Abstract
Autosomal dominant Hyper IgE syndrome (AD-HIES), a rare immune deficiency affecting fewer than one per million people, is caused by heterozygous deleterious mutations in STAT3. STAT3 signaling plays crucial roles in basic cellular functions affecting broad aspects of cellular homeostasis. Accordingly, in addition to immunological deficits, patients experience severe multisystem non-immunological features. Human induced pluripotent stem cells (hiPSC) are well established as in vivo disease models for various human pathologies. We describe the generation of iPSC from three AD-HIES patients. These iPSCs express pluripotency markers, differentiate into three germ layers, have normal karyotype and similar genome identity to parental cells.
Keywords: iPSC, Autosomal dominant Hyper IgE syndrome (AD-HIES), STAT3
1. Resource utility
The AD-HIES hiPSC lines and their derivatives could be used for in vitro disease modeling to better elucidate the pathophysiology of AD-HIES. These cells could help clarify the effects of the STAT3 mutation in developmental processes as well as differentiation and proliferation of various cell types in vitro.
2. Resource Details
Hyperimmunoglobulin syndromes (HIES) consist of a group of rare primary immune deficiency disorders characterized by elevated immunoglobulin E levels, eczema, and recurrent pulmonary and skin infections (Hashmi et al., 2017). Both autosomal dominant and recessive patterns of inheritance have been reported in HIES with autosomal dominant (AD-HIES) patterns being more common. In addition to immunodeficiency, patients experience severe non-immunological features including skeletal, connective tissue and vascular abnormalities, poor post-infection healing and subsequent pulmonary failure (Freeman and Holland, 2010). AD-HIES is caused by dominant negative mutations in the signal transducer and activator of the transcription factor 3 (STAT3) gene (Holland et al., 2007; Minegishi et al., 2007). Despite the identified genetic cause of the disease, the exact mechanisms of the AD-HIES pathologies are not well understood, preventing the development of specific therapies and limiting treatment options to general antimicrobial prophylaxis and multidisciplinary care.
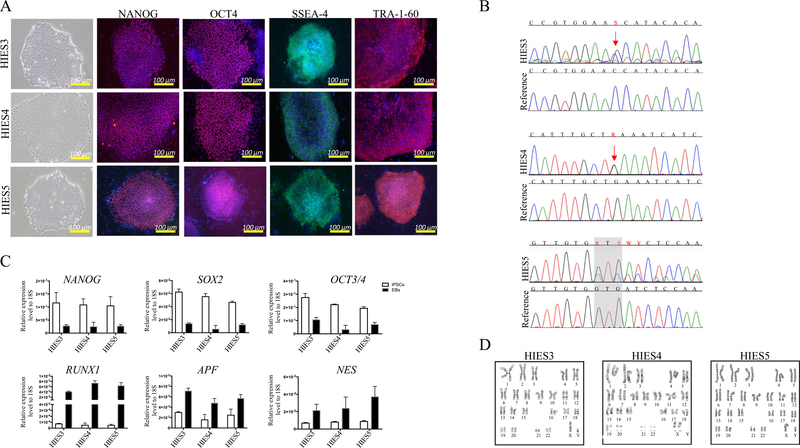
In our study, skin punch biopsy samples from 3 patients with AD-HIES were collected at the NIH Clinical Center in addition to a complete medical history and physical exam including clinical onset, demographics and other phenotype specific details (Table 1). As with standard control iPSCs lines, AD-HIES iPSCs were able to maintain pluripotency over 60 passages as determined by morphological observations and the expression of typical pluripotency markers, OCT4, NANOG, TRA-1–60 and SSEA4 (Fig 1A). Genotyping of hiPSC identified either an SH2 domain or a DNA binding domain mutation in all the clones (total of 3) shown in Fig 1B. To test the differentiation potential of the three cell lines, we performed embryoid body formation assays that drive the cells toward differentiation into the three primary germ layers in vitro. The expression of the endoderm (AFP), ectoderm (NESTIN) and mesoderm (RUNX1) signature genes was determined with real-time (RT) PCR, which showed upregulated expression levels in AD-HIES lines after differentiation (Fig 1C). All generated cell lines demonstrated chromosomal stability and a normal karyotype (Fig 1D). Table 2 summarizes the characterization and validation of AD-HIES hiPSCs. In addition, short tandem repeat (STR) profiles confirmed that all iPSC lines matched their parental fibroblasts completely in 15 amplified STR loci (supplement 1). All cultures were routinely tested for mycoplasma contamination and were found to be negative (supplement 2). To the best of our knowledge, this is the first published study to generate iPSC lines from AD-HIES patients.
Table 1.
Summary of three AD-HIES patients with a STAT3 mutation
| iPSC line names | Abbreviation in figures | Gender | Age (years) | Ethnicity | Genotype of locus | Mutation | Protein Mutation | Disease |
|---|---|---|---|---|---|---|---|---|
| NIHTVBi011-A | HIES3 | M | 24 | European-American | STAT3, 17q21.2 | 1915 C-G | P639A | AD-HIES |
| NIHTVBi012-A | HIES4 | F | 56 | European-American | STAT3, 17q21.2 | 1954 G-A | E652K | AD-HIES |
| NIHTVBi013-A | HIES5 | M | 49 | European-American | STAT3, 17q21.2 | 1387delGTG | del V463 | AD-HIES |
Figure 1.
Table 2.
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Phase-contrast microscope | Normal | Figure 1A |
| Phenotype | Qualitative analysis (immunofluorescence staining) | Expression of pluripotency markers: OCT4, NANOG, SSEA4 and TRA-1–60 | Figure 1A |
| Quantitative analysis (RT-qPCR) | Expression of pluripotency markers: NANOG, SOX2, and OCT3/4 | Figure 1C | |
| Genotype | Karyotype (G-banding) and resolution | 46,XX or 46,XY; resolution 450–500 bands | Figure 1D |
| Identity | Microsatellite PCR OR STR analysis | Not performed | N/A |
| 15 sites tested, 100% match | Supplementary file | ||
| Mutation analysis (IF APPLICABLE) | DNA sequencing | STAT3 mutation | Figure 1B |
| Southern blot OR WGS | Not performed | N/A | |
| Microbiology and virology | Mycoplasma testing by luminescence | Negative | Supplementary file |
| Differentiation potential | EB formation assay | Differentiating cells are expression of RUNX1, AFP, and NES; iPSCs were able to differentiate into three germ layers | Figure 1C |
| Donor screening (OPTIONAL) | HIV1 + HIV2, hepatitis B virus, hepatitis C virus | Not performed | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | Not performed | N/A |
| HLA tissue typing | Not performed | N/A |
3. Materials and methods
3.1. Subjects and study approval
Unrelated patients with clinical evidence of AD-HIES and confirmed STAT3 mutations were consented to NHLBI IRB approved study 10-H-0126.
3.2. Derivation of patient-specific fibroblasts
Fibroblasts lines were generated from punch skin biopsies as previously described (Jin et al., 2016). Briefly, the sample was cut into 1 mm pieces and digested for 1h at 37°C in 0.1% Collagenase Type II/0.25 U/ml Dispase (both from Thermo Fisher Scientific)/PBS solution. The pieces were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 20% fetal bovine serum (FBS) and 1% penicillin-streptomycin. After 3–4 weeks, fibroblast outgrowths from the explants were passaged and cultured in DMEM containing 10% FBS and 1% penicillin-streptomycin.
3.3. Generation and culture of human iPSCs from human fibroblasts
Fibroblasts from AD-HIES patients were reprogrammed with the Human STEMCCA Cre-Excisable Constitutive Polycistronic (OKSM) Lentivirus reprogramming kit (Millipore). iPSC colonies were collected 21 days post-transduction, maintained in full E8 medium at 37°C with 5% CO2 and passaged with 0.5mM EDTA every 3–5 days (Jin et al., 2016).
3.4. Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde for 10 minutes, permeabilized with 0.3% Triton X-100 for 30 min and then blocked with donkey 5% serum. They were then incubated with primary antibodies NANOG, OCT4, SSEA4, and TRA-1–60 (Table 3) overnight at 4°C, washed and incubated with fluorophore-tagged secondary antibodies for 1 h at room temperature. After PBS wash, DAPI was added for nuclear staining before capturing images with a fluorescence microscope (Zeiss).
Table 3.
Reagents. Antibodies used for immunocytochemistry
| Antibody | Dilution | Company | Cat# | RRID | |
|
| |||||
| Primary antibodies | Rabbit anti-OCT4 | 1:100 | Cell Signaling Technology | 2750 | AB_823583 |
| Mouse anti-NANOG | 1:100 | Cell Signaling Technology | 4893 | AB_10548762 | |
| Mouse anti-SSEA4 | 1:100 | MilliporeSigma | MAB4304 | AB_177629 | |
| Mouse anti-TRA-1–60 | 1:150 | MilliporeSigma | MAB4360 | AB_2119183 | |
| Secondary antibodies | Alexa Fluor 594 Donkey anti-rabbit | 1:300 | Life Technologies | A21207 | AB_141637 |
| Alexa Fluor 594 Donkey anti-mouse | 1:300 | Life Technologies | A21203 | AB_141633 | |
| Alexa Fluor 488 Donkey anti-mouse | 1:300 | Life Technologies | A21202 | AB_141607 | |
| Alexa Fluor 555 Goat anti-mouse | 1:300 | Life Technologies | A21426 | AB_2535847 | |
|
| |||||
| Primers used for RT-qPCR and PCR | |||||
|
| |||||
| Target | Forward/reverse primer (5’–3’) | ||||
| NANOG | AGG GAA ACA ACC CAC TTC T/CCT TCT GCG TCA CAC CAT T | ||||
| SOX2 | CCC AGC AGA CTT CAC ATG T/CCT CCC ATT TCC CTC GTT TT | ||||
| AFP | AGC TTG GTG GAT GAA AC/CCC TCT TCA GCA AAG CAG AC | ||||
| NESTIN | GCG TTG GAA CAG AGG TTG GA/TGG GAG CAA AGA TCC AAG AC | ||||
| RUNX1 | CTG CCC ATC GCT TTC AAG GT/GCC GAG TAG TTT TCA TTG CC | ||||
| STAT3 P82 | TGG CCA CGC TGG GCA TTC TTT CCA CTA T/CTC AGT AGA CAT GGC CCA AAT GAA CAG CCC TAT G | ||||
| STAT3 P19/P77 | |||||
| AGT AAG GAG CGG GAG CGG GCC ATC TTG/TCC TCC CAC CTC AGC CTC CCA AGT AGC TAC GAC TA | |||||
3.5. Embryoid Body (EB) formation
EB formation was performed as described with minor modifications. Briefly, iPSCs were dissociated into small clumps by EDTA/PBS incubation for 5 minutes and transferred into ultra-low attachment plates (Corning) to allow for self-aggregation. Cells were cultured at 37C, 20% CO2 in KnockOut DMEM (ThermoFisher Scientific) supplemented with 10% FBS. The media was replaced every other day and EBs were analyzed at differentiation day 10.
3.6. Gene Expression Analysis
Endogenous mRNA expression levels of genes were determined by RT-PCR in iPSCs and EB at differentiation day 10. RT-PCR was performed with SYBR Green premix (Bio-rad) and mRNA copy number was quantified relative to 18 S. All assays were run in duplicate and primers are shown in Table 3. Data are represented as the average ± SEM relative to the mRNA levels found in fibroblasts of each sample. The specificity of the amplified PCR products was confirmed by analysis of the melting curve.
3.7. Karyotyping assay
To exclude introduction of chromosomal abnormalities during the iPSC generation and propagation procedure, cell karyotype was evaluated by the WiCell Research Institute using G-banding metaphase karyotype analysis every 10 cell passages starting at passage 10 and counting 20 metaphase spreads (supplement 3).
3.8. DNA sequencing and STR
Genomic DNA was isolated by using DNeasy Blood & Tissue Kit (Qiagen). To amplify the corresponding mutation position in STAT3, PCR was performed with specific primers (Table 3). Following purification, the PCR products were sent to Eurofins Scientific for sequencing. STR analysis was performed by WiCell Research Institute, which generated a STR profile via the Promega Powerplex® 16 System to verify STR polymorphisms for 15 loci plus amelogenin in genomic DNA extracted from iPSCs and their parental fibroblasts.
3.9. Mycoplasma detection
To ensure iPSC cultures were mycoplasma free, media were collected after culturing for 48 h and tested with the MycoAlert™ Mycoplasma Detection Kit (Lonza).
Supplementary Material
Resource Table.
| Unique stem cell lines identifier | NIHTVBi011-A |
| NIHTVBi012-A | |
| NIHTVBi013-A | |
| Alternative names of stem cell lines | AD-HIES3 |
| AD-HIES4 | |
| AD-HIES5 | |
| Institution | National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Bethesda, Maryland, USA |
| Contact information of distributor | Manfred Boehm; boehmm@nhlbi.nih.gov and Guibin Chen; chengb@nhlbi.nhi.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | Dermal fibroblasts |
| Clonality | Clonal cell lines |
| Method of reprogramming | Lentiviral vectors containing the transcription factors Oct4, Klf4, Sox2 and c-MYC |
| Multiline rationale | Lines derived from the five patients |
| Gene modification | Yes |
| Type of modification | Hereditary |
| Associated disease | None |
| Gene/locus | STAT3, 17q21.2 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | September 2012 |
| Cell line repository/bank | N/A |
| Ethical approval | National Institutes of Health Ethics Committee (Approval Number: 10-H-0126) |
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.scr.2019.101586.
References
- Freeman AF, Holland SM, 2010. Clinical manifestations of hyper IgE syndromes. Dis. Markers 29, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashmi H, Mohebbi M, Mehravaran S, Mazloumi M, Jahanbani-Ardakani H, Abtahi SH, 2017. Hyperimmunoglobulin E syndrome: Genetics, immunopathogenesis, clinical findings, and treatment modalities. J. Res. Med. Sci 22, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SM, Deleo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns DB, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Whitney AR, Voyich JM, Musser JM, Woellner C, Schaeffer AA, Puck JM, Grimbacher B, 2007. STAT3 mutations in the hyper-IgE syndrome. N. Eng. J. Med 357 (16), 1608–1619. [DOI] [PubMed] [Google Scholar]
- Jin H, St Hilaire C, Huang Y, Yang D, Dmitrieva NI, Negro A, Schwartzbeck R, Liu Y, Yu Z, Walts A, Davaine JM, Lee DY, Donahue D, Hsu KS, Chen J, Cheng T, Gahl W, Chen G, Boehm M, 2016. Increased activity of TNAP compensates for reduced adenosine production and promotes ectopic calcification in the genetic disease ACDC. Sci. Signal 9, pp. ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, Metin A, Karasuyama H, 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448 (2007), 1058–1062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.