences of Proteins of Immunological Interest (U.S. Dept. of Health and Human Services, Public Health Service, National Institutes of Health, 1991). [Google Scholar]
- 42.Lefranc MP et al. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Developmental and comparative immunology 27, 55–77 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Kong WP et al. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proceedings of the National Academy of Sciences of the United States of America 103, 15987–15991 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei CJ et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329, 1060–1064 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Muller R, Wienands J & Reth M The serine and threonine residues in the Ig-alpha cytoplasmic tail negatively regulate immunoreceptor tyrosine-based activation motif-mediated signal transduction. Proceedings of the National Academy of Sciences of the United States of America 97, 8451–8454 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heizmann B, Reth M & Infantino S Syk is a dual-specificity kinase that self-regulates the signal output from the B-cell antigen receptor. Proceedings of the National Academy of Sciences of the United States of America 107, 18563–18568 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sulzer B & Perelson AS Immunons revisited: binding of multivalent antigens to B cells. Molecular immunology 34, 63–74 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Ekiert DC et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]