Abstract
SOX9 is a pivotal transcription factor in chondrocytes, a lineage essential in skeletogenesis. Its mandatory role in transactivating many cartilage-specific genes is well established, whereas a pioneer role in lineage specification, which along with transactivation defines master transcription factors, remains elusive. Abundant, but yet incomplete evidence exists that intricate molecular networks control SOX9 activity during the multi-step chondrogenesis pathway. They include a highly modular genetic regulation, posttranscriptional and posttranslational modifications, and varying sets of functional partners. Fully uncovering SOX9 actions and regulation is fundamental to explain mechanisms underlying many diseases that directly or indirectly affect SOX9 activity and to design effective disease treatments. We here review current knowledge, highlight recent discoveries, and propose new research directions to answer remaining questions.
Introduction
Cartilage, a distinctive and essential tissue in vertebrates, is built and maintained by a unique cell type, the chondrocyte. Chondrocytes derive in embryogenesis from multipotent skeletal progenitor cells, as do osteoblasts and a few other cell types. Upon lineage specification, chondrogenic cells condense and undergo early differentiation to establish cartilage primordia that constitute the primary skeleton of the embryo [1]. They then remodel the tissues into growth plates or permanent cartilage. Growth plates are temporary structures that ensure skeletal elongation and the progressive replacement of cartilage by bone. They are made of layers of chondrocytes at sequential stages of differentiation: early-stage cells proliferating in columns, and terminally maturing prehypertrophic and hypertrophic cells. The latter eventually die or participate in endochondral ossification through osteoblastic transdifferentiation. In joints and in the respiratory and auditory systems, chondrocytes mature into quiescent, lowly anabolic cells that ensure permanent cartilage homeostasis throughout life. Elaborate molecular networks control chondrocyte specification and differentiation [1–3]. They mediate their effects at the genetic level through specific sets of transcription factors, of which SOX9 is central [4]. SOX9 belongs to a family of twenty SRY-related HMG box-containing (SOX) proteins, most of which contribute to cell type specification and differentiation in discrete lineages [5]. The first clue that SOX9 is essential in development, including chondrogenesis, came with the discovery that heterozygous mutations occurring within and around SOX9 cause Campomelic Dysplasia (CMPD), a severe skeleton malformation syndrome often associated with XY sex reversal, and milder skeletal dysplasias, namely acampomelic campomelic dysplasia (ACMPD) and Pierre Robin Sequence (PRS) [6–9]. Studies in animal models and molecular studies have uncovered many aspects of SOX9’s specific roles and modes of actions and regulation in development and diseases, but many questions remain. It is thus a good time to review current knowledge, underline recent discoveries, and set priorities for future studies.
Roles and partners of SOX9
Lineage-tracing approaches using mouse Sox9Cre and Sox9CreER knockin alleles have shown that many progenitor cell types express Sox9 and give rise to more cell types than those currently known to be SOX9-dependent [10,11]. In the skeletal system, these cells include osteoblasts, tenocytes and synovial fibroblasts, but solid evidence is still lacking that SOX9 is needed in skeletal progenitors and in other progeny than chondrocytes. Whole transcriptome profiling assays (RNA-seq) detected few changes between Sox9wild-type and Sox9null mouse embryo limb bud skeletogenic cells, and whole epigenome and targetome assays (ChIP-seq) showed that SOX9 helps, but is not required to, remove transrepression marks and deposit active promoter and enhancer marks at chondrocyte-specific genes [12]. These findings are consistent with a contributing rather than mandatory pioneer role of SOX9 in setting a chondrogenic chromatin landscape, but more work is needed to identify the chondrogenic pioneers that act before or with SOX9 and thereby answer the important question of whether SOX9 is a master chondrogenic factor, i.e., a transcription factor involved in both the epigenetic and the transcriptional governance of the chondrocyte lineage. FOX transcription factors, including FOXC, FOXF, and FOXP proteins, are chondrogenic pioneer candidates: they are expressed before and at the onset of chondrogenesis; they are necessary for proper chondrogenesis; FOX-binding sites are enriched near SOX9-binding sites on chondrocyte enhancers; and several family members are pioneers in non-chondrogenic processes [13–17].
Precartilaginous condensation is the first morphologically recognizable step in chondrogenesis. It is absolutely dependent upon SOX9 [18] (Figure 1), but its effector and other regulatory mechanisms remain largely elusive. Recent effort using Sox9 mutant mouse limb buds have indicated that SOX9 upregulates genes for specific autocrine and paracrine regulatory factors and for cytoskeleton and actin network components [12]. These candidate genes for the implementation of precartilaginous condensation warrant full validation.
Figure 1.
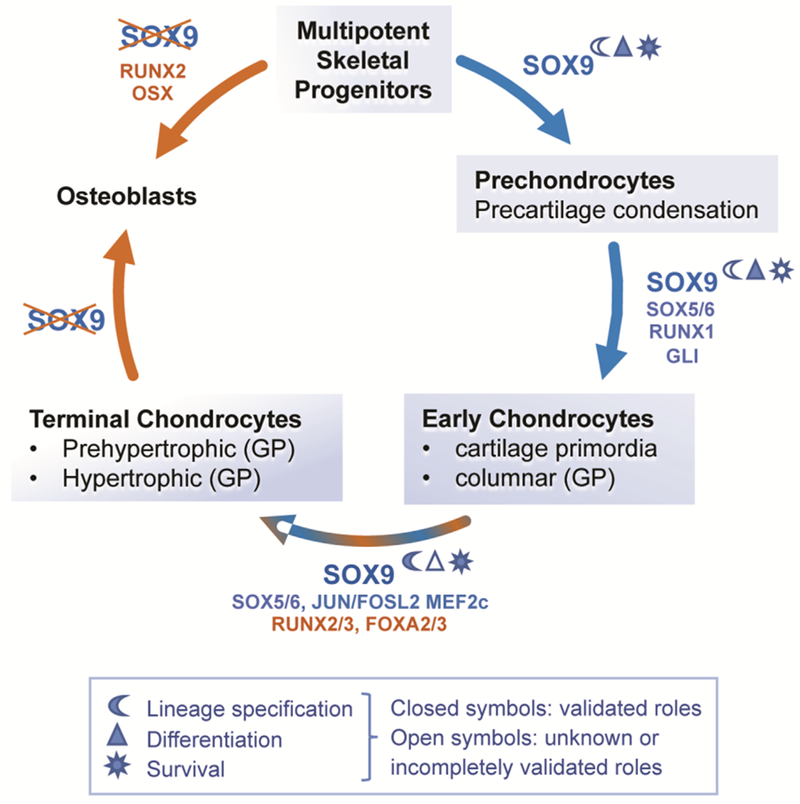
Current knowledge and gaps in knowledge regarding SOX9’s main roles in skeletal progenitors, chondrocytes and osteoblasts. SOX9 is expressed from the multipotent skeletal progenitor stage until chondrocyte hypertrophic differentiation (blue shading of boxes) and has been shown to be involved in chondrocyte lineage specification, differentiation, and survival. Repression of SOX9 is required for osteoblast differentiation from progenitor cells and terminal chondrocytes. Confirmed and candidate co-factors of SOX9 are indicated (blue) as well as other factors involved in specific cell differentiation stages (brown).
Overt chondrogenesis occurs as condensed prechondrocytes differentiate into early-stage chondrocytes, cells whose main activity is to produce cartilage extracellular matrix. This step requires SOX9 and SOX5/SOX6 [18,19]. SOX5 and SOX6 are structurally and functionally very similar to one another, but not to SOX9, and act redundantly [19–22]. They cooperatively bind with SOX9 on active enhancers and super-enhancers associated with hundreds of cartilage-specific genes, and thereby potentiate SOX9’s ability to transactivate [14,15]. Occasional binding to the promoters of ubiquitously expressed genes and to transcriptionally repressed gene loci suggest that the trio may also upregulate and inhibit more genes than those specifically expressed in early chondrocytes [15,23]. The RUNT-domain factor RUNX1 also contributes to overt chondrogenesis, and was recently shown to physically interact with the SOX trio and enhance activation of chondrocyte-specific genes [24]. This finding is in line with the abundance of RUNX binding sites near SOX trio motifs in chondrocyte enhancers [14,15]. ChIP-seq and transactivation (reporter) assays in chondrocytes should be considered to fully validate this RUNX1/SOX partnership. Of note, this role of RUNX1 is distinct from that of RUNX2 and RUNX3, which drive growth plate chondrocyte maturation [1].
The SOX trio remains required to maintain chondrocyte at the early differentiation stage as they proliferate in columns in growth plates and maintain permanent cartilage homeostasis. This activity may involve cooperativity between SOX9 and the Hedgehog pathway-dependent GLI transcription factors [25]. Despite being still highly expressed in prehypertrophic chondrocytes, the SOX trio delays progression to this stage and inhibits rather than activates prehypertrophic marker genes. The three SOX RNAs abruptly vanish and the SOX9 protein (SOX5/6 have not been tested) slowly decays as chondrocytes undergo overt hypertrophy [26]. SOX9 is required at this stage to prevent cell apoptosis, to express to at least some of the early hypertrophic markers, and to prevent chondrocytes from converting to the osteoblast lineage [19,20,26–28]. These activities namely involve keeping RUNX2 and canonical WNT signaling levels in check. Besides SOX5/SOX6, co-factors of SOX9 likely include the JUN and FOSL2 AP1 factors [29]. Cooperativity between GLI, JUN/FOSL2 and SOX9 is based on frequent binding sites for these factors near those of SOX9 in chondrocyte enhancers [14,15] and in vivo evidence of the expression and importance of the factors in chondrogenesis [25,29]. In contrast, SOX9 may compete with FOXA2 to delay terminal chondrocyte maturation [25]. Complementary studies are warranted to fully validate these findings and possibly identify additional factors.
SOX9 structure/function
Reaching deep understanding of the molecular actions of SOX9 and how diseases may disrupt them requires detailed knowledge of the protein’s structure/function attributes. Such information is still being gathered. Like all twenty SOX proteins, SOX9 features an SRY-related high-mobility-group (HMG) domain (Figure 2A) [5]. This domain penetrates the DNA minor groove and binds motifs matching or resembling C[A/T]TTG[A/T][A/T]. This property combines with DNA bending and protein interaction abilities to establish transcriptional complexes. This domain also harbors the nuclear import and export signals of the protein. Within the SOX family, SOX9 forms the SOXE group with SOX8 and SOX10 because the three proteins share a higher degree of identity with one another in the HMG domain than with other SOX proteins and also feature group-specific domains. One of these domains (DIM) confers preferential binding t targets at sites containing inverted SOX motifs separated by 3–4 nucleotides (Figure 2B). Data were recently provided that DIM may promote protein homodimerization, not through DIM:DIM interactions, as long thought, but through DIM:HMG interactions [30]. Consistent with tight structures and essential functions of these domains, all CMPD-causing SOX9 missense mutations occur in the HMG and DIM domains (Angelozzi and Lefebvre, in press).
Figure 2.
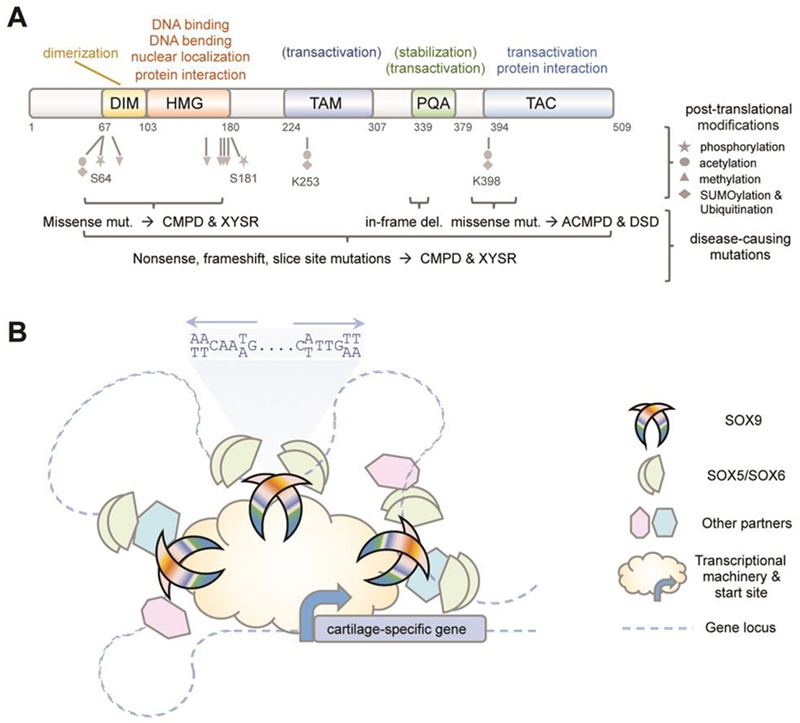
Current knowledge of SOX9 protein’s modes of action and regulation. (A) Domain organization of the protein, with indication of functions validated in vivo (no parentheses), functions identified only in vitro (in parentheses), and post-translational modifications identified only in vitro (grey). The location of mutations (mut.) causing diseases as severe as campomelic dysplasia (CMPD), XY sex reversal (XYSR), acampomelic CMPD (ACMPD) and disorders of sex development (DSD) are indicated (brackets). (B) Schematic of the main mode of action of SOX9 in chondrocytes. SOX9 binds as a homodimer to pairs of inverted SOX recognition sites on multiple enhancers associated with cartilage-specific genes. It functionally interacts with SOX5/SOX6 and other co-factors that bind to nearby sites on the enhancers. SOX9 uses its transactivation domains to contact transcriptional co-activators and basal transcriptional machinery components and thereby induce gene transactivation.
The C-terminus of SOX9 has been known to contain a potent transactivation domain, called TAC (transactivation domain at the C-terminus), ever since nonsense mutations located before or within TAC were found to cause CMPD [31] (Figure 2A). By definition, transactivation domains contact the promoter-based basal transcriptional machinery directly or via transcriptional co-activators. Accordingly, TAC can interact with the transcriptional co-activators CBP and P300 [15,32] and MED25 (Mediator Complex Subunit 25) [33]. In contrast, its interaction with β-catenin inhibits transactivation by both proteins and induces their degradation [34]. TAC is about 100-residue-long, but the residues critical for transactivation and other protein interactions remain undeciphered. Very recently, another transactivation domain was identified in the middle of SOX9 (TAM) [35]. Both TAM and TAC have intrinsic transactivation capability and the two domains synergize to activate chondrocyte-specific genes and reporters in vitro. The minimal sequence required for TAM activity is 40-residue long. It is predicted to form a protein-binding pocket, but its preys remain unknown. These data, along with demonstration that SOX10 TAM is essential in mouse development [36], strongly suggest that SOX9 decisively involves TAM in its chondrogenic actions. Finally, vertebrates have evolved a unique domain in SOX9 [35,37]. This domain of up to 45 residues is called PQA because it contains only prolines, glutamines and alanines. It helps stabilize SOX9 and facilitates transactivation in vitro, but lacks intrinsic transactivation capability. Only two missense mutations in TAC and one microdeletion in PQA have been reported to be pathogenic so far, and all resulted in disorders of sex development (DSD) with ACMPD or no skeletal defects [38,39]. The mildness of the diseases may be due to the mutation types or to the fact that, unlike HMG and DIM, TAM, TAC and PQA are less structured and can therefore better tolerate point variants. This finding should not undermine the likelihood that each domain may be critical for SOX9’s chondrogenic and other activities and thus deserves further investigations.
SOX9 post-translational modifications
The chondrogenic activities of SOX9 depend not only on functional partners’ availability and protein integrity, but also on post-translational modifications (Figure 2A). Several have been detected and linked to cartilage development and diseases, but their functional impact in vivo remains unvalidated [40]. Among them, phosphorylation of S64 and S181 has received much attention. It was shown to increase SOX9 transcriptional activity in vitro and proposed to help delay chondrocyte maturation downstream of PTHrP signaling in a PKA (cAMP-dependent protein kinase A)-dependent manner [41,42]. S64 and S181 phosphorylation may also be achieved by other kinases downstream of other pathways. For instance, it was proposed to occur and promote SOX9 SUMOylation downstream of BMP and canonical WNT signaling during neural crest delamination [43]. Further, SHP2, a Src homology domain-containing tyrosine phosphatase and key player in RAS/MAPK signaling, was recently found to be critical for proper skeletal patterning and growth [44]. It was shown to block SOX9 phosphorylation by PKA and hence subsequent SUMOylation. Of note, gain-of-function mutations in PTPN11 (encoding SHP2) cause Noonan and LEOPARD syndromes, which include skeletal dysplasia, whereas loss-of-function mutations cause metachondromatosis. Ubiquitination, acetylation and deacetylation of SOX9 were also suggested to be decisive. Recently, DDRGK1 mutations were shown to cause Shohat-type spondyloepimetaphyseal dysplasia [45]. DDRGK1, a component of the UFM1 conjugation pathway, was found to inhibit SOX9 ubiquitination and subsequent proteasomal degradation. Its SOX9 residue target remains uncharacterized, but be K398 or other residues previously shown to be ubiquitinated [46]. Sirtuins are NAD+-dependent lysine deacetylases involved in many processes, including chondrogenesis. Sirt1+/− mice are growth-delayed and develop osteoarthritis precociously, and Sirt1−/− pups neonatally with severe skeletal malformations [47], whereas Sirt7−/− mice are resistant to osteoarthritis [48]. SIRT1 may promote chondrogenesis by increasing SOX9 nuclear localization [49]. It can deacetylate SOX9, but the affected residues remain unknown. In contrast, SIRT7 suppresses SOX9 activity in vitro, but appears unable to deacetylate SOX9, leaving open its mechanism of action [48]. Altogether, these studies point to posttranslational mechanisms that could be paramount in chondrogenesis and thus call for more investigations to fully validate them and likely to uncover many more.
SOX9 gene regulation
The specific spatiotemporal pattern of the SOX9 RNA implies that SOX9 regulation starts at the gene and RNA levels. Many pathways have been shown to control SOX9 expression in chondrogenesis: Hedgehog, PTHrP, BMP, TGFβ, FGF, non-canonical WNT and hypoxia signaling induce or upregulate it, whereas canonical WNT, NOTCH, retinoic acid and inflammatory pathways decrease it [1,2]. Their importance is illustrated by the fact that mutations in various components of these pathways cause skeletal dysplasias. An example is achondroplasia, due to activating mutations in FGFR3 [50]. Focusing on the SOX9 proximal region to explain changes in SOX9 expression, these studies have provided in vitro evidence that HIF-1α (hypoxia) [51], SMAD (BMP) [52], RBPjk/NICD (NOTCH) [53], STAT3 (many cytokine and growth factor pathways) [54], and YAP1/TEAD (Hippo) [55] bind specific motifs in this region (Figure 3). While proximal regulatory elements may indeed critically modulate SOX9 expression, it is worth noting that the SOX9 promoter is insufficient to drive reporter gene expression in chondrocytes and other SOX9-expressing cells in transgenic mice [56,57]. Thus, distant elements may also mediate the effects of signaling pathways.
Figure 3.
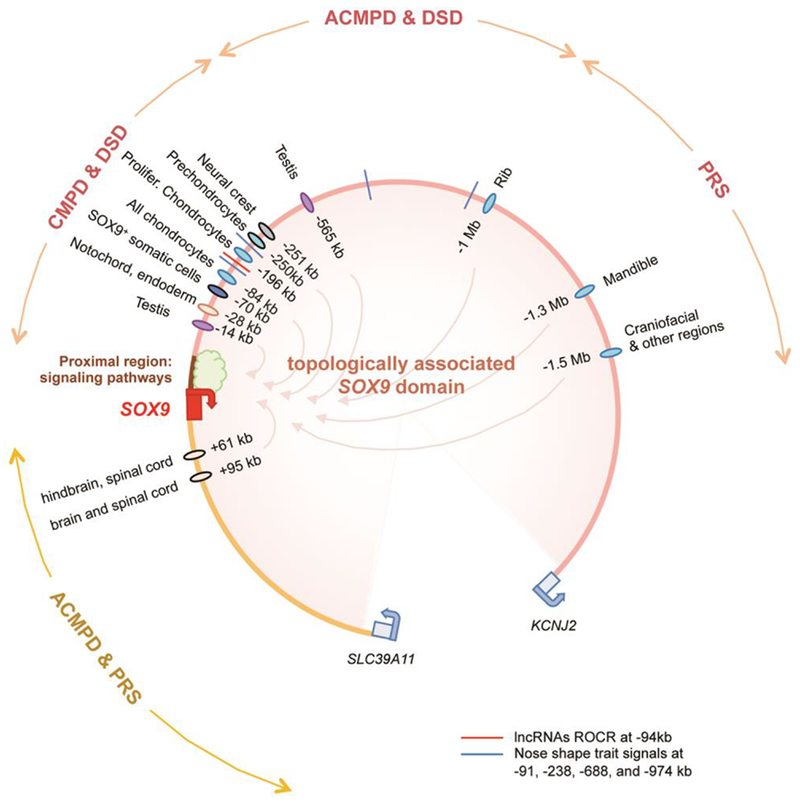
Current knowledge of the complex modular organization of SOX9 cis-acting elements. The domain topologically associated with SOX9 spans 1.9 Mb upstream and 0.5 Mb downstream of the gene. The SOX9 proximal region is believed to be a hub for signaling pathways. The distal regions house at least a dozen and likely more enhancers driving gene expression in various tissues, chondrocyte differentiation stages, and cartilage anatomical sites. The upstream region also houses ROCR, SOX9-regulating IncRNAs. Chromosomal translocations, microdeletions and duplications in the distal regions cause chondrodysplasias (CMPD, ACMPD, and PRS) and disorders of sex development (DSD) with a severity generally proportional to their proximity to the SOX9 gene body.
The discovery that translocations and microdeletions occurring far around SOX9 cause CMPD, ACM PD, PRS or DSDs provided the first clue years ago that SOX9 cis-acting elements are numerous and spread over large distances (Figure 3). Delineation of the SOX9 topologically associated domain to the 2-Mb gene desert around SOX9 has confirmed this notion [58]. To date, a dozen enhancers have been delineated throughout this domain and have been shown to be active in multiple or discrete tissues, chondrocyte-specific stages, or anatomical sites [8,56,57,59–61]. Moreover, a recent GWAS study linked nose shape traits in humans to four loci located between 91 and 974 kb upstream of SOX9 and containing active promoter/enhancer marks [62]. Together, current knowledge supports the notion that SOX9 is modularly controlled, likely by more enhancers than currently known, and that variants in enhancers may contribute to skeletal pattern diversity among humans as well as among vertebrate species.
Identifying the transcription factors that control SOX9 enhancers is a difficult task still in its infancy. SOX9 can positively regulate several of its gene enhancers [57,59], leading to the important concept that pathways affecting SOX9 protein stability may secondarily affect SOX9 gene expression through this feedback loop. There is also evidence that SOX5/SOX6 contribute to the activity of several SOX9 enhancers [59], that STAT3 targets a rib-specific enhancer [61], and that MSX1, a homeodomain transcription factor involved in limb and craniofacial development, controls a mandible-specific SOX9 enhancer [8]. Further, very recent ChIP-seq assays for PITX1, a bicoid-class homeodomain transcription factor required for hindlimb development, have uncovered a dozen of PITX1-bound limb enhancers upstream and downstream of SOX9, consistent with Sox9 downregulation in Pitx1−− mouse embryos [63].
Finally, several miRNAs and IncRNAs have been shown to affect SOX9 RNA level or translation during chondrogenesis in vitro [40]. Direct roles for miRNAs are supported by conserved binding sites in the SOX9 3’ untranslated region. Recently, the IncRNAs referred to as ROCR (regulator of chondrogenesis RNA) were located 94 kb upstream of SOX9 and found critical for SOX9 expression and successful chondrogenesis of human mesenchymal stem cells [64]. In all cases, in vivo validation remains to be provided.
Summary and outlook
Ever since SOX9 mutations were linked to campomelic dysplasia, substantial research efforts have been devoted towards uncovering the ins and outs of the gene in the chondrocyte lineage. To date, the roles of SOX9 at the multiple steps of chondrocyte differentiation are fairly well understood, although more work should still be done to assess the impact of SOX9 binding to many genes with known or yet-unknown roles in cartilage. Less understood, despite a potential for breakthroughs, is the impact of SOX9 expression on progenitor cells. Further, key mechanisms involved in SOX9 regulation at the gene, RNA and protein level have been uncovered, but have also led us to humbly acknowledge that the SOX molecular network is likely far more sophisticated than currently appreciated. Important goals for the future are to continue to dissect the many nuts and bolts of SOX9 actions and regulation. Definitive knowledge will come with the use of cutting-edge approaches in vitro and in vivo, including the Crispr/Cas technology to genetically modify transcribed and cis-acting regions of SOX9, its targets, co-factors and regulators. New findings are promised to be insightful for many areas of fundamental science, for better grasping the mechanisms underlying a large number of cartilage diseases and for designing better strategies to treat these diseases.
Acknowledgments
We gratefully acknowledge the financial support of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01 grants AR068308 and AR072649 to V.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published in the last two years, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Kozhemyakina E, Lassar AB, Zelzer E: A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development 2015, 142:817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samsa WE, Zhou X, Zhou G: Signaling pathways regulating cartilage growth plate formation and activity. Semin Cell Dev Biol 2017, 62:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rux D, Decker RS, Koyama E, Pacifici M: Joints in the appendicular skeleton: Developmental mechanisms and evolutionary influences. Curr Top Dev Biol 2019, 133:119–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CF, Samsa WE, Zhou G, Lefebvre V: Transcriptional control of chondrocyte specification and differentiation. Semin Cell Dev Biol 2017, 62:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamachi Y, Kondoh H: Sox proteins: regulators of cell fate specification and differentiation. Development 2013, 140:4129–4144. [DOI] [PubMed] [Google Scholar]
- 6.Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al. : Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 1994, 372:525–530. [DOI] [PubMed] [Google Scholar]
- 7.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al. : Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 1994, 79:1111–1120. [DOI] [PubMed] [Google Scholar]
- 8.Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, et al. : Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet 2009, 41:359–364. [DOI] [PubMed] [Google Scholar]
- 9.Yip RKH, Chan D, Cheah KSE: Mechanistic insights into skeletal development gained from genetic disorders. Curr Top Dev Biol 2019, 133:343–385. [DOI] [PubMed] [Google Scholar]
- 10.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, et al. : Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A 2005, 102:14665–14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono N, Ono W, Nagasawa T, Kronenberg HM: A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol 2014, 16:1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.**.Liu CF, Angelozzi M, Haseeb A, Lefebvre V: SOX9 is dispensable for the initiation of epigenetic remodeling and the activation of marker genes at the onset of chondrogenesis. Development 2018, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used RNA-seq and ChIP-seq assays on Sox9 control and null mouse limb buds to identify genes involved in precartilaginous condensation and the epigenetic and transcriptional roles of SOX9 at the onset of chondrogenesis.
- 13.Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL: The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 1998, 93:985–996. [DOI] [PubMed] [Google Scholar]
- 14.Liu CF, Lefebvre V: The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res 2015, 43:8183–8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohba S, He X, Hojo H, McMahon AP: Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep 2015, 12:229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao H, Zhou W, Yao Z, Wan Y, Cao J, Zhang L, Zhao J, Li H, Zhou R, Li B, et al. : Foxp½/4 regulate endochondral ossification as a suppresser complex. Dev Biol 2015, 398:242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.**.Xu P, Balczerski B, Ciozda A, Louie K, Oralova V, Huysseune A, Crump JG: Fox proteins are modular competency factors for facial cartilage and tooth specification. Development 2018, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uncovers that FOXC and FOXF members of the FOX family of transcription factors are required to allow SOX9 to activate chondrogenic genes in zebrafish embryos. The data support the notion that multiple FOX proteins overlap in the various skeletogenic sites to exert pioneer transcription factor functions at the onset of chondrogenesis.
- 18.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B: The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 2002, 16:2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V: The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell 2001, 1:277–290. [DOI] [PubMed] [Google Scholar]
- 20.Smits P, Dy P, Mitra S, Lefebvre V: Sox5 and Sox6 are needed to develop and maintain source, columnar, and hypertrophic chondrocytes in the cartilage growth plate. J Cell Biol 2004, 164:747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung UI: The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum 2004, 50:3561–3573. [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre V: Roles and regulation of SOX transcription factors in skeletogenesis. Curr Top Dev Biol 2019, 133:171–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung VY, Gao B, Leung KK, Melhado IG, Wynn SL, Au TY, Dung NW, Lau JY, Mak AC, Chan D, et al. : SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet 2011, 7:e1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.*.Yano F, Ohba S, Murahashi Y, Tanaka S, Saito T, Chung UI: Runx1 contributes to articular cartilage maintenance by enhancement of cartilage matrix production and suppression of hypertrophic differentiation. Sci Rep 2019, 9:7666. [DOI] [PMC free article] [PubMed] [Google Scholar]; The analysis of Runx1-null mice and data from in vitro assays demonstrate that RUNX1 enhances chondrogenesis likely through direct physical and functional interaction with the SOX5/6/9 trio in the governance of early chondrocyte differentiation.
- 25.*.Tan Z, Niu B, Tsang KY, Melhado IG, Ohba S, He X, Huang Y, Wang C, McMahon AP, Jauch R, et al. : Synergistic co-regulation and competition by a SOX9-GLI-FOXA phasic transcriptional network coordinate chondrocyte differentiation transitions. PLoS Genet 2018, 14:e1007346. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tan and collaborators generated a growth plate differential gene expression library and available ChIP-seq datasets from various tissues to suggest synergistic and antagonistic interactions between SOX9 and other transcription factors known to have important roles in the multi-step differentiation process of growth plate chondrocytes.
- 26.Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, Lefebvre V: Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell 2012, 22:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.*.Lui JC, Yue S, Lee A, Kikani B, Temnycky A, Barnes KM, Baron J: Persistent Sox9 expression in hypertrophic chondrocytes suppresses transdifferentiation into osteoblasts. Bone 2019, 125:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]; Transgenic mice overexpressing SOX9 in chondrocytes provide consolidating evidence that SOX9 inhibits the transdifferentiation of terminal chondrocytes into osteoblastic cells during endochondral ossification.
- 28.Ikegami D, Akiyama H, Suzuki A, Nakamura T, Nakano T, Yoshikawa H, Tsumaki N: Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development 2011, 138:1507–1519. [DOI] [PubMed] [Google Scholar]
- 29.He X, Ohba S, Hojo H, McMahon AP: AP-1 family members act with Sox9 to promote chondrocyte hypertrophy. Development 2016, 143:3012–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YH, Jankowski A, Cheah KS, Prabhakar S, Jauch R: SOXE transcription factors form selective dimers on non-compact DNA motifs through multifaceted interactions between dimerization and high-mobility group domains. Sci Rep 2015, 5:10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudbeck P, Schmitz ML, Baeuerle PA, Scherer G: Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nat Genet 1996, 13:230–232. [DOI] [PubMed] [Google Scholar]
- 32.Tsuda M, Takahashi S, Takahashi Y, Asahara H: Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem 2003, 278:27224–27229. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura Y, Yamamoto K, He X, Otsuki B, Kim Y, Murao H, Soeda T, Tsumaki N, Deng JM, Zhang Z, et al. : Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat Commun 2011, 2:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, et al. : Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev 2004, 18:1072–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.**.Haseeb A, Lefebvre V: The SOXE transcription factors-SOX8, SOX9 and SOX10-share a bi-partite transactivation mechanism. Nucleic Acids Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides evidence that SOX9 and its closest relatives use two synergistic domains to achieve potent transactivation. One domain, located at the C-terminus, was previously known as the transactivation domain of SOX9, whereas the other, located in the middle of the protein, was not known to be functional in SOX9.
- 36.Schreiner S, Cossais F, Fischer K, Scholz S, Bosl MR, Holtmann B, Sendtner M, Wegner M: Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development 2007, 134:3271–3281. [DOI] [PubMed] [Google Scholar]
- 37.McDowall S, Argentaro A, Ranganathan S, Weller P, Mertin S, Mansour S, Tolmie J, Harley V: Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J Biol Chem 1999, 274:24023–24030. [DOI] [PubMed] [Google Scholar]
- 38.Katoh-Fukui Y, Igarashi M, Nagasaki K, Horikawa R, Nagai T, Tsuchiya T, Suzuki E, Miyado M, Hata K, Nakabayashi K, et al. : Testicular dysgenesis/regression without campomelic dysplasia in patients carrying missense mutations and upstream deletion of SOX9. Mol Genet Genomic Med 2015, 3:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen SY, Lin SJ, Tsai LP, Chou YY: Sex-reversed acampomelic campomelic dysplasia with a homozygous deletion mutation in SOX9 gene. Urology 2012, 79:908–911. [DOI] [PubMed] [Google Scholar]
- 40.Lefebvre V, Dvir-Ginzberg M: SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect Tissue Res 2017, 58:2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W, Zhou X, Lefebvre V, de Crombrugghe B: Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9’s ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol Cell Biol 2000, 20:4149–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang W, Chung UI, Kronenberg HM, de Crombrugghe B: The chondrogenic transcription factor Sox9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc Natl Acad Sci U S A 2001, 98:160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu JA, Wu MH, Yan CH, Chau BK, So H, Ng A, Chan A, Cheah KS, Briscoe J, Cheung M: Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc Natl Acad Sci U S A 2013, 110:2882–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.*.Zuo C, Wang L, Kamalesh RM, Bowen ME, Moore DC, Dooner MS, Reginato AM, Wu Q, Schorl C, Song Y, et al. : SHP2 regulates skeletal cell fate by modifying SOX9 expression and transcriptional activity. Bone Res 2018, 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that the tyrosine dephosphatase SHP2 is required to downregulate the SOX9 protein level in skeletal progenitors and chondrocytes and thereby achieve proper skeletogenesis in mice. SHP2 is proposed to inhibit SOX9 SUMOylation via PKA signaling.
- 45.*.Egunsola AT, Bae Y, Jiang MM, Liu DS, Chen-Evenson Y, Bertin T, Chen S, Lu JT, Nevarez L, Magal N, et al. : Loss of DDRGK1 modulates SOX9 ubiquitination in spondyloepimetaphyseal dysplasia. J Clin Invest 2017, 127:1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]; Egunsola and colleagues identify loss-of-function mutations in DDRGK1 as the cause of a severe skeletal dysplasia. They provide experimental evidence that DDRGK1 stabilizes the SOX9 protein by directly inhibiting its ubiquination and subsequent proteasomal degradation.
- 46.Akiyama H, Kamitani T, Yang X, Kandyil R, Bridgewater LC, Fellous M, Mori-Akiyama Y, de Crombrugghe B: The transcription factor Sox9 is degraded by the ubiquitin-proteasome system and stabilized by a mutation in a ubiquitin-target site. Matrix Biol 2005, 23:499–505. [DOI] [PubMed] [Google Scholar]
- 47.Dvir-Ginzberg M, Mobasheri A, Kumar A: The Role of Sirtuins in Cartilage Homeostasis and Osteoarthritis. Curr Rheumatol Rep 2016, 18:43. [DOI] [PubMed] [Google Scholar]
- 48.Korogi W, Yoshizawa T, Karim MF, Tanoue H, Yugami M, Sobuz SU, Hinoi E, Sato Y, Oike Y, Mizuta H, et al. : SIRT7 is an important regulator of cartilage homeostasis and osteoarthritis development. Biochem Biophys Res Commun 2018. [DOI] [PubMed] [Google Scholar]
- 49.Bar Oz M, Kumar A, Elayyan J, Reich E, Binyamin M, Kandel L, Liebergall M, Steinmeyer J, Lefebvre V, Dvir-Ginzberg M: Acetylation reduces SOX9 nuclear entry and ACAN gene transactivation in human chondrocytes. Aging Cell 2016, 15:499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou ZQ, Ota S, Deng C, Akiyama H, Hurlin PJ: Mutant activated FGFR3 impairs endochondral bone growth by preventing SOX9 downregulation in differentiating chondrocytes. Hum Mol Genet 2015, 24:1764–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E: HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development 2007, 134:3917–3928. [DOI] [PubMed] [Google Scholar]
- 52.Pan Q, Yu Y, Chen Q, Li C, Wu H, Wan Y, Ma J, Sun F: Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J Cell Physiol 2008, 217:228–241. [DOI] [PubMed] [Google Scholar]
- 53.Kohn A, Rutkowski TP, Liu Z, Mirando AJ, Zuscik MJ, O’Keefe RJ, Hilton MJ: Notch signaling controls chondrocyte hypertrophy via indirect regulation of Sox9. Bone Res 2015, 3:15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall MD, Murray CA, Valdez MJ, Perantoni AO: Mesoderm-specific Stat3 deletion affects expression of Sox9 yielding Sox9-dependent phenotypes. PLoS Genet 2017, 13:e1006610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.*.Goto H, Nishio M, To Y, Oishi T, Miyachi Y, Maehama T, Nishina H, Akiyama H, Mak TW, Makii Y, et al. : Loss of Mob1a/b in mice results in chondrodysplasia due to YAP1/TAZ-TEAD-dependent repression of SOX9. Development 2018, 145. [DOI] [PubMed] [Google Scholar]; This study shows that MOB1a/b, which are adaptors increasing LATS kinase activity in the Hippo pathway, are required for mouse postnatal growth. This activity may involve positive contribution to SOX9 expression through binding of the transcriptional co-activators YAP1 and TAZ to the SOX9 promoter.
- 56.Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G: Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol 2006, 291:382–397. [DOI] [PubMed] [Google Scholar]
- 57.Mead TJ, Wang Q, Bhattaram P, Dy P, Afelik S, Jensen J, Lefebvre V: A far-upstream (−70 kb) enhancer mediates Sox9 auto-regulation in somatic tissues during development and adult regeneration. Nucleic Acids Res 2013, 41:4459–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schopflin R, Kraft K, Kempfer R, Jerkovic I, Chan WL, et al. : Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 2016, 538:265–269. [DOI] [PubMed] [Google Scholar]
- 59.Yao B, Wang Q, Liu CF, Bhattaram P, Li W, Mead TJ, Crish JF, Lefebvre V: The SOX9 upstream region prone to chromosomal aberrations causing campomelic dysplasia contains multiple cartilage enhancers. Nucleic Acids Res 2015, 43:5394–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonen N, Futtner CR, Wood S, Garcia-Moreno SA, Salamone IM, Samson SC, Sekido R, Poulat F, Maatouk DM, Lovell-Badge R: Sex reversal following deletion of a single distal enhancer of Sox9. Science 2018, 360:1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.**.Mochizuki Y, Chiba T, Kataoka K, Yamashita S, Sato T, Kato T, Takahashi K, Miyamoto T, Kitazawa M, Hatta T, et al. : Combinatorial CRISPR/Cas9 Approach to Elucidate a Far-Upstream Enhancer Complex for Tissue-Specific Sox9 Expression. Dev Cell 2018, 46:794–806 e796. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study involves a novel high-throughput screening approach to identify a SOX9 far-upstream enhancer specifically active in rib cartilage. This is the first SOX9 enhancer whose deletion in the mouse is found to lead to a chondrodysplasia phenotype.
- 62.**.Cha S, Lim JE, Park AY, Do JH, Lee SW, Shin C, Cho NH, Kang JO, Nam JM, Kim JS, et al. : Identification of five novel genetic loci related to facial morphology by genome-wide association studies. BMC Genomics 2018, 19:481. [DOI] [PMC free article] [PubMed] [Google Scholar]; Searching for genetic loci determining face morphological traits in Koreans, Cha and collaborators identify four signals in the SOX9 upstream region associated with nose shape traits. These signals locate at sites featuring active enhancer marks, suggesting direct implication in SOX9 regulation.
- 63.**.Wang JS, Infante CR, Park S, Menke DB: PITX1 promotes chondrogenesis and myogenesis in mouse hindlimbs through conserved regulatory targets. Dev Biol 2018, 434:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang and colleagues combined ChIP-seq and RNA-seq assays to identify PITX1 as an transcriptional regulator of cartilage development in hindlimbs acing in part by binding to limb-specific enhancers in the far-upstream regulatory region of SOX9. PITX1 is thereby one of the first transcription factors proposed to control SOX9 expression in specific sites.
- 64.**.Barter MJ, Gomez R, Hyatt S, Cheung K, Skelton AJ, Xu Y, Clark IM, Young DA: The long non-coding RNA ROCR contributes to SOX9 expression and chondrogenic differentiation of human mesenchymal stem cells. Development 2017, 144:4510–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is first to identify lncRNAs in the upstream domain of SOX9 that are involved in the expression of the gene. These ROCR IncRNAs are shown to significantly promote SOX9 expression and thereby chondrogenesis of human mesenchymal stem cells in vitro.


