Abstract
Objective:
A study was undertaken to identify the gene underlying DYT4 dystonia, a dominantly inherited form of spasmodic dysphonia combined with other focal or generalized dystonia and a characteristic facies and body habitus, in an Australian family.
Methods:
Genome-wide linkage analysis was carried out in 14 family members followed by genome sequencing in 2 individuals. The index patient underwent a detailed neurological follow-up examination, including electrophysiological studies and magnetic resonance imaging scanning. Biopsies of the skin and olfactory mucosa were obtained, and expression levels of TUBB4 mRNA were determined by quantitative real-time polymerase chain reaction in 3 different cell types. All exons of TUBB4 were screened for mutations in 394 unrelated dystonia patients.
Results:
The disease-causing gene was mapped to a 23cM region on chromosome 19p13.3-p13.2 with a maximum multipoint LOD score of 5.338 at markers D9S427 and D9S1034. Genome sequencing revealed a missense variant in the TUBB4 (tubulin beta-4; Arg2Gly) gene as the likely cause of disease. Sequencing of TUBB4 in 394 unrelated dystonia patients revealed another missense variant (Ala271Thr) in a familial case of segmental dystonia with spasmodic dysphonia. mRNA expression studies demonstrated significantly reduced levels of mutant TUBB4 mRNA in different cell types from a heterozygous Arg2Gly mutation carrier compared to controls.
Interpretation:
A mutation in TUBB4 causes DYT4 dystonia in this Australian family with so-called whispering dysphonia, and other mutations in TUBB4 may contribute to spasmodic dysphonia. Given that TUBB4 is a neuronally expressed tubulin, our results imply abnormal microtubule function as a novel mechanism in the pathophysiology of dystonia.
Hereditary whispering dysphonia, later designated DYT4, was first described by Parker in 1985 in an Australian family with 20 affected family members.1 DYT4 dystonia follows an autosomal dominant mode of inheritance with seemingly high penetrance based on lack of unaffected obligate carriers and a high number of affected individuals in large sibships. The phenotype is clinically characterized by craniocervical dystonia with prominent spasmodic dysphonia and shows variable expressivity in affected family members. The dystonia frequently generalizes and is at least partially responsive to alcohol and propranolol.2 When recently re-evaluating this family, we identified a total of 191 family members including 7 newly affected individuals. For the first time, additional characteristic clinical features beyond the motor phenotype were described, that is, a thin face and body habitus that completely cosegregated with the DYT4 motor phenotype.2
Genetic analyses ruled out mutations in and/or linkage to DYT1, THAP1 (DYT6), DYT7, DYT13, and PRKRA (DYT16).2–5 Compound heterozygous mutations in the ATP7B gene explain Wilson disease in 2 family members. However, these mutations did not segregate with the DYT4 phenotype.2 To elucidate the genetic basis of DYT4 dystonia, we performed a genome-wide linkage analysis followed by genome sequencing and identified a novel mutation in the TUBB4 gene as the likely genetic cause of whispering dysphonia combined with other dystonia and a characteristic facies and body habitus in the original Australian DYT4 family. Screening of about 400 unrelated dystonia patients revealed another missense mutation in a patient with segmental dystonia including spasmodic dysphonia.
Subjects and Methods
Study Participants and Clinical Studies
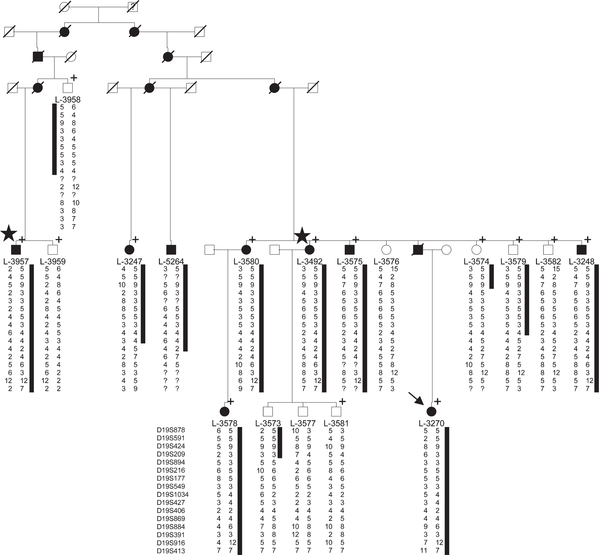
The study was approved by the local ethics committee, and all participants gave informed consent. DNA samples were available for 18 family members including 9 affected individuals (Fig 1). The index patient (L-3270) underwent a detailed neurological follow-up investigation, an electroencephalographic (EEG) examination, nerve conduction studies, and 3 Tesla brain magnetic resonance imaging (MRI) examinations. Skin and olfactory mucosal biopsies were taken according to standard protocols.6
FIGURE 1:
Simplified pedigree of the Australian DYT4 family including only those individuals with available DNA (L-number) and their ancestors. The shared haplotype on chromosome 19 is indicated by a black bar. Genotypes at the indicated markers are given as numbers; a question mark symbolizes that genotyping did not work. Individuals marked with a plus sign were included in the genome-wide linkage analysis; in those marked with a star, genome sequencing was performed. DYT4 dystonia-affected family members are labeled with black and unaffected with white symbols, and presumably affected ones with question marks. The proband is indicated by an arrow. Slashes indicate deceased family members. Circles = females; squares = males.
As disease controls, we included 394 unrelated dystonia patients. Of these, 124 had spasmodic dysphonia, and the remaining 270 were selected based on an age at onset of ≤30 years and/or a positive family history. These patients had a mean age at onset of 31.4 ± 14.1 (range = 7–87) years, 47.2% were male, and a positive family history was known for 27.4%. Types of dystonia included generalized dystonia (n = 49), segmental dystonia (n = 75; including 20 with spasmodic dysphonia), spasmodic dysphonia (n = 104), cervical dystonia (n = 61), musician’s dystonia (n = 61; including 11 with dystonia of the embouchure), blepharospasm (n=30), and writer’s cramp (n = 14). They were recruited at different movement disorder centers in Lübeck, Hamburg, Kassel, and Hannover (Germany), Amsterdam (the Netherlands), Belgrade (Serbia), Toronto (Canada), and New York (USA) and clinically diagnosed and characterized by experienced movement disorder specialists. In addition, 500 unrelated, healthy controls were collected as part of a population-based epidemiological study (EPIPARK) at the University of Lübeck (Germany), and movement disorders were excluded by a diagnostic interview.7
Linkage Analysis and Genome Sequencing
We performed a genome-wide linkage analysis in 14 family members (including 8 affected DYT4 patients) using the Illumina (San Diego, CA) HumanHap 610-Quad SNP chip. Samples were processed at ATLAS Biolabs (Berlin, Germany). Due to the large size of the family, statistical linkage analysis was performed for 2 branches of the pedigree separately using Allegro 1.2c within EasyLinkage Plus 5.08.8 Five regions with an additive LOD score >2 underwent fine-mapping with microsatellite markers. Statistical linkage analysis within the linked region after fine-mapping was performed using Simwalk2 within EasyLinkage Plus 5.08.
Next, genome sequencing was carried out in 2 definitely affected distant relatives (L-3492 and L-3957; see Fig 1) using the service at Knome (Boston, MA). For validation, we chose those variants that were (1) located within the linked region on chromosome 19; (2) shared among the 2 investigated patients; (3) absent from 5 unrelated individuals sequenced together with the 2 patients; (4) considered to change the protein sequence; and (5) absent from the National Heart, Lung, and Blood Institute Exome Sequencing Project (publically available at the exome variant server [EVS] at http://evs.gs.washington.edu/EVS/. For filtering, we used knomeDISCOVERY data filtering software (Knome).
Variants fulfilling these criteria were resequenced by Sanger sequencing and tested for segregation in all available family members. The frequency of the TUBB4 variant was determined by sequencing of 1,000 control chromosomes. Finally, the entire coding region including 4 exons and exon/intron boundaries of the TUBB4 gene was sequenced in 394 disease controls. Primer sequences are listed in Supplementary Table S1.
RNA Extraction and Quantitative Polymerase Chain Reaction
RNA extraction and quantitative polymerase chain reaction (PCR) were performed independently at 2 sites. Whereas blood samples were investigated at the University of Lübeck (Germany), RNA from blood, skin, and olfactory cells was measured at the University of Sydney (Australia). Total RNA was extracted from lymphoblasts from the only available heterozygous mutation carrier and 13 unrelated healthy controls using the PaxGene blood RNA isolation kit (PreAnalytiX, Hombrechtikon, Switzerland) according to the manufacturer’s instructions. In addition, we extracted RNA from fibroblast and primary olfactory cell cultures from the same heterozygous mutation carrier and a related but unaffected control using the RNeasy Mini kit (QIAGEN, Valencia, CA). RNA was reverse-transcribed into cDNA with the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA) or Superscript III first-strand synthesis kit for real-time PCR (Invitrogen, Carlsbad, CA). TUBB4 expression was investigated by quantitative PCR with exon-spanning primers (see Supplementary Table S1). Expression was quantified by real-time PCR on the LightCycler 480 (Roche Diagnostics, Mannheim, Germany) using the LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche Diagnostics) or on the Rotor-gene 6000 (Corbett Life Science, Mortlake, Australia) using the Quanti-Tect SYBR green PCR kit (QIAGEN). GAPDH and ACTB served as reference genes (see Supplementary Table S1). All measurements were performed at least in duplicate, and at least 2 independent PCR runs were analyzed. To determine potential differences in RNA levels, we used the Mann–Whitney U test.
Results
Clinical Follow-up, Electrophysiological, and MRI Studies of the Proband
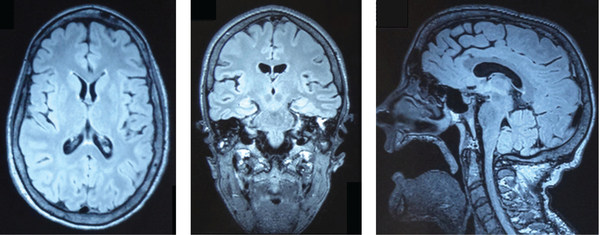
The detailed medical history of the 31-year-old index patient is summarized in the Supplementary Information. She presented with the characteristic facies and body habitus of DYT4 dystonia patients (Fig 2, left panels) and the “hobby horse” type gait (see Fig 2, right panel). Her EEG was contaminated by prominent muscle artifacts secondary to her cranial, facial, and cervical dystonia but was otherwise unremarkable. Nerve conduction studies were mildly abnormal in keeping with a mild demyelinating and axonal, predominantly sensory peripheral neuropathy. Like many of the affected family members, she had often used and occasionally abused alcohol, as it initially provided significant relief from her dystonic symptoms. In the past 2 years, she had significantly increased her alcohol consumption, which had further compromised her postural stability and may have contributed to her mild sensory peripheral neuropathy. A 3T brain MRI scan demonstrated a structurally normal brain. Specifically, no mass lesion, cerebral hemorrhage, or cerebral infarct were present. There was no evidence of a neural migration disorder such as gray matter heterotopia (Fig 3).
FIGURE 2:
Recent photographs of the index patient showing the characteristic facial features of atrophy most prominent in the jaw and lower facial muscle producing a triangular appearance of the lower face (left panels). The index patient and several other members of the family also exhibit bilateral eyelid ptosis. All observed affected family members have a generally thin body habitus from childhood until well before the obvious onset of DYT4 features, which invariably commences with spasmodic dysphonia. Poor dentition and eventually edentulousness is common in the more severely affected family members, especially those with extrusional tongue movements (left, middle panel). Synkinetic movements of the levator palpebrae superioris, ocular, and jaw muscles are triggered by jaw opening (left, lower panel). The characteristic dystonic “hobby horse” gait of DYT4 dystonia is seen in the right panel. [Color figure can be viewed in the online issue, which is available at www.annalsofneurology.org.]
FIGURE 3:
Cranial 3T magnetic resonance imaging with representative fluid attenuated inversion recovery sequences in the axial (left), coronal (middle), and midline sagittal plane (right). Brain parenchyma and in particular the deep and cortical gray and white matter exhibited normal patterning. The subcutaneous tissues of the inferior face, predominantly comprising the muscles of facial expression, are atrophic, and the patient is edentulous. The muscles of mastication were of normal volume, and an incidental, small, left mastoid effusion was present.
Identification of a Mutation in TUBB4
Genome-wide linkage analysis resulted in 5 regions with LOD scores >2. By fine-mapping, 4 candidate regions were excluded (Supplementary Table S2), and linkage of DYT4 was established to a 23cM region on chromosome 19p13.3-p13.2. Haplotypes were shared in a 6.2 Mb region from the telomere to marker D19S427 (see Fig 1) by all affected and up to 4 unaffected family members. The maximum multipoint LOD score reached 5.338 for markers D9S1034 and D9S427.
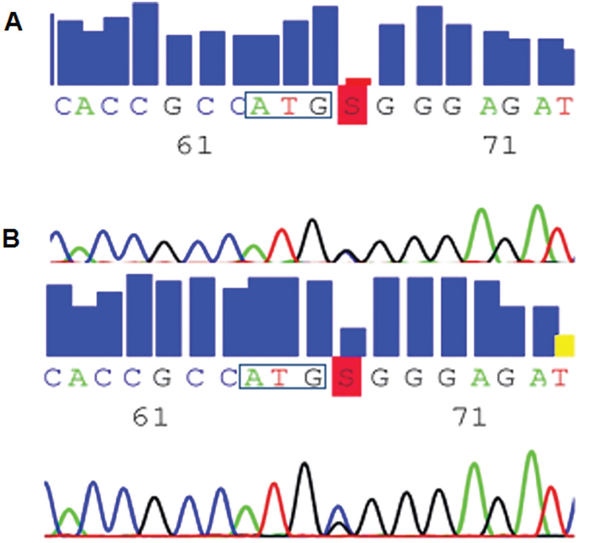
Genome sequencing had a mean coverage of 48×, and about 90% of the genome was called (Table 1). About 4.5 million mismatches to the target reference sequence were found in both patients, about 10% of which were novel (see Table 1). Of these variants, only 2 substitutions within the linked region were predicted to alter a protein sequence and were shared by both investigated patients. Furthermore, they were absent in 5 unrelated individuals from the same sequencing batch. This filtering step was used to filter out sequencing artifacts that tend to be present in different samples. The 2 candidate missense variants in the TUBB4 (tubulin beta-4; c.4C>G; Arg2Gly; Fig 4A) and DOT1L (DOT1-like, histone H3 methyltransferase; c.2723 G>A; Arg908His) genes were confirmed by Sanger sequencing and present in all 9 affected family members. Both variants were absent in 1,000 control chromosomes and not listed in any database. Notably, the mutation in TUBB4 was also absent in all healthy family members, whereas the DOT1L variant was found in 4 of 9 unaffected family members. All of these individuals were older than 42 years, which is the latest observed age of onset for DYT4.2 In addition, the EVS lists only 7 different missense variants (at amino acid positions 32, 90, 125, 251, 315, 434, and 444) in single individuals and 1 carrier of a nonsense mutation (at position 344) in the 444–amino acid protein TUBB4 in about 6,500 individuals (4,300 of European and 2,200 of African American back-ground). Assuming that all these variants are true, only 0.12% of individuals carry a TUBB4 variant, underlining that the TUBB4 protein is highly conserved, tolerating only little variation. In contrast, almost 100 rare and frequent missense variants have been described for the 1,537–amino acid protein DOT1L in a comparable number of sequenced individuals.
TABLE 1.
Statistics of Genome Sequencing in 2 DYT4 Patients
| L-3492 | L-3957 | |
|---|---|---|
| Mean coverage depth | 48.2 | 48 |
| Proportion of genome covered | 89.6% | 90.0% |
| Total sites called reference-mismatching | 4,589,260 | 4,532,122 |
| Novel single nucleotide substitutions | 433,888 | 427,112 |
| Synonymous | 9,410 | 9,061 |
| Missense | 8,192 | 7,772 |
| Nonsense | 53 | 63 |
| Read-through | 16 | 16 |
| Splice region (≤5b from splice site) | 443 | 536 |
| Insertions/deletions (Indel) | 999,101 | 1,709,685 |
FIGURE 4:
Novel mutation in the TUBB4 gene. The electropherograms show the sequence of exon 1 of TUBB4 around the start codon (boxed) for Patient L-3270. The c.4C>G mutation is highlighted by the letter S in red. (A) At the DNA level, the mutation is heterozygous with equal peak heights for both alleles. (B) At the RNA level, the wild-type allele is apparently more abundant than the mutant allele, prompting real-time quantitative polymerase chain reaction experiments.
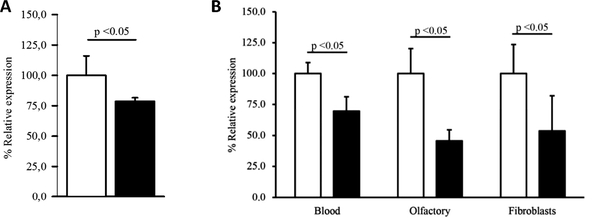
The Arg2Gly mutation in TUBB4 is predicted to be deleterious by in-silico analyses with PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/; possibly damaging [score 0.876]) and SIFT (http://sift.jcvi.org/; probably damaging [score 0.996]). Because substitutions of the Arg2 to any other amino acid were shown to result in loss of the autoregulated instability of TUBB4 mRNA,9 we analyzed mutant and wild-type TUBB4 mRNA levels. Sequence analysis of cDNA reversely transcribed from blood mRNA from a heterozygous mutation carrier suggested that the wild-type allele of TUBB4 was more abundant than the mutant allele (see Fig 4B), prompting us to perform quantitative real-time PCR in lymphoblasts and in cultures of the index patient’s fibroblasts and primary olfactory cells. By this, we demonstrated that the amount of TUBB4 in the heterozygous mutation carrier is significantly reduced compared to controls in all 3 different cell types (p < 0.05; Fig 5).
FIGURE 5:
Expression levels of wild-type and mutant TUBB4. (A) Quantitative real-time polymerase chain reaction (PCR) done at the University of Lübeck revealed a lower level of TUBB4 in the available heterozygous mutation carrier (filled bar) compared to the mean of 13 unrelated individuals without TUBB4 mutations (open bar). GAPDH was used as a reference gene. The expression level for the controls was set to 1. Standard deviation is indicated. All measurements were performed in duplicate, and all samples were analyzed at least twice. (B) Quantitative PCR was also performed at the University of Sydney in an independently extracted blood sample (Blood) as well as in fibroblasts and human primary olfactory cell cultures (Olfactory) from the same mutation carrier (filled bars) and a related but unaffected control (open bar). The expression level for the control was set to 1, and ACTB was used as a reference gene. Standard deviation is indicated, and all measurements were performed at least in duplicate.
Screening of 394 unrelated dystonia patients for mutations in TUBB4 revealed 1 additional missense variant (Ala271Thr) in a 71-year-old female patient with spasmodic dysphonia and oromandibular dystonia and dyskinesia. Age at onset was at 60 years; symptoms were progressive and interfered with speaking and eating. Her mother had had dyskinesia around the mouth from age 70 years, had died at age 76 years, and was therefore not available for mutation analysis. The Ala271Thr variant was predicted to affect protein function by SIFT (score 0.02) and to be disease-causing by Mutationtaster (http://www.mutationtaster.org/; p = 0.9996).
Discussion
DYT4 dystonia has long remained among the few members of the DYT list lacking both locus assignment and gene identification.10 We here provide comprehensive linkage and genome sequencing combined with clinical and functional data, suggesting TUBB4 as the gene causing DYT4 dystonia in the original Australian whispering dysphonia family.
After a candidate gene approach,2 linkage analysis, and genome sequencing, we were left with 2 candidate genes for DYT4 dystonia, that is, TUBB4 and DOT1L. However, the variant in DOT1L was also found in about half of the unaffected family members, who all exceeded the latest reported age at onset in the family. In addition, both rare and frequent missense variants are spread all over the DOT1L protein (EVS). Because protein-changing variants in TUBB4 are extremely rare (8 in 6,500 individuals), and the Arg2Gly variant was exclusively found in affected family members, we consider this to be the disease-causing mutation in the Australian DYT4 family and the substitution in DOT1L to be a likely private but disease-unrelated variant. Interestingly, DYT4 dystonia seems to differ with respect to its high level of penetrance from other dominantly inherited dystonias, such as DYT1 or DYT6 dystonia, in which penetrance is as low as 30%.11
There are 3 additional lines of evidence supporting TUBB4 as the true disease-causing gene of DYT4 dystonia. First, although the DOT1L gene is ubiquitously expressed,12 expression of TUBB4 is mostly restricted to the brain, with only low expression levels detected in selected other tissues, including testis, cardiomyocytes, and blood.12,13 By far the highest TUBB4 expression levels are present in fetal brain, suggesting an important role of this gene in brain development. Although TUBB4 expression is also abundant in all adult brain regions tested, the highest levels were found in the amygdala, followed by the hypothalamus, thalamus, and prefrontal cortex.12 This expression pattern is of potential interest, as it may relate to some of the additional, nonmotor signs observed in DYT4 dystonia, including psychiatric features1 and weight loss2.
Second, the novel TUBB4 mutation is situated within a highly conserved sequence in all beta-tubulins, which had been functionally investigated decades before the newly observed link to DYT4 dystonia. The stability of beta-tubulin mRNAs is autoregulated by their own translation product.9 The underlying mechanism includes cotranslational recognition of the amino-terminal beta-tubulin tetrapeptide Met-Arg-Glu-Ile (MREI) after its emergence from the ribosome and activation of an RNAse that degrades the polysome-bound mRNA. Notably, a variant within this MREI motif was found neither in TUBB4 nor in any other human TUBB gene in data from the EVS. In vitro mutations of this motif, especially at Arg2, the very codon mutated in the DYT4 family, were shown to abolish the autoregulatory effect.9 When investigating RNA levels of the index patient, we detected a consistent decrease of mutant TUBB4 RNA levels compared to wild-type in our index patient in all 3 different cell types studied, lending functional support to our genetic data. Thus, the disease mechanism of the TUBB4 mutation probably includes reduced levels of TUBB4 rather than affecting its functionality. Of note, there is an apparent discrepancy between our and the previously published data; whereas our in vivo data revealed reduced levels of mutant mRNA compared to wild-type using 2 different methods in 3 different cell types, the previous study reported autoregulated instability,9 which would result in stabilization, that is, increased levels of the mutant transcript. Notably, our data are based on a single patient only and not yet confirmed in independent samples. Thus, it is also possible that the reduction of TUBB4 RNA levels in our patient is not related to the TUBB4 mutation. Alternatively, the findings by Yen et al,9 which are based on a highly artificial assay, may not adequately reflect the in-vivo situation. At present, we can only speculate on the reason for this discrepancy, and further research will be necessary, including testing of additional patients and detailed in-vitro studies.
Third, by screening about 400 unrelated dystonia patients, we identified a second novel missense variant (Ala271Thr) in TUBB4. Interestingly, this variant was detected in 1 of 124 patients with spasmodic dysphonia (0.8%). Affected family members were not available to test for segregation, but pathogenicity is supported by prediction programs and a positive family history in the mother. This mutation is located outside the MREI motif and may have a different disease mechanism compared to Arg2Gly. RNA of the mutation carrier for expression analysis was not available.
The function of proteins encoded by previously identified DYT dystonia genes has implicated a number of different disease mechanisms underlying dystonias, such as dysfunction of the nuclear envelope caused by a mutation in TorsinA in DYT1 dystonia,14,15 transcriptional dysregulation due to mutations in THAP1 in DYT6 dystonia,16,17 and disturbed neurotransmitter synthesis (GCH1 mutations in dopa-responsive dystonia, DYT5).18 Identification of mutations in TUBB4, coding for a tubulin, the major constituent of microtubules, emphasizes another perspective to the pathophysiology of dystonia. Notably, a link between dystonia and the cytoskeleton has previously been reported by identification of kinesin light chain 1 and nesprin 3 as interactors of TorsinA.15,19
A spectrum of neurological conditions characterized by abnormal migration and differentiation of neurons and of disturbed axon guidance has recently been linked to mutations in genes encoding the α- and β-tubulin subtypes TUBA1A, TUBA8, TUBB2B, and TUBB3, all of which are thought to coassemble into neuronal microtubules. Associated phenotypes include nervous system malformations with different types of cortical malformations, defects in commissural fiber tracts, and degeneration of motor and sensory axons.20 Mutations in TUBB3, another β-tubulin with a similar expression pattern13 to TUBB4, are localized outside the highly conserved MREI sequence involved in autoregulation. These mutations are thought to disrupt microtubule dynamics and to perturb the interaction of microtubules with proteins that use the microtubule system. This may produce defective axonal transport of growth materials and signals necessary for axon outgrowth.21 Similar to patients with TUBB3 mutations and congenital ocular motility defects,22 the proband of the DYT4 family had evidence of ptosis, impaired extraocular eye movements, and possibly synkinetic movements of the levator palpebrae superioris, ocular, and jaw muscles. Although there were no associated neuroimaging abnormalities, it is possible that patients with TUBB4 mutations may also have clinical manifestations indicative of aberrant innervation due to axon guidance defects similar to those observed in patients with TUBB3 mutations.
Intriguingly, a role of TUBB4 for synaptic function has recently been suggested, as microarray gene profiling and electron microscopic stereology revealed lower expression of TUBB4 and other genes in the dorsolateral prefrontal cortex of subjects with major depressive disorder and a corresponding lower number of synapses.23
Although there is strong evidence that the mutation in TUBB4 is the cause of DYT4 dystonia, a limitation of our study is that the method of genome sequencing never provides complete coverage of every single base pair, especially in GC-rich and repetitive sequences. Also, we focused our analysis on novel, protein-changing variants under the assumption of an autosomal dominant mode of inheritance with high penetrance. It is possible, albeit unlikely, that other (noncoding or rare) variants cause the DYT4 phenotype. The finding of a second mutation in an unrelated patient with spasmodic dysphonia supports a pathogenic role for TUBB4, but further evidence including segregation of TUBB4 mutations in independent families and detailed functional studies will help to further establish TUBB4 as the gene causing DYT4 dystonia. Furthermore, the (full-blown) phenotype of DYT4 as in the Australian family may be overall very rare.2
In summary, we provide strong evidence supporting the causative role of a mutation in TUBB4, altering a highly conserved and functionally important amino acid, in DYT4 dystonia. Other mutations in TUBB4 may contribute to spasmodic dysphonia. Our results imply a neuronally expressed tubulin, and thus, abnormal microtubule function as a novel mechanism in the pathophysiology of dystonia. Future studies may be directed at (1) screening larger patient cohorts, especially individuals with spasmodic dysphonia or a combination of generalized dystonia with thin face and body habitus, for mutations in TUBB4; and (2) performing additional functional investigations of mutant TUBB4 to further support pathogenicity of (the) TUBB4 mutation(s).
Supplementary Material
Acknowledgment
This work was supported by grants from the Bachmann Strauss Dystonia and Parkinson Foundation (to C.K. and K.L.), intramural funds from the University of Lübeck (SPP Genetics), the Hermann and Lilly Schilling Foundation (to C.K.), the Australian Brain Foundation (to C.M.S., R.A.W., and C.K.), and the National Health and Medical Research Council of Australia (to K.R.K. and C.M.S.).
We thank the members of the DYT4 family for their participation and support of this study.
Footnotes
Potential Conflicts of Interest
K.L., grants/grant pending, German Research Foundation. R.A.W.: grants/grants pending, Brain Foundation; travel expenses, Medtronics, Novartis. M.K.: grants/grants pending, German Research Foundation. F.J.K.: grants/grants pending, German Research Foundation. K.R.K.: travel expenses, Movement Disorders Society; Dora Lush NHMRC Postgraduate Scholarship, National Health and Medical Research Council. D.A.-F.: grants/grants pending, University Medical Center Giessen and Marburg (UKGM). E.A.: grants/grants pending, German Research Foundation; speaking fees, Merz, Allergan, Ipsen Pharma, Eisai; royalties, Music, Motor Control and the Brain (Oxford University Press). A.M.: grants/grants pending, Deutsche Forschungsgemeinschaft, European Science Foundation, Else Kröner-Fresenius-Stiftung; speaking fees, Pharm Allergan, Ipsen, Merz Pharmaceuticals; travel expenses, Pharm Allergan, Ipsen, Merz Pharmaceuticals. V.K.: board membership, Boehringer Ingelheim, Pfizer; grants/grants pending, project #175090 MNT Serbia; speaking fees, Novartis, Alkaloid Skoplje, Boehringer, Libra, Abbott Laboratories, GlaxoSmithKline, Pfizer; travel expenses, Boehringer Ingelheim. L.J.O.: patents, royalties, Athena Diagnostics. A.P.M.L.: consultancy, Biovail, Ceregene, Novartis, Merck Serono, Solvay, Teva, Abbott Laboratories, Allon Therapeutics, AstraZeneca, Eisai, GSK, MSD, BMS; grants/grants pending, Ontario Problem Gambling Research Centre, Canadian Institutes of Health Research, Michael J. Fox Foundation, National Parkinson Foundation; royalties, Saunders, Wiley-Blackwell, Johns Hopkins Press, Cambridge University Press. M.A.J.T.: grants/grants pending, Prinses Beatrix Fonds Stichting Wetenschapsfonds Dystonie Vereniging STW Technology Society (program: Perspective - NeuroSIPE Nuts-Ohra); dystonia nurse, Ipsen. C.K.: consultancy, Centogene; grants/grants pending, German Research Foundation, European Foundation, BMBF; speaking fees, AAN Annual Meeting.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Parker N Hereditary whispering dysphonia. J Neurol Neurosurg Psychiatry 1985;48:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilcox RA, Winkler S, Lohmann K, Klein C. Whispering dysphonia in an Australian family (DYT4): a clinical and genetic reappraisal. Mov Disord 2011;26:2404–2408. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad F, Davis MB, Waddy HM, et al. Evidence for locus heterogeneity in autosomal dominant torsion dystonia. Genomics 1993;15:9–12. [DOI] [PubMed] [Google Scholar]
- 4.Djarmati A, Schneider SA, Lohmann K, et al. Mutations in THAP1 (DYT6) and generalised dystonia with prominent spasmodic dysphonia: a genetic screening study. Lancet Neurol 2009;8:447–452. [DOI] [PubMed] [Google Scholar]
- 5.Jarman PR, del Grosso N, Valente EM, et al. Primary torsion dystonia: the search for genes is not over. J Neurol Neurosurg Psychiatry 1999;67:395–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JS, Mehta P, Cooper AA, et al. Pathogenic effects of novel mutations in the P-type ATPase ATP13A2 (PARK9) causing Kufor-Rakeb syndrome, a form of early-onset parkinsonism. Hum Mutat 2011;32:956–964. [DOI] [PubMed] [Google Scholar]
- 7.Kasten M, Hagenah J, Graf J, et al. Cohort profile: a population-based cohort to study non-motor symptoms in parkinsonism (EPI-PARK). Int J Epidemiol . 2012. December 19. [Epub ahead of print]; doi: 10.1093/ije/dys202. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann K, Lindner TH. easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics 2005;21:3565–3567. [DOI] [PubMed] [Google Scholar]
- 9.Yen TJ, Machlin PS, Cleveland DW. Autoregulated instability of beta-tubulin mRNAs by recognition of the nascent amino terminus of beta-tubulin. Nature 1988;334:580–585. [DOI] [PubMed] [Google Scholar]
- 10.Marras C, Lohmann K, Lang A, Klein C. Fixing the broken system of genetic locus symbols: Parkinson disease and dystonia as examples. Neurology 2012;78:1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruggemann N, Klein C. Genetics of primary torsion dystonia. Curr Neurol Neurosci Rep 2010;10:199–206. [DOI] [PubMed] [Google Scholar]
- 12.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 2004;101:6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leandro-Garcia LJ, Leskela S, Landa I, et al. Tumoral and tissue-specific expression of the major human beta-tubulin isotypes. Cytoskeleton (Hoboken) 2010;67:214–223. [DOI] [PubMed] [Google Scholar]
- 14.Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci U S A 2004;101:847–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nery FC, Zeng J, Niland BP, et al. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci 2008;121:3476–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser FJ, Osmanoric A, Rakovic A, et al. The dystonia gene DYT1 is repressed by the transcription factor THAP1 (DYT6). Ann Neurol 2010;68:554–559. [DOI] [PubMed] [Google Scholar]
- 17.Gavarini S, Cayrol C, Fuchs T, et al. Direct interaction between causative genes of DYT1 and DYT6 primary dystonia. Ann Neurol 2010;68:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichinose H, Ohye T, Takahashi E, et al. Hereditary progressive dystonia with marked diurnal fluctuation caused by mutations in the GTP cyclohydrolase I gene. Nat Genet 1994;8:236–242. [DOI] [PubMed] [Google Scholar]
- 19.Kamm C, Boston H, Hewett J, et al. The early onset dystonia protein torsinA interacts with kinesin light chain 1. J Biol Chem 2004;279:19882–19892. [DOI] [PubMed] [Google Scholar]
- 20.Tischfield MA, Engle EC. Distinct alpha- and beta-tubulin isotypes are required for the positioning, differentiation and survival of neurons: new support for the ‘multi-tubulin’ hypothesis. Biosci Rep 2010;30:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh KK, Tsai LH. MicroTUB(B3)ules and brain development. Cell 2010;140:30–32. [DOI] [PubMed] [Google Scholar]
- 22.Tischfield MA, Baris HN, Wu C, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell 2010;140:74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang HJ, Voleti B, Hajszan T, et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med 2012;18:1413–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.