Abstract
The SRY-related (SOX) transcription factor family pivotally contributes to determining cell fate and identity in many lineages. Since the original discovery that SRY deletions cause sex reversal, mutations in half of the twenty human SOX genes have been associated with rare congenital disorders, henceforward called SOXopathies. Mutations are generally de novo, heterozygous and inactivating, revealing gene haploinsufficiency, but other types, including duplications, have been reported too. Missense variants primarily target the HMG domain, the SOX hallmark that mediates DNA binding and bending, nuclear trafficking, and protein-protein interactions. We here review key clinical and molecular features of SOXopathies and discuss the prospect that the disease family likely involves more SOX genes and larger clinical and genetic spectrums than currently appreciated.
Keywords: developmental disorder, genetic variant, human disease, mutation, SOX, SRY
DEFINING SOXOPATHIES
A seminal discovery was made in 1990 with the cloning of SRY (see glossary), the gene that occupies and defines the sex-determining region on the Y chromosome and whose inactivation underlies disorders of sex development (DSDs) [1]. SRY encodes a transcription factor with a high-mobility-group (HMG)-type DNA-binding domain. This discovery prompted a search for close relatives, with the vision that SRY-related HMG box (SOX)-containing genes would also have critical roles. Major cloning efforts and completion of genome sequencing projects made it clear by the turn of the 21st century that humans and most mammals possess nineteen SOX genes in addition to SRY [2]. Both forward and reverse genetic approaches have uncovered pivotal functions for most SOX genes, such that it is now well recognized that the SOX family exerts master roles in many developmental, physiological and pathological processes by governing cell type-specific genetic programs in both stem/progenitor cells and highly specialized cell types [3]. Thanks to major advances in genetic testing procedures, mutations in half of the SOX genes have been associated to date with congenital diseases. Several of these associations were made very recently, and the numbers of reported pathogenic mutations have been increasing exponentially over the years (Figure 1). SOX mutation-driven diseases affect various processes, but most are developmental disorders and due to de novo alterations inactivating one SOX allele. We henceforward refer to them as SOXopathies, just as RASopathies, for instance, are due to mutations in components of the RAS/MAPK pathway [4] and collagenopathies are primarily due to mutations in collagen genes [5]. We here review these diseases clinically and genetically and in the context of current knowledge of SOX functions. While focusing on developmental disorders due to germline mutations in SOX genes, we also briefly discuss other diseases, such as cancers, which may be triggered or influenced by somatic mutations in SOX genes or by factors altering SOX gene or protein activities. We end with a discussion on the perspective that SOXopathies likely involve more SOX genes and exhibit larger clinical and genetic spectrums than currently known.
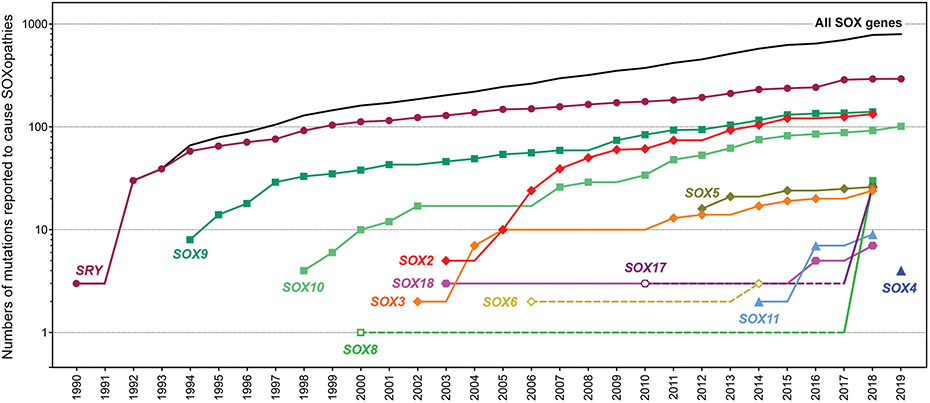
Figure 1.
Timeline of SOXopathy discovery. Cumulative graph showing the numbers of distinct pathogenic alterations identified within and around SOX genes over time. Closed symbols and plain lines represent validated gene-disease associations, whereas open symbols and dotted lines represent suggested associations. Links made through GWAS are not included because of undefined variants and numbers.
SHARED AND UNIQUE FEATURES OF SOX PROTEINS AND GENES
SOX proteins, like TCF/LEF proteins, share significant identity in their DNA-binding domain with HMGB proteins (Figure 2A). The latter are ubiquitous chromatin architectural factors that run in SDS-PAGE with a high mobility [6], whereas SOX and TCF/LEF proteins are classical transcription factors expressed in discrete cell types. The HMG domain forms three α-helices that fold into an L-shaped structure, penetrates the minor groove of DNA, and sharply bends DNA (Figure 2B). Key residues responsible for these properties and for nuclear trafficking are conserved among HMGB, SOX, and TCF/LEF proteins, but the degree of residue identity is much higher within than across families (Figure 2A). Differences account for DNA sequence specificities and bending angles. SOX factors preferentially bind motifs matching or resembling C[A/T]TTG[A/T][A/T]. DNA bending is critical for transcriptional activity, likely by facilitating enhanceosome assembly [7]. As further described later, missense variants in many HMG-domain residues cause SOXopathies, showing how important the domain and many of its residues are.
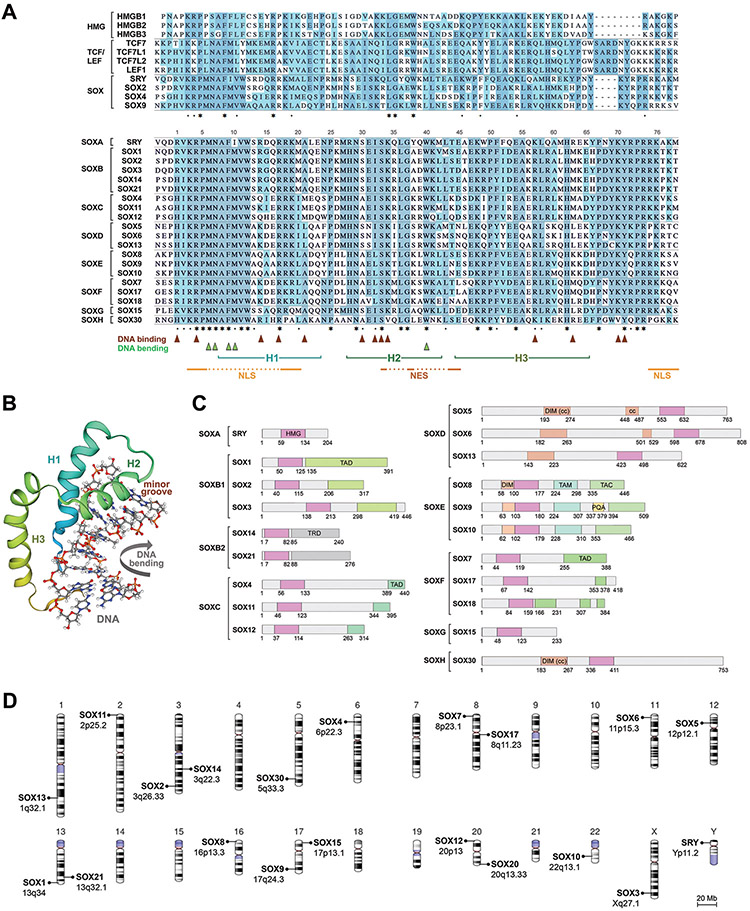
Figure 2.
Shared and distinctive features of SOX proteins and genes.
A. Alignment of the HMG domain sequences (including 3 flanking residues on each side) of the human HMGB and TCF/LEF proteins with those of a few SOX proteins (top) and all human SOX proteins (bottom) highlights full conservation (greyish blue) and semi-conservation (cyan blue) of specific residues. Residues involved in DNA binding, DNA bending, α-helices, nuclear localization signals (NLS), and nuclear export signal (NES) are indicated.
B. 3D solution NMR structure of the human SRY HMG domain complexed to DNA shows that the HMG domain is characterized by three α-helices (H1 to H3 from the N- to the C-terminus) that position themselves into an L-shape, contact DNA exclusively in the minor groove, and force bending of the DNA helix. This schematic was generated by SWISS-MODEL according to [94].
C. Domain structure organization of the human SOX proteins. HMG, HMG domain; TAD, transactivation domain; TRD, transrepression domain; DIM, homodimerization domain; cc, coiled coil; TAM, middle transactivation domain; TAC, C-terminal transactivation domain; PQA, PQA-rich domain.
D. Chromosomal distribution of the human SOX genes.
The SOX family comprises eight groups, SOXA to SOXH (Figure 2A and 2C). SOX proteins share almost 100% identity in the HMG domain with same-group relatives, but only about 50% with other-group members. They also share significant identity with same-group members outside the HMG domain, especially in functional domains, which include homodimerization, transactivation and transrepression domains, but share virtually no identity with other-group members outside the HMG domain. One would expect that missense variants would cause diseases even if located outside the HMG domain, but as described later, only a few cases have been described so far.
SRY is located on the Y chromosome in a region ancestrally related to a segment of the X chromosome containing SOX3. The other SOX genes are spread across autosomal chromosomes (Figure 2D). Same-group SOX genes have identical exon-intron structures. The SOXA, SOXB and SOXC genes are made of a single exon, whereas SOXD genes comprise at least 15 coding exons and multiple 5’ untranslated ones, and SOX5 and SOX6 are spread over hundreds of kb. The other SOX genes are small and feature 2 to 5 exons. Regardless of body size, most SOX genes are separated from coding neighbors by dozens to thousands of kb. These flanking regions typically house multiple enhancers that underlie complex modes of gene regulation. Accordingly, mutations in these regions have been shown in multiple cases to cause diseases. The expression pattern of each SOX gene is unique, typically including several cell types, but overlaps with that of same-group members, allowing the genes to exert additive or redundant functions. This property implies that inactivating mutations often cause disease only in processes where key roles of a gene cannot be compensated by those of a co-expressed close relative.
SOXOPATHIES REVEAL KEY ROLES FOR HUMAN SOX GENES DURING AND BEYOND DEVELOPMENT
SRY
To date, several hundreds of distinct SRY mutations have been reported to cause disease, more than for any other SOX gene, likely because SRY is a master determinant of sex determination (Figure 3), is present at only one copy, and has no SOXA relative to share its functions with. Most SRY mutations cause XY sex reversal (Key Table) [8, 9]. They include full or partial gene deletions as well as point mutations affecting protein integrity [10, 11]. Disease-causing missense variants have been identified in almost every HMG-domain residue, but rarely outside this domain [12]. This is explained by the fact that SRY has no functional domain other than its HMG motif. SRY translocations from the Y to the X chromosome also cause DSDs. In these cases, individuals carrying SRY on an X chromosome develop as males (XX sex reversal), and individuals with an SRY-depleted Y chromosome develop as females (XY sex reversal) [13]. Mouse models have confirmed and explained the master role of SRY in sex determination: XY mice lacking Sry develop as females, and XX mice carrying an Sry transgene develop as males [14, 15]. Sry is transiently expressed in the embryonic gonad and its main role is to activate Sox9, which then activates other male sex differentiation genes, including Sox8 [16].
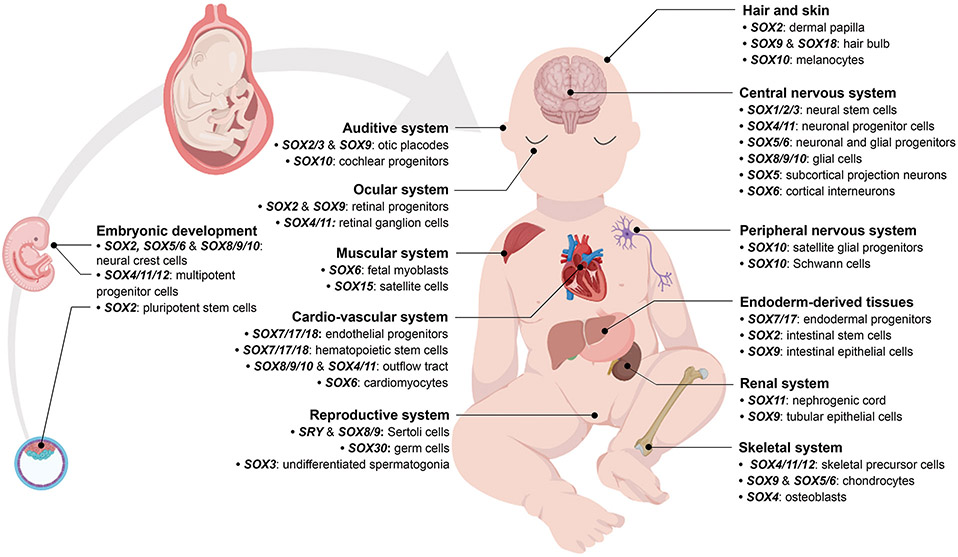
Figure 3.
Examples of key roles of SOX genes in development derived primarily from experiments in vitro and in animal models. Drawings were created using BioRender.
Key Table.
Currently known SOXopathies and types of mutations involved
| Group | Gene | Disease | Type of mutations | ||||||
|---|---|---|---|---|---|---|---|---|---|
| T | Del | Dup | Inv | Ms | Ns/Fs | ||||
| HMG | Other | ||||||||
| SOXA | SRY | Disorder of sex development (DSD) | ✓ | ✓ | - | - | ✓ | ✓ | ✓ |
| SOXB1 | SOX1 | - | - | - | - | - | - | - | - |
| SOX2 | Anophthalmia/microphthalmia syndrome | - | ✓ | - | - | ✓ | ✓ | ✓ | |
| SOX3 | Mental retardation with panhypopituitarism, X-linked | - | - | ✓ | ✓ | ✓ | ✓ | - | |
| Septo-optic dysplasia (SOD) syndrome | - | - | ✓ | - | - | ✓ | - | ||
| XX male sex reversal | ✓ | ✓ | ✓ | - | - | - | - | ||
| SOXB2 | SOX14 | - | - | - | - | - | - | - | - |
| SOX21 | - | - | - | - | - | - | - | - | |
| SOXC | SOX4 | Neurodevelopmental syndrome with mild dysmorphism | - | - | - | - | ✓ | - | - |
| SOX11 | Coffin-Siris syndrome-like syndrome (CSSLS) | - | ✓ | - | - | ✓ | - | ✓ | |
| SOX12 | - | - | - | - | - | - | - | - | |
| SOXD | SOX5 | Lamb-Shaffer syndrome | ✓ | ✓ | - | - | - | - | ✓ |
| SOX6 | [Craniosynostosis and craniofacial dysostosis] | ✓ | - | - | - | - | ✓ | - | |
| SOX13 | - | - | - | - | - | - | - | - | |
| SOXE | SOX8 | [Alpha-thalassemia/mental retardation syndrome (ATR-16)] | - | ✓ | - | - | - | - | - |
| Disorder of sex development (DSD) | - | - | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| SOX9 | Campomelic dysplasia (CD) | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | |
| Acampomelic dysplasia (ACD) | ✓ | ✓ | - | - | ✓ | ✓ | - | ||
| Disorder of sex development (DSD) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Isolated Pierre Robin Sequence (PRS) | ✓ | ✓ | - | - | - | - | - | ||
| SOX10 | Waardenburg-Hirschsprung syndrome | - | ✓ | - | - | ✓ | ✓ | ✓ | |
| Peripheral demyelinating neuropathy, central dysmyelination, Waardenburg syndrome, and Hirschsprung disease (PCWH) | - | ✓ | - | - | ✓ | - | ✓ | ||
| Kallmann syndrome | - | - | - | - | ✓ | ✓ | ✓ | ||
| SOXF | SOX7 | - | - | - | - | - | - | - | - |
| SOX17 | [Congenital anomalies of the kidney and urinary tract (CAKUT)] | - | - | - | - | - | ✓ | - | |
| Pulmonary arterial hypertension and congenital heart failure (PAH-CHD) | - | - | - | - | ✓ | ✓ | ✓ | ||
| SOX18 | hypotrichosis-lymphedema-telangiectasia-renal defect syndrome (HLTRS) | - | - | - | - | ✓ | - | ✓ | |
| SOXG | SOX15 | - | - | - | - | - | - | - | - |
| SOXH | SOX30 | - | - | - | - | - | - | - | - |
Del, deletion; Dup, duplication; Fs, frameshift mutation; Inv, inversion; Ms, missense mutation within (HMG) or outside (Other) the HMG domain; Ns; nonsense mutation; T, translocation. Unconfirmed diseases are listed in square brackets.
SOXE genes
The SOXE genes, SOX8, SOX9 and SOX10, were next after SRY to be associated with diseases. They encode transcriptional activators with critical functions in many processes.
Mutations inactivating one SOX9 allele were first shown in 1994 to cause Campomelic Dysplasia (CD) [17, 18]. The disease owes its name to the bending (campo) of limbs (melic), one of many features of this neonatally lethal skeletal dysplasia. The few individuals that have survived to adulthood presented such clinical features as mental retardation and hearing loss in addition to short stature and generalized skeletal malformations [19]. SOX9 is a master regulator of chondrogenesis [20]. It is highly expressed in skeletal progenitor cells and throughout chondrocyte differentiation, and activates most chondrocyte-specific genes [21, 22]. Its heterozygous inactivation in mice reproduces human CD and its homozygous inactivation precludes chondrogenesis [23, 24]. Non-skeletal defects of CD patients reflect important functions of SOX9 in other processes, but based on data from homozygous mutant mice, they reveal only the “tip of the iceberg” regarding SOX9 functions. As indicated earlier, SOX9 is also a master of sex determination. Two-thirds of XY CD patients are sex reversed, and 17q duplications that include SOX9 cause XX sex reversal [25]. In mice, Sox9 homozygous inactivation causes XY sex reversal, as so does Sox9 heterozygous inactivation in a Sox8-null background [26, 27]. More than a hundred different mutations affecting SOX9 have been shown to cause disease. They are described in depth in BOX 1 as a paradigm of the wide spectrums of mutations and diseases than can be associated with a SOX gene. In brief, CD with XY sex reversal is due to de novo heterozygous SOX9 mutations that delete the gene body, translocate most of the upstream regulatory region, or preclude expression of a functional protein. Missense variants are almost always located in the HMG and homodimerization domains, the latter allowing high-affinity binding of SOX9 to pairs of DNA recognition sites. Microdeletions and translocations occurring far-upstream or downstream of SOX9 cause milder diseases, namely acampomelic dysplasia, Pierre Robin sequence (PRS), and DSD without skeletal dysplasia, while duplications of specific upstream regions have been shown to cause XX sex reversal. While nonsense mutations affecting the C-terminal transactivation domain cause CD and XY sex reversal, proving the critical role of this domain, missense mutations in this domain only cause testicular dysgenesis. The reason is likely that transactivation domains are intrinsically disordered and may thus tolerate missense variants better than the highly structured HMG and dimerization domains.
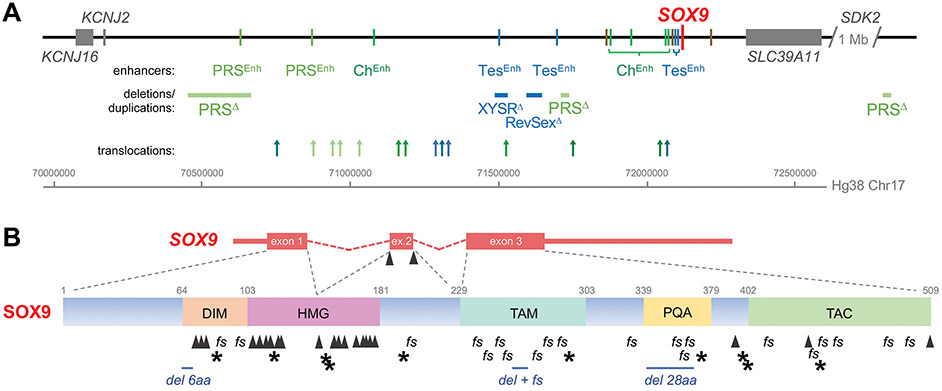
Box 1. A variety of mutations can cause SOXopathies: an example from SOX9.
Many mutations within and around SOX9 have been associated with disease. The gene itself is small (5.4 kb), but embedded within a 2-Mb-long region lacking any other coding gene. This region constitutes the SOX9 topologically associated domain (TAD), i.e., a higher-order chromatin interaction structure controlling SOX9 expression (Figure IA) [95]. It comprises many enhancers driving SOX9 expression in Sertoli cells, chondrocytes or other cell types, and translocations, deletions, duplications, and point variants at various locations within this TAD underlie skeletal dysplasias and DSD phenotypes with various degrees of severity [1, 20, 27, 96-100]. Mutations within the SOX9 gene body have also been associated with SOXopathies (Figure IB). Nonsense variants result in truncated proteins retaining partial activity or completely inactive. Frameshift variants result in shorter or longer proteins with altered activities. Nonsense and frameshift mutations are widely distributed along the coding sequence, whereas missense variants are largely restricted to splice sites and to the dimerization, DNA-binding or transactivation domains. Most missense mutations and in-frame deletions in the dimerization and HMG domains and in splice sites cause severe disease (CD with XY sex reversal), whereas missense variants outside these regions cause mild genitalia defects without skeletal abnormalities.
Figure I.
Types and distributions of pathogenic mutations in the SOX9 locus. A) SOX9 locus and flanking genes on 17q24.3, including enhancers primarily active in chondrocytes (ChEnh, green bars), Sertoli cells (TesEnh, blue bars), embryonic mandibular region (PRSEnh, light green bars) and other cell types/tissues (brown bars); microdeletions causing Pierre Robin sequence (PRSΔ) and XY sex reversal (XYSRΔ); a duplication causing XX sex reversal (RevSexΔ); and translocations causing campomelic dysplasia (dark green arrows), acampomelic dysplasia (lighter green arrows), Pierre Robin sequence (light green arrows), XY or XX sex reversal (blue arrows), or skeletal dysplasia and XY sex reversal (teal arrows). B) SOX9 exon/intron and protein structures, including pathogenic microdeletions (del) and nonsense variants (*), frameshift variants (fs) and missense and splice variants (Δ).
SOX8 inactivation was initially proposed to contribute to mental retardation in Alpha-Thalassemia/Mental Retardation (ATR-16), a syndrome due to deletions or unbalanced translocations within a 1-Mb 16p13.3 region that includes SOX8 [28]. However, this proposition remains unvalidated. Recently, genome rearrangements just upstream of SOX8 and missense variants within and outside the HMG domain were identified in males and females with a DSD spectrum that included oligozoospermia, azoospermia, primary ovary deficiency and XY sex reversal [29]. Noteworthily, mental retardation was not reported. These findings establish the importance of human SOX8 in sex determination, like mouse Sox8. Sox8-null mice are viable and leaner than normal [30]. Sex determination is unaffected unless, as reported earlier, the mice are also Sox9 heterozygous null. Sox8-null males, however, become infertile early in adulthood [31].
Heterozygous mutations inactivating SOX10 cause various neurocristopathies: Waardenburg disease, characterized by hearing loss and pigmentation defects; the Hirschsprung intestinal disorder; PCWH, which comprises Peripheral demyelinating neuropathy, Central demyelinating leukodystrophy, and Waardenburg and Hirschsprung disease [32, 33]; and Kallmann syndrome, a form of hypogonadism characterized by delayed or absent puberty and olfactory defects [34]. The diseases are reminiscent of the phenotypes of mice carrying spontaneous Sox10 inactivating mutationa (e.g., Sox10DOM) or a Sox10 null allele at the heterozygous state (megacolon and pigmentation defect) or homozygous state (namely, lack of peripheral nervous system glia and disrupted differentiation of oligodendroglia) [35-37]. Many aspects of these diseases reflect the fact that SOX10 is essential to specify neural crest cells, controls the development of various neural crest derivatives, including Schwann cells, cardiac crest cells, sensory neurons and melanocytes, and is also essential for the development of oligodendrocytes from neuroectoderm [38, 39].
SOXB genes
The SOXB group comprises SOXB1 (SOX1, SOX2, and SOX3) and SOXB2 genes (SOX14 and SOX21), originally described as encoding transcriptional activators and repressors, respectively. Recent studies, however, have shown that this functional distinction between the two subgroups may not be as strict as initially proposed, as SOX2, for instance, represses as many genes as it activates in neural stem cells [40]. All SOXB genes are expressed in progenitor cells from early development and most highly in the nervous system.
Expansions of a polyalanine tract within the SOX3 coding sequence were shown in 2002 to cause X-linked mental retardation, short stature due to growth hormone deficiency, and occasionally facial dysmorphism and complete panhypopituitarism [41]. These alterations cause protein aggregation, and thus loss of function. Other mutations, either reducing or increasing SOX3 dosage, may cause variants of septic-optic dysplasia (SOD), a highly heterogeneous disease that includes optic nerve hypoplasia, corpus callosum and septum pellucidum agenesis, and panhypopituitarism due to pituitary hypoplasia [42]. Furthermore, unique rearrangements in the SOX3 regulatory region, which likely led to ectopic expression of SOX3 in the developing gonad, were reported in patients with XX male sex reversal [43]. Consistent with these diseases, Sox3-null mice have profound growth insufficiency, weakness, craniofacial abnormalities, hypopituitarism, and midline CNS defects [44]. They do not have sex reversal, but both males and females show severely reduced fertility.
SOX2 is well known for its master roles in specification, differentiation and maintenance of pluripotent embryonic stem cells and other progenitor cell types [45]. Various kinds of heterozygous loss-of-function mutations were first associated in 2003 with anophtalmia or microphtalmia syndromes often including craniofacial and other skeletal abnormalities, developmental delay, learning difficulties, esophageal atresia, sensori-neural hearing loss and genital abnormalities [42, 46]. In agreement with these data, Sox2-null mice die in early embryogenesis from failure to form pluripotent epiblast [47]; mice with Sox2 hypomorphic mutations display a spectrum of eye and other malformations [48]; and Sox2+/− mice show impaired development of the hypothalamo-pituitary and reproductive axes [49].
It remains unknown whether SOX1, SOX14 and SOX21 mutations cause diseases. It also unknown whether mouse Sox14 is critical. In contrast, Sox1-null mice have microphthalmia and cataract [50] and suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability [51], and Sox21-null mice are small, for unexplained reasons [52], and show cyclic alopecia, explained by master roles for SOX21 in hair shaft cuticle differentiation [53]. One can thus predict that SOXopathies may soon be revealed for these genes.
SOXF genes
SOX7, SOX17, and SOX18 compose the SOXF group. They encode transcriptional activators that have been shown in animal models to be pivotal in several developmental processes, including cardiogenesis, vasculogenesis and angiogenesis (SOX7, SOX17 and SOX18), lymphangiogenesis and hair follicle development (SOX18), and hemangioblastogenesis, definitive endoderm and gastro-intestinal system formation (SOX17) [54-60].
First described in 2003, SOX18 loss-of-function mutations cause Hypotrichosis-Lymphedema-Telangiectasia syndrome (HLTS), i.e., sparse hair, absence of eyebrows and eyelashes, lymphatic edema, and peripheral vein anomalies [61]. Following this report, one patient developed renal failure and additional patients with Hypotrichosis-Lymphedema-Telangiectasia-Renal Defect syndrome (HLTRS) were found to carry pathogenic SOX18 variants [62]. Some variants were heterozygous and nonsense, truncating SOX18 before or within its transactivation domains. It was proposed, but not tested, that the mutant protein could dominant-negatively affect the wild-type protein. Other variants were homozygous missense and found in consanguineous families, in which heterozygotes were unaffected. They constitute the first and so far, only cases of recessive SOXopathy.
SOX17 variants were described in 2010 in patients with congenital anomalies of the kidney and urinary tract (CAKUT) [63]. Several patients were carrying a missense variant located in a region of unknown function and causing excessive accumulation of SOX17 protein in vitro. It was later found in an individual that did not have CAKUT disease [64]. Other patients also had missense variants outside the HMG domain of unknown functional impact. These SOX17 variants could generate risk rather than causative alleles for CAKUT. Very recently, SOX17 heterozygous variants linked to pulmonary arterial hypertension and congenital heart disease (PAT-CHD). Two studies reported frameshift, nonsense and missense variants, the latter affecting highly conserved residues in the HMG domain or transactivation/β-catenin-binding domain [65, 66]. Several alterations segregated with PAH in families. Further, genome-wide association studies found common genetic variations associated with PAH in a critical enhancer upstream of SOX17 [67].
Mutations in SOX7 have not been firmly linked to a disease yet, but recurrent microdeletions of 8p23.1 that include SOX7 and GATA4 confer a high risk of congenital diaphragmatic hernia (CDH) and cardiac defects [68]. CDH is partially penetrant in Sox7+/− and Gata4+/− mice, suggesting that combined haploinsufficiency of SOX7 and GATA4 may cause CDH.
SOXD genes
The SOXD group comprises SOX5, SOX6, and SOX13. These genes encode proteins that homodimerize through coiled-coil domains and bind target genes preferentially to pairs of SOX sites. SOX5 and SOX6 are closer to one another than to SOX13, and control several developmental processes. They help either in transactivation or in transrepression depending on the cell context. Sox5 and Sox6 single-null mice are born with discrete skeletal malformations, and double-null fetuses die in utero with a severe chondrodysplasia [69]. This is explained by cooperation of SOX5 and SOX6 with SOX9 in activating the chondrocyte program [20, 21]. In contrast, these SOXD proteins inhibit transactivation by SOXC, SOXE or other factors in neocorticogenesis (SOX5), oligodendrogenesis (SOX5 and SOX6), myogenesis (SOX6), erythropoiesis (SOX6), and melanogenesis (SOX5) [70-75].
In 2006, a child with craniosynostosis and other dysostosis features was found to carry a balanced translocation (t(9; 11)(q33;p15) disrupting SOX6 (11p15) [76]. Another child, with a 9q32-q34 deletion, had a similar phenotype but no craniosynostosis, and a third child, who inherited a missense variant from his unaffected mother, only had craniosynostosis. The variant was located in an N-terminal region of SOX6 of unknown function. These cases concur that SOX6 mutations might cause craniosynostosis, but this possibility needs validation.
In 2012, de novo heterozygous translocations and microdeletions disrupting SOX5 were reported in patients with global developmental delay, intellectual disability, hypotonia, autistic-like features, and mild facial dysmorphism and skeletal malformations [77]. The disorder was named Lamb-Shaffer syndrome and additional loss-of-function variants, including nonsense ones, were subsequently reported in other patients [78].
SOX13 is expressed in several tissues, including kidney, pancreas, lung, liver, and spinal cord. Its inactivation and overexpression in the mouse have revealed that it promotes gammadelta T cell development while opposing alphabeta T cell differentiation [79], and adds to SOX5 and SOX6 to control the development of mouse spinal cord oligodendrocytes [80]. To date, however, no human disease has yet been associated with mutations in SOX13.
SOXC genes
SOX4, SOX11, and SOX12 form the SOXC group. They encode transcriptional activators, of which SOX11 is the strongest, and SOX12 the weakest. They are expressed in many progenitor cell types and critically control cell survival and fate determination in response to various signaling pathways [81, 82] . Sox4-null mice die in utero from heart malformation and Sox11-null mice die at birth with abnormalities in the heart, skeleton, and multiple internal organs, whereas Sox12-null mice are healthy throughout development and adulthood under regular conditions [83-85]. Mouse conditional knockouts have uncovered redundant roles for Sox4 and Sox11 in many developmental processes from early organogenesis, including neurogenesis, skeletogenesis and outflow tract formation [86-88].
In 2014, two de novo heterozygous missense variants in the SOX11 HMG box were linked to a Coffin-Siris syndrome-like syndrome (CSSLS) characterized by intellectual disability, growth deficiency, facial dysmorphism and hypoplasia of the fifth digit [89]. The variant proteins were unable to bind DNA. Consolidating the notion of SOX11 haploinsufficiency, more de novo heterozygous mutations were later reported in patients with similar features [90]. They included SOX11-containing 2p25 deletions, a nonsense variant and additional HMG-domain missense variants.
Very recently, four de novo heterozygous missense variants in the SOX4 HMG box were identified in patients with intellectual disability and mild facial and digit dysmorphism [91]. Resemblance to CSSLS is consistent with combined roles for mouse Sox4 and Sox11 in many processes. Interestingly, the patients’ variants were nonfunctional in vitro, whereas all twelve variants listed in gnomAD, a database of control individuals, were functional. Thus, while many HMG-domain variants have been reported in SOX genes to cause diseases, this finding calls for caution in interpreting diagnostic data as it implies that not every such variant should be considered pathogenic.
To date, SOX12 has not been linked to a disease yet. Sox12-null mice were recently found to show impaired regulatory T cell/lymphocyte differentiation during colitis [92]. This first finding of an important role for mouse Sox12 in vivo should encourage studies to link human SOX12 variants to SOXopathies.
SOXG and SOXH genes
Although SOX15 and SOX30 are classified as SOXG and SOXH genes, respectively, SOX15 shares recent ancestry with SOXB1 genes and SOX30 with SOXD genes. Neither gene has been linked to a human disease yet, but important roles have been shown for their mouse orthologs. Sox15-null mice develop normally and have an unremarkable adult life except for a reduced ability to regenerate skeletal muscle in response to a crush injury [93]. This weakness is explained by the expression of Sox15 in satellite cell-derived myoblasts and its involvement in myogenic determination. Sox30-null mice look normal too, but males are sterile, due to a block of spermiogenesis at the round spermatid stage [40]. Based on these data, it is tempting to speculate that mutations in human SOX15 and SOX30 underlie yet-to-be-uncovered SOXopathies.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
The study of SOX genes and discovery of SOXopathies have provided seminal information on genetic, cellular and molecular mechanisms underlying fundamental processes from early development onwards. With all the current information, we can tentatively provide a unified view of SOXopathy disease features. Indeed, most SOXopathies are rare developmental syndromes. Based on findings in animals, most SOXopathies only show “the tip of icebergs” regarding crucial involvement of SOX genes in human processes. Intellectual disability, disorders of sex development, and skeletal and cardiovascular malformations are common, but defects in virtually every system have been reported. Additionally, most SOXopathies result from de novo heterozygous loss-of-function mutations, and thus reveal gene haploinsufficiency. Of course, there are exceptions. For example, SRY loss-of-function variants fully reveal SRY functions because SRY has no close relative and is expressed from a single allele; Reduced fertility due to SOX8 mutations are adult rather than developmental diseases; and SOX3 and SOX9 duplications as well as SOX18 missense variants outside the HMG domain have been associated with diseases. All cases, however, reflect the importance of proper gene dosage to achieve normalcy.
To date, the discovery of SOXopathies is merely mid-way completed. Many important questions remain unanswered (see Outstanding Questions). Half of the SOX genes are still disease-orphan and more disease associations may remain unknown for the other half. One might think that the remaining diseases are benign, otherwise they would be known by now, but this argument is easy to counter since SOX17 was linked to pulmonary arterial hypertension and congenital heart failure only in the last year. In addition to developmental disorders, some still-elusive SOXopathies may arise only with increasing age and in specific contexts, such as cancer, tissue repair and immune response (BOX 2). Further research is also needed to better understand the cellular and molecular basis of SOXopathies, and in particular, the issue of disease penetrance and severity. For this, we need to learn how to distinguish pathogenic from risk and benign variants. To explain diseases and develop therapeutic strategies, we also have to further characterize the factors that functionally interact with SOX genes and proteins. All new knowledge will undoubtedly be very valuable to inform genetic counseling and to better understand and treat many other diseases, including those in which SOX genes may intervene abnormally even if intact.
OUTSTANDING QUESTIONS BOX.
Are all twenty human SOX genes involved in SOXopathies? Are SOXopathies primarily developmental disorders or do they also include a broad range of adult-onset diseases? How broad is the spectrum of diseases associated with any single SOX gene? Are diseases the only outcome of SOX gene variants or are there any phenotypic advantages conferred by some rare or common SOX gene variants to human carriers?
Are pathogenic SOX missense and nonsense variants primarily resulting in null alleles? Do some confer reduced, increased, dominant-negative, or ectopic activity?
Have SOX genes acquired new functions or new expression level requirements during evolution that could explain why several SOXopathies are detected in humans but not in mice upon heterozygous inactivation of some SOX genes? In particular, as many SOX genes are required for brain development, has the evolution of human brain-specific features relied on regulatory changes in SOX gene dosage and expression pattern?
What are the treatment options for SOXopathies? Is gene therapy an option? Are SOX proteins druggable? When should therapies be initiated?
Box 2. SOX genes and non-developmental diseases.
Many types of diseases implicate SOX genes, but are not due to germline SOX mutations and therefore do not classify as SOXopathies. Cancers are the great majority of them [101]. In fact, all SOX genes have been shown to be dysregulated in at least one tumor type. Deregulation can occur at the genetic level, or at the epigenetic, transcriptional, translational and post-translational levels, resulting in either increased or decreased SOX activities. SOX factors being master determinants of cell fate, their deregulation can cause drastic changes in cell stemness, survival, proliferation, migration, and differentiated activities. SOX genes can be either tumor repressors or promoters depending on tumor types and environment.
Among other adult-onset and degenerative diseases, single nucleotide polymorphisms within and around SOX4 correlate with moderate risks for osteoporosis and reduced expression of SOX4 in bone correlates with postmenopausal osteoporosis [102, 103]. A significant association exists between SOX5 variants and a familial form of late-onset Alzheimer’s disease [104]. Also, a single nucleotide polymorphism in SOX8 was identified as a genuine multiple sclerosis susceptibility locus [105], a finding consistent with the importance of mouse Sox8 in oligodendrocyte myelination [106]. If confirmed, these polymorphisms within or around SOX4, SOX5 and SOX8 could classify these disease forms as SOXopathies. Additionally, SOX5, SOX6, and SOX9 downregulation [107] and SOX4 and SOX11 upregulation correlate with cartilage degeneration in osteoarthritic patients [108]. Also, SOX2 downregulation is seen in brain sections from Alzheimer’s patients, which is consistent with neurodegeneration features resembling Huntington’s and Alzheimer’s disease described in mice with Sox2 deficiency [109].
Autoimmune diseases are another class of disorders worth mentioning. Like other transcription factors, several SOX proteins are inclined to generate pathogenic autoimmune responses. For instance, SOX13 was initially discovered in humans as an autoantigen in type 1 diabetes [110] and later in primary biliary cirrhosis [111], and SOX9 and SOX10 are vitiligo autoantigens in autoimmune polyendocrine syndrome type I [112].
HIGHLIGHTS.
SOXopathies are rare severe disorders resulting from mutations in the SOX genes. They have been associated to date with half of the twenty SOX family members and the numbers of genes involved and pathogenic variants are still on the rise.
Most SOXopathies result in developmental defects and are syndromic, including such severe defects as sex reversal, intellectual disability, skeletal dysmorphism, and cardiovascular anomalies.
SOXopathies can be caused by many types of gene alterations, and most mutations are de novo, heterozygous and loss-of-function, thus exposing gene haploinsufficiency.
Missense variants are almost exclusively located in the HMG domain, a distinctive and multifunctional feature of all SOX proteins.
ACKNOWLEGMENTS
We are grateful to patients, families and friends who have motivated and supported our research on SOXopathies. This work was supported by funding from the National Institutes of Health NIAMS (R01 AR068308 and R01 AR72649), Harper’s Quest and the LAMSHF Syndrome Research Fund.
GLOSSARY
- De novo
refers to gene deletions or variants detected in a child, but not in the biological parents.
- DNA bending
ability of the HMG domain to induce a strong bend of the DNA helix. This property is believed to have an architectural role in the formation of enhanceosomes (complexes of transcription factors bound to enhancer sequences).
- Dominant negative
refers to a heterozygous mutation that results in a variant protein that negatively interferes with the activity of the wild-type protein.
- Haploinsufficiency
term referring to a gene located on an autosomal chromosome that is unable to fully achieve its normal functions when one of its alleles carries a loss-of-function mutation.
- HMG domain
DNA-binding domain originally identified in HMGB proteins, which are members of the superfamily of non-histone chromatin proteins that exhibit high mobility in SDS-PAGE. This domain also characterizes the SOX and TCF/LEF families.
- SOX
acronym for SRY-related HMG box-containing gene or protein. SOX proteins share at least 50% similarity in the HMG domain with SRY and with one another. The SOX family counts 19 members in addition to SRY in humans and most mammals.
- SOX motif
DNA sequence specifically recognized and bound by the HMG domain of SOX proteins. This motif corresponds to or resembles the C[A/T]TTG[A/T][A/T] sequence. Interactions occur at the level of A/T pairs in the minor groove of the DNA helix.
- SRY
gene located in the sex-determining region of the Y chromosome. SRY is the founder (first identified member) of the SOX family.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gonen N and Lovell-Badge R, The regulation of Sox9 expression in the gonad. Curr Top Dev Biol, 2019. 134: p. 223–252. [DOI] [PubMed] [Google Scholar]
- 2.Schepers GE, Teasdale RD, and Koopman P, Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell, 2002. 3(2): p. 167–70. [DOI] [PubMed] [Google Scholar]
- 3.Kamachi Y and Kondoh H, Sox proteins: regulators of cell fate specification and differentiation. Development, 2013. 140(20): p. 4129–44. [DOI] [PubMed] [Google Scholar]
- 4.Tajan M, et al. , The RASopathy Family: Consequences of Germline Activation of the RAS/MAPK Pathway. Endocr Rev, 2018. 39(5): p. 676–700. [DOI] [PubMed] [Google Scholar]
- 5.Jobling R, et al. , The collagenopathies: review of clinical phenotypes and molecular correlations. Curr Rheumatol Rep, 2014. 16(1): p. 394. [DOI] [PubMed] [Google Scholar]
- 6.Hock R, et al. , HMG chromosomal proteins in development and disease. Trends Cell Biol, 2007. 17(2): p. 72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou L, Srivastava Y, and Jauch R, Molecular basis for the genome engagement by Sox proteins. Semin Cell Dev Biol, 2017. 63: p. 2–12. [DOI] [PubMed] [Google Scholar]
- 8.Berta P, et al. , Genetic evidence equating SRY and the testis-determining factor. Nature, 1990. 348(6300): p. 448–50. [DOI] [PubMed] [Google Scholar]
- 9.Berkovitz GD, et al. , The role of the sex-determining region of the Y chromosome (SRY) in the etiology of 46,XX true hermaphroditism. Hum Genet, 1992. 88(4): p. 411–6. [DOI] [PubMed] [Google Scholar]
- 10.Cameron FJ and Sinclair AH, Mutations in SRY and SOX9: testis-determining genes. Hum Mutat, 1997. 9(5): p. 388–95. [DOI] [PubMed] [Google Scholar]
- 11.Harley VR, Clarkson MJ, and Argentaro A, The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9 [SRY-related high-mobility group (HMG) box 9]. Endocr Rev, 2003. 24(4): p. 466–87. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. , Identification of a novel mutation (Ala66Thr) of SRY gene causes XY pure gonadal dysgenesis by affecting DNA binding activity and nuclear import. Gene, 2018. 651: p. 143–151. [DOI] [PubMed] [Google Scholar]
- 13.McElreavey K and Cortes LS, X-Y translocations and sex differentiation. Semin Reprod Med, 2001. 19(2): p. 133–9. [DOI] [PubMed] [Google Scholar]
- 14.Lovell-Badge R and Robertson E, XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development, 1990. 109(3): p. 635–46. [DOI] [PubMed] [Google Scholar]
- 15.Koopman P, et al. , Male development of chromosomally female mice transgenic for Sry. Nature, 1991. 351(6322): p. 117–21. [DOI] [PubMed] [Google Scholar]
- 16.She ZY and Yang WX, Sry and SoxE genes: How they participate in mammalian sex determination and gonadal development? Semin Cell Dev Biol, 2017. 63: p. 13–22. [DOI] [PubMed] [Google Scholar]
- 17.Foster JW, et al. , Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature, 1994. 372(6506): p. 525–30. [DOI] [PubMed] [Google Scholar]
- 18.Wagner T, et al. , Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell, 1994. 79(6): p. 1111–20. [DOI] [PubMed] [Google Scholar]
- 19.Mansour S, et al. , The phenotype of survivors of campomelic dysplasia. J Med Genet, 2002. 39(8): p. 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefebvre V and Dvir-Ginzberg M, SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect Tissue Res, 2017. 58(1): p. 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CF and Lefebvre V, The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res, 2015. 43(17): p. 8183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohba S, et al. , Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep, 2015. 12(2): p. 229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi W, et al. , Sox9 is required for cartilage formation. Nat Genet, 1999. 22(1): p. 85–9. [DOI] [PubMed] [Google Scholar]
- 24.Akiyama H, et al. , The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev, 2002. 16(21): p. 2813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn A and Koopman P, The molecular genetics of sex determination and sex reversal in mammals. Semin Reprod Med, 2012. 30(5): p. 351–63. [DOI] [PubMed] [Google Scholar]
- 26.Chaboissier MC, et al. , Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development, 2004. 131(9): p. 1891–901. [DOI] [PubMed] [Google Scholar]
- 27.Barrionuevo F, et al. , Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol Reprod, 2006. 74(1): p. 195–201. [DOI] [PubMed] [Google Scholar]
- 28.Pfeifer D, et al. , The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics, 2000. 63(1): p. 108–16. [DOI] [PubMed] [Google Scholar]
- 29.Portnoi MF, et al. , Mutations involving the SRY-related gene SOX8 are associated with a spectrum of human reproductive anomalies. Hum Mol Genet, 2018. 27(7): p. 1228–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sock E, et al. , Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol Cell Biol, 2001. 21(20): p. 6951–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrionuevo F, et al. , Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol, 2009. 327(2): p. 301–12. [DOI] [PubMed] [Google Scholar]
- 32.Pingault V, et al. , SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet, 1998. 18(2): p. 171–3. [DOI] [PubMed] [Google Scholar]
- 33.Inoue K, et al. , Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet, 2004. 36(4): p. 361–9. [DOI] [PubMed] [Google Scholar]
- 34.Pingault V, et al. , Loss-of-function mutations in SOX10 cause Kallmann syndrome with deafness. Am J Hum Genet, 2013. 92(5): p. 707–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Southard-Smith EM, Kos L, and Pavan WJ, Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet, 1998. 18(1): p. 60–4. [DOI] [PubMed] [Google Scholar]
- 36.Britsch S, et al. , The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev, 2001. 15(1): p. 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stolt CC, et al. , Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev, 2002. 16(2): p. 165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldin CE and LaBonne C, SoxE factors as multifunctional neural crest regulatory factors. Int J Biochem Cell Biol, 2010. 42(3): p. 441–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weider M and Wegner M, SoxE factors: Transcriptional regulators of neural differentiation and nervous system development. Semin Cell Dev Biol, 2017. 63: p. 35–42. [DOI] [PubMed] [Google Scholar]
- 40.Han F, et al. , Epigenetic regulation of sox30 is associated with testis development in mice. PLoS One, 2014. 9(5): p. e97203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laumonnier F, et al. , Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet, 2002. 71(6): p. 1450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelberman D and Dattani MT, Genetics of septo-optic dysplasia. Pituitary, 2007. 10(4): p. 393–407. [DOI] [PubMed] [Google Scholar]
- 43.Sutton E, et al. , Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest, 2011. 121(1): p. 328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss J, et al. , Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol, 2003. 23(22): p. 8084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar A and Hochedlinger K, The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell, 2013. 12(1): p. 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slavotinek A, Genetics of anophthalmia and microphthalmia. Part 2: Syndromes associated with anophthalmia-microphthalmia. Hum Genet, 2018. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S and Cui W, Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J Stem Cells, 2014. 6(3): p. 305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taranova OV, et al. , SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev, 2006. 20(9): p. 1187–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelberman D, et al. , Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest, 2006. 116(9): p. 2442–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishiguchi S, et al. , Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev, 1998. 12(6): p. 776–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malas S, et al. , Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience, 2003. 119(2): p. 421–32. [DOI] [PubMed] [Google Scholar]
- 52.Cheung LYM, Okano H, and Camper SA, Sox21 deletion in mice causes postnatal growth deficiency without physiological disruption of hypothalamic-pituitary endocrine axes. Mol Cell Endocrinol, 2017. 439: p. 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiso M, et al. , The disruption of Sox21-mediated hair shaft cuticle differentiation causes cyclic alopecia in mice. Proc Natl Acad Sci U S A, 2009. 106(23): p. 9292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lilly AJ, Lacaud G, and Kouskoff V, SOXF transcription factors in cardiovascular development. Semin Cell Dev Biol, 2017. 63: p. 50–57. [DOI] [PubMed] [Google Scholar]
- 55.Francois M, Koopman P, and Beltrame M, SoxF genes: Key players in the development of the cardio-vascular system. Int J Biochem Cell Biol, 2010. 42(3): p. 445–8. [DOI] [PubMed] [Google Scholar]
- 56.Nakajima-Takagi Y, et al. , Role of SOX17 in hematopoietic development from human embryonic stem cells. Blood, 2013. 121(3): p. 447–58. [DOI] [PubMed] [Google Scholar]
- 57.Seguin CA, et al. , Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell, 2008. 3(2): p. 182–95. [DOI] [PubMed] [Google Scholar]
- 58.Lange AW, et al. , Sox17 is required for normal pulmonary vascular morphogenesis. Dev Biol, 2014. 387(1): p. 109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfister S, et al. , Sox17-dependent gene expression and early heart and gut development in Sox17-deficient mouse embryos. Int J Dev Biol, 2011. 55(1): p. 45–58. [DOI] [PubMed] [Google Scholar]
- 60.Pennisi D, et al. , Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet, 2000. 24(4): p. 434–7. [DOI] [PubMed] [Google Scholar]
- 61.Irrthum A, et al. , Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am J Hum Genet, 2003. 72(6): p. 1470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moalem S, et al. , Hypotrichosis-lymphedema-telangiectasia-renal defect associated with a truncating mutation in the SOX18 gene. Clin Genet, 2015. 87(4): p. 378–82. [DOI] [PubMed] [Google Scholar]
- 63.Gimelli S, et al. , Mutations in SOX17 are associated with congenital anomalies of the kidney and the urinary tract. Hum Mutat, 2010. 31(12): p. 1352–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Combes P, et al. , Relevance of SOX17 variants for hypomyelinating leukodystrophies and congenital anomalies of the kidney and urinary tract (CAKUT). Ann Hum Genet, 2012. 76(3): p. 261–7. [DOI] [PubMed] [Google Scholar]
- 65.Zhu N, et al. , Rare variants in SOX17 are associated with pulmonary arterial hypertension with congenital heart disease. Genome Med, 2018. 10(1): p. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graf S, et al. , Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat Commun, 2018. 9(1): p. 1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhodes CJ, et al. , Genetic determinants of risk in pulmonary arterial hypertension: international genome-wide association studies and meta-analysis. Lancet Respir Med, 2019. 7(3): p. 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wat MJ, et al. , Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet, 2012. 21(18): p. 4115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smits P, et al. , The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell, 2001. 1(2): p. 277–90. [DOI] [PubMed] [Google Scholar]
- 70.Lai T, et al. , SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron, 2008. 57(2): p. 232–47. [DOI] [PubMed] [Google Scholar]
- 71.Kwan KY, et al. , SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A, 2008. 105(41): p. 16021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Azim E, et al. , SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat Neurosci, 2009. 12(10): p. 1238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hagiwara N, Yeh M, and Liu A, Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dyn, 2007. 236(8): p. 2062–76. [DOI] [PubMed] [Google Scholar]
- 74.Dumitriu B, et al. , Sox6 cell-autonomously stimulates erythroid cell survival, proliferation, and terminal maturation and is thereby an important enhancer of definitive erythropoiesis during mouse development. Blood, 2006. 108(4): p. 1198–207. [DOI] [PubMed] [Google Scholar]
- 75.Stolt CC, et al. , The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic Acids Res, 2008. 36(17): p. 5427–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tagariello A, et al. , Balanced translocation in a patient with craniosynostosis disrupts the SOX6 gene and an evolutionarily conserved non-transcribed region. J Med Genet, 2006. 43(6): p. 534–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamb AN, et al. , Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum Mutat, 2012. 33(4): p. 728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nesbitt A, et al. , Exome sequencing expands the mechanism of SOX5-associated intellectual disability: A case presentation with review of sox-related disorders. Am J Med Genet A, 2015. 167A(11): p. 2548–54. [DOI] [PubMed] [Google Scholar]
- 79.Melichar HJ, et al. , Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science, 2007. 315(5809): p. 230–3. [DOI] [PubMed] [Google Scholar]
- 80.Baroti T, et al. , Sox13 functionally complements the related Sox5 and Sox6 as important developmental modulators in mouse spinal cord oligodendrocytes. J Neurochem, 2016. 136(2): p. 316–28. [DOI] [PubMed] [Google Scholar]
- 81.Lefebvre V and Bhattaram P, SOXC Genes and the Control of Skeletogenesis. Curr Osteoporos Rep, 2016. 14(1): p. 32–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kavyanifar A, Turan S, and Lie DC, SoxC transcription factors: multifunctional regulators of neurodevelopment. Cell Tissue Res, 2018. 371(1): p. 91–103. [DOI] [PubMed] [Google Scholar]
- 83.Schilham MW, et al. , Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature, 1996. 380(6576): p. 711–4. [DOI] [PubMed] [Google Scholar]
- 84.Sock E, et al. , Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol, 2004. 24(15): p. 6635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoser M, et al. , Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol, 2008. 28(15): p. 4675–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bhattaram P, et al. , Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun, 2010. 1: p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bergsland M, et al. , Sequentially acting Sox transcription factors in neural lineage development. Genes Dev, 2011. 25(23): p. 2453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paul MH, et al. , Cardiac outflow tract development relies on the complex function of Sox4 and Sox11 in multiple cell types. Cell Mol Life Sci, 2014. 71(15): p. 2931–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsurusaki Y, et al. , De novo SOX11 mutations cause Coffin-Siris syndrome. Nat Commun, 2014. 5: p. 4011. [DOI] [PubMed] [Google Scholar]
- 90.Hempel A, et al. , Deletions and de novo mutations of SOX11 are associated with a neurodevelopmental disorder with features of Coffin-Siris syndrome. J Med Genet, 2016. 53(3): p. 152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zawerton A, et al. , De Novo SOX4 Variants Cause a Neurodevelopmental Disease Associated with Mild Dysmorphism. Am J Hum Genet, 2019. 104(2): p. 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tanaka S, et al. , Sox12 promotes T reg differentiation in the periphery during colitis. J Exp Med, 2018. 215(10): p. 2509–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee HJ, et al. , Sox15 is required for skeletal muscle regeneration. Mol Cell Biol, 2004. 24(19): p. 8428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murphy EC, et al. , Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J Mol Biol, 2001. 312(3): p. 481–99. [DOI] [PubMed] [Google Scholar]
- 95.Franke M, et al. , Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature, 2016. 538(7624): p. 265–269. [DOI] [PubMed] [Google Scholar]
- 96.Mochizuki Y, et al. , Combinatorial CRISPR/Cas9 Approach to Elucidate a Far-Upstream Enhancer Complex for Tissue-Specific Sox9 Expression. Dev Cell, 2018. 46(6): p. 794–806 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Symon A and Harley V, SOX9: A genomic view of tissue specific expression and action. Int J Biochem Cell Biol, 2017. 87: p. 18–22. [DOI] [PubMed] [Google Scholar]
- 98.Croft B, et al. , Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9. Nat Commun, 2018. 9(1): p. 5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao B, et al. , The SOX9 upstream region prone to chromosomal aberrations causing campomelic dysplasia contains multiple cartilage enhancers. Nucleic Acids Res, 2015. 43(11): p. 5394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gonen N, et al. , Sex reversal following deletion of a single distal enhancer of Sox9. Science, 2018. 360(6396): p. 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grimm D, et al. , The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol, 2019. [DOI] [PubMed] [Google Scholar]
- 102.Duncan EL, et al. , Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet, 2011. 7(4): p. e1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jemtland R, et al. , Molecular disease map of bone characterizing the postmenopausal osteoporosis phenotype. J Bone Miner Res, 2011. 26(8): p. 1793–801. [DOI] [PubMed] [Google Scholar]
- 104.Li A, et al. , Silencing of the Drosophila ortholog of SOX5 leads to abnormal neuronal development and behavioral impairment. Hum Mol Genet, 2017. 26(8): p. 1472–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.International Multiple Sclerosis Genetics, C., et al. , MANBA, CXCR5, SOX8, RPS6KB1 and ZBTB46 are genetic risk loci for multiple sclerosis. Brain, 2013. 136(Pt 6): p. 1778–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Turnescu T, et al. , Sox8 and Sox10 jointly maintain myelin gene expression in oligodendrocytes. Glia, 2018. 66(2): p. 279–294. [DOI] [PubMed] [Google Scholar]
- 107.Lee JS and Im GI, SOX trio decrease in the articular cartilage with the advancement of osteoarthritis. Connect Tissue Res, 2011. 52(6): p. 496–502. [DOI] [PubMed] [Google Scholar]
- 108.Takahata Y, et al. , Sox4 is involved in osteoarthritic cartilage deterioration through induction of ADAMTS4 and ADAMTS5. FASEB J, 2019. 33(1): p. 619–630. [DOI] [PubMed] [Google Scholar]
- 109.Sarlak G and Vincent B, The Roles of the Stem Cell-Controlling Sox2 Transcription Factor: from Neuroectoderm Development to Alzheimer's Disease? Mol Neurobiol, 2016. 53(3): p. 1679–1698. [DOI] [PubMed] [Google Scholar]
- 110.Kasimiotis H, et al. , Sex-determining region Y-related protein SOX13 is a diabetes autoantigen expressed in pancreatic islets. Diabetes, 2000. 49(4): p. 555–61. [DOI] [PubMed] [Google Scholar]
- 111.Fida S, et al. , Autoantibodies to the transcriptional factor SOX13 in primary biliary cirrhosis compared with other diseases. J Autoimmun, 2002. 19(4): p. 251–7. [DOI] [PubMed] [Google Scholar]
- 112.Hedstrand H, et al. , The transcription factors SOX9 and SOX10 are vitiligo autoantigens in autoimmune polyendocrine syndrome type I. J Biol Chem, 2001. 276(38): p. 35390–5. [DOI] [PubMed] [Google Scholar]