Abstract
Background
Oral squamous cell carcinoma (OSCC) is the most frequent oral malignancy. Recent studies have revealed that long non-coding RNA (lncRNA) PVT1 plays important roles in the pathogenesis of various cancers. However, the functional roles of PVT1 in OSCC progression and cisplatin resistance have not been elucidated.
Material and Methods
In this study, PVT1 expression level in cisplatin-sensitive and cisplatin-resistant OSCC tissues and cell lines was determined using qRT-PCR. Gain-of-function and loss-of-function assays were performed to explore the biological roles of PVT1 in OSCC cell proliferation and cisplatin resistance. Western blot, luciferase reporter assay and bioinformatics analysis were employed to investigate the underlying mechanism of PVT1 in OSCC progression.
Results
Here, we found that PVT1 was frequently up-regulated in cisplatin-resistant tissues and cell lines and strongly correlated with worse overall survival. Functional studies showed that PVT1 promoted OSCC cell proliferation and cisplatin resistance. Mechanistic investigation revealed that PVT1 could positively regulate HIF1a expression via its competing endogenous RNA (ceRNA) activity on miR-194-5p. In addition, miR-194-5p conversely correlated with PVT1 and HIF1a expression in OSCC samples. More importantly, HIF1a knock-down or miR-194-5p overexpression reversed PVT1-induced promotion of OSCC cell proliferation and cisplatin resistance.
Conclusion
Our results indicated that PVT1 functions as an oncogene involved in OSCC cell proliferation and cisplatin-resistance and may serve as a novel therapeutic target for OSCC treatment.
Keywords: oral squamous cell carcinoma, miR-194-5p, HIF1a, cisplatin resistance
Introduction
Long non-coding RNAs (lncRNAs) are a class of transcribed RNAs with a length of more than 200 nucleotides that are commonly lack of protein-coding potential.1 Accumulating evidence suggests that lncRNAs are involved in multiple biological processes, including tumor initiation, metastasis, autophagy and chemoresistance.2 Dysregulated lncRNAs have been frequently observed in various diseases, including cancer, and have also been identified as valuable biomarkers for diagnosis and prognosis.3 Recently, several lncRNAs were reported to be capable of encoding for small peptide, such as LINC00961, LOC100507537 and LINC00948.4–6
Emerging evidence demonstrates that lncRNAs can act as a competing endogenous RNA (ceRNA) to regulate gene expression through decoying microRNAs (miRNAs).7 Plasmacytoma variant translocation 1 (PVT1) is a 1957 bp long intergenic non-coding RNA and located on human chromosome 8q24.21.8 Previous studies have shown that PVT1 is an important oncogenic lncRNA and highly expressed PVT1 correlates with poor outcomes in various types of cancer, including glioma, colorectal cancer, melanoma, small cell lung cancer and gastric cancer.9 Silence of PVT1 was reported to significantly inhibit proliferation, invasion and epithelial-mesenchymal transition via reducing miR-16-5p in renal cell carcinoma cells.10 In ovarian cancer, PVT1 up-regulated SOX2 expression to promote cell proliferation and invasion.11 These findings support a crucial clinical value for PVT1 in the development of useful biomarker and targeted therapy.
Oral squamous cell carcinoma (OSCC) is the sixth most common cancer worldwide with low cure rate and high mortality.12 Despite the substantial improvements in diagnosis and treatment approaches, 5-year survival rate still ranges from 45% to 50%.13 Cisplatin chemotherapy is commonly used for OSCC treatment, however the emergence of chemoresistance limits its long-term curative effect.14 In recent studies, up-regulated PTV1 was broadly implicated in multidrug resistance, including 5-fluorouracil, gemcitabine and cisplatin,15–17 yet the roles of PVT1 in OSCC progression and cisplatin resistance still remain largely unknown.
In this study, we first examined the expression profile of PVT1 in OSCC patients and found PVT1 was frequently up-regulated in cisplatin-resistant patients and strongly correlated with worse overall survival. Mechanically, PVT1 increased HIF1a expression to promote proliferation and cisplatin resistance via down-regulating miR-194-5p. Taken together, these results provide a novel mechanism for PVT1 in OSCC.
Materials and Methods
Patients and Samples
A total of 83 OSCC patients were enrolled at Binzhou Medical University Hospital between May 2010 and August 2015. The patients were treated with cisplatin prior to surgery. The patients’ responses were classified as cisplatin-sensitive or cisplatin-resistant according to the Response Evaluation Criteria in Solid Tumors of World Health Organization. The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Binzhou Medical University Hospital (BMUH-2017-093). Written informed consent was obtained from all participants.
Cell Culture
Human OSCC cell lines SCC9 and CAL27, and HEK-293T cells were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in DMEM (Gibico) with 10% FBS (Gibico) at 37°C under 5% CO2. Stable cisplatin-resistant cell lines SCC9-R and CAL27-R were established by continuous treatment with gradually increasing concentrations of cisplatin. To maintain the resistance, cisplatin (5 μM for SCC9-R and 7.5 μM for CAL27-R) was routinely added to the medium every other day before the experiments.
Oligonucleotide and Plasmid Transfection
miR-194-5p mimics, miRNA negative control (miRNA-NC), pcDNA-PVT1, si-PVT1, si-HIF1a and non-specific siRNA (si-NC) were designed and chemically synthesized by GenePharma (GenePharma, Shanghai, China). A concentration of 50 nM oligonucleotide or plasmid was transfected into cells using Lipofectamine 2000. After 48 h transfection, the cells were harvested for further experiments.
Cell Viability Assay
Cells were seeded at a density of 2000 cells/mL per well into a 96-well plate overnight and treated with different concentrations of cisplatin (2, 4, 6, 8, and 10 µM) for 48 hrs. Then, 10 µL CCK-8 solution (Dojindo, Kyushu, Japan) was added to each well. After incubating for 2 h at 37°C, the absorbance at 450 nm was measured with a microplate reader (Biotek, Winooski, VT, USA). The IC50 value was calculated by the relative survival curve.
Cell Survival Assay
Cells were seeded at a density of 2000 cells/mL per well in 96-well plates with 100 µL DMEM. CCK-8 assay was applied to measure cell proliferation daily for a consecutive 5 days as mentioned above.
Colony Formation Assay
A total of 500 cells/mL per well were seeded into 6-well plates and exposed to cisplatin for 24 hrs. Then, the drugs were washed away and the medium was replaced every 4 days. After 14 days, colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, Steinheim, Germany) for 15 min, and the total number of visible colonies were counted.
qRT-PCR Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). For miR-194-5p, cDNA was synthesized using PrimeScript miRNA cDNA Synthesis Kit (TaKaRa, Shiga, Japan) and U6 served as an internal control. For PVT1 and HIF1a, cDNA was synthesized using PrimeScript RT Reagent Kit (TaKaRa) and β-actin served as an internal control. qRT-PCR assay was performed using SYBR Premix Ex Taq (TaKaRa) according to the instructions. The relative expression levels were calculated using 2−ΔΔCT method.
Western Blot
Cells were lysed in RIPA buffer and 30 μg of total protein lysates were separated by 10% SDS-PAGE, transferred to PVDF membranes (Millipore), blocked with 5% non-fat milk for 60 min at room temperature, incubated with antibodies against HIF1a (1:1000, Abcam) at 4°C overnight followed by horseradish peroxidase-labeled secondary antibody for 60 min at room temperature. β-actin served as the loading controls.
Luciferase Reporter Assay
The putative binding sequence of miR-194-5p in wild type (WT) or mutant (Mut) PVT1 and HIF1a 3ʹUTR (WT-PVT1, Mut-PVT1, WT-HIF1a 3ʹUTR and Mut-HIF1a 3ʹUTR) were synthesized by GenePharma and subcloned were into the pmirGLO vector (Promega, Madison, WI, USA). HEK-293T cells were co-transfected WT or Mut reporter with miRNA mimics or miRNA-NC. After 48 h transfection, luciferase activity was determined using a Dual Luciferase Reporter Assay Kit (Promega).
Statistical Analysis
The mean ± standard deviation (SD) was used to show quantitative data from at least three independent experiments. One-way analysis of variance was applied to compare the difference between two groups. All tests were done with GraphPad Prism 5 software (GraphPad Software, Inc.). A p value < 0.05 was considered to indicate a significant difference.
Results
PVT1 Is Up-Regulated and Associated with Poor Prognosis and Cisplatin Resistance in OSCC
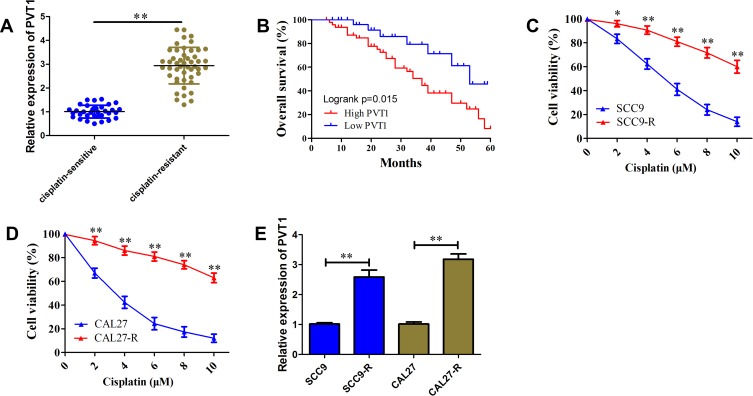
To investigate the functional role of PVT1 in the development of resistance to cisplatin, we first determined its expression profile in OSCC samples using qRT-PCR. Results showed that PVT1 expression level in cisplatin-resistant patients was higher than cisplatin-sensitive patients (Figure 1A). Besides, an unfavorable overall survival was observed in OSCC patients with high PVT1 expression (Figure 1B, P=0.015). To further validate the correlation of PVT1 with cisplatin resistance, two cisplatin-resistant cell lines SCC9-R and CAL27-R were generated. CCK-8 results showed SCC9-R (IC50= 13.56±0.87 µM) and CAL27-R (IC50= 16.43±1.08 µM) cells were more resistant to clinically doses of cisplatin (1–10 µM) compared with their parental cells (Figure 1C and D). We observed the PVT1 expression level was also significantly up-regulated 2.58-fold and 3.18-fold in SCC9-R and CAL27-R cells, respectively (Figure 1E). These results indicate that PVT1 may promote OSCC progression through regulating cisplatin resistance.
Figure 1.
PVT1 is up-regulated and associated with poor prognosis and cisplatin resistance in OSCC. (A) The level of PVT1 in cisplatin-resistant OSCC tissues and cisplatin-sensitive OSCC tissues. (B) Kaplan-Meier analysis of overall survival in OSCC patients according to PVT1 level. (C) CCK-8 assay analysis of cisplatin-resistance for SCC9-R cells and its parallel SCC9 cells. (D) CCK-8 assay analysis of cisplatin-resistance for CAL27-R cells and its parallel CAL27 cells. (E) The level of PVT1 in SCC9-R and CAL27-R cells and their parallel cells. *P<0.05, **P<0.01.
PVT1 Enhances OSCC Cell Proliferation and Cisplatin Resistance
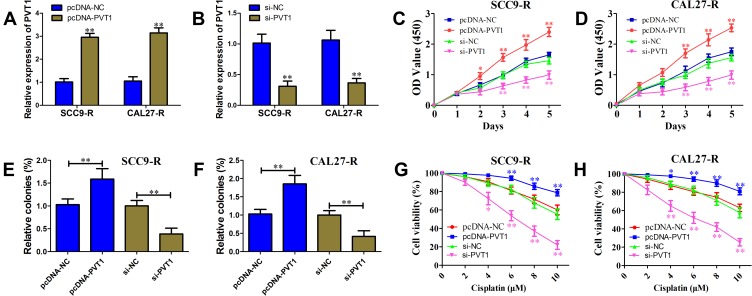
To directly observe the effects of PVT1 on the OSCC progression, gain-of-function and loss-of-function assays were conducted in SCC9-R and CAL27-R cells. As shown in Figure 2A and B, qRT-PCR assay was first performed to validate the efficiency of overexpression and knock-down. In both SCC9-R and CAL27-R cells, CCK-8 assay results showed overexpression of PVT1 significantly inhibited the proliferation compared with negative control, but an opposite effect was observed for silence of PVT1 (Figure 2C and D). Colony-formation assays confirmed that cells transfected with PVT1 siRNA have fewer colonies, and vice versa (Figure 2E and F). In both SCC9-R and CAL27-R cells, PVT1 overexpression exhibited greatly enhanced resistance to cisplatin compared with negative control (Figure 2G and H). Conversely, PVT1 knockdown prominently enhanced OSCC cell susceptibility to cisplatin.
Figure 2.
PVT1 enhances OSCC cell proliferation and cisplatin resistance. (A) Effect of pcDNA-PVT1 on PVT1 level in SCC9-R and CAL27-R cells. (B) Effect of si-PVT1 on PVT1 level in SCC9-R and CAL27-R cells. (C and D) Cell survival rate of SCC9-R (C) and CAL27-R (D) cells transfected with pcDNA-NC, pcDNA-PVT1, si-NC or si-PVT1 simultaneously under cisplatin pressure examined with CCK-8 assay. (E and F) Colony formation ability of SCC9-R (E) and CAL27-R (F) cells transfected with pcDNA-NC, pcDNA-PVT1, si-NC or si-PVT1 simultaneously under cisplatin pressure examined with colony formation assay. (G and H) Cell proliferation ability of SCC9-R (G) and CAL27-R (H) cells transfected with pcDNA-NC, pcDNA-PVT1, si-NC or si-PVT1 simultaneously under cisplatin pressure examined with CCK-8 assay. *P<0.05, **P<0.01.
PVT1 Positively Correlates HIF1a
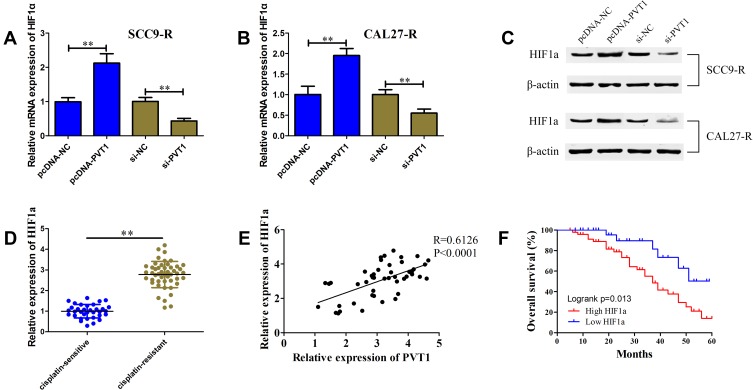
According to previous studies, HIF1a was closely related to the OSCC cell proliferation and cisplatin resistance. We speculated that PVT1 might affect OSCC cell proliferation and cisplatin resistance through regulating HIF1a expression. qRT-PCR and Western blot were first performed to determine whether PVT1 could regulate HIF1a expression. We observed that both PVT1 overexpression and PVT1 knockdown could positively affect HIF1a mRNA and protein levels in SCC9-R and CAL27-R cells (Figure 3A–C). We also evaluated the profile of HIF1a in OSCC samples and found that HIF1a in cisplatin-resistant patients increased 2.78-fold compared with cisplatin-sensitive patients (Figure 3D). Pearson analysis revealed a strong positive correlation of HIF1a with PVT1 in OSCC samples (Figure 3E; R=0.6126; P<0.0001). Kaplan-Meier survival analysis demonstrated that high mRNA expression level of HIF1a was significantly associated with poor outcome (Figure 3F, P=0.013).
Figure 3.
PVT1 positively correlates HIF1a. (A and B) The HIF1a mRNA level in SCC9-R (A) and CAL27-R (B) cells transfected with pcDNA-NC, pcDNA-PVT1, si-NC or si-PVT1 detected using qRT-PCR. (C) The HIF1a protein level in SCC9-R and CAL27-R cells transfected with pcDNA-NC, pcDNA-PVT1, si-NC or si-PVT1 detected using Western blot. (D) The HIF1a level in cisplatin-resistant OSCC tissues and cisplatin-sensitive OSCC tissues. (E) Pearson correlation analysis between PVT1 and HIF1a in OSCC tissues. (F) Kaplan-Meier analysis of overall survival in OSCC patients according to HIF1a level. **P<0.01.
PVT1 Up-Regulated HIF1a via Interaction with miR-194-5p
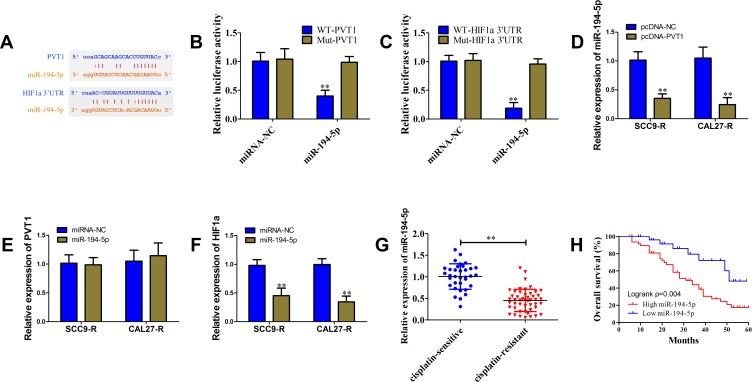
To elucidate the possible mechanism through which PVT1 promotes HIF1a expression, bioinformatics analysis including starBase, TargetScan and microRNA.org was used and miR-194-5p was identified as candidate miRNA that could bind to PVT1 and HIF1a 3ʹUTR (Figure 4A). The vectors containing WT or Mut putative binding sequences of HIF1a and PVT1 3ʹUTR were constructed. In HEK-293T cells, miR-194-5p mimics decreased the luciferase activity of WT vectors, but not Mut vectors (Figure 4B and C). As shown in Figure 4D–F, overexpression of PVT1 could reduce miR-194-5p level, but miR-194-5p mimics only down-regulated HIF1a expression and had no significant effect on PVT1 expression in SCC9-R and CAL27-R cells. We also observed a down-regulated level of miR-194-5p in cisplatin-resistant OSCC patients (Figure 4G). In addition, low expression level of miR-194-5p predicted a poor outcome (Figure 4H, P=0.004).
Figure 4.
PVT1 up-regulated HIF1a via interaction with miR-194-5p. (A) The putative binding sites of miR-194-5p in the PVT1 and HIF1a 3ʹUTR predicted with starBase v3.0. (B and C) Luciferase reporter assay in HEK-293T cells, co-transfected of WT or Mut of PVT1 (B) and HIF1a 3ʹUTR (C), with miR-194-5p mimics or miRNA-NC. (D) The miR-194-5p level in SCC9-R and CAL27-R cells transfected with pcDNA-PVT1 or pcDNA-NC determined using qRT-PCR. (E) The PVT1 level in SCC9-R and CAL27-R cells transfected with miR-194-5p mimics or miRNA-NC determined using qRT-PCR. (F) The HIF1a level in SCC9-R and CAL27-R cells transfected with miR-194-5p mimics or miRNA-NC determined using qRT-PCR. (G) The miR-194-5p level in cisplatin-resistant OSCC tissues and cisplatin-sensitive OSCC tissues. (H) Kaplan-Meier analysis of overall survival in OSCC patients according to miR-194-5p level. **P<0.01.
PVT1 Promotes Cell Proliferation and Cisplatin Resistance via Up-Regulation of HIF1a
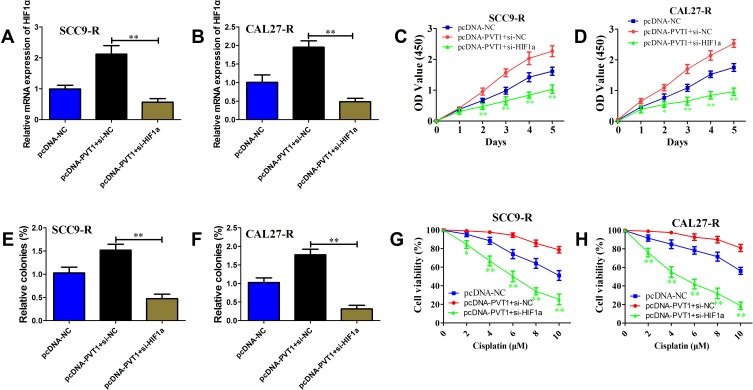
To further determine the role of HIF1a in PVT1-dependent OSCC cell proliferation and cisplatin resistance, specific HIF1a siRNA or non-specific siRNA was co-transfected with pcDNA-PVT1 in SCC9-R and CAL27-R cells. The level of HIF1a was obviously reduced in cells transfected with si-HIF1a compared with si-NC (Figure 5A and B). As shown in Figure 5C and D, knockdown of HIF1a could abolish the growth promotion of OSCC cells induced by PVT1 overexpression. Colony-formation assay results revealed that silence of HIF1a could decrease the number of colonies induced by PVT1 up-regulation (Figure 5E and F). In addition, the increased cisplatin resistance in both SCC9-R and CAL27-R cells was impaired via HIF1a knockdown (Figure 5G and H). These findings suggest that PVT1 promotes OSCC proliferation and cisplatin resistance at least partly via increasing HIF1a expression.
Figure 5.
PVT1 promotes cell proliferation and cisplatin resistance via up-regulation of HIF1a. (A and B) The HIF1a mRNA level in SCC9-R (A) and CAL27-R (B) cells co-transfected pcDNA-NC or pcDNA-PVT1 with si-NC or si-HIF1a detected using qRT-PCR. (C and D) Cell survival rate of SCC9-R (C) and CAL27-R (D) cells co-transfected pcDNA-NC or pcDNA-PVT1 with si-NC or si-HIF1a examined with CCK-8 assay. (E and F) Colony formation ability of SCC9-R (E) and CAL27-R (F) cells co-transfected pcDNA-NC or pcDNA-PVT1 with si-NC or si-HIF1a examined with colony formation assay. (G and H) Cell proliferation ability of SCC9-R (G) and CAL27-R (H) cells co-transfected pcDNA-NC or pcDNA-PVT1 with si-NC or si-HIF1a simultaneously examined with CCK-8 assay. *P<0.05, **P<0.01.
miR-194-5p Reverses PVT1-Induced OSCC Cell Proliferation and Cisplatin Resistance
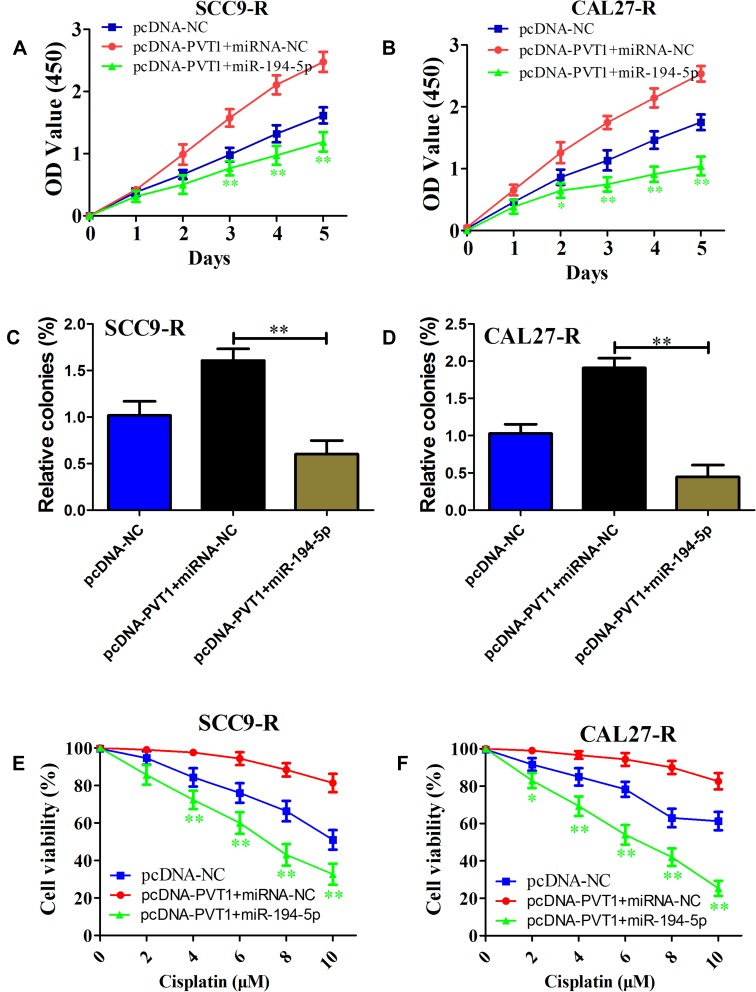
To further explore the potential role of miR-194-5p in PVT1-induced OSCC cell proliferation and cisplatin resistance, miR-194-5p mimics or miRNA negative control was co-transfected with pcDNA-PVT1 in SCC9-R and CAL27-R cells. As expected, miR-194-5p overexpression inhibited PVT1 overexpression-induced cell proliferation (Figure 6A and B). As shown in Figure 6C and D, we observed that miR-194-5p mimics significantly suppressed colony formation compared with miRNA negative control. In line with these findings, the promotion of cisplatin resistance-modulated by PVT1 up-regulation was reversed when cisplatin-resistant cells were introduced into miR-194-5p mimics (Figure 6E and F). These data suggest that miR-194-5p is an important downstream of PVT1 to promote OSCC proliferation and cisplatin resistance.
Figure 6.
miR-194-5p reverses PVT1-induced OSCC cell proliferation and cisplatin resistance. (A and B) Cell survival rate of SCC9-R (A) and CAL27-R (B) cells co-transfected pcDNA-NC or pcDNA-PVT1 with miRNA-NC or miR-194-5p mimics examined with CCK-8 assay. (C and D) Colony formation ability of SCC9-R (C) and CAL27-R (D) cells co-transfected pcDNA-NC or pcDNA-PVT1 with miRNA-NC or miR-194-5p mimics examined with colony formation assay. (E and F) Cell proliferation ability of SCC9-R (E) and CAL27-R (F) cells co-transfected pcDNA-NC or pcDNA-PVT1 with miRNA-NC or miR-194-5p mimics examined with CCK-8 assay. *P<0.05, **P<0.01.
Discussion
Since first approved by FDA for the treatment of testicular and bladder cancer in 1978, cisplatin has been widely used for the treatment of solid tumors, including ovary, testes, and the head and neck.18 However, many patients develop resistance to cisplatin-based therapy which results in a high rate of non-response and relapse, especially in ovarian cancer.19 Chemoresistance has become the main limitation to the clinical usefulness of cisplatin as an anticancer drug and hence, understanding the molecular mechanisms of cisplatin resistance will help to the development of more efficient anticancer therapies.
In this study, we identified that PVT1 was up-regulated in OSCC cisplatin-resistant tissues and cell lines compared with cisplatin-sensitive tissues and cell lines, and high PVT1 level was significantly associated with poor overall survival. We also found that PVT1 overexpression enhanced OSCC cell proliferation, colony formation and resistance to cisplatin. These findings suggest that PVT1 functions as an oncogene in OSCC. Further investigations identified HIF1a is a crucial downstream gene of PVT1. We observed that high expression level of HIF1a in OSCC cisplatin-resistant tissues predicted a worse outcome.
HIF1a gene encodes the alpha subunit of transcription factor HIF1 that controls cellular and systemic homeostatic response to hypoxia.20 HIF1 can activate the transcription of dozens of genes and many of them have been identified to be involved in cell metabolism, cell survival and angiogenesis.21 Up-regulated HIF1a has been observed in several cancers, including esophageal, rectal, hepatocellular and nasopharyngeal carcinoma.22 Previous studies demonstrated HIF-1α was associated with OSCC overall survival and disease-free survival, and involved in tumor angiogenesis, growth, and radiochemotherapy.23,24 Further studies are needed to determine the underlying mechanism of how HIF1a regulates the downstream genes or signal pathways to promote OSCC proliferation and cisplatin resistance.
miR-194-5p was reported to be down-regulated in hypopharyngeal carcinoma, acute myeloid leukemia, bladder cancer and glioblastoma and functioned as a tumor suppressor.25–27 Interestingly, Yang et al found that miR‑194‑5p was up-regulated in breast cancer tissues and silence of miR-194-5p inhibited cell proliferation and metastasis via targeting SOX17.28 Meanwhile, decreased miR-194-5p was implicated in sunitinib resistance in human renal cell carcinoma and paclitaxel resistance in ovarian cancer.29,30 These studies revealed a complex function and molecular basis of miR-194-5p, yet its role in OSCC still remains largely unknown.
We identified that miR-194-5p as a candidate miRNA that could directly target PVT1 and HIF1a 3ʹUTR. We also observed miR-194-5p was down-regulated in cisplatin-resistant OSCC tissues and low miR-194-5p level was related to unfavorable prognosis. Moreover, PVT1 overexpression obviously reduced miR-194-5p level and miR-194-5p mimics down-regulated HIF1a expression. Overexpression of miR-194-5p was observed to partly relieve the promotion of cisplatin resistance-modulated by PVT1. These data indicate that PVT1 acted as a ceRNA through interacting with miR-194-5p and indirectly up-regulating HIF1a. Except for ceRNA mechanism, lncRNAs can also execute biological functions as archetypes of signals, guides, and scaffolds. PVT1 drives triple-negative breast cancer tumorigenesis through binding with KLF5 and increases its stability via BAP1.31 In Cholangiocarcinoma, PVT1 regulates histone methylation of the promoter of ANGPTL4 to promote tumor growth and migration by binding to epigenetic modification complexes (PRC2).32
Conclusion
The present study for the first time revealed that PVT1 was up-regulated in OSCC cisplatin-resistant tissues and cell lines, and high PVT1 level was associated with poor prognosis. Furthermore, we also found that PVT1 promoted OSCC cell proliferation and cisplatin-resistance through miR-194-5p/HIF1a axis. PVT1 might be used as a new biomarker and therapeutic target for OSCC patients with cisplatin-resistance.
Acknowledgments
This work was supported by grants from the Science and Technology Development Plan Project of Binzhou (2014ZC0111), the Scientific Research Foundation for the Excellent Middle-Aged and Young Scientists of Shandong Province (BS2013YY056) and the Science and Technology Development Plan Project of Jinan (2013-60).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 3.Li H, Liu C, Lu Z, et al. Upregulation of the long non-coding RNA SPRY4-IT1 indicates a poor prognosis and promotes tumorigenesis in ovarian cancer. Biomed Pharmacother. 2017;88:529–534. doi: 10.1016/j.biopha.2017.01.037 [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto A, Pasut A, Matsumoto M, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541(7636):228–232. doi: 10.1038/nature21034 [DOI] [PubMed] [Google Scholar]
- 5.Nelson BR, Makarewich CA, Anderson DM, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351(6270):271–275. doi: 10.1126/science.aad4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. 2018;17(1):119. doi: 10.1186/s12943-018-0870-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carramusa L, Contino F, Ferro A, et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol. 2007;213(2):511–518. doi: 10.1002/(ISSN)1097-4652 [DOI] [PubMed] [Google Scholar]
- 9.Cui M, You L, Ren X, et al. Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun. 2016;471(1):10–14. doi: 10.1016/j.bbrc.2015.12.101 [DOI] [PubMed] [Google Scholar]
- 10.Ren Y, Huang W, Weng G, Cui P, Liang H, Li Y. LncRNA PVT1 promotes proliferation, invasion and epithelial-mesenchymal transition of renal cell carcinoma cells through downregulation of miR-16-5p. Onco Targets Ther. 2019;12:2563–2575. doi: 10.2147/OTT.S190239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou MF, Ling J, QY W, Zhang CX. Long non-coding RNA PVT1 functions as an oncogene in ovarian cancer via upregulating SOX2. Eur Rev Med Pharmacol Sci. 2018;22(21):7183–7188. doi: 10.26355/eurrev_201811_16251 [DOI] [PubMed] [Google Scholar]
- 12.Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin. 2015;65(5):401–421. doi: 10.3322/caac.v65.5 [DOI] [PubMed] [Google Scholar]
- 13.Bloebaum M, Poort L, Böckmann R, Kessler P. Survival after curative surgical treatment for primary oral squamous cell carcinoma. J Craniomaxillofac Surg. 2014;42(8):1572–1576. doi: 10.1016/j.jcms.2014.01.046 [DOI] [PubMed] [Google Scholar]
- 14.Yamauchi K, Yang M, Hayashi K, et al. Induction of cancer metastasis by cyclophosphamide pretreatment of host mice: an opposite effect of chemotherapy. Cancer Res. 2008;68(2):516–520. doi: 10.1158/0008-5472.CAN-07-3063 [DOI] [PubMed] [Google Scholar]
- 15.Du P, Hu C, Qin Y, et al. LncRNA PVT1 mediates antiapoptosis and 5-fluorouracil resistance via increasing Bcl2 expression in gastric cancer. J Oncol. 2019;2019:9325407. doi: 10.1155/2019/9325407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun ZY, Jian YK, Zhu HY, Li B. lncRNAPVT1 targets miR-152 to enhance chemoresistance of osteosarcoma to gemcitabine through activating c-MET/PI3K/AKT pathway. Pathol Res Pract. 2019;215(3):555–563. doi: 10.1016/j.prp.2018.12.013 [DOI] [PubMed] [Google Scholar]
- 17.Ping G, Xiong W, Zhang L, Li Y, Zhang Y, Zhao Y. Silencing long noncoding RNA PVT1 inhibits tumorigenesis and cisplatin resistance of colorectal cancer. Am J Transl Res. 2018;10(1):138–149. [PMC free article] [PubMed] [Google Scholar]
- 18.Prestayko AW, D’Aoust JC, Issell BF, Crooke ST. Cisplatin (cis-diamminedichloroplatinum II). Cancer Treat Rev. 1979;6(1):17–39. doi: 10.1016/S0305-7372(79)80057-2 [DOI] [PubMed] [Google Scholar]
- 19.Ozols RF. Ovarian cancer: new clinical approaches. Cancer Treat Rev. 1991;18(Suppl):A:77–83. doi: 10.1016/0305-7372(91)90027-W [DOI] [PubMed] [Google Scholar]
- 20.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masoud GN, Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5(5):378–389. doi: 10.1016/j.apsb.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu GQ, Tang YL, Li L, et al. Hypoxia inducible factor 1α and hypoxia inducible factor 2α play distinct and functionally overlapping roles in oral squamous cell carcinoma. Clin Cancer Res. 2010;16(19):4732–4741. doi: 10.1158/1078-0432.CCR-10-1408 [DOI] [PubMed] [Google Scholar]
- 24.Sasabe E, Zhou X, Li D, et al. The involvement of hypoxia-inducible factor-1alpha in the susceptibility to gamma-rays and chemotherapeutic drugs of oral squamous cell carcinoma cells. Int J Cancer. 2007;120(2):268–277. doi: 10.1002/ijc.22294 [DOI] [PubMed] [Google Scholar]
- 25.Dell’Aversana C, Giorgio C, D’Amato L, et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia. 2017;31(11):2315–2325. doi: 10.1038/leu.2017.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Sun G, Wang C, Guo W, Tang Q, Wang M. MiR-194-5p inhibits cell migration and invasion in bladder cancer by targeting E2F3. J BUON. 2018;23(5):1492–1499. [PubMed] [Google Scholar]
- 27.Zhang Z, Lei B, Wu H, Zhang X, Zheng N. Tumor suppressive role of miR-194-5p in glioblastoma multiforme. Mol Med Rep. 2017;16(6):9317–9322. doi: 10.3892/mmr.2017.7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F, Xiao Z, Zhang S. Knockdown of miR-194-5p inhibits cell proliferation, migration and invasion in breast cancer by regulating the Wnt/β-catenin signaling pathway. Int J Mol Med. 2018;42(6):3355–3363. doi: 10.3892/ijmm.2018.3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yumioka T, Osaki M, Sasaki R, et al. Lysosome-associated membrane protein 2 (LAMP-2) expression induced by miR-194-5p downregulation contributes to sunitinib resistance in human renal cell carcinoma cells. Oncol Lett. 2018;15(1):893–900. doi: 10.3892/ol.2017.7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura K, Sawada K, Miyamoto M, et al. Downregulation of miR-194-5p induces paclitaxel resistance in ovarian cancer cells by altering MDM2 expression. Oncotarget. 2019;10(6):673–683. doi: 10.18632/oncotarget.v10i6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang J, Li Y, Sang Y, et al. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37(34):4723–4734. doi: 10.1038/s41388-018-0310-4 [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, Zhang M, Liu J, et al. Long non-coding RNA PVT1 promotes cell proliferation and migration by silencing ANGPTL4 expression in cholangiocarcinoma. Mol Ther Nucleic Acids. 2018;13:503–513. doi: 10.1016/j.omtn.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]