Abstract
In many human-altered ecosystems, organisms are increasingly faced with more diverse and complex environmental stressors and pollutant mixtures, to which the adaptations necessary to survive exposure are likely to be numerous and varied. Improving our understanding of the molecular mechanisms that underlie complex polygenic adaptations in natural settings requires significant toxicological, biochemical, physiological, and genomic data rarely available for non-model organisms. Here, we build upon two decades of study of adaptation to anthropogenic pollutants in a population of Atlantic killifish (Fundulus heteroclitus) that inhabits the creosote-contaminated Atlantic Wood Industries Superfund (AW) site on the Elizabeth River, Virginia in the United States. To better understand the genotypes that underlie previously characterized resistance to PCBs and PAHs, we performed Restriction site-Associated DNA sequencing (RADseq) on killifish from AW and two relatively clean reference sites (King’s Creek–KC, and Mains Creek–MC). Across the genome, we analyzed over 83,000 loci and 12,000 single nucleotide polymorphisms (SNPs). Shared across both comparisons of killifish from polluted (AW) and relatively unpolluted (KC and MC) sites, we found eight genomic regions with smoothed FST values significantly (p<0.001) elevated above background. Using the recently published F. heteroclitus reference genome, we identified candidate genes in these significant regions involved in the AHR pathway (e.g. AIP, ARNT1c), as well as genes relating to cardiac structure and function. These genes represent both previously characterized and potentially novel molecular adaptations involved with various aspects of resistance to these environmental toxins.
Keywords: AIP, ARNT, Elizabeth River, PAH, RADseq
1. Introduction
Human civilization has been responsible for polluting and altering the environment in which all creatures must live at an ever increasing magnitude and rate of change. In this changing world, an organism’s ability to acclimate and ultimately evolve resistance to new baselines will be decisive. Molecular techniques are now readily available to help understand the forces and mechanisms underlying rapid evolutionary change in wild populations. There may be no better natural experiment in which to explore rapid evolution of resistance to lethally toxic aquatic pollution than the well-studied Atlantic killifish, Fundulus heteroclitus, populations that inhabit highly polluted, and often Superfund-listed, sites along the east coast of the United States. Superfund sites are those that have been identified by the US Environmental Protection Agency as candidates for cleanup due to contamination by hazardous waste that poses a risk to human or environmental health.
The Atlantic killifish is a highly euryhaline, eurythermal, and euryoxic estuarine minnow native to the east coast of North America from Newfoundland to mid Florida with a subspecies split near New Jersey (Dunson et al., 1993; Gonzalez et al., 1989; Nordlie, 2006; Smith and Able, 2003; Stierhoff et al., 2003; Wood and Marshall, 1994). Its small home range of approximately 600 m (Lotrich, 1975) and non-dispersive spawning predisposes it to local adaptation and makes it an excellent teleost model of adaptation (Burnett et al., 2007). Several populations of killifish adapted to live in estuaries polluted with polycyclic aromatic hydrocarbons (PAHs) (e.g. Elizabeth River (ER), Virginia), polychlorinated biphenyls (PCBs) (e.g. New Bedford Harbor, Massachusetts; Newark Bay, New Jersey), and dioxins (e.g., Newark Bay, New Jersey) have been described and studied for more than 20 years (Di Giulio and Clark, 2015; Meyer and Di Giulio, 2003; Nacci et al., 2010; Nacci et al., 1999).
The story of ER killifish adaptation to PAHs is complicated by both extremely high and complex pollutant exposure. For example, the killifish subpopulation that inhabits the Atlantic Wood Industries Superfund site (AW) on the Southern Branch of the ER lives with sediments that contain up to 500 μg/g dry sediment total PAH (Vogelbein and Unger, 2003) [122.6 μg/g dry sediment total PAH in Clark et al. (2013)], some of the highest concentrations of environmental PAHs ever measured (Walker et al., 2004). Pollutants at this site are primarily the result of several decades of adjacent creosote wood industry that operated until 1990 (Di Giulio and Clark, 2015) and consist of a complex mixture of PAHs including benzo[b,k,j]fluoranthene, chrysene, fluoranthene, and benzo[a]pyrene, among others (Clark et al., 2013). PAHs and their metabolites are differentially known to be carcinogenic (Luch, 2005), immunosuppressive (Kaplan et al., 2013), teratogenic (Billiard et al., 2008), aryl hydrocarbon receptor (AHR) agonists (Billiard et al., 2002; Schmidt and Bradfield, 1996) and/or cytochrome P450 (CYP) inhibitors (Wassenberg and Di Giulio, 2004a, b). Thus, the adaptations necessary to survive exposure to such diverse and complex mixtures of PAHs are likely to be numerous and varied, so as to address both the individual and combined effects of multiple PAHs and other toxicants. We expect that the complete adaptation story for populations exposed to complex pollutant mixtures is likely far more multifaceted than adaptation to single source pollutants, for example the case of Hudson River tomcod where a single mutation to the AHR gene explains much of the adaptation to PCB pollution (Wirgin et al., 2011).
The AW killifish were originally studied by Hargis et al. (1989) and Vogelbein et al. (1990) who reported high incidences of cancer, particularly in the liver. Further studies of AW killifish have generally shown a lack of up-regulation of CYP1s in response to common inducing agents (Van Veld and Westbrook, 1995) and a depression of the AHR pathway (Meyer et al., 2003b). These adaptations appear to be associated with the fatal cardiac teratogenesis that occur in fish embryos exposed to PAHs. Other findings in adapted fish from the ER show generally upregulated antioxidant defenses (Meyer et al., 2003a), increased resistance to the development of liver tumors (though a higher incidence of liver tumors in the field) (Wills et al., 2010), increased DNA repair capabilities (Jung et al., 2009), and cross resistance to dioxin-like compounds and several pesticides despite those compounds not being present in high concentrations at AW (Clark and Di Giulio, 2012; Meyer and Di Giulio, 2002). Importantly, several of these adaptations appear to be at least in part heritable (Clark et al., 2014; Meyer and Di Giulio, 2002; Meyer and Di Giulio, 2003; Meyer et al., 2002; Nacci et al., 2010; Ownby et al., 2002).
Attempting to describe the genetic mechanisms that underlie such a suite of adaptations in a natural population exposed to complex pollutant mixtures can be extremely challenging. Yet, a growing toolkit of ecological and evolutionary genomic techniques increasingly enables genome-wide scans for molecular adaptation. For example, restriction-site associated DNA sequencing (RADseq) enables the simultaneous discovery and genotyping of thousands of SNPs across the genomes of non-model organisms through sequencing short DNA fragments that flank restriction enzyme cut sites (Andrews et al., 2016; Baird et al., 2008; Davey et al., 2011). Such reduced-representation genome approaches provide a systematic way to reduce genome complexity, enabling genome-wide sequencing at a population scale within a limited budget. Though whole genome re-sequencing provides higher genomic resolution (e.g., Jones et al., 2012), such approaches still come at a higher cost per individual for lower read depth and require greater computational resources to process very large datasets (Ellegren, 2014). In fact, genotyping-by-sequencing approaches such as RADseq do not require a reference whole genome sequence, though physiological and ecological interpretation of the resulting data is greatly enhanced by annotation information. Some studies have recently used RADseq in systems with and without reference genomes to identify regions of the genome that show significant deviation among natural populations and which are therefore likely under selection, putatively as a result of variation in habitat and/or exposure to stressors (Cammen et al., 2015; Hohenlohe et al., 2010).
Recent collaborative efforts to improve the genomic resources for Atlantic killifish have dramatically increased the potential power of genome-wide studies for molecular adaptation in this non-model system (Burnett et al., 2007). In particular, the recent sequencing and annotation of the genome of a single northern subspecies killifish from Maine by the Killifish Genome Consortium (Reid et al., 2017; Reid et al., 2016) facilitates a genome-enabled RADseq approach to investigate the complex physiological adaptations observed in response to PAH exposure across ER populations. Here we use RADseq to compare genomic variation of killifish collected from a polluted Superfund site (AW) to killifish collected from two relatively unpolluted nearby sites (Kings Creek [KC] and Mains Creek [MC]). Using multiple comparisons between polluted and unpolluted sites provides higher power to detect regions of the genome that are truly associated with adaptation to PAHs and reduce the impact of false positives. We present several candidate regions showing signs of selection and discuss the genes within those regions in the context of two decades of molecular, toxicological, and phenotypic data on these populations.
2. Materials and Methods
2.1. Samples
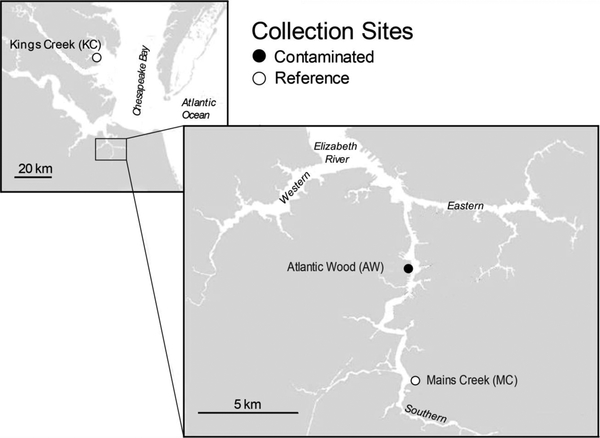
Atlantic killifish, Fundulus heteroclitus, were collected from three sites in Virginia: the Atlantic Wood Industries Superfund site (AW; 36°48’27.2”N, 76°17’38.1”W) on the ER; Mains Creek (MC; 36°45’13.5”N, 76°24’58.9”W) on the ER; and King’s Creek (KC; 37°18’16.2”N, 76°24’58.9”W) located off of Mobjack Bay near the mouth of the York River (Figure 1). The AW and KC sites have been the source of killifish for dozens of published studies (reviewed in Di Giulio and Clark, 2015), while MC was first used in Clark et al. (2013). Recent measurements of the sum of selected PAH concentrations from the three study sites found AW to be far more polluted (122,665 ± 16,854 ng/g dry sediment) than either reference site (KC: 526 ± 624 ng/g; MC: 186 ± 201 ng/g) (Clark et al., 2013). Accordingly, killifish from AW are the most phenotypically resistant (adapted) to PAH exposure and killifish from KC are the most susceptible. Killifish from MC display intermediate and variable resistance (Clark et al., 2013).
Figure 1.
Map of collection sites: the Atlantic Wood Industries Superfund site (AW) and Mains Creek (MC) on the Elizabeth River, VA; and Kings Creek (KC) located near the mouth of the York River, VA.
Fish were collected in wire mesh minnow traps in Summer and Fall 2012 and transported live in coolers to Duke University. After two weeks in a circulating artificial seawater system, 32 fish from each site were cold anesthetized and euthanized by decapitation. Muscle samples were isolated, flash frozen in liquid nitrogen, and stored at −80°C. Sub-samples were subsequently taken and stored at room temperature in 95% ethanol until DNA isolation. The collection, care, and euthanasia techniques used herein were approved by the Duke University Institutional Animal Care & Use Committee (A184–13-07).
2.2. Genome-wide Sequencing
Double digest RADseq libraries were prepared using the protocol described in Peterson et al. (2012) with minor modifications. High quality genomic DNA was isolated from approximately 20 mg of muscle sample using a Wizard® Plus SV Miniprep DNA Purification kit (Promega, Madison, WI) with suction manifold, following the manufacturer’s instructions. DNA concentrations were quantified using a Qubit 2.0 (Life Technologies, Calsbad, CA) fluorometer with manufacturer-provided broad range standards. To prepare RADseq libraries, 500 ng of DNA from each sample was double digested with 10 U each of SBfI-HF® and MspI (New England Biolabs, Ipswich, MA) at 37 °C for 3 h. Digested DNA was purified using 1.5x volume AMPure (Agencourt, Beverly, MA) magnetic beads. Thirty-two custom P1-SBfI oligo adapters with unique 6 bp barcodes were ligated to DNA fragments from each of the respective 32 samples from each site. Two custom P2-MspI oligo adapters were annealed to the other end of each fragment. Ligation reactions consisted of 6.25 nM P1 adapter, 0.625 μM P2 adapter, 1.25 mM rATP and 200 U concentrated T4 DNA ligase (New England Biolabs), incubated at room temperature for 2 h. Further ligation was prevented using a heat kill step of 65 °C for 10 min, followed by slow (2%) ramp to room temperature.
The 32 labeled samples from each site were subsequently pooled and purified using 1.5x volume AMPure (Agencourt) magnetic beads and 2 μl of each DNA pool was amplified using 1x Phusion® High-Fidelity PCR Master Mix (New England Biolabs) and 1 μM of a modified Solexa© amplification primer mix (P1-forward: 5’-AATGATACGGC GACCACCG*A-3’ ; P2-reverse: 5’-CAAGCAGAAGACG GCATACG*A-3’). PCR was run as follows: 30 s 98 °C, 30 s 58 °C, 1 min 72 °C, 25 cycles. Following magnetic bead purification of the amplified product, RAD libraries were quantified and sequenced. AW and KC libraries were sequenced together in a single Illumina Hi-Seq 2500 lane by the Duke University Center for Genomic and Computational Biology sequencing center (50 bp single-end run). The MC library was sequenced with other killifish libraries at a later date under identical settings.
2.3. SNP Genotyping
We used Stacks version 1.21 (Catchen et al., 2011) to identify and genotype SNPs in the RAD sequences. Reads were first filtered for quality using a minimum average read score of 10 across 8 bp sliding windows and reads with sequencing errors in the barcode or restriction enzyme site were removed. Reads were then demultiplexed by collection site using P2 index sequences and by individual within collection site using P1 barcode sequences. We used Bowtie v0.12.9 (Langmead et al., 2009) to map all remaining reads to the F. heteroclitus genome assembly 3.0.2 (GCF_000826765.1), accepting only alignments with less than three mismatches and a single best match to the genome. For each individual, all reads that mapped to the same genomic location were considered a “stack” and SNPs were detected using a maximum likelihood framework in Stacks (Catchen et al., 2013). All stacks were combined into a single catalog of loci, allowing up to two mismatches at a locus between individuals. A minimum read depth of 10 reads per locus per individual was required for all further analyses.
2.4. Population Statistics
We used Stacks to calculate kernel-smoothed average pairwise AMOVA FST (Weir, 1996) between sampling sites for each SNP genotyped in at least 75% of the individuals in two populations and with a sample-wide minor allele frequency greater than or equal to 0.1. Smoothed values were calculated across a sliding window of 150 kb along the genome scaffolds. Bootstrap resampling (10,000 replicates) of variable sites across the genome was used to test if a kernel-smoothed FST value was significantly different from the average FST observed across the genome (Catchen et al., 2013).
2.5. Candidate Genes
We further evaluated candidate genes located on scaffolds with smoothed pairwise FST values that were significantly greater than the average across the genome (P<0.001) in both pairwise comparisons of polluted and clean sites (AW-KC and AW-MC), using the F. heteroclitus genome assembly and annotation loaded into CLC Genomics Workbench version 9.1 (Qiagen Bioinformatics, Redwood City, CA, USA). Within significant scaffolds, we identified candidate genes within 300 kb (Hohenlohe et al., 2010) of non-smoothed FST values greater than 0.1 in either comparison. Non-smoothed FST values were used to increase the fine-scale resolution of differentiation across the scaffold. A non-smoothed FST value of 0.1 was used as the threshold for significance in our analysis as it is greater than three standard deviations above the mean (e.g. Proestou et al., 2014). All genes located at least partly within the given range were included in the list of candidates, regardless of whether the genes were named or functions known. The putative function of un-named genes (annotated as LOC1059*****) that were located within candidate scaffold regions were further described by the annotated RNA product in the killifish genome assembly (3.0.2), by comparison with highly similar sequences identified in NCBI’s nucleotide collection (nr/nt) database using the online blastn tool, or by referral to recently published papers (especially Reid et al., 2016 supplemental). We refer to these as “gene-like” and initially use the locus name when the gene is not fully annotated.
Because this assembly (3.0.2) of the killifish genome consists of 10,180 scaffolds with an N50 of 1,252,252 bp, many of the candidate gene regions were on scaffolds that were either too small to encompass the complete ± 300kb range or near the edge of a large scaffold so that one arm of the range was truncated at the scaffold edge. It is therefore likely that additional candidate genes may be found by reanalyzing these data with future versions of the killifish genome that are composed of fewer larger scaffolds. Furthermore, we acknowledge that SNPs on small scaffolds may also have truncated sliding windows across which to calculate smoothed FST values, which may artificially inflate smoothed values for these loci. On the other hand, large scaffolds may have been selected by our filtering system if multiple non-overlapping genomic regions were significant across multiple pairwise comparisons; however, the subsequent analysis of non-smoothed FST values would not be equally biased.
3. Results
RAD sequencing was used to identify potential genetic adaptations in killifish to highly polluted habitat at an EPA Superfund site on the Elizabeth River, VA using multiple pairwise comparisons between fish collected at a highly contaminated creosote storage site (AW) and two relatively PAH/PCB-free reference sites (KC and MC). Illumina HiSeq sequencing of 32 individual fish from each site generated over 43 million reads per collection site. An average of 76.4% of the raw reads (post-quality filtering) from each of 96 individuals mapped to single locations across the genome. The number of mapped reads per individual ranged from 123,978 to 8,941,786 with an average of 1,330,973 (±1,143,420 SD) and no significant difference among study sites (AW: 1,047,547 ±471,931; KC: 1,065,297 ±351,031; MC: 1,880,165 ±1,797,027).
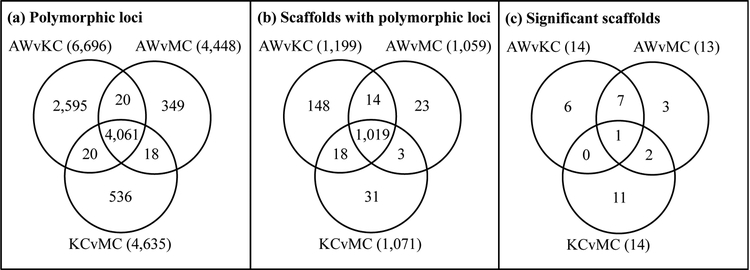
Sequencing identified 83,144 loci, of which 7,599 loci contained at least one SNP matching the filters described above (12,071 SNPs total). These filtered loci had an average read depth of 70 sequences per RAD tag per individual when missing data were excluded. Over half of these polymorphic loci (4,061; 53.4%) were sequenced to sufficient depth in all three populations (Figure 2A). These SNPs were distributed across 1,256 of the 10,180 scaffolds in the F. heteroclitus genome version 3.0.2 (Figure 2B).
Figure 2.
Number of (a) polymorphic loci and (b) Fundulus genome scaffolds that were evaluated in each pairwise comparison between collection sites (abbreviations as in Figure 1). Further, (c) the subset of these scaffolds that contained at least one region with significantly greater differentiation (smoothed FST, P<0.001) than the average across the genome.
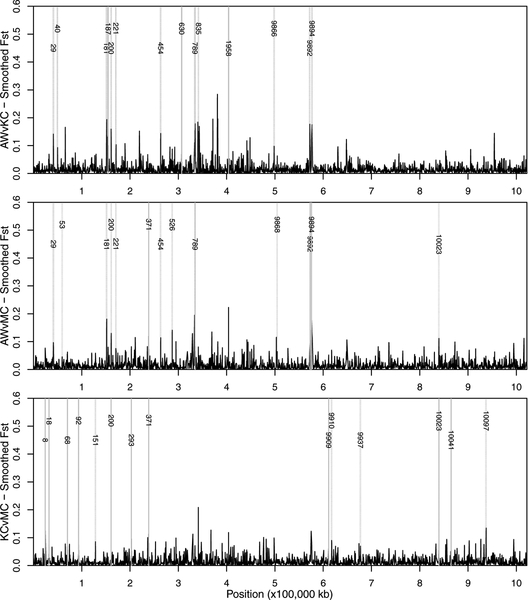
The filtered SNPs were used to identify genomic scaffolds showing high levels of differentiation in two pairwise comparisons between clean (KC and MC) and polluted (AW) sample sites (Figure 3). For each pairwise comparison, we identified 13–14 scaffolds that contained at least one region with significantly greater differentiation (P<0.001) than the average across the genome. In order to minimize the number of false positives, we searched for candidate genes only on scaffolds with significant smoothed FST values in both comparisons between clean and polluted sites (AW vs KC and AW vs MC). We identified eight such candidate scaffolds, of which only one (scaffold 200) was also significant in the pairwise comparison between the two clean sites (KC vs. MC) (Figure 2C). Each of these eight scaffolds (Table 1) contained multiple SNPs with non-smoothed FST values greater than 0.1, around which multiple candidate genes were identified (Figures 4 & 5) and are discussed further below.
Figure 3.
Smoothed pairwise FST values across Fundulus scaffolds (ordered by scaffold number, not size) for comparisons of Atlantic Wood (AW), Mains Creek (MC), and Kings Creek (KC). Shaded vertical bars highlight genomic regions with significantly greater genetic differentiation than the average across the genome within each pairwise comparison. Numbers on these shaded bars correspond to scaffold with significant differentiation.
Table 1.
Summary of the eight scaffolds with smoothed FST values greater than background at p<0.001 for both the AW-KC and AW-MC comparison.
| AW-KC | AW-MC | |||||
|---|---|---|---|---|---|---|
| Scaffold | Size (bp) | Candidate Region (bp) | p-value | Candidate Region (bp) | p-value | Genes of Interest |
| 29 | 822750 | 153745–822750 | 0.0001 | 153737–822750 | 0.0001 | ITGB5 |
| 181 | 540100 | 0–540100 | 0.0001 | 0–540100 | 0.0001 | GSTT1 |
| 200 | 483985 | 0–483985 | 0.0001 | 0–483985 | 0.0001 | TTN |
| 221 | 439259 | 0–439259 | 0.0001 | 0–439259 | 0.001 | CYB5 |
| 454 | 240234 | 0–240234 | 0.0001 | 0–240234 | 0.0001 | NEBL |
| 789 | 3477790 | 900901–2318446 | 0.0001 | 144707–1767624 | 0.001 | NCOA2, KCNB2 |
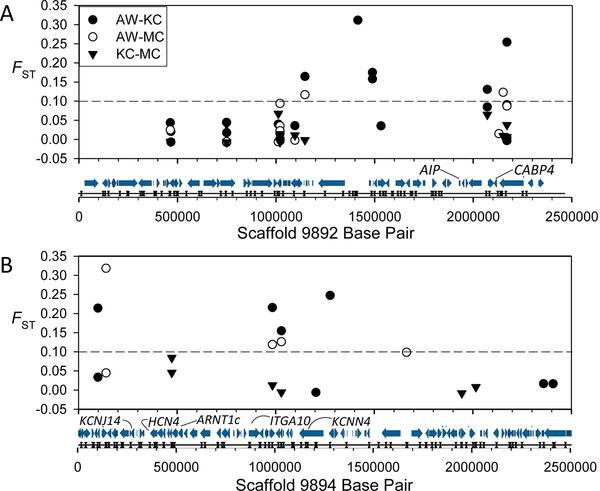
| 9892 | 2472794 | 1115291–1832508 | 0.0001 | 1830869–2471427 | 0.001 | AIP, CABP4 |
| 9894 | 2500692 | 0–398276 683455–1576579 |
0.001 | 0–1328585 | 0.0001 | ARNT1c, KCNJ14, HCN4, KCNN4, ITGA10 |
Figure 4.
Scaffolds 9892 (A) and 9894 (B) showing FST values of SNPs identified within pairwise comparisons. Below the x-axis are gene models (arrows) and Sbf1 restriction sites (opposing triangles) from Fundulus heteroclitus genome version 3.0.2 with mentioned genes labeled.
Figure 5.
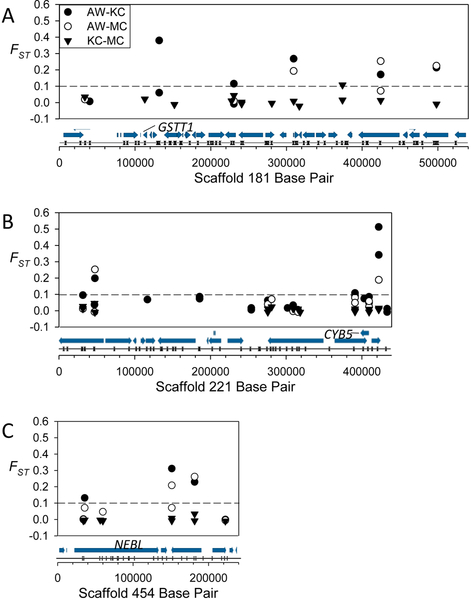
Scaffolds 181 (A), 221 (B) and 454 (C) showing FST values of SNPs identified within pairwise comparisons. Below the x-axis are gene models (arrows) and Sbf1 restriction sites (opposing triangles) from Fundulus heteroclitus genome version 3.0.2 with mentioned genes labeled.
4. Discussion
Our ability to fully understand the molecular mechanisms that underlie complex, polygenic adaptations in natural settings is often limited by both sampling and technological constraints. Few natural study systems enable replicate study designs and confounding environmental conditions often complicate our interpretations of adaptation to single or multiple stressors (Menzel and Case, 1977). Furthermore, non-model organisms, in which we are most interested in studying adaptation in natural settings, typically lack the genomic resources necessary for detailed interpretations of observed genetic diversity. Here, we overcome some of these challenges in the well-studied, non-model system Fundulus heteroclitus, using comparisons of a single contaminated site to multiple natural control sites to explore the evolution of resistance to anthropogenic pollutants. Data generated using RADseq, a high-resolution genotyping-by-sequencing approach, were interpreted following current best practices (Catchen et al., 2017; Lowry et al. 2017a,b; McKinney et al., 2017), within the context of an annotated reference genome and decades of evidence from complementary field and laboratory experiments.
By comparing killifish collected from a well-studied pollution-adapted population to killifish collected at two reference sites, we identified multiple locations across the Fundulus genome that are associated with PAH resistance. The overlap in scaffolds that were found to be significantly differentiated in both pairwise comparisons of polluted and non-polluted sites (but not between the two non-polluted sites, Figure 2C) is strong evidence for a genetic basis to the impressive evolution of PAH resistance in killifish and should address concerns regarding the risk of false positives in SNP outlier analyses (Narum and Hess, 2011). These findings offer additional support for our prior understanding of the heritability of these adaptations (Clark et al., 2014; Nacci et al., 2010), as well as offer further insights into which underlying physiological pathways and processes are involved, and potentially identify novel pathway linkages. Within the unique context provided by both decades of toxicological research (reviewed by Di Giulio and Clark, 2015) as well as genomic resources released within the past year (Reid et al., 2017; Reid et al., 2016), we are further able to describe the physiological relevance of these outlier regions. Many of our findings match findings recently reported by Reid et al. (2016) who conducted low-coverage whole genome re-sequencing of four paired resistant and sensitive killifish populations including AW and KC. However, our findings are not completely overlapping, and each approach identified candidate genes that were not found using the alternate approach. An analysis of the two studies thus provides broader insights into the comparability and utility of reduced-representation and re-sequencing genomic approaches. While we acknowledge the limitations of RADseq highlighted previously by Lowry et al. (2017a,b), including a lack of knowledge on linkage disequilibrium for our study species and potential alternate hypotheses (e.g., neutral, demographic processes) that could explain some of the observed variation, we argue that the observed agreement in results with recent whole genome re-sequencing efforts provides greater confidence in the utility of our RADseq approach to identify signatures of adaptation. The present RADseq study used different fish collected independently from Reid et al. (2016) and form part of a larger effort to describe the landscape genomics of killifish within the ER.
Among the candidate genes found in scaffold regions showing significantly elevated smoothed FST, we see three major pathways and processes that could play a role in the resistant phenotype: the AHR pathway, cardiac structure, and cardiac function. We acknowledge that our selection of candidate genes from large significantly differentiated genomic regions is biased by our current understanding of the biology of our study system. Any of the genes located between significantly elevated FST values could hypothetically be a target of selection for PAH resistance. However, we choose to highlight and discuss the subset of these genes for which we predict the greatest likelihood of their involvement in the adapted phenotype based on extensive background knowledge of the biology, toxicology, and physiology of our study system. The systematically random genome-wide nature of RADseq analysis removes the bias inherent in targeted studies (like Proestou et al. (2014) and Reitzel et al. (2014)), or those using microarrays enriched in cardiac genes (Jayasundara et al., 2015), but re-introduces it on a much more restricted collection of genes produced from relatively blind analysis.
4.1. AHR Pathway
The aryl hydrocarbon receptor (AHR) pathway has long been a target of study in adapted killifish from the ER as well as other chronically PAH- and PCB-polluted sites along the Atlantic coast. The AHR pathway is one of the primary means of metabolizing PAHs and one of the primary ways relatively benign PAHs are activated into more reactive and toxic PAH metabolites. Activation of this pathway can lead to the transcription of several xenobiotic metabolizing enzymes, including several cytochrome p450s (CYPs) and glutathione s-transferases (GSTs) (Nebert et al., 2000). The recalcitrance of AW killifish to upregulate CYP1a following treatment with AHR-agonist PAHs was one of the original hallmarks of the adapted phenotype that suggested involvement of the AHR pathway (Van Veld & Westbrook, 1995). More recently, Clark et al. (2010) showed that morpholino knockdown of AHR2 (actually AHR2a), but not AHR1 (actually AHR1a), was protective against PAH exposure in embryos of killifish collected from the clean KC site. That work and other recent reports (e.g. Reid et al., 2016) have suggested that it is adaptation of AHR2 itself, or co-factors directly affecting the activity of AHR2, that is central to PAH (and dioxin-like compounds, DLC) resistance, rather than genes regulated by the AHR pathway.
We found two canonical AHR complex members in our candidate gene regions, the aryl hydrocarbon receptor interacting protein (AIP, also known as XAP-2 or ARA9) and the aryl hydrocarbon receptor nuclear translocator 1 (LOC105928492, ARNT1c-like) (Table 1, Figure 4); one gene known to influence the AHR pathway, the Nuclear Receptor Coactivator 2 (NCOA2); and two genes controlled, at least in part, by AHR pathway activation and that play a direct role in xenobiotic metabolism, GST theta 1 (LOC105916512, GSTT1-like), and cytochrome B5 (LOC105917072; CYB5-like) (Figure 5). These results agree well with those of Reid et al. (2016) who found AIP, ARNT1c, and GSTT1 as candidate genes in significant regions.
AIP and ARNT bind to AHR in the cytoplasm and nucleus, respectively, and the latter enables AHR to act as a transcription factor that binds DNA at xenobiotic response elements (XREs). Since both AIP and ARNT interact with the pathway before and during the transcription step, mutations in these genes could affect the activity of the entire pathway. NCOA2 is a transcriptional coregulator that can interact with and regulate the AHR pathway (Tsai et al., 2015) and may play a role in heart failure in other systems (Yu et al., 2013). Several other studies have highlighted the likelihood of members of the AHR-ARNT complex as targets of selection and searched for evidence within this pathway (Reitzel et al., 2014). Candidate gene studies of AHR-related genes in several other adapted killifish populations in New England (USA) have found SNPs with significantly different allele frequencies between resistant and susceptible populations in AHR1, AHR2 (Hahn et al., 2004; Proestou et al., 2014) and the aryl hydrocarbon receptor repressor (AHRR) (Reitzel et al., 2014). Although both ARNT1c and AIP (ARA9) were sequenced by Proestou et al. (2014), no significant difference in SNP allele frequencies were reported in either case. Likewise, contrary to Proestou et al. (2014), the genomic neighborhoods around AHR1a-AHR2a, AHR1b-AHR2b, or AHRR were not identified as candidate genes in our study. No SNPs passed our filtering on scaffold 217, the location of the AHR1b-AHR2b tandem, and only a few SNPs with non-significant FST values were identified on scaffolds with the AHR1a-AHR2a tandem and with AHRR.
Two major differences between our findings and those of Reid et al. (2016) in relation to the AHR pathway is the absence in our data of any strong signs of selection around AHR2a and CYP1a. Reid et al. (2016) noted that AW-KC especially shows strong selection of a critical deletion in the tandem paralogs AHR2a and AHR1a. Since our data filtering criteria require markers to be found in both populations of a given pairwise comparison, deletions like this would not be detected. Our data do include two SNPs with significant non-smoothed FST values on the scaffold with CYP1a (scaffold 9923) from each of our pairwise comparisons. However, smoothed FST values were not significant in either comparison and so the scaffold was not considered in our selection of candidate genes. The recalcitrance to induce CYP1a in adapted killifish makes it a logical target of adaptation, even if the knockdown of CYP1a is not protective against DLC cardiotoxicity and in fact exacerbates PAH toxicity (Matson et al., 2008). Several other studies have also found significant differentiation at SNPs in CYP1a genes between killifish populations from contaminated and uncontaminated sites (Proestou et al., 2014; Williams and Oleksiak, 2008, 2011).
The two xenobiotic metabolizing enzyme candidate genes we identified are a GST, of the theta subfamily (also reported by Reid et al., 2016), and cytochrome b5 (CYB5). GSTs generally facilitate xenobiotic excretion through linkage to the tripeptide glutathione, while CYB5 functions as an electron carrier that helps minimize the incomplete oxidation of xenobiotics by CYPs (Hildebrandt and Estabrook, 1971) and thus reduces the production of superoxide radicals. Carney et al. (2006)) reported no change in GSTT1 expression and a non-significant increase in CYB5 expression in zebrafish dosed with TCDD, so they do not appear to play a direct role in DLC cardiotoxicity. Rather, GSTT1 may play a role in the carcinogenicity of PAHs and help explain some of the lesser studied aspects of the adapted phenotype, such as a resistance to DNA damage and cancer (Jung et al., 2009; Wills et al., 2010). The toxicities, and specifically carcinogenicities, of PAHs are usually a function of their metabolites, rather than the parent compounds themselves. Thus, individual variations in the expression of these xenobiotic metabolizing enzymes could be a significant factor in how and how well an individual handles exposure to anthropogenic pollutants like PAHs (Henderson et al., 2014). Association studies of GSTs and cancer in other systems (Chen et al., 1996; Chen et al., 2006) suggest that adaptations to the GSTT1 gene could play a role in minimizing the carcinogenicity of PAH metabolites, and help explain the increased resistance to DNA damage (Jung et al., 2009; Wills et al., 2010) and liver cancers observed in the adapted AW killifish population (Wills et al., 2010). Interestingly, deletion of the GSTT1 gene in humans confers some resistance to PAH-induced DNA damage (Garte et al., 2007). Although several toxicological studies of GST activity and gene expression have been performed in relationship to PAH toxicity/resistance in fish (Bello et al., 2001; Garner and Di Giulio, 2012; Paetzold et al., 2009; Van Tiem and Di Giulio, 2011), none has studied the theta 1 variant. Further exploration of the roles CYB5 and GSTT1 play in adapted killifish should be interesting.
Several of these candidate genes also play important and potentially relevant roles beyond their contributions to the AHR pathway. For example, ARNT is also involved in activating responses to hypoxic conditions, and therefore, changes to ARNT gene sequence could secondarily affect hypoxic tolerance. Killifish, in particular among teleosts, show great tolerance to environmental extremes including hypoxia (Stierhoff et al., 2003). Yet, co-exposure to certain anthropogenic pollutants and hypoxic conditions can negatively impact the activation of each pathway (Matson et al., 2008; Prasch et al., 2004; Vorrink and Domann, 2014), resulting in additive or synergistic toxicity (Fleming and Di Giulio, 2011) that can have both individual and ecological level impacts (e.g., negative reproductive consequences (Hedgpeth and Griffitt, 2016)). Furthermore, certain CYB5 variants in humans result in heritable methemoglobinemia type IV (Hegesh et al., 1986), a condition that causes a buildup of methemoglobin, and a subsequent reduction in oxygen carrying capacity. Similar outcomes of CYB5 gene changes in AW killifish could contribute to the physiological costs of the Elizabeth River adaptations reported in (Jayasundara et al., 2017).
4.2. Cardiac Structure and Function
Until only recently, it was poorly understood whether and how AHR activation, specifically, leads to developmental cardiac abnormalities in embryonic fish. Structurally, exposed fish embryo hearts resemble hearts with dilated cardiomyopathy. Insights into the mechanisms underlying this toxicity were provided by (Lanham et al., 2014), who showed that TCDD (dioxin)- induced AHR activation in zebrafish cardiomyocites leads directly to most of the cardiac abnormalities classic to dioxin (and PAH) toxicity. Yet, mechanisms independent of the AHR pathway have also been shown to cause PAH cardiotoxity (e.g. Incardona and Scholz, 2016). In order to become fully resistant to chronic PAH toxicity, fish might therefore need both AHR-dependent and AHR-independent adaptations. Since cardiac teratogenesis is a classic toxic phenotype, genes involved with developmental cardiac structure and cardiac function, which are often closely linked, seem likely candidates for adaptation primarily or secondarily involved with PAH resistance.
Within our candidate gene regions, we found two key cardiac and muscle genes that could affect cardiac structural development under polluted conditions, the genes encoding the giant molecular spring protein titin (TTN) and cardiac-specific nebulette (NEBL) (Figure 5C). Polymorphisms in both TTN and NEBL are associated with dilated cardiomyopathy (Arimura et al., 2000; Purevjav et al., 2010). In striated muscle, titin functions to anchor myosin to the Z-disc and is responsible for the elastic nature of muscle. In zebrafish, two titin orthologs exist, ttna and ttnb, with ttna being the earliest gene expressed in the developing cardiac sarcomere and required for proper sarcomere assembly (Seeley et al., 2007). It should be noted that TTN exists on the one scaffold (200) that showed significant differences in all three pair-wise comparisons (Table 1, Figure 2C). Nebulette, which is found in the Z-discs of cardiac muscle (Moncman and Wang, 1995), binds and regulates the stability of actin thin filaments and plays an essential role in maintaining proper structure and function of cardiac sarcomeres (Moncman and Wang, 2002). Since mutations in these genes are known to cause dilated cardiomyopathy, any mutations in AW killifish would need to instill resistance, for example through increased expression or alternative splicing to a more resistant isoform. To our knowledge, this is the first suggestion that nebulette could play a role in pollution resistance in killifish.
In addition to the evident structural deformities, PAHs are also known to cause cardiac arrhythmias, bradycardia (slow heart rate), and reduced contractility in fish (Brette et al., 2014; Carls et al., 1999; Heintz et al., 1999; Incardona et al., 2014). This functional toxicity is attributed to PAH interference with various ion channels, leading to atrio-ventricular conduction block (Brette et al., 2017). Complex PAH mixtures of weathered crude oil directly block delayed rectifying potassium channels and disrupt calcium cycling in developing tuna embryos (Brette et al., 2014; Brette et al., 2017). AHR activation by TCDD exposure in zebrafish results in differential expression of several ion channels and ion regulatory genes (Carney et al., 2006). Not only are ion channels crucial to proper cardiac function, they are also necessary for normal developmental cardiac structure. Certain mutations in zebrafish ion channel genes, specifically the Na+/Ca2+ exchanger (NCX1) and Ca2+ pump (SERCA2) result in structural abnormalities similar to PCB and PAH exposure (Frey et al., 2000). Within our eight candidate regions, there exist several potassium channels covering many portions of the cardiac action potential, including the delayed rectifier potassium channel (KCNB2), the inward rectifying potassium channel (KCNJ14), the potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (LOC10598494; HCN4-like), and the small conductance calcium-activated potassium channel protein 2 (LOC10598426; KCNN4-like) (Figure 4B). KCNB2 was also reported as a candidate gene under resequencing analysis (Reid et al., 2016), along with KCNC3, which we do not report. Unlike the mutations that cause structural abnormalities (Frey et al., 2000), other mutations could result in ion channels that remain functional under polluted conditions.
A final major class of proteins that is crucial to proper cardiac structure and function, and which was identified within our candidate regions (IGTB5 and IGTA10) (Figure 4B), is the integrins. Integrins are transmembrane heterodimer (α and β) glycoprotein receptors that function as transmembrane mechanotransducers linking the extracellular matrix to the intracellular actin cytoskeleton. These proteins play essential roles in cell adhesion, cell migration and proliferation, and cell signaling (reviewed by Israeli-Rosenberg et al. (2014). In relation to adapted killifish, we see three possible roles for integrins: (1) development of proper cardiac and vascular structures (Le Gat et al., 2001); (2) controlling Ca2+ and K+ ion channels (Gui et al., 2010); and (3) influencing tumor progression and metastasis (Desgrosellier and Cheresh, 2010).
5. Conclusion
The wealth of toxicological, biochemical, gene expression, and knockdown studies of the several resistant killifish populations studied for the past twenty years provides the key background context needed to interpret genome-wide studies such as this one. Our ability to interpret biological significance of molecular findings is further greatly enhanced by the recently released Fundulus heteroclitus annotated genome. Thus, these natural experiments of pollution adaptation are ideal for the application of genome-wide approaches, such as RADseq, to gain significant understanding of the genotypes underlying the phenotypes and mechanisms of toxicity and resistance. In fact, they could be useful natural experiments on which to test multiple traditional and cutting-edge genomic methodologies to determine better approaches for understanding lesser known systems or species.
One benefit of employing genome-wide studies as opposed to candidate gene studies is the ability to identify novel and less obvious mechanisms that can ultimately expand our understanding of the pathways involved. Here we have used RADseq to identify regions of the genome showing signs of selection in killifish from one highly polluted site at AW in relation to one historical control site at KC and a relatively clean site at MC. Our conservative results suggest at least eight genomic regions show significant signs of selection. These regions contain several genes that make biological sense in a resistant animal including canonical AHR pathway members AIP and ARNT1c; genes associated with the AHR pathway such as GSTT1, CYB5 and NCOA2; genes related to cardiac structure including titin and nebulette; and ion channel and integrin genes related to cardiac function. Many of our findings are consistent with prior knowledge from other toxicological and molecular work, and in agreement with findings recently reported from a low-coverage whole genome re-sequencing study (Reid et al. 2016). Considering these findings together, we propose a scenario in which AHR-related genetic adaptations directly decrease the functionality of the pathway and thus reduce (or prevent) bioactivation of PAHs and consequently reduce direct toxicity. Other complementary genetic adaptations could help the heart, specifically, become more resistant to direct and indirect effects of PAHs or could be compensatory adaptations to overcome challenges introduced by AHR-related mutations.
Acknowledgements
We would like to thank members of the Di Giulio lab for assistance collecting and processing fish and tissue samples and especially to Dr. N. Jayasundara for review and advice and to S. Blinebry for RAD library prep assistance. This work was funded by the National Institutes of Environmental Health and Superfund Research Program (P42-ES-10356). The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the US EPA.
Footnotes
Data Accessibility
The killifish genome assembly version 3.0.2 can be found on NCBI (GCF_000826765.1). All sequencing reads can be found on the NCBI Sequence Read Archive (XXXXXXX).
References
- Andrews K, Good JM, Miller MR, Luikart G, Hohenlohe PA, 2016. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet 17, 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura T, Nakamura T, Hiroi S, Satoh M, Takahashi M, Ohbuchi N, Ueda K, Nouchi T, Yamaguchi N, Akai J, Matsumori A, Sasayama S, Kimura A, 2000. Characterization of the human nebulette gene: a polymorphism in an actin-binding motif is associated with nonfamilial idiopathic dilated cardiomyopathy. Hum. Genet 107, 440–451. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA, 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PloS One 3, e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME, 2001. Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: In vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicol. Sci 60, 77–91. [DOI] [PubMed] [Google Scholar]
- Billiard SM, Hahn ME, Franks DG, Peterson RE, Bols NC, Hodson PV, 2002. Binding of polycyclic aromatic hydrocarbons (PAHs) to teleost aryl hydrocarbon receptors (AHRs). Comp. Biochem. Physiol. B: Biochem. Mol. Biol 133, 55–68. [DOI] [PubMed] [Google Scholar]
- Billiard SM, Meyer JN, Wassenberg DM, Hodson PV, Di Giulio RT, 2008. Nonadditive effects of PAHs on early vertebrate development: mechanisms and implications for risk assessment. Toxicol. Sci 105, 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brette F, Machado B, Cros C, Incardona JP, Scholz NL, Block BA, 2014. Crude Oil Impairs Cardiac Excitation-Contraction Coupling in Fish. Science 343, 772–776. [DOI] [PubMed] [Google Scholar]
- Brette F, Shiels HA, Galli GLJ, Cros C, Incardona JP, Scholz NL, Block BA, 2017. A Novel Cardiotoxic Mechanism for a Pervasive Global Pollutant. Sci. Rep 7 DOI: 10.1038/srep41476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gómez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, MacLatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Bees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL, 2007. Fundulus as the premier teleost model in environmental biology: opportunities for new insights using genomics. Comp. Biochem. Physiol. D: Genomics Proteomics 2, 257–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammen KM, Schultz TF, Rosel PE, Wells RS, Read AJ, 2015. Genomewide investigation of adaptation to harmful algal blooms in common bottlenose dolphins (Tursiops truncatus). Mol. Ecol 24, 4697–4710. [DOI] [PubMed] [Google Scholar]
- Carls MG, Rice SD, Hose JE, 1999. Sensitivity of fish embryos to weathered crude oil: Part I. Low-level exposure during incubation causes malformations, genetic damage, and mortality in larval Pacific herring (Clupea pallasi). Environ. Toxicol. Chem 18, 481–493. [Google Scholar]
- Carney SA, Prasch AL, Heideman W, Peterson RE, 2006. Understanding dioxin developmental toxicity using the zebrafish model. Birth Defects Res. A 76, 7–18. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe PA, Cresko WA, Postlethwait JH, 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3 1, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Hohenlohe PA, Bassham S, Amores A, Cresko WA, 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol 22, 3124–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Hohenlohe PA, Bernatchez L, Funk WC, Andrews KR, Allendorf FW, 2017. Unbroken: RADseq remains a powerful tool for understanding the genetics of adaptation in natural populations. Mol. Ecol. Resour 17, 362–365. [DOI] [PubMed] [Google Scholar]
- Chen HW, Sandler DP, Taylor JA, Shore DL, Liu E, Bloomfield CD, Bell DA, 1996. Increased risk for myelodysplastic syndromes in individuals with glutathione transferase theta 1 (GSTT1) gene defect. Lancet 347, 295–297. [DOI] [PubMed] [Google Scholar]
- Chen YW, Bai Y, Yuan J, Chen WH, Sun JY, Wang H, Liang HS, Guo L, Yang XB, Tan H, Su YG, Wei QY, Wu TC, 2006. Association of Polymorphisms in AhR, CYP1A1, GSTM1, and GSTT1 genes with levels of DNA damage in peripheral blood lymphocytes among coke-oven workers. Cancer Epidem. Biomar 15, 1703–1707. [DOI] [PubMed] [Google Scholar]
- Clark BW, Bone AJ, Di Giulio RT, 2014. Resistance to teratogenesis by F1 and F2 embryos of PAH-adapted Fundulus heteroclitus is strongly inherited despite reduced recalcitrance of the AHR pathway. Environ. Sci. Poll. Res. Int 21, 13898–13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BW, Cooper EM, Stapleton HM, Di Giulio RT, 2013. Compound- and mixture-specific differences in resistance to PAHs and PCB-126 among Fundulus heteroclitus subpopulations throughout the Elizabeth River estuary (Virginia, USA). Environ. Sci. Tech 47, 10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BW, Di Giulio RT, 2012. Fundulus heteroclitus adapted to PAHs are cross-resistant to multiple insecticides. Ecotoxicology 21, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BW, Matson CW, Jung D, Di Giulio RT, 2010. AHR2 mediates cardiac teratogenesis of polycylic aromatic hydrocarbons and PCB-126 in Atlantic killifish (Fundulus heteroclitus). Aquat. Toxicol 99, 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML, 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet 12, 499–510. [DOI] [PubMed] [Google Scholar]
- Desgrosellier JS, Cheresh DA, 2010. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio RT, Clark BW, 2015. The Elizabeth River story: a case study in evolutionary toxicology. J. Toxicol. Environ. Heal. B 18, 259–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunson WA, Fricano P, Sadinski WJ, 1993. Variation in tolerance to abiotic stresses among sympatric salt marsh fish. Wetlands 13, 16–24. [Google Scholar]
- Ellegren H, 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol 29, 51–63. [DOI] [PubMed] [Google Scholar]
- Fleming CR, Di Giulio RT, 2011. The role of CYP1A inhibition in the embryotoxic interactions between hypoxia and polycyclic aromatic hydrocarbons (PAHs) and PAH mixtures in zebrafish (Danio rerio). Ecotoxicology 20, 1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey N, McKinsey TA, Olson EN, 2000. Decoding calcium signals involved in cardiac growth and function. Nat. Med 6, 1221–1227. [DOI] [PubMed] [Google Scholar]
- Garner LVT, Di Giulio RT, 2012. Glutathione transferase pi class 2 (GSTp2) protects against the cardiac deformities caused by exposure to PAHs but not PCB-126 in zebrafish embryos. Comp. Biochem. Physiol. C: Toxicol. Pharmacol 155, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garte S, Taioli E, Raimondi S, Paracchini V, Binkova B, Sram RJ, Kalina I, Popov TA, Singh R, Farmer PB, 2007. Effects of metabolic genotypes on intermediary biomarkers in subjects exposed to PAHS: Results from the EXPAH study. Mutat. Res.- Fund. Mol. M 620, 7–15. [DOI] [PubMed] [Google Scholar]
- Gonzalez RJ, Mason CH, Dunson WA, 1989. Anomalous tolerance to low pH in the estuarine killifish Fundulus heteroclitus. Comp. Biochem. Physiol. C: Comp. Pharmacol 94, 169–172. [Google Scholar]
- Gui PC, Chao JT, Wu X, Yang Y, Davis GE, Davis MJ, 2010. Coordinated Regulation of Vascular Ca2+ and K+ Channels by Integrin Signaling, in: Becchetti A, Arcangeli A (Eds.), Integrins and Ion Channels: Molecular Complexes and Signaling, pp. 69–79. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Franks DG, Merson RR, 2004. Aryl hydrocarbon receptor polymorphisms and dioxin resistance in Atlantic killifish (Fundulus heteroclitus). Pharmacogenetics 14, 131–143. [DOI] [PubMed] [Google Scholar]
- Hargis WG Jr., Zwerner DE, Thoney DA, Kelly KL, Warinner III JE, 1989. Neoplasms in mummichogs from the Elizabeth River, Viriginia. J. Aquat. Anim. Heal 1, 165–172. [Google Scholar]
- Hedgpeth BM, Griffitt RJ, 2016. Simultaneous exposure to chronic hypoxia and dissolved polycyclic aromatic hydrocarbons results in reduced egg production and larval survival in the sheepshead minnow (Cyprinodon variegatus). Environ. Toxicol. Chem 35, 645–651. [DOI] [PubMed] [Google Scholar]
- Hegesh E, Hegesh J, Kaftory A, 1986. Congenital Methemoglobinemia with a Deficiency of Cytochrome-B5. New Eng. J. Med 314, 757–761. [DOI] [PubMed] [Google Scholar]
- Heintz RA, Short JW, Rice SD, 1999. Sensitivity of fish embryos to weathered crude oil: Part II. Increased mortality of pink salmon (Oncorhynchus gorbuscha) embryos incubating downstream from weathered Exxon Valdez crude oil. Environ. Toxicol. Chem 18, 494–503. [Google Scholar]
- Henderson CJ, McLaren AW, Wolf CR, 2014. In Vivo Regulation of Human Glutathione Transferase GSTP by Chemopreventive Agents. Cancer Res. 74, 4378–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt A, Estabrook RW, 1971. Evidence for pariticpation of cytochrome B5 in hepatic microsomal mixed-function oxidation reactions. Arch. Biochem. Biophys 143, 66-+. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA, 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet 6, e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona JP, Gardner LD, Linbo TL, Brown TL, Esbaugh AJ, Mager EM, Stieglitz JD, French BL, Labenia JS, Laetz CA, Tagal M, Sloan CA, Elizur A, Benetti DD, Grosell M, Block BA, Scholz NL, 2014. Deepwater Horizon crude oil impacts the developing hearts of large predatory pelagic fish. P. Natl. Acad. Sci. USA 111, E1510–E1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona JP, Scholz NL, 2016. The influence of heart developmental anatomy on cardiotoxicity-based adverse outcome pathways in fish. Aquat. Toxicol 177, 515–525. [DOI] [PubMed] [Google Scholar]
- Israeli-Rosenberg S, Manso AM, Okada H, Ross RS, 2014. Integrins and Integrin-Associated Proteins in the Cardiac Myocyte. Circ. Res 114, 572–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasundara N, Fernando PW, Osterberg JS, Cammen KM, Schultz TF, Di Giulio RT, 2017. Cost of Tolerance: Physiological Consequences of Evolved Resistance to Inhabit a Polluted Environment in Teleost Fish Fundulus heteroclitus. Environ. Sci. Tech 51, 8763–8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasundara N, Garner LV, Meyer JN, Erwin KN, Di Giulio RT, 2015. AHR2-Mediated Transcriptomic Responses Underlying the Synergistic Cardiac Developmental Toxicity of PAHs. Toxicol. Sci 143, 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, Birney E, Searle S, Schmutz J, Grimwood J, Dickson MC, Myers RM, Miller CT, Summers BR, Knecht AK, Brady SD, Zhang H, Pollen AA, Howes T, Amemiya C, Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team, Lander ES, Di Palma F, Lindblad-Toh K, Kingsley DM, 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Cho Y, Collins LB, Swenberg JA, Di Giulio RT, 2009. Effects of benzo[a]pyrene on mitochondrial and nuclear DNA damage in Atlantic killifish (Fundulus heteroclitus) from a creosote-contaminated and reference site. Aquat. Toxicol 95, 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BLF, Sulentic CEW, Holsappel MP, Kaminski NE, 2013. Toxic responses of the immune system, in: Klaasen CD (Ed.), Casarett and Doull’s toxicology: The basic science of toxicology. McGraw Hill, New York, NY, pp. 559–638. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S, 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanham KA, Plavicki J, Peterson RE, Heideman W, 2014. Cardiac Myocyte-Specific AHR Activation Phenocopies TCDD-Induced Toxicity in Zebrafish. Toxicol. Sci 141, 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gat L, Bonnel S, Gogat K, Brizard M, Van Den Berghe L, Kobetz A, Gadin S, Dureau P, Dufier JL, Abitbol M, Menasche M, 2001. Prominent beta-5 gene expression in the cardiovascular system and in the cartilaginous primordiae of the skeleton during mouse development. Cell Commun. Adhes 8, 99–112. [DOI] [PubMed] [Google Scholar]
- Lotrich VA, 1975. Summer home range and movements of Fundulus heteroclitus (Pisces: Cyprinodontidae) in a tidal creek. Ecology 56, 191–198. [Google Scholar]
- Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, Storfer A, 2017a. Responsible RAD: Striving for best practices in population genomic studies of adaptation. Mol. Ecol. Resour 17, 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, Storfer A, 2017b. Breaking RAD: an evaluation of the utility of restriction site-associated DNA sequencing for genome scans of adaptation. Mol. Ecol. Resour 17, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luch A, 2005. The carcinogenic effects of polycylic aromatic hydrocarbons. Imperial College Press, London, UK. [Google Scholar]
- Matson CW, Clark BW, Jenny MJ, Fleming CR, Hahn ME, Di Giulio RT, 2008. Development of the morpholino gene knockdown technique in Fundulus heteroclitus: A tool for studying molecular mechanisms in an established environmental model. Aquat. Toxicol 87, 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney GJ, Larson WA, Seeb LW, Seeb JE, 2017. RADseq provides unprecedented insights into molecular ecology and evolutionary genetics: comment on Breaking RAD by Lowry et al. (2016). Mol. Ecol. Resour 17, 356–361. [DOI] [PubMed] [Google Scholar]
- Menzel DW, Case J, 1977. Concept and design - Controlled ecosystem pollution experiment. Bull. Mar. Sci 27, 1–7. [Google Scholar]
- Meyer J, Di Giulio R, 2002. Patterns of heritability of decreased EROD activity and resistance to PCB 126-induced teratogenesis in laboratory-reared offspring of killifish (Fundulus heteroclitus) from a creosote-contaminated site in the Elizabeth River, VA, USA. Mar. Environ. Res 54, 621–626. [DOI] [PubMed] [Google Scholar]
- Meyer J, Di Giulio RT, 2003. Heritable adaptation and fitness costs in killfish (Fundulus heteroclitus) inhabiting a polluted estuary. Ecol. Appl 13, 490–503. [Google Scholar]
- Meyer J, Nacci DE, Di Giulio RT, 2002. Cytochrome P4501A (CYP1A) in killifish (Fundulus heteroclitus): heritability of altered expression and relationship to survival in contaminated sediments. Toxicol. Sci 68, 69–81. [DOI] [PubMed] [Google Scholar]
- Meyer J, Smith JD, Winston GW, Di Giulio RT, 2003a. Antioxidant defenses in killifish (Fundulus heteroclitus) exposed to contaminated sediments and model prooxidants: short-term and heritable responses. Aquat. Toxicol 65, 377–395. [DOI] [PubMed] [Google Scholar]
- Meyer J, Wassenberg DM, Karchner SI, Hahn ME, Di Giulio RT, 2003b. Expression and inducibility of aryl hydrocarbon receptor pathway genes in wild-caught killifish (Fundulus heteroclitus) with different contaminant-exposure histories. Environ. Toxicol. Chem 22, 2337–2343. [DOI] [PubMed] [Google Scholar]
- Moncman CL, Wang K, 1995. Nebulette - A 107 KD Nebulin-Like Protein in Cardiac-Muscle. Cell Motil. Cytoskel 32, 205–225. [DOI] [PubMed] [Google Scholar]
- Moncman CL, Wang K, 2002. Targeted disruption of nebulette protein expression alters cardiac myofibril assembly and function. Exp. Cell Res 273, 204–218. [DOI] [PubMed] [Google Scholar]
- Nacci DE, Champlin D, Jayaraman S, 2010. Adaptation of estuarine fish Fundulus heteroclitus (Atlantic killifish) to polychlorinated biphenyls (PCBs). Estuaries Coasts 33, 853–864. [Google Scholar]
- Nacci DE, Coiro L, Champlin D, Jayaraman S, McKinney R, Gleason TR, Munns WR Jr., Specker JL, Cooper KR, 1999. Adapations of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Mar. Biol 134, 9–17. [Google Scholar]
- Narum SR, Hess JE, 2011. Comparison of F-ST outlier tests for SNP loci under selection. Mol. Ecol. Resour 11, 184–194. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP, 2000. Role of the aromatic hydrocarbon receptor and Ah gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol 59, 65–85. [DOI] [PubMed] [Google Scholar]
- Nordlie FG, 2006. Physiochemical environments and tolerances of cyprinodontoid fishes found in estuaries and salt marshes of eastern North America. Rev. Fish Biol. Fish 16, 51–106. [Google Scholar]
- Ownby DR, Newman MC, Mulvey M, Vogelbein WK, Unger MA, Arzayus LF, 2002. Fish (Fundulus heteroclitus) populations with different exposure histories differ in tolerance of creosote-contaminated sediments. Environ. Toxicol. Chem 21, 1897–1902. [PubMed] [Google Scholar]
- Paetzold SC, Ross NW, Richards RC, Jones M, Hellou J, Bard SM, 2009. Up-regulation of hepatic ABCC2, ABCG2, CYP1A1 and GST in multixenobiotic-resistant killifish (Fundulus heteroclitus) from the Sydney Tar Ponds, Nova Scotia, Canada. Mar. Environ. Res 68, 37–47. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE, 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PloS One 7, e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch AL, Andreasen EA, Peterson RE, Heideman W, 2004. Interactions between 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and hypoxia signaling pathways in zebrafish: Hypoxia decreases responses to TCDD in zebrafish embryos. Toxicol. Sci 78, 68–77. [DOI] [PubMed] [Google Scholar]
- Proestou DA, Flight P, Champlin D, Nacci D, 2014. Targeted approach to identify genetic loci associated with evolved dioxin tolerance in Atlantic Killifish (Fundulus heteroclitus). BMC Evol. Biol 14 DOI: 10.1186/1471-2148-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purevjav E, Varela J, Morgado M, Kearney DL, Li H, Taylor MD, Arimura T, Moncman CL, McKenna W, Murphy RT, Labeit S, Vatta M, Bowles NE, Kimura A, Boriek AM, Towbin JA, 2010. Nebulette Mutations Are Associated With Dilated Cardiomyopathy and Endocardial Fibroelastosis. J. Am. Coll. Cardiol 56, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid NM, Jackson CE, Gilbert D, Minx P, Montague MJ, Hampton TH, Helfrich LW, King BL, Nacci DE, Aluru N, Karchner SI, Colbourne JK, Hahn ME, Shaw JR, Oleksiak MF, Crawford DL, Warren WC, Whitehead A, 2017. The Landscape of Extreme Genomic Variation in the Highly Adaptable Altantic Killifish. Genome Biol. Evol 9, 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid NM, Proestou DA, Clark BW, Warren WC, Colbourne JK, Shaw JR, Karchner SI, Hahn ME, Nacci D, Oleksiak MF, Crawford DL, Whitehead A, 2016. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science 354, 1305–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Karchner SI, Franks DG, Evans BR, Nacci D, Champlin D, Vieira VM, Hahn ME, 2014. Genetic variation at aryl hydrocarbon receptor (AHR) loci in populations of Atlantic killifish (Fundulus heteroclitus) inhabiting polluted and reference habitats. BMC Evol. Biol 14 DOI: 10.1186/1471-2148-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JV, Bradfield CA, 1996. Ah receptor signaling pathways. Annual Review of Cell and Dev. Biol 12, 55–89. [DOI] [PubMed] [Google Scholar]
- Seeley M, Huang W, Chen ZY, Wolff WO, Lin XY, Xu XL, 2007. Depletion of zebrafish titin reduces cardiac contractility by disrupting the assembly of Z-discs and A-bands. Circul. Res 100, 238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KJ, Able KW, 2003. Dissolved oxygen dynamics in salt marsh pools and its potential impacts on fish assemblages. Mar. Ecol. Prog. Ser 258, 223–232. [Google Scholar]
- Stierhoff KL, Targett TE, Grecay PA, 2003. Hypoxia tolerance of the mummichog: the role of access to the water surface. J. Fish Biol 63, 580–592. [Google Scholar]
- Tsai CH, Li CH, Liao PL, Cheng YW, Lin CH, Huang SH, Kang JJ, 2015. NcoA2-Dependent Inhibition of HIF-1 alpha Activation Is Regulated via AhR. Toxicol. Sci 148, 517–530. [DOI] [PubMed] [Google Scholar]
- Van Tiem LA, Di Giulio RT, 2011. AHR2 knockdown prevents PAH-mediated cardiac toxicity and XRE- and ARE-associated gene induction in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol 254, 280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veld PA, Westbrook DJ, 1995. Evidence for depression of cytochrome P4501A in a population of chemically resistant mummichog (Fundulus heteroclitus). Environ. Sci 3, 221–234. [Google Scholar]
- Vogelbein W, Fournie JW, Van Veld PA, Huggett RJ, 1990. Hepatic neoplasms in the mummichog Fundulus heteroclitus from a creosote-contaminated site. Cancer Res. 50, 5978–5986. [PubMed] [Google Scholar]
- Vogelbein W, Unger MA, 2003. The Elizabeth River monitoring program 2001–2002: Association between mummichog liver histopathology and sediment chemical contamination Final report to the Viriginia Department of Environmental Quality, Gloucester Point, VA. [Google Scholar]
- Vorrink SU, Domann FE, 2014. Regulatory crosstalk and interference between the xenobiotic and hypoxia sensing pathways at the AhR-ARNT-HIF1 alpha signaling node. Chem-Biol. Interact 218, 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Dickhut RM, Chisholm-Brause C, 2004. Polcyclic aromatic hydrocarbons in a highly industrialized urban estuary: inventories and trends. Environ. Toxicol. Chem 23, 2655–2664. [DOI] [PubMed] [Google Scholar]
- Wassenberg DM, Di Giulio RT, 2004a. Synergistic embryotoxicity of polycyclic aromatic hydrocarbon aryl hydrocarbon receptor Agonists with cytochrome P4501A inhibitors in Fundulus heteroclitus. Environ. Health Perspect 112, 1658–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenberg DM, Di Giulio RT, 2004b. Teratogenesis in Fundulus heteroclitus embryos exposed to a creosote-contaminated sediment extract and CYP1A inhibitors. Mar. Environ. Res 58, 163–168. [DOI] [PubMed] [Google Scholar]
- Weir B, 1996. Population structure, Genetic Data Analysis II. Sinauer Associates, Sunderland, MA, pp. 161–201. [Google Scholar]
- Williams LM, Oleksiak MF, 2008. Signatures of selection in natural populations adapted to chronic pollution. BMC Evol. Biol 8 DOI: 10.1186/1471-2148-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Oleksiak MF, 2011. Ecologically and evolutionarily important SNPs identified in natural populations. Mol. Biol. Evol 28, 1817–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills LP, Jung D, Koehrn K, Zhu S, Willett KL, Hinton DE, DI Giulio RT, 2010. Comparative chronic liver toxicity of benzo[a]pyrene in two populations of the Atlantic killifish (Fundulus heteroclitus) with different exposure histories. Environ. Health Perspect 118, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Roy NK, Loftus M, Chambers RC, Franks DG, Hahn ME, 2011. Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River. Science 331, 1322–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CM, Marshall WS, 1994. Ion balance, acid-base regulation, and chloride cell function in the common killifish, Fundulus heteroclitus: a euryhaline estuarine teleost. Estuaries 17, 34–52. [Google Scholar]
- Yu B, Barbalic M, Brautbar A, Nambi V, Hoogeveen RC, Tang WH, Mosley TH, Rotter JI, deFilippi CR, O’Donnell CJ, Kathiresan S, Rice K, Heckbert SR, Ballantyne CM, Psaty BM, Boerwinkle E, Consortium CA, 2013. Association of Genome-Wide Variation With Highly Sensitive Cardiac Troponin-T Levels in European Americans and Blacks A Meta-Analysis From Atherosclerosis Risk in Communities and Cardiovascular Health Studies. Circ-Cardiovasc. Gene 6, 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]