Abstract
Background
Acinetobacter baumannii is a nosocomial pathogen of critical importance due to the increasing numbers of antibiotic-resistant isolates. Colonies can have a smooth or matt appearance, but also exhibit slimy, mucoid growth, with the latter being increasingly isolated in patients in recent years.
Methods
We isolated 60 A. baumannii strains from altogether 56 patients and found that all patients were infected by mucoid strains, with four patients having also matt phenotypes in addition to the mucoid ones. The morphology of the colonies and capsules was observed. The antibiotics susceptibilities were tested, and the biofilm formation ability was determined by crystal violet staining. The whole-genome sequencing (WGS) was performed on all the strains, and then the core genome multilocus sequence typing (cgMLST) and drug resistance gene analysis were performed. Finally, a part of isolates were selected to test virulence in a Galleria mellonella model.
Results
We observed much larger capsules in the mucoid strains compared to the matt isolates. But the mucoid phenotype did not correlate with the amount of biofilm produced by the strain. Almost all mucus-type A. baumannii were multi-drug resistant isolates, containing various antibiotic resistance genes. The main ST types of mucoid-type A.baumannii were ST191 and ST195, of which ST191 isolates were more virulence, while ST195 isolates were weaker.
Conclusion
The mucoid A. baumannii had resistance to most antibiotics and some strains had high virulence, which should be paid attention in clinical.
Keywords: Acinetobacter baumannii, capsule, biofilm formation, mucoid phenotype
Introduction
Acinetobacter baumannii has become an important nosocomial pathogen causing a range of infections, including skin and soft tissue infections, infections of the urinary tract, endocarditis and meningitis.1,2 Due to its ability to acquire various antibiotic resistance, more than 80% of Acinetobacter species are defined as multidrug-resistant (MDR), and without treatment options result in high mortality rates, while presenting a huge healthcare burden in general.3,4
Acinetobacter baumannii isolates have been found to exhibit both, mucoid and non-mucoid (matt) phenotypes. In case of Pseudomonas aeruginosa, during the course of an acute early infection establishing itself as chronic infection, a previously matt phenotype can “switch” to a mucoid one which shows reduced growth rate concurrent with a changed metabolic pathway and over-expression of alginate.5 The mucoid Ps. aeruginosa isolates secrete a variety of substances including various polysaccharides to form a biofilm, which protects it from antibiotics and contributes to persistence in the host as well as to virulence in general.6,7 In Klebsiella pneumonia, hypermucoviscous K.pneumonia isolates are considered to be hypervirulent.8 In A. baumannii, the K locus cluster is responsible for capsular biosynthesis,9 with several genes involved in the regulation process, including itrA and wzc.10 When these genes are non-functional, the non-encapsulated mutants are quickly inactivated and killed by serum.11 We previously reported that matt A. baumannii are able to evolve to a mucoid phenotype in vivo.12 In this work, we aimed to define the molecular basis of mucoid A. baumannii formation, but also analysed antibiotic resistance and virulence of mucoid isolates.
Materials and Methods
Ethics Statement
The study was carried out in accordance with the principles of the Declaration of Helsinki and was approved by the Research Ethics Committee of Hangzhou General Hospital of Chinese People’s Armed Police which waived the need for consent, as the patient information was anonymized and deidentified prior to analysis.
Patients and Bacteria Strains
60 A. baumannii isolates from 56 hospitalized patients in Chinese People’s Armed Police Forces, Hangzhou during 2014–2018, were collected. After obtaining single colonies, all isolates were identified as A. baumannii by 16S rRNA analysis (primers: forward, 5ʹ-ACATCGTTTACGGCATGGACT-3ʹ, and reverse, 5ʹ- TGCCGCGTGTGTGAAGAA-3ʹ). ATCC19606 and MDR-ZJ0613 were taken as reference isolate in this study.
Capsule Thickness Measuring
Bacterial capsules were detected using the India ink method.10 Briefly, bacterial colonies and India ink were mixed in 0.85% NaCl solution. And the bacteria were observed and photographed under light microscopy (1000×). Ten capsules were measured for each isolate by image J software (https://imagej.nih.gov/ij/).
WGS and Core Genome Multilocus Sequence Typing (cgMLST)
The genomic DNA of 60 isolates were extracted using a QIAamp DNA Mini kit (Qiagen, Valencia, CA, USA) following the manufacturer’s recommendations, and were sequenced at Zhejiang Tianke (Hangzhou, China) on an Illumina HiSeq2000 platform (Illumina, San Diego, CA, United States) following a pair-end 2×150 bp protocol. De novo assembled using CLC Genomic Workbench 8.5.1 (Qiagen, Aarhus, Denmark). Phylogenetic analyses were performed based on validated A. baumannii cgMLST version 1.0 (2390 target genes), using Ridom SeqSphere+ software (Ridom GmbH, Munster, Germany).14
Antimicrobial Susceptibility Testing
The Minimal Inhibitory Concentrations (MICs) of colistin, tigecycline and eravacycline were determined by the broth microdilution method with cation-adjusted Mueller-Hinton broth (CAMHB), and MICs of the other antibiotics, including ceftazidime, cefepime, cefoperazone/sulbactam, ciprofloxacin, amikacin, gentamicin, imipenem, meropenem, were detected by the agar dilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI-M100-S26). The results of tigecycline and eravacycline were interpreted according to EUCAST breakpoints, and the breakpoint of eravacycline was referred to tigecycline.
Growth Rate Determination
Four independent cultures per strain were grown overnight and diluted to 1:100 in MH broth before aliquots were placed into a flat-bottom 100-well plate with three replicates each. The plate was incubated at 37°C with agitation while the OD600 of each culture was determined every 5 min for 20hrs using a Bioscreen C MBR machine (Oy Growth Curves Ab Ltd., Finland). The growth rate was determined by applying a linear regression to the exponential growth rate data using an R script.15
Biofilm Assay
Biofilm assays of each isolate were performed in triplicates as described previously,16 with slight modifications. Overnight bacterial cultures were diluted by fresh Luria-Bertani (LB) medium until the bacterial suspension reached an OD600 of 0.5. Two hundred milliliters of the cell number-adjusted bacterial cultures were transferred to 96-well polystyrene plates, and incubated for 72 h at 37°C. Cells were washed three times with phosphate-buffered saline (PBS), and stained with 0.1% crystal violet for 20 min at room temperature. Wells were washed again and dried naturally. The bound dye was solubilized with 200 μL of 95% ethanol and quantified by measuring the absorbance at 540 nm. Three wells containing LB without bacteria served as a negative control. Strong biofilm-forming ability was defined that OD540 of the isolate was more than ATCC19606. Weak biofilm-forming ability was defined with OD540 between ATCC19606 and negative control. If OD540 of the isolate was less than the negative control, it was defined as no biofilm-forming ability.
In vivo Virulence Study Using G. mellonella
Virulence was evaluated in the insect model G. mellonella as described previously.17 Briefly, groups of 10 larvae (~200 mg; Yuejiayin, Tianjin, China) were stored in the dark at 4°C prior to use. To determine the virulence of isolates, every larva was infected with 1 × 106 CFU bacteria (n = 10), and 10 larvae injected with 10 μL PBS as a negative control. Survival was monitored up to 72hrs post-infection at 37°C. Experiments were performed in triplicate and re-tested when the difference between the two experiments was >15%. Data from all experiments were combined to calculate the mean values of percent survival values.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software Inc., La Jolla CA, USA). Independent samples Student's t-test was used to determine differences between mucoid and matt A. baumannii in capsule thickness, growth rate and biofilm-forming ability. Survival rates were evaluated using Kaplan Meier survival curves and analyzed with the log-rank (Mantel-Cox) method. Chi-squared or Fisher’s exact tests were used to determine differences in the number or percentage of isolates between isolates. Differences were considered significant at P < 0.05.
Results
Clinical Parameters of Patients Infected with A. baumannii
The clinical data of the 56 patients are shown in Table 1. The median age of the patients was 61.68 ±14.29 years, with 60.7% male patients. The average hospitalization time was 90.4 ±82.5 days with 41.1% having been admitted to an Intensive Care Unit (ICU). The most prevalent comorbidities were cerebrovascular diseases followed by hypertension and diabetes. A 89.3% of the patients underwent invasive procedures, with 30.4% undergoing an operation.
Table 1.
Baseline Characteristics of Patients and Antimicrobial Treatments
| Characteristics | Total (n=56) |
|---|---|
| Demographics | |
| Age (years, Mean ± SD) | 61.68±14.29 |
| Male (n, %) | 34 (60.7%) |
| Hospitalization time (days, Mean ± SD) | 90.4±82.5 |
| Comorbidities (n, %) | |
| Hypertension | 29(51.8%) |
| Diabetes mellitus | 11(19.7%) |
| Cerebrovascular diseases | 46 (82.1%) |
| Solid tumors | 4 (7.1%) |
| Hematonosis | 2 (3.6%) |
| Chronic obstructive pulmonary disease | 5 (8.9%) |
| Chronic renal diseases | 3 (5.4%) |
| Invasive procedures a (n, %) | 50 (89.3%) |
| Operationb (n, %) | 17 (30.4%) |
| ICU admission (n, %) | 23 (41.1%) |
| Antibiotic treatmentc (n, %) | |
| β-lactam | 34 (60.7%) |
| β-lactamase inhibitor combination | 41 (73.2%) |
| Carbapenem | 36 (64.3%) |
| Glycopeptides | 24 (42.9%) |
| Aminoglycosides | 22 (39.3%) |
| Tetracyclines | 14 (25.0%) |
| Quinolones | 15 (26.8%) |
| Fosfomycin | 10 (17.9%) |
| Macrolides | 8 (14.3%) |
| Colistin | 2 (3.6%) |
| Othersd | 12 (21.4%) |
| Clinical outcome (n, %) | |
| Cure | 24 (42.9%) |
| Invalid | 19 (33.9%) |
| Deterioration | 10 (17.9%) |
| Death | 3 (5.4%) |
Notes: aInvasive procedure: including endotracheal intubation or incision, central venous catheterization, arterial catheterization, and hemodialysis tube, angiography or embolization, PICCO, ≥2 punctures (thoracic puncture, abdominal puncture, lumbar puncture, bone marrow puncture). bOperation: including operations during hospitalization and 1 month before admission. cAntibiotic treatment: uses of antibiotics within half a year prior to isolation. dOthers: linezolid,8 sulfonamides.4
The previous uses of antibiotics half a year prior to isolation of the microbial strains were recorded. The antibiotics were classified into 11 categories, with more than 50% of the patients having used β-lactam, β-lactamase inhibitor combination or carbapenem antibiotics. The clinical outcome of the patients during the duration of the study found only 42.9% of the patients fully cured, while the medical condition of approx. 18% deteriorated, and 3 died during the duration of the study, one of the bacterial infections.
The Majority of the A. baumannii Isolates Display a Mucoid Phenotype
From the 56 patients, we isolated 60 A. baumannii strains and found that all patients were infected by mucoid strains, with four patients having also matt phenotypes in addition to the mucoid ones. The colonies of mucoid A. baumannii appeared moist, irregular round, with an elevated surface. Adjacent colonies often formed fused associations on agar plates (Figure 1). In contrast to this, matt A. baumannii colonies had a smaller, round appearance with distinct edges.
Figure 1.
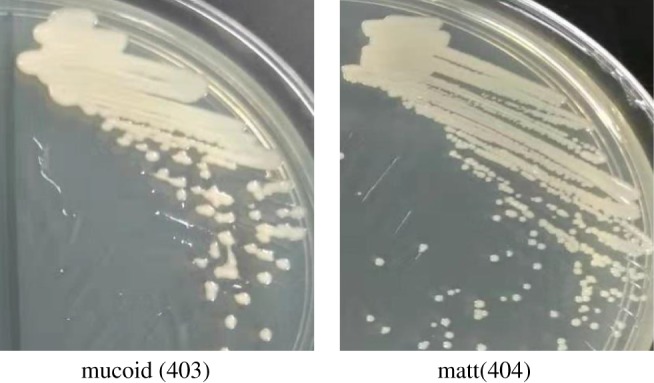
Colony morphology of the A. baumannii isolates on MH plates. Isolate 403 was mucoid with moist, irregular round colonies, and isolate 404 was matt with round and neat edge colonies.
Mucoid A. baumannii Isolates Display Thicker Capsules
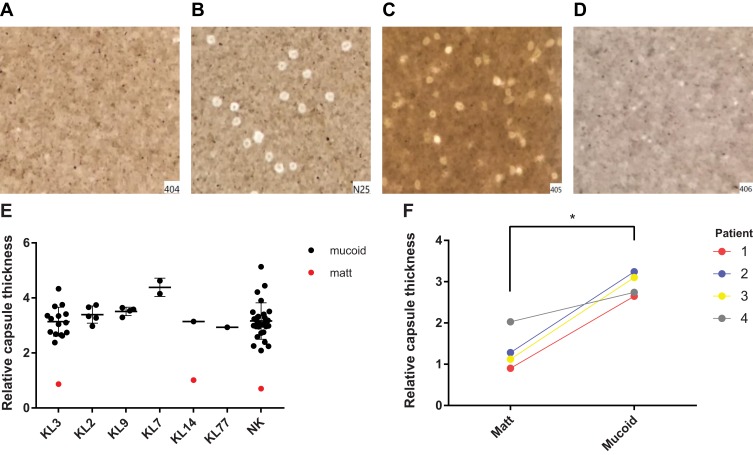
After determining the matt and mucoid phenotypes of the isolates, we then analysed the capsular types of the 60 strains. Here, we found that the capsular types of 29 strains were so-far unknown, and the remaining 31 strains belonged to six capsular types; 17 strains belong to the type KL3, five strains to KL2, four strains to KL9, two strains to KL14, two strains to KL7, and one strain to KL77. We then visualized the capsules of mucoid and matt A. baumannii isolates by India ink staining, and observed by light microscopy that matt A. baumannii had much thinner capsules compared to mucoid isolates (Figure 2A–D). The capsule thickness was determined by measuring the diameter of the cell halos (including the cells), with the capsule thickness of the reference strain MDR-ZJ06 (matt phenotype) as a comparison. The average capsule thickness of mucoid A. baumannii isolates was determined to be 3.24 um ± 0.60, more than 2.5 times that of the matt phenotype (p < 0.001). Interestingly, we found that the capsule thickness of KL7 isolates was significantly higher than that of other capsular types (p < 0.05). In contrast to this observation, no differences among the rest of the capsular types were observed (Figure 2E). In the four patients where we isolated both, matt and mucoid phenotypes, the capsule thickness of mucoid-type A. baumannii was also significantly higher than that of the matt isolate (p < 0.05) (Figure 2F).
Figure 2.
Capsule thickness. (A-D) (A) Matt isolate 404; (B) mucoid isolate N25; (C, D) mucoid isolate 405 and matt isolate 406, isolated from the same patient. (E) The average capsule thickness of different capsular types strains. Mucoid A baumannii isolate was more than 2.5 times that of the matt phenotype (p < 0.001); (F) Capsule thickness of 8 isolates, isolated from 4 patients, one matt isolate and one mucoid isolate were isolated from each patient. *p < 0.05; ***p < 0.0001.
Antibiotic Resistance Did Not Correlate with Capsule Thickness
The MICs of all isolates for 11 antibiotics were determined (Table 2). The resistance rate to cephalosporins, quinolones and carbapenems antibiotics was up to 96.7%, followed by aminoglycoside antibiotics (amikacin of 73.3% and gentamicin of 83.3%). The susceptibility to cefoperazone/sulbactam, tigecycline and colistin was lower, with resistance rates of 15.0%, 35.0% and 3.3%, respectively. The novel antibiotic eravacycline, which belongs to the tetracycline class, had the highest antibacterial activity to mucoid A. baumannii with a resistance rate of only 1.7%. And the 4 matt isolates (isolate 402, 404, 406 and 408) had similar resistances, which were only sensitive to colistin and eravacycline.
Table 2.
Antimicrobial Susceptibility in A. Baumannii
| Antibiotics | Number of Isolates | Resistance Rate (%) | ||
|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||
| CAZ | 1 | 1 | 58 | 96.7 |
| FEP | 1 | 1 | 58 | 96.7 |
| CIP | 1 | 1 | 58 | 96.7 |
| IMP | 1 | 1 | 58 | 96.7 |
| MEN | 1 | 1 | 58 | 96.7 |
| GEN | 4 | 6 | 50 | 83.3 |
| AK | 16 | 0 | 44 | 73.3 |
| TGC | 4 | 35 | 21 | 35.0 |
| SCF | 12 | 39 | 9 | 15.0 |
| CST | 58 | 0 | 2 | 3.3 |
| ERA | 46 | 13 | 1 | 1.7 |
Abbreviations: CAZ, ceftazidime; FEP, cefepime; CIP, ciprofloxacin; IPM, imipenem; MEM, meropenem; GEN, gentamicin; AK, amikacin; TGC, tigecycline; SCF, cefoperazone/sulbactam; CST, colistin; ERA, eravacycline.
Genomic Data, Sequence Types and AMR Genes of the Isolates
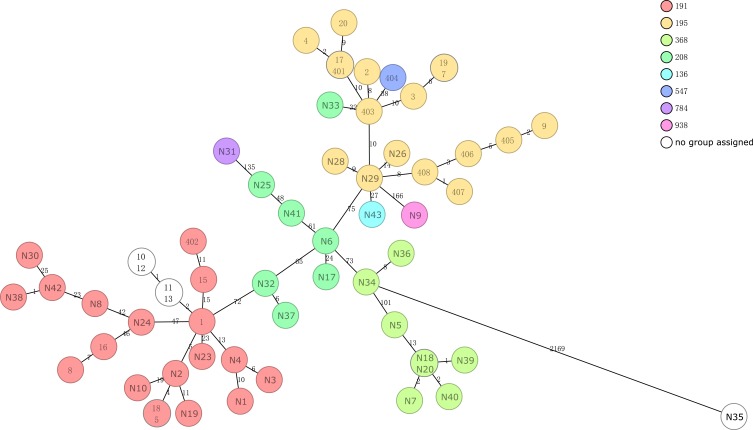
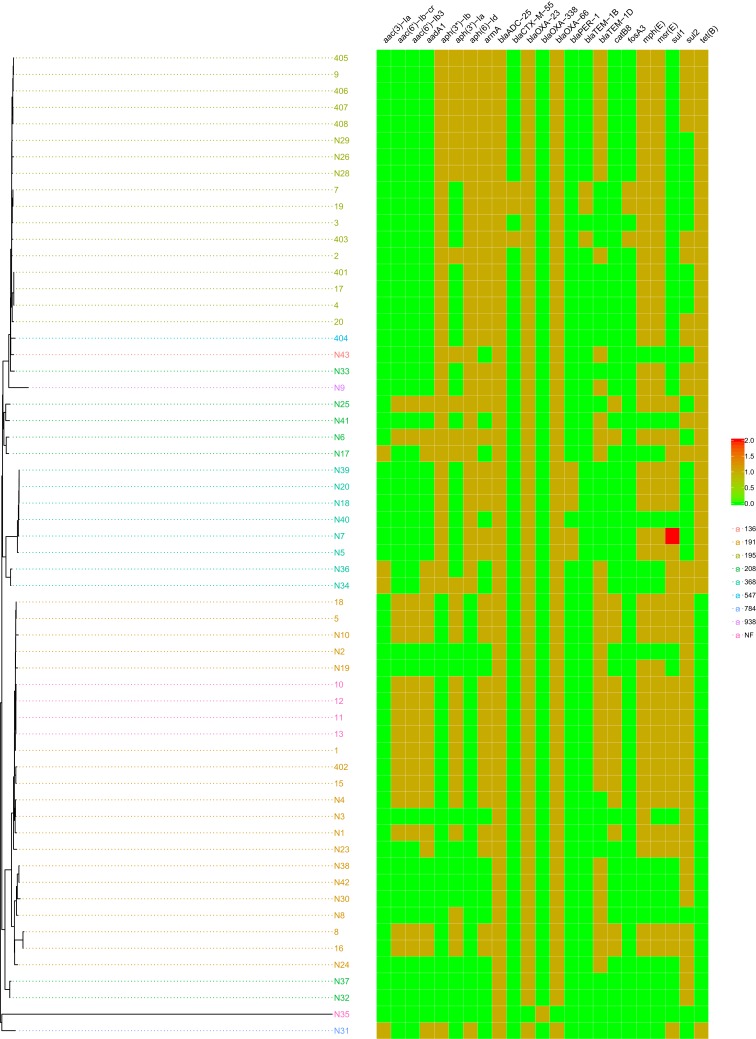
According to the analysis by cgMLST, the 60 isolates belonged to eight different sequence types (ST), including five new types (Figure 3). The main ST of mucoid A. baumannii were ST191 and ST19, followed by ST 368, 208, 136, 547, 784 and 938. Having obtained the whole-genome data, we were able to determine the occurrence of resistance genes and the mutations that confer resistance to the isolates (Figure 4). There were only two resistance genes in the isolate N35, which was susceptible to all 11 antibiotics (Table 2). The isolate N32 and N37 contained four resistance genes, blaADC-25, blaOXA-66, blaOXA-23, and sul2, which confer resistance to β-lactam and carbapenem antibiotics. The remaining 57 isolates all contained more than five resistance genes. The copy number of resistance genes in all isolates was 1, except that isolate N7 contained double copy of the sul2 gene. The mutation in adeN or adeR appeared in 30 isolates. The gene adeR is involved in the regulation of efflux pump AdeABC and adeN regulates efflux pump AdeIJK, which both contribute to tigecycline resistances. The isolates contained the mutation all not susceptible to tigecycline (Table 3).
Figure 3.
Minimum spanning tree based on cgMLST analysis. Each circle represents an allelic profile, i.e. genotype, based on sequence analysis of up to 2390 target genes. The numbers on the connecting lines illustrate the numbers of target genes with different alleles.
Figure 4.
Resistance genes. The isolate N35 only had two resistance genes, which was susceptible to all 11 antibiotics (Table 2). The isolate N32 and N37 contained four resistance genes, and the remaining 57 isolates all contained more than five resistance genes.
Table 3.
adeN and adeR Mutation Conferred Tigecycline Resistance
| Isolates | TGC(mg/L) | Mutation |
|---|---|---|
| 1 | 8 | adeN ISAba1 299 |
| 5 | 8 | adeN ISAba1 299 |
| 10 | 8 | adeN ISAba1 299 |
| 11 | 4 | adeN ISAba1 299 |
| 12 | 8 | adeN ISAba1 299 |
| 13 | 8 | adeN ISAba1 299 |
| 15 | 4 | adeN ISAba1 299 |
| 18 | 8 | adeN ISAba1 299 |
| 402 | 8 | adeN ISAba1 299 |
| N1 | 4 | adeN ISAba1 299 |
| N2 | 4 | adeN ISAba1 299 |
| N3 | 4 | adeN ISAba1 299 |
| N4 | 4 | adeN ISAba1 299 |
| N5 | 4 | adeN IS26 277 |
| N6 | 4 | adeN ISAba1 299 |
| N7 | 8 | adeN IS26 277 |
| N10 | 4 | adeN ISAba1 299 |
| N17 | 4 | adeN ISAba1 299 |
| N18 | 4 | adeN IS26 277 |
| N19 | >32 | adeN ISAba1 299 |
| N20 | 8 | adeN IS26 277 |
| N23 | 16 | adeN ISAba1 299 |
| N25 | 8 | adeN ISAba1 288 |
| N31 | 16 | adeR D23N N115T |
| N32 | 8 | adeN ISAba1 299 |
| N34 | 8 | adeN ISAba1 299 |
| N36 | 8 | adeN ISAba1 299 |
| N37 | 8 | adeN ISAba1 299 |
| N39 | 8 | adeN IS26 277 |
| N40 | 4 | adeN IS26 277 |
| N41 | 4 | adeN ISAba1 288 |
Single Nucleotide Polymorphism (SNP) Analysis
To investigate possible molecular mechanisms for the appearance of mucoid or matt phenotypes, we analysed the SNPs of the double isolates that were obtained from one patient. The isolates 405 and 406 were obtained from one patient, and 407 and 408 were obtained from a second patient. All four isolates belonged to ST195. According to the sequence comparison of isolate 405 (mucoid), isolate 406 (matt) revealed two mutations in gene ptk (Table 4). The other two isolates, 407 (mucoid) and 408 (matt), also showed one mutation in the same gene, ptk (Table 5). This result indicated that gene ptk might be a key determinant for matt versus mucoid phenotypes during capsule biosynthesis, an observation that was consistent with previous studies.10,12
Table 4.
SNPs Analysis of Isolate 405 (Mucoid) and 406 (Matt)
| Gene | Type | Product | Effect |
|---|---|---|---|
| sppA | SNP | Putative signal peptide peptidase SppA | missense_variant c.415A>G p.Lys139Glu |
| ftsI | SNP | Peptidoglycan D,D-transpeptidase FtsI | missense_variant c.1589G>A p.Arg530His |
| tadA | SNP | tRNA-specific adenosine deaminase | missense_variant c.109T>C p.Ser37Pro |
| ptk | SNP | Tyrosine-protein kinase | missense_variant c.2000A>G p.Asp667Gly |
| missense_variant c.1628C>T p.Pro543Leu | |||
| ppx | SNP | Exopolyphosphatase | missense_variant c.1441A>G p.Thr481Ala |
| rapZ | del | RNase adapter protein RapZ | frameshift_variant c.688delA p.Met230fs |
| ibaG | SNP | Acid stress protein IbaG | missense_variant c.65A>G p.Asn22Ser |
| – | SNP | Hypothetical protein | missense_variant c.443C>T p.Pro148Leu |
| – | del | Putative TonB-dependent receptor | frameshift_variant c.1297delT p.Trp433fs |
Table 5.
SNPs Analysis of Isolate 407 (Mucoid) and 408 (Matt)
| Gene | Type | Product | Effect |
|---|---|---|---|
| nimR_1 | SNP | HTH-type transcriptional regulator NimR | missense_variant c.578A>G p.His193Arg |
| rpsA | SNP | 30S ribosomal protein S1 | missense_variant c.410G>A p.Gly137Asp |
| ptk | SNP | Tyrosine-protein kinase | missense_variant c.833T>A p.Leu278Gln |
| fhuA_4 | SNP | Ferrichrome-iron receptor | synonymous_variant c.273A>G p.Leu91Leu |
| – | del | Hypothetical protein | frameshift_variant c.3000delA p.Val1001fs |
No Correlation Between Biofilm Formation and Mucoid/Matt Phenotypes of A. baumannii Isolates
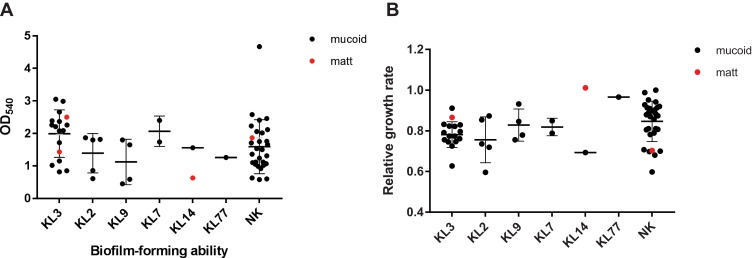
Previously, it was reported that matt A. baumannii isolates produced more biofilms than mucoid ones. Therefore, we determined the biofilm-forming ability of all strains by crystal violet staining, which can be quantified by measuring the optical density at 540 nm, OD540. As reference served the type strain ATCC19606. While the reference strain was set to 1 ± 0.18, the negative control with media only, gave a value of 0.25 ±0.07. Three isolates formed no biofilms with OD540 values lower than the negative control. Fifty (83.3%) isolates showed weak biofilm-forming ability with OD540 values between the negative control and the reference strain (0.63 ± 0.22). The remaining seven isolates (11.7%) formed extensive biofilms with OD540 values significantly higher than the control strain ATCC19606 (1.23 ± 0.30). While a significant difference between the groups with a different biofilm-forming ability (p < 0.05) was observed, no significant difference was observed between the mucoid A. baumannii and the matt isolates (0.68 ± 0.11, 0.66±0.13, p > 0.05), and also no differences between different capsular types (Figure 5A).
Figure 5.
Biofilm-formating ability and relative growth rate. (A) Biofilm-forming ability of different capsular types. (B) Relative growth rate of different capsular type strains, taking the isolate 1 as reference. Both biofilm-forming ability and relative growth rate had no significant differences between matt and mucoid isolates.
Abbreviation: NK, unknown capsular types.
Mucoid and Matt A. baumannii Isolates Show Similar Growth Rates
Previously as the study reported that mucoid A. baumannii isolates display a slower growth rate than their matt counterparts.18 We, therefore, measured the growth rates of all isolates. Taking the growth rate of isolate 1 (a mucoid strain) as a reference, the relative growth rates of all isolates were calculated. Our results showed that there was no significant difference between mucoid A. baumannii and matt A. baumannii (p > 0.05) in the relative growth rate, and no differences between different capsular types (Figure 5B).
No Correlation in Virulence with Matt versus Mucoid A. baumannii in the G. mellonella Model
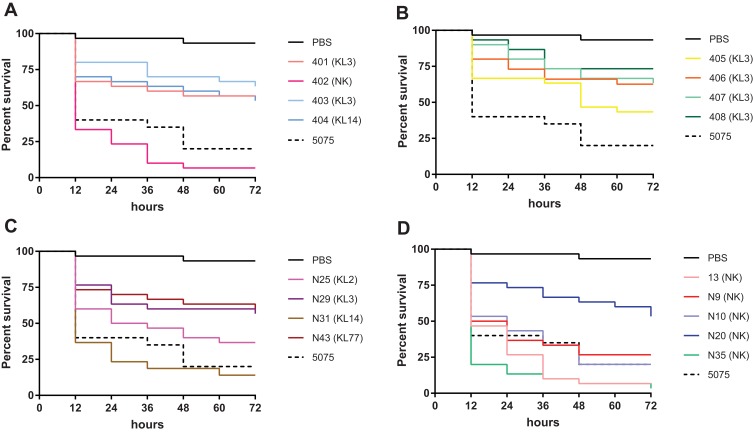
According to ST and resistance genes of all isolates, we selected the isolate 13, N9, N10, N20, N25, N29, N31, N35, N43, and the two double isolates (matt and mucoid together; 401–408), a total of 17 isolates for virulence test in G. mellonella (Figure 6A–D). The virulence of 17 clinical isolates varied widely, with an average 72 h survival rate ranging from 3.33% to 73.33%. The percent survival of the isolate 402, 405, 13, N9, N10, N25, N31 and N35 was less than 50%, among which, the percent survival of 402, 13 and N35 was lower than 10%. The isolates 401, 403, 404, 406, 407, 408, N20, N29 and N43 displayed a lower virulence level, with survival rates at 72 hrs of higher than 50%. Interestingly, when testing the double isolate that was obtained from one patient, the virulence of the matt isolate (402) was significantly higher than the mucoid one (401, p < 0.0001). In must be mentioned that 401 belongs to ST191, while 402 belongs to ST195 which makes it difficult to conclude whether virulence is influenced by the matt or mucoid phenotype of A. baumannii. An indication that it might the phenotypes might not correlate with virulence is the data we obtained from the other two double isolates, obtained from a single patient each (403, 404 and 407, 408). When comparing these isolates, no difference in virulence could be observed. We also were able to determine that the isolates of the capsule-type KL3 generally displayed a lower virulence level, while isolates N31 (KL14) and N25 (KL2) had a higher virulence level. Interestingly, we found highly virulent isolates with so-far unknown capsular types, such as isolate 13 and N35.
Figure 6.
Percent survival of G. mellonella infected with A. baumannii. (A) 401 (mucoid, ST195, KL3) and 402 (matt, ST191, NK) isolated from one patient, and (mucoid, ST195, KL3) and 404 (matt, ST547, KL14) isolated from one patient; (B) 403 (mucoid) and 404 (matt) isolated from one patient, and (mucoid) and 404 (matt) isolated from one patient, all 4 isolates belong to ST195, KL3; (C, D) all isolates were mucoid.
Abbreviation: NK, unknown capsular types.
Discussion
In 1998 the first mucoid A. baumannii strains in China were reported, which were isolated from the sputum of patients with chronic bronchitis.19 As the isolation of mucoid A. baumannii strains is comparably rare in the clinical setting, reports about their isolation rate are lacking to date. We observed that the quantity of mucoid A. baumannii isolates is increasing in clinical care. Mucoid bacteria are often associated with chronic infections, with two major factors possibly contributing to the long persistence in the host: higher levels of antibiotic resistance and stronger attachment due to the mucoid nature of the bacterial colonies.20,21 In our study, we could demonstrate that the capsules of the mucoid A. baumannii isolates were significantly thicker than that of matt A. baumannii isolates. In contrast to previous reports,18 we could not conclude that matt cells form more biofilm than mucoid ones, as we observed no significant differences in biofilm-forming ability between the phenotypes. In addition, our data showed that the growth rate between the different phenotypes was highly similar.
Our data showed that up to 77% of patients had used more than three categories of antibiotics prior to isolation of the bacteria, which may have led to the observed high resistance rates to major antibiotics in mucoid A. baumannii. Notably, the resistance rate of tigecycline had reached up to 35.0%. Whole-genome sequencing revealed that 30 isolates harbored tigecycline resistance mutations in adeN and one isolate had a mutation in adeR. Other mutations were observed which might confer resistance, although more experiments are required to fully understand the resistance mechanism.
The major sequence types (ST) in this study were ST191 and ST195, which were previously reported as the major STs carrying carbapenem-resistance gene blaOXA-23 in the carbapenem-resistance A. baumannii in China.22 Consistently, all ST191 and ST195 isolates in our study carrying a single copy of blaOXA-23 were shown to be resistant to carbapenem antibiotics. Interestingly, the survival rate of two ST191 isolates (402 and N10) was similar to AB5057, a model highly virulent A. baumannii strain,23 but the survival rate of six ST195 isolates (401, 405–408, N29) was significantly higher than AB5075 (p < 0.05). These results indicate that ST191 might belong to the more virulent isolates whereas ST195 might be regarded as isolates with comparably lower virulence. It is noteworthy that the six ST195 isolates all belonged to KL3. Infections with these isolates and another KL3 isolate (403) had significantly higher survival rates compared to AB5075 (p < 0.05), which indicates that the capsule type KL3 might be less virulent. For the other types, more data are required to be able to correlate capsule type with the virulence of the strain.
When performing the virulence test in G. mellonella, we could not see any correlation between virulence and the matt or mucoid phenotype. While this test is usually a good indicator of virulence, the larva model might not be the best test of the human pathogen. Therefore, more data and possibly other tests in higher organisms are required to understand virulence levels of the isolates and a possible correlation with the mucoid or matt phenotype.
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (31670135, 31770142, 81702041, 81871614 and 81861138054), and the Zhejiang Province Medical Platform Backbone Talent Plan (2016KYA158). Xiaoting Huab and Zhihui Zhoub should be considered co-corresponding authors.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Munoz-Price LS, Weinstein RA. Acinetobacter infection. N Engl J Med. 2008;358:1271–1281. doi: 10.1056/NEJMra070741 [DOI] [PubMed] [Google Scholar]
- 2.McConnell MJ, Actis L, Pachón J. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev. 2013;37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x [DOI] [PubMed] [Google Scholar]
- 3.Karageorgopoulos DE, Falagas ME. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis. 2008;8:751–762. doi: 10.1016/S1473-3099(08)70279-2 [DOI] [PubMed] [Google Scholar]
- 4.Karaiskos I, Giamarellou H. Multidrug-resistant and extensively drug-resistant gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother. 2014;15:1351–1370. doi: 10.1517/14656566.2014.914172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sousa AM, Pereira MO. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs-a review. Pathogens. 2014;3(3):680–703. doi: 10.3390/pathogens3030680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiens JR, Vasil AI, Schurr MJ, Vasil ML. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. mBio. 2014;5(1):e01010–13. doi: 10.1128/mBio.01010-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Germoni LAP, Bremer PJ, Lamont IL. The effect of alginate lyase on the gentamicin resistance of Pseudomonas aeruginosa in mucoid biofilms. J Appl Microbiol. 2016;121:126–135. doi: 10.1111/jam.2016.121.issue-1 [DOI] [PubMed] [Google Scholar]
- 8.Fung CP, Chang FY, Lee SC, et al. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut. 2002;50:420–424. doi: 10.1136/gut.50.3.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenyon JJ, Hall RM. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One. 2013;8:e62160. doi: 10.1371/journal.pone.0062160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisinger E, Isberg RR. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015;11(2):e1004691. doi: 10.1371/journal.ppat.1004691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tipton KA, Chin C-Y, Farokhyfar M, Weiss DS, Rather PN. Role of capsule in resistance to disinfectants, host antimicrobials, and desiccation in acinetobacter baumannii. Antimicrob Agents Chemother. 2018;62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua X, Zhou Z, Yang Q, et al. Evolution of Acinetobacter baumannii in vivo: international clone II, more resistance to ceftazidime, mutation in ptk. Front Microbiol. 2017;8(1256). doi: 10.3389/fmicb.2017.01256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Zhang T, Yu D, et al. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob Agents Chemother. 2011;55:4506–4512. doi: 10.1128/AAC.01134-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins PG, Prior K, Harmsen D, Seifert H. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One. 2017;12(6):e0179228. doi: 10.1371/journal.pone.0179228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Team R. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 16.Lee HW, Koh YM, Kim J, et al. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Inf. 2008;14:49–54. doi: 10.1111/j.1469-0691.2007.01842.x [DOI] [PubMed] [Google Scholar]
- 17.Blum G, Hörtnagl C, Jukic E, et al. New insight into amphotericin B resistance in Aspergillus terreus. Antimicrob Agents Chemother. 2013;57:1583–1588. doi: 10.1128/AAC.01283-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempf M, Eveillard M, Deshayes C, et al. Cell surface properties of two differently virulent strains of Acinetobacter baumannii isolated from a patient. Can J Microbiol. 2012;58:311–317. doi: 10.1139/w11-131 [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Kong H. Mucoid-type Acinetobacter baumannii was detected from the sputum of the old chronic bronchitis. Shanghai Med Lab. 1998;1:6. [Google Scholar]
- 20.Min KB, Lee K-M, Oh YT, Yoon SS. Nonmucoid conversion of mucoid Pseudomonas aeruginosa induced by sulfate-stimulated growth. FEMS Microbiol Lett. 2014;360:157–166. doi: 10.1111/fml.2014.360.issue-2 [DOI] [PubMed] [Google Scholar]
- 21.Tuchscherr L, Medina E, Hussain M, et al. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med. 2011;3:129–141. doi: 10.1002/emmm.v3.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning NZ, Liu X, Bao CM, et al. Molecular epidemiology of bla OXA-23 -producing carbapenem-resistant Acinetobacter baumannii in a single institution over a 65-month period in north China. BMC Infect Dis. 2017;17:14. doi: 10.1186/s12879-016-2110-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs AC, Thompson MG, Black CC, et al. AB5075, a highly virulent isolate of acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio. 2014;5(3):e01076–14. doi: 10.1128/mBio.01076-14 [DOI] [PMC free article] [PubMed] [Google Scholar]