Abstract
Background
Increasing evidence has suggested that multiple long non-coding RNAs (lncRNAs) act key regulatory functions in the pathogenesis of bladder cancer. This study aimed to determine the expression and clinical significance of lncRNA ROR1 antisense RNA 1 (ROR1-AS1) from patients with bladder cancer, and to explore the potential role and mechanism underlying ROR1-AS1-related cancer progression.
Methods
Real time quantitative PCR (RT-qPCR) was conducted to detected the expression levels of ROR1-AS1 and miR-504 in bladder cancer samples and cell lines. Chi-square test was used for correlation analysis. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) and wound scratch assays were applied to assesses the effects of ROR1-AS1 overexpression and knockdown on bladder cancer cell growth and migration in vitro, respectively. The prognosis of bladder cancer patients was evaluated by survival curves with Kaplan-Meier method. The regulatory mechanism of ROR1-AS1 on miR-504 was confirmed by bioinformatics analysis and luciferase reporter gene assay.
Results
ROR1-AS1 levels were obviously upregulated in bladder cancer tissues than matched normal bladder tissues. High expression of ROR1-AS1 was remarkably correlated with higher histological grade, advanced tumor stage, and positive lymph node metastasis. High ROR1-AS1 expression was markedly correlated with shorter overall survival of bladder cancer patients. Moreover, knockdown of ROR1-AS1 notably repressed T24 and 5637 cell growth and migration. ROR1-AS1 directly bound with miR-504 and act as a molecular sponge to decrease miR-504 expression. Silencing of miR-504 partly abrogated ROR1-AS1 knockdown-induced inhibitory effects on bladder cancer cell growth and migration.
Conclusions
Our data demonstrated that increased ROR1-AS1 promotes cell growth and migration of bladder cancer via regulation of miR-504, indicating ROR1-AS1 may be used as a prognostic biomarker and therapeutic target for bladder cancer.
Introduction
Bladder cancer is the fourth most common diagnosed malignancy, and is one of the most expensive malignancies to treatment in men worldwide [1]. According to the Global Cancer Statistics in 2017, approximately 440,000 new cases are diagnosed with bladder cancer, and among those patients about 130,000 cases died of cancer [2]. Patients with invasive bladder cancer are commonly treated with radical cystectomy and urinary diversion; however, those patients usually have a poor prognosis [3,4]. At present, because of the poor understanding of pathological mechanisms in the progression of bladder cancer, the effective treatment for this cancer is very limited [5,6]. Therefore, it’s essential to identify new sensitive and promising therapeutic targets for bladder cancer.
Long non-coding RNA (lncRNA) is a class of non-coding RNA which surpasses 200 nucleotides in length, usually exhibiting little or no coding potential [7]. Increasing data have demonstrated that lncRNAs play a crucial role in modulating various cellular biological processes, including growth and differentiation, apoptosis, malignant metastasis, and epithelial-mesenchymal transition [8–10]. According to their location on the human genome, lncRNAs can be placed into five broad categories: sense, antisense, bidirectional, intronic, and intergenic. Recent studies have identified that some antisense lncRNAs participate in the tumorigenesis and carcinogenesis of diverse human cancers, these lncRNAs can also be used as potential and effective biomarkers for cancer therapies [11,12]. For instance, ABHD11 antisense RNA1 acts as an oncogene and a potential target for antitumor therapies in ovarian cancer [13]. Long intergenic non-protein coding RNA 1133 inhibits breast cancer cell invasion and metastasis by negatively regulating SRY-box transcription factor 4 expression via enhancer of zeste 2 polycomb repressive complex 2 subunit [14]. SMAD5 antisense RNA 1 functions as a miR-106a-5p sponge to promote epithelial mesenchymal transition in nasopharyngeal carcinoma [15]. Additional, DLX6 antisense RNA 1 promotes cell growth and invasiveness in bladder cancer via modulating the miR-223-HSP90B1 axis [16].
ROR1 antisense RNA 1 (ROR1-AS1) is a novel found lncRNA, which locates at human genome 1p31.3 position. Hu and his team [17] in 2017 reported that ROR-AS1 is involved in regulation of gene transcription via associating with polycomb repressive complex 2 complex, and may serve as a new biomarker in patients with mantle cell lymphoma. Recently, several studies indicated that ROR1-AS1 can enhance colorectal cancer metastasis and tumorigenesis [18,19]. MicroRNAs (miRs) act as tumor suppressive or oncogenic factors and are major posttranscriptional gene regulators in diverse cancers. LncRNAs can associate with miRs to impact cell biological behaviors by acting as ceRNAs by competitively binding common miRs [19]. Despite the above-mentioned knowledge, how ROR1-AS1 participated in the progression of bladder cancer remains unclear. The present study aimed to explore the functional role and downstream target miRs of ROR1-AS1 in the progression of bladder cancer. We firstly analyzed the expression and clinical significances of ROR1-AS1 in patients with bladder cancer. Then, we performed in vitro loss-of-function and gain-of-function experiments to determine the potential function of ROR1-AS1 in bladder cancer cell growth and migration. Finally, luciferase reporter gene assay and rescue experiments were applied to confirm the regulatory mechanism of ROR1-AS1 on miR-504. Our study highlighted a key role of ROR1-AS1/miR-504 axis in the progression of bladder cancer.
Materials and methods
Clinical specimens and cell culture
This research was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (Approval No. 2018070), and the written informed consents were received from all patients in accordance with the 1964 Helsinki declaration and its later amendments. This retrospective study included 65 cases of bladder cancer patients who underwent surgery at Department of Urology, First Affiliated Hospital of Nanchang University between 09/2011 and 05/2017. Human bladder tissues and adjacent matched normal bladder tissues (≥ 3 cm away from the edge of cancer) from all patients were collected between 09/2011 and 05/2017, and instantly frozen in liquid nitrogen after operation until further use. The clinicopatholigcal information were recorded and summarized by patient’s attending physician. The grade of bladder cancer was assessed according to the World Health Organization 2004 Grading System [20] and the stage was classified according to the modified tumor–node–metastasis (TNM) cancer staging system (UICC, 2002) [21]. The research was carried out at First Affiliated Hospital of Nanchang University during 01/2018-08/2019.
Five human bladder cancer cells (T24, 5637, J82, 253J and RT4) and a normal bladder epithelial cell (SV-HUC-1) were directly purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in DMEM medium (Thermo Fisher Scientific, Waltham, MA, USA) and 10% FBS (HyClone, Logan, UT, USA) with an incubator containing 5% CO2 at 37°C. The SV-HUC-1 cell were cultured in F-12K medium (Thermo Fisher Scientific) plus 10% FBS and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin).
RNA isolation and real time quantitative PCR (RT-qPCR)
Total RNA from bladder cancer tissues (size at 2 mm3) or cells (number at 2 × 106) was obtained by using TRIzol reagent (Thermo Fisher Scientific). Each RNA (1 μg) was reversed transcribed to complementary DNA by utilizing a PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Beijing, China), following the manufacturer’s instructions. After that, qPCR was performed with a SYBR Premix ExTaq II kit (Takara) on an ABI 7900 Real Time PCR system (Applied Biosystems, Foster City, CA, USA) to quantify the relative the expression of ROR1-AS1 and miR-504. The qPCR cycling conditions: one denaturation step of 10 min at 95°C; 40 cycles, with one cycle consisting of 10 sec at 95°C, 30 sec at 58°C, and 30 sec at 72°C. The primer sequences were shown in Table 1. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and small nuclear RNA U6 (U6) genes were served as the internal controls. The 2-ΔΔCt method was utilized to analyze the data and calculate the relative expression of each gene [22]. The experiments were performed in triplicate and repeated three times.
Table 1. The primer sequences used for real time quantitative PCR (RT-qPCR).
| Gene symbol | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| ROR1-AS1 | GACGAAACACTGGAA | GTCTGATTTGGTAGCTT |
| GAPDH | CCAAAATCAAGTGGGGC | TGATGGCATGGACTGTGGT |
| miR-504 | GCTGCTGTTGGGAGACC | GCCCTCTGTATGGGAAAC |
| U6 | CTCGCTTCGGCAGCACATA | ACGCTTCACGAATTTGCGT |
Transfection
The specific short hairpin RNA (shRNA) targeting ROR1-AS1 (shRNA-ROR1-AS1, 5’-GUAGGGAAGUACAAUUUUUGAGUUA-3’) and the corresponding non-specific shRNA (shRNA-NC, 5’-UGCAUCCUACTAGAUGGCCUGUAA-3’) were obtained from GenePharma Biotechnology (Suzhou, China). The miR-504 inhibitor (5’-CAGACGUGAGAUAGACAUAAGAA-3’) and inhibitor NC (5’-AUUCCCGAAUCCAAUAGUGCAU-3’) were synthesized in RiboBio Biotechnology (Guangzhou, China). T24 and 5637 cells were cultured in 6-well plates to get 80% confluence and then transfected with the indicated amounts of shRNA-ROR1-AS1 or shRNA-NC and miR-504 inhibitor or inhibitor NC by using lipofectamine 2000 transfection reagent (Thermo Fisher Scientific), according to the specific instructions. The transfection efficiency was analyzed after 48 h transfection by RT-qPCR.
MTT assay
MTT (3-(4,5-dimethylthiazol-2yl)-2,5-diphenyl tetrazolium bromide) assay was applied to detect cell proliferation ability. T24 and 5637 cells were seeded into 96-well culture plate 24 h prior to transfection with shRNA-ROR1-AS1 or shRNA-NC and miR-504 inhibitor or inhibitor NC. After 0, 24, 48 or 72 h transfection, 12 μl of MTT solution (TIANGEN Biotechnology, Beijing, China) at 5 mg/ml concentration was added to each well, and incubated for 3 h at 37°C. Then, the supernatants were discarded, and 120 μl of DMSO (TIANGEN Biotechnology) was used to solubilize the crystals for 20 min at 37°C. Data of absorbance were measured at a wavelength of 480 nm (with 590 nm as the reference wavelength) using a Synergy™ HT Multi-Mode Microplate Reader (Biotek, Winooski, VT, USA) at the indicated time points.
Wound scratch assay
Wound scratch assay was performed to evaluate cell migration capacity. Briefly, T24 and 5637 cells transfected with shRNA-ROR1-AS1 or shRNA-NC and miR-504 inhibitor or inhibitor NC were seeded into six-well culture plates and cultured with DMEM medium and 10% FBS to form a tight monolayer. Scraped lines were created with 100 μl sterile pipette tips, and the cell debris was removed with PBS and the remaining cells were incubated for 24 h at 37°C with no serum-containing DMEM medium. The migrated distances of the growing edge on the monolayer were observed by using a ECLIPSE TS100 light microscope (Nikon Corporation, Tokyo, Japan) under a 200× microscope field at 0~24 h after being wounded.
Luciferase reporter gene assay
A target prediction tool, starBase v2.0 (http://starbase.sysu.edu.cn/starbase2/), was used to predict potential miRs target of ROR1-AS1, and we found ROR1-AS1 could potentially bind with miR-504 sequence. The ROR1-AS1 transcript 1 containing the binding sites and non-binding sites for miR-504 sequence were synthesized and inserted into a psiCHECK2 reporter vector (Promega, WI, USA) to get the ROR1-AS1-WT and ROR1-AS1-MUT reporter vectors, respectively. Subsequently, bladder cancer cells were seeded into 24-well plates and transfected with the ROR1-AS1-WT and ROR1-AS1-MUT vector, together with miR-504 inhibitor or inhibitor NC using Lipofectamine 2000, according to the manufacturer’s instructions. 24 h after the cell transfection, the luciferase activity was determined by using a dual-luciferase reporter assay (Promega, WI, USA). The relative Renilla luciferase activities were normalized to Firefly luciferase activities, which was served as an internal control for transfection efficiency.
Statistical analysis
The statistical analyses were performed using SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Data were presented as mean ± SD (standard deviation) from the three independent experiments. The expression differences between bladder cancer tissues and adjacent matched normal bladder tissues were analyzed using paired student’s t-test. Chi-square test was used for correlation analysis. Kaplan-Meier method was utilized for analysis of prognosis. CCK-8 assay were analyzed using ANOVA analysis followed by post hoc testing. P values were two-sided and a two-tailed value of p<0.05 was considered to be a statistically significant difference.
Results
ROR1-AS1 expression is upregulated in patients with bladder cancer and associates with malignant clinicopatholigcal features
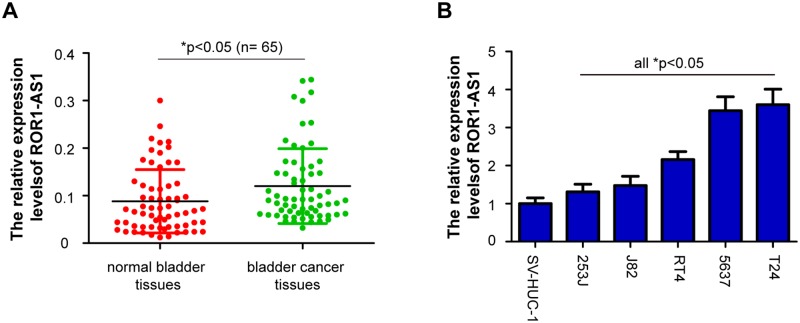
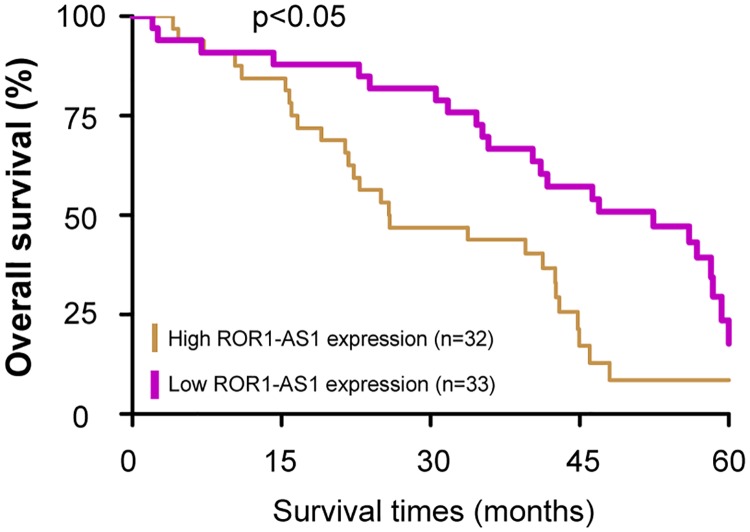
The relative expression levels of ROR1-AS1 was determined by RT-qPCR in a total of 65 cases of bladder cancer patients. Compared with adjacent matched normal bladder tissues, the ROR1-AS1 levels were notably upregulated in bladder cancer tissues (Fig 1A, p<0.05). Subsequently, we analyzed the expression levels of ROR1-AS1 in multiple bladder cancer cell lines (T24, 5637, J82, 253J and RT4). ROR1-AS1 expression was also increased significantly in the five bladder cancer cells compared with the normal bladder epithelial cell (SV-HUC-1) (Fig 1B, p<0.05). Furthermore, the correlations between ROR1-AS1 expression and clinicopathologic variables was evaluated. The 65 pairs of bladder cancer patients were divided into two groups based on the median value of relative ROR1-AS1 expression level: low (n = 33) and high (n = 32) ROR1-AS1 expression groups. As shown in Table 2, the high ROR1-AS1 expression was notably correlated with advanced tumor stage (p = 0.005), higher tumor grade (p = 0.045), and positive lymph node metastasis (p = 0.015). But not correlated with patient’s sex (p = 0.097), age (p = 0.199), smoking status (p = 0.590) and tumor size (p = 0.505). Moreover, patients with high ROR1-AS1 expression had a significantly shorter survival times compared with those patients with low ROR1-AS1 expression (Fig 2, p<0.05). These data indicated that upregulation of ROR1-AS1 may be involved in the progression of bladder cancer.
Fig 1. The expression of lncRNA ROR1-AS1 is upregulated in bladder cancer samples and cell lines.
(A) Real time quantitative PCR (RT-qPCR) analysis of ROR1 antisense RNA 1 (ROR1-AS1) expression in 65 cases of pairs bladder cancer tissues and adjacent matched normal bladder tissues. (B) ROR1-AS1 expression was determined by RT-qPCR in five bladder cancer cell lines (T24, 5637, J82, 253J and RT4) and a normal bladder epithelial cell line (SV-HUC-1). Data were expressed as the mean ± SD (n = 3). *p < 0.05.
Table 2. Correlation between ROR1 antisense RNA 1 (ROR1-AS1) expression and different clinicopathologic features in 65 cases of bladder cancer patients.
| Clinicopathologic features | No. | ROR1-AS1 | p | |
|---|---|---|---|---|
| High (n, %) | Low (n, %) | |||
| Age (years) | 0.199 | |||
| <55 | 19 | 7 (10.8) | 12 (18.5) | |
| ≥55 | 46 | 25 (38.5) | 21 (32.3) | |
| Sex | 0.097 | |||
| Male | 38 | 22 (33.8) | 16 (24.6) | |
| Female | 27 | 10 (15.4) | 17 (26.2) | |
| Smoking status | 0.590 | |||
| No | 14 | 6 (9.2) | 8 (12.3) | |
| Yes | 51 | 26 (40.0) | 25 (38.5) | |
| Size | 0.505 | |||
| <3 cm | 25 | 11 (16.9) | 14 (21.5) | |
| ≥3 cm | 40 | 21 (32.3) | 19 (29.2) | |
| Grade | 0.045* | |||
| G1-G2 | 22 | 7 (10.8) | 15 (23.1) | |
| G3 | 43 | 25 (38.5) | 18 (27.7) | |
| Stage | 0.005* | |||
| T1-T2 | 21 | 5 (7.7) | 16 (24.6) | |
| T3 | 44 | 27 (41.5) | 17 (26.2) | |
| Lymph node metastasis | 0.015* | |||
| No | 26 | 8 (12.3) | 18 (27.7) | |
| Yes | 39 | 24 (36.9) | 15 (23.1) | |
*p<0.05.
Fig 2. Kaplan-Meier overall survival curves by ROR1-AS1 expression.
Patients with high ROR1-AS1 expression exhibited a significantly shorter survival times compared with those patients with low ROR1-AS1 expression.
ROR1-AS1 knockdown suppresses the proliferation and migration of bladder cancer cells
To examine the potential roles of ROR1-AS1 in the proliferation and migration of bladder cancer, T24 and 5637 cells were chose and treated with shRNA-ROR1-AS1 or shRNA-NC using MTT and wound scratch assays. As presented in Fig 3A, ROR1-AS1 siRNA transfected T24 cells significantly decreased ROR1-AS1 expression levels, and knockdown of ROR1-AS1 inhibited T24 cell growth and migration (p<0.05). Meanwhile, shRNA-ROR1-AS1 treated cells downregulated ROR1-AS1 expression in 5637 cells, and arrested 5637 cell proliferation and migration (Fig 3B, p<0.05). These data demonstrated that ROR1-AS1 contributes to cell proliferation and migration in bladder cancer.
Fig 3. The effects of ROR1-AS1 knockdown on bladder cancer cells proliferation and migration.
(A) The T24 cells were transfected with shRNA-ROR1-AS1 or shRNA-NC, and ROR1-AS1 expression levels were analyzed by RT-qPCR after 48 h transfection. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) and wound scratch assays were applied to determine T24 cells growth and migration in vitro. (B) shRNA-ROR1-AS1 treated cells downregulated ROR1-AS1 expression in 5637 cells. ROR1-AS1 knockdown obviously suppressed 5637 cell proliferation and migration. Data from at least three independent experiments, shown as the mean ± SD. *p < 0.05.
ROR1-AS1 acts as a molecular sponge to decrease miR-504 expression
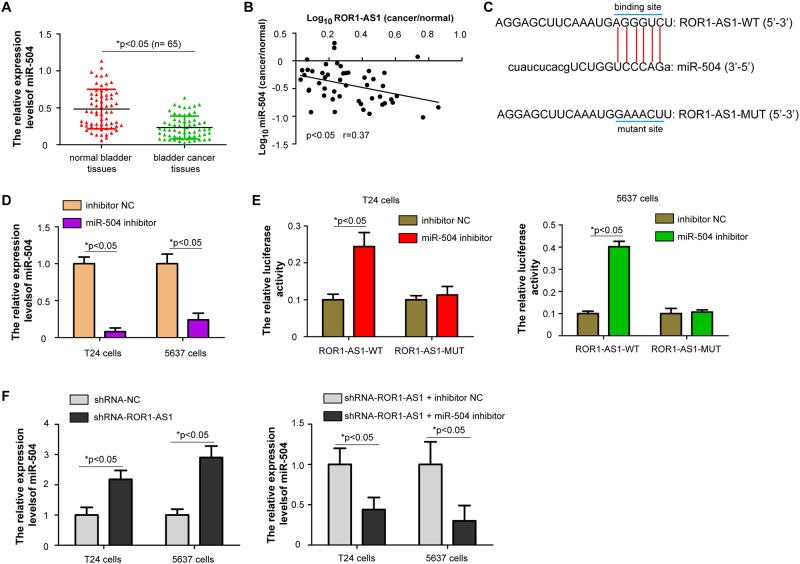
Using bioinformatics software starBase 2.0, we found that ROR1-AS1 contained a binding sites for miR-504 sequence. RT-qPCR found that miR-504 was markedly downregulated in bladder cancer tissues than matched normal bladder tissues (Fig 4A, p<0.05). Pearson’s correlation analysis showed that miR-504 expression was negatively correlated with ROR1-AS1 expression in bladder cancer samples (Fig 4B, p<0.05). The potential binding sites for miR-504, ROR1-AS1-WT and ROR1-AS1-MUT were constructed in luciferase reporter gene vectors (Fig 4C). Results showed that transfection of miR-504 inhibitor in T24 and 5637 cells significantly suppressed miR-504 expression (Fig 4D, p<0.05). Knockdown of miR-504 increased the luciferase activity of ROR1-AS1-WT vector, but not ROR1-AS1-MUT vector in T24 and 5637 cells (Fig 4E, p<0.05). Transfection of shRNA-ROR1-AS1 into bladder cancer cells showed upregulation of miR-504 expression, but co-transfection with shRNA-ROR1-AS1 and miR-504 inhibitor reversed these effects (Fig 4F, p<0.05). These findings indicated that ROR1-AS1 bind with miR-504 and acts as a molecular sponge to decrease miR-504 expression.
Fig 4. The regulatory effects of ROR1-AS1 on miR-504.
(A) RT-qPCR analysis of miR-504 expression in 65 cases of bladder cancer samples. (B) The correlation of miR-504 and ROR1-AS1 expression in bladder cancer samples was evaluated by Pearson’s correlation analysis. (C) Sequences of ROR1-AS1 binding sites within the miR-504. (D) The miR-504 expression in T24 and 5637 cells was detected by RT-qPCR after transfection with miR-504 inhibitor or inhibitor NC. (E) Luciferase assay was measured 24 h post transfection in bladder cancer cells, which were co-transfected with ROR1-AS1-WT and ROR1-AS1-MUT vectors, together with miR-504 inhibitor or inhibitor NC. (F) Transfection of shRNA-ROR1-AS1 into bladder cancer cells showed upregulation of miR-504 expression, but co-transfection with shRNA-ROR1-AS1 and miR-504 inhibitor reversed these effects. Data were presented as the mean ± SD (n = 3). *p < 0.05.
Inhibition of miR-504 partly abrogates ROR1-AS1 knockdown-induced inhibitory effects on bladder cancer cell growth and migration
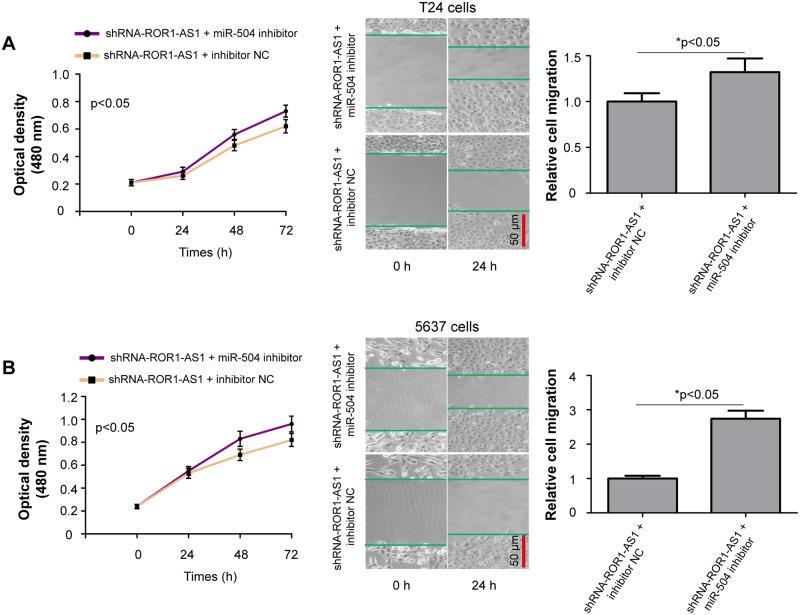
Due to the negatively regulation of ROR1-AS1 on miR-504 in bladder cancer cells, we speculated that the role of ROR1-AS1 in regulating bladder cancer cell proliferation and migration was mediated by sponging miR-504 expression. We transfected miR-504 inhibitor or inhibitor NC into the shRNA-ROR1-AS1 treated T24 and 5637 cells, and functional rescue experiments were performed. Interesting, we found that ROR1-AS1 knockdown mediated inhibitory effects on bladder cancer cell proliferation and migration were partially reversed by co-transfection with shRNA-ROR1-AS1 and miR-504 inhibitor (Fig 5A and 5B, p<0.05).
Fig 5. ROR1-AS1 promotes bladder cancer cells proliferation and migration by sponging miR-504.
(A) MTT and wound scratch assays were applied to determine T24 cells growth and migration in vitro after co-transfection with shRNA-ROR1-AS1 and miR-504 inhibitor or inhibitor NC. (B) Transfection of miR-504 inhibitor partly abrogated ROR1-AS1 knockdown-induced inhibitory effects on 5637 cell growth and migration. Data were expressed as the mean ± SD (n = 3). *p < 0.05.
Discussion
As an emerging hotspot in the investigation of human cancer, disregulation of lncRNAs in bladder cancer have been well studied. Some lncRNAs in bladder cancer can be utilized as potential therapeutic targets [23,24]. For example, Li et al [25] found that deleted in lymphocytic leukemia 1 is upregulated in bladder cancer tissues and patients with high DLEU1 expression exhibits a shorter survival time, and DLEU1 increases cell proliferation, invasion, and cisplatin resistance of bladder cancer cells. Miao et al [26] showed that long intergenic non-protein coding RNA 612 facilitates the proliferation and invasion capacity of bladder cancer cells, suggesting that the lncRNA may act as a potential biomarker and therapeutic target. Gao et al [27] revealed that ZEB1-AS1/miR-200b/FSCN1 axis may serve as a potential target for molecular therapies of bladder cancer. Though there are many researches focusing on the regulation of lncRNAs on bladder cancer, there are still some potential mechanism need to be explored. To the best of our knowledge, this is the first report to illustrate the role of ROR1-AS1 in bladder cancer. Moreover, we demonstrated that increased ROR1-AS1 promotes the proliferation and migration of bladder cancer by sponging miR-504, and suggesting ROR1-AS1 may be used as a prognostic biomarker and therapeutic target for bladder cancer treatment.
Previous studies have indicated that antisense lncRNAs including those located antisense to cancer-related genes were participated in complicate and accurate gene-net of human cancers [28]. ROR1-AS1 is a novel antisense lncRNAs, which was first identified in mantle cell lymphoma, and could mediate tumor growth by histone modification through enhancer of zeste 2 polycomb repressive complex 2 subunit [17]. Recent studies have demonstrated that deregulation of ROR1-AS1 contributes to carcinogenesis. In colorectal cancer, Wang et al [29] found that ROR1-AS1 promotes cell metastasis and proliferation via inducing Wnt/β-catenin signaling pathway. However, the function and molecular mechanism of ROR1-AS1 in bladder cancer are largely unknown until now. In this study, we identified that ROR1-AS1 expression was increased in bladder cancer tissues and cell lines. Furthermore, the high ROR1-AS1 expression levels were closely correlated with higher histological grade, advanced tumor stage, and positive lymph node metastasis in patients with bladder cancer, indicating that upregulation of ROR1-AS1 represents an aggressive phenotypes of bladder cancer. More importantly, series of functional experiments further showed that knockdown of ROR1-AS1 expression inhibited cell growth and migration of bladder cancer cells in vitro. These results are consistent with prior reports suggesting a oncogenic role of ROR1-AS1 in the progression of bladder cancer.
A better understanding of the mechanisms underlying the pathogenesis of bladder cancer is key for improvement of anticancer therapy. Increasing evidence has exhibited that lncRNAs compete for miRNAs response elements (MREs) with the driver genes strongly relevant to tumor progression by acting as a ceRNA [30]. To further identify the potential mechanism of ROR1-AS1 affects bladder cancer cell proliferation and migration, we predicted and chose miR-504 as a potential sponge of ROR1-AS1 using bioinformatic analysis. Recently, investigations demonstrated the tumor-suppressive role of miR-504 in hypopharyngeal squamous cell carcinoma and liver cancer [31,32]. Additional, Ye et al [33] showed that miR-504 is downregulated in non-small cell lung cancer tissues and can inhibit cell proliferation, invasion, and epithelial-mesenchymal transition by targeting lysyl oxidase like 2 expression. Similarly, Liu et al [34] found that miR-504 represses mesenchymal phenotype of glioblastoma by directly targeting the frizzled class receptor 7-mediated Wnt-β-catenin signaling pathway. However, role of miR-504 in bladder cancer is still unknown. In the present study, we first identified the relationship between miR-504 and ROR1-AS1 in bladder cancer samples. Results revealed that miR-504 expression was downregulated and negatively correlated with ROR1-AS1 expression. Moreover, luciferase reporter gene assay demonstrated that ROR1-AS1 acts as a molecular sponge to decrease miR-504 expression. Functional rescue experiments confirmed that ROR1-AS1 promotes cell growth and migration of bladder cancer via regulation of miR-504. Consistent with previous findings, we have validated that downregulated miR-504 acts as a tumor suppressor in bladder cancer.
In conclusion, our data suggested that upregulation of ROR1-AS1 promotes bladder cancer cells proliferation and migration by regulating miR-504. Our findings contribute to a better understanding of the importance of ROR1-AS1/miR-504 axis in bladder cancer progression, and provide a promising of lncRNA-based targeted approach for bladder cancer treatment. Further researches should also be conducted to illustrate the detailed molecular mechanism of ROR1-AS1 in other urinary tumors.
Abbreviations
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- LncRNAs
long non-coding RNAs
- MicroRNAs
miRs
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
- ROR1-AS1
ROR1 antisense RNA 1
- RT-qPCR
real time quantitative PCR
Data Availability
All relevant data are within the manuscript.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Dobruch J, Daneshmand S, Fisch M, Lotan Y, Noon AP, Resnick MJ, et al. (2016) Gender and Bladder Cancer: A Collaborative Review of Etiology, Biology, and Outcomes. Eur Urol. 69(2): 300–310. 10.1016/j.eururo.2015.08.037 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. (2017) Cancer Statistics, 2017. CA Cancer J Clin. 67(1): 7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 3.Maraver A, Fernandez-Marcos PJ, Cash TP, Mendez-Pertuz M, Duenas M, Maietta P, et al. (2015) NOTCH pathway inactivation promotes bladder cancer progression. J Clin Invest. 125(2): 824–830. 10.1172/JCI78185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alifrangis C, McGovern U, Freeman A, Powles T, Linch M. (2019) Molecular and histopathology directed therapy for advanced bladder cancer. Nat Rev Urol. 16(8): 465–483. 10.1038/s41585-019-0208-0 [DOI] [PubMed] [Google Scholar]
- 5.Kim WJ, Bae SC. (2008) Molecular biomarkers in urothelial bladder cancer. Cancer Sci. 99(4): 646–652. 10.1111/j.1349-7006.2008.00735.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty A, Dasari S, Long W, Mohan C. (2019) Urine protein biomarkers for the detection, surveillance, and treatment response prediction of bladder cancer. Am J Cancer Res. 9(6): 1104–1117. [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet. 10(3): 155–159. 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 8.Moran VA, Perera RJ, Khalil AM. (2012) Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 40(14): 6391–6400. 10.1093/nar/gks296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, et al. (2013) Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 20(7): 908–913. 10.1038/nsmb.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. (2016) Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 7(9): e2389 10.1038/cddis.2016.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. (2017) Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 77(15): 3965–3981. 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi Y, Wang D, Wang J, Yu W, Yang J. (2019) Long Non-Coding RNA in the Pathogenesis of Cancers. Cells. 8(9): 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng XY, Jiang XY, Yong JH, Xie H, Yuan J, Zeng D, et al. (2019) lncRNA ABHD11-AS1, regulated by the EGFR pathway, contributes to the ovarian cancer tumorigenesis by epigenetically suppressing TIMP2. Cancer Med. 8(16): 7074–7085. 10.1002/cam4.2586 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Song Z, Zhang X, Lin Y, Wei Y, Liang S, Dong C. (2019) LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J Cell Mol Med. 23(11): 7554–7565. 10.1111/jcmm.14625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng YJ, Zhao JY, Liang TS, Wang P, Wang J, Yang DK, et al. (2019) Long noncoding RNA SMAD5-AS1 acts as a microRNA-106a-5p sponge to promote epithelial mesenchymal transition in nasopharyngeal carcinoma. FASEB J. 33(11): 12915–12928. 10.1096/fj.201900803R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang C, Xu L, He W, Dai J, Sun F. (2019) Long noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle. 18(23): 3288–3299. 10.1080/15384101.2019.1673633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu G, Gupta SK, Troska TP, Nair A, Gupta M. (2017) Long non-coding RNA profile in mantle cell lymphoma identifies a functional lncRNA ROR1-AS1 associated with EZH2/PRC2 complex. Oncotarget. 8(46): 80223–80234. 10.18632/oncotarget.17956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao T, Maierdan SL, Lv C. (2019) ROR1-AS1 promotes tumorigenesis of colorectal cancer via targeting Wnt/beta-catenin. Eur Rev Med Pharmacol Sci. 23(3 Suppl): 217–223. 10.26355/eurrev_201908_18650 [DOI] [PubMed] [Google Scholar]
- 19.Wang FZ, Zhang MQ, Zhang L, Zhang MC. (2019) Long non-coding RNA ROR1-AS1 enhances colorectal cancer metastasis by targeting miR-375. Eur Rev Med Pharmacol Sci. 23(16): 6899–6905. 10.26355/eurrev_201908_18729 [DOI] [PubMed] [Google Scholar]
- 20.Pellucchi F, Freschi M, Ibrahim B, Rocchini L, Maccagnano C, Briganti A, et al. (2011) Clinical reliability of the 2004 WHO histological classification system compared with the 1973 WHO system for Ta primary bladder tumors. J Urol. 186(6): 2194–2199. 10.1016/j.juro.2011.07.070 [DOI] [PubMed] [Google Scholar]
- 21.Oosterlinck W, Lobel B, Jakse G, Malmstrom PU, Stockle M, Sternberg C. (2002) Guidelines on bladder cancer. Eur Urol. 41(2): 105–112. 10.1016/s0302-2838(01)00026-4 [DOI] [PubMed] [Google Scholar]
- 22.Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 3(6): 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 23.Gulia C, Baldassarra S, Signore F, Rigon G, Pizzuti V, Gaffi M, et al. (2017) Role of Non-Coding RNAs in the Etiology of Bladder Cancer. Genes (Basel). 8(11): 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taheri M, Omrani MD, Ghafouri-Fard S. (2018) Long non-coding RNA expression in bladder cancer. Biophys Rev. 10(4): 1205–1213. 10.1007/s12551-017-0379-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Shi B, Dong F, Zhu X, Liu B, Liu Y. (2019) Long Non-coding RNA DLEU1 Promotes Cell Proliferation, Invasion, and Confers Cisplatin Resistance in Bladder Cancer by Regulating the miR-99b/HS3ST3B1 Axis. Front Genet. 10: 280 10.3389/fgene.2019.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao L, Liu HY, Zhou C, He X. (2019) LINC00612 enhances the proliferation and invasion ability of bladder cancer cells as ceRNA by sponging miR-590 to elevate expression of PHF14. J Exp Clin Cancer Res. 38(1): 143 10.1186/s13046-019-1149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao R, Zhang N, Yang J, Zhu Y, Zhang Z, Wang J, et al. (2019) Long non-coding RNA ZEB1-AS1 regulates miR-200b/FSCN1 signaling and enhances migration and invasion induced by TGF-beta1 in bladder cancer cells. J Exp Clin Cancer Res. 38(1): 111 10.1186/s13046-019-1102-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinescu S, Ignat S, Lazar AD, Constantin C, Neagu M, Costache M. (2019) Epitranscriptomic Signatures in lncRNAs and Their Possible Roles in Cancer. Genes (Basel). 10(1): 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Zheng W, Zhang L, Li K. (2019) Long non-coding RNA ROR1-AS1 incudes tumorigenesis of colorectal cancer by affecting Wnt/beta-catenin signaling pathway. Biosci Rep. 39(11): BSR20191453 10.1042/BSR20191453 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. (2019) Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 234(7): 10080–10100. 10.1002/jcp.27941 [DOI] [PubMed] [Google Scholar]
- 31.Quan H, Li B, Yang J. (2018) MicroRNA-504 functions as a tumor suppressor in hepatocellular carcinoma through inhibiting Frizzled-7-mediated-Wnt/beta-catenin signaling. Biomed Pharmacother. 107: 754–762. 10.1016/j.biopha.2018.07.150 [DOI] [PubMed] [Google Scholar]
- 32.Kikkawa N, Kinoshita T, Nohata N, Hanazawa T, Yamamoto N, Fukumoto I, et al. (2014) microRNA-504 inhibits cancer cell proliferation via targeting CDK6 in hypopharyngeal squamous cell carcinoma. Int J Oncol. 44(6): 2085–2092. 10.3892/ijo.2014.2349 [DOI] [PubMed] [Google Scholar]
- 33.Ye MF, Zhang JG, Guo TX, Pan XJ. (2018) MiR-504 inhibits cell proliferation and invasion by targeting LOXL2 in non small cell lung cancer. Biomed Pharmacother. 97: 1289–1295. 10.1016/j.biopha.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 34.Liu Q, Guan Y, Li Z, Wang Y, Liu Y, Cui R. (2019) miR-504 suppresses mesenchymal phenotype of glioblastoma by directly targeting the FZD7-mediated Wnt-beta-catenin pathway. J Exp Clin Cancer Res. 38(1): 358 10.1186/s13046-019-1370-1 [DOI] [PMC free article] [PubMed] [Google Scholar]