Abstract
Background:
A major roadblock to reducing the mortality of colorectal cancer (CRC) is prompt detection and treatment and a simple blood test is likely to have higher compliance than all of the current methods. The purpose of this report is to examine the utility of a mass spectrometry based blood serum protein biomarker test for detection of CRC.
Materials and Methods:
Blood was drawn from individuals (n=213) prior to colonoscopy or from patients with non-metastatic colorectal cancer (n=50) prior to surgery.
Proteins were isolated from the serum of patients using targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS). We designed a machine-learning statistical model to assess these proteins.
Results:
When considered individually, over 70% of the selected biomarkers showed significance by Mann-Whitney testing for distinguishing cancer-bearing cases from cancer-free cases. Using machine-learning methods, peptides derived from EGFR and LRG1 were consistently identified as highly predictive for detecting colorectal cancer from cancer-free cases. A five-marker panel consisting of LRG1, EGFR, ITIH4, HPX, and SOD3 performed the best with 70% specificity at over 89% sensitivity (AUC=0.86) in the validation set. For distinguishing regional from localized cancers, cross-validation within the training set showed that a panel of four proteins consisting of CD44, GC, CRP, and ITIH3 yielded the highest performance (AUC=0.75).
Conclusions:
The minimally invasive blood biomarker panels identified here could serve as screening/detection alternatives for colorectal cancer in a human population, and potentially useful for staging of existing cancer.
Keywords: Colorectal neoplasms, biomarkers, proteomic
INTRODUCTION
Colorectal cancer has the second highest incidence and mortality of cancers diagnosed in the United States(1, 2). Early stages of the disease are highly curable and can be detected in asymptomatic patients through a variety of primary screening procedures(2). The most common methods include endoscopic, fecal, and radiologic screening(3). However, approximately 40% of the recommended screening population does not adhere to screening guidelines, which results in late-stage disease diagnosis and increased rates of morbidity and mortality(4, 5). Available screening methods are often expensive and require laborious bowel preparation (endoscopy, CT colonography), or require the physical handling of stool by the patient (gFOBT, FIT, multitarget stool DNA testing)(5, 6). In order to increase adherence to colorectal cancer screening, the United States Preventative Task Force (USPSTF) has recently changed its guidelines for colorectal cancer screening to include any effective test to screen for colorectal cancer in patients between the ages of 50 and 75(7). These changes have the capability to increase screening compliance in cases where patients cannot afford or access a particular screening method by providing more screening options(8, 9). The new guidelines also indicate the need for developing screening methods that would increase screening choices and may subsequently improve compliance.
Colorectal cancer morbidity and mortality rates are also affected by complications arising from colorectal surgery. Up to 35% of colorectal surgery patients suffer from post-operative complications, which have been shown to translate to a worse quality of life, poor oncologic outcomes, additional complications, and overall higher risk for mortality(10). Localized surgical techniques such as endoscopic and/or minimally-invasive resection could reduce the incidence of patient morbidity and mortality if regional lymph node status could be more accurately predicted(11). Preoperative lymph node status is typically assessed using CT and MR with low accuracy(12, 13). Perhaps in the future serum studies will be harnessed in tandem with imaging modalities will provide a pristine picture of nodal disease burden. In addition, neoadjuvant therapy of advanced colon cancer may provide benefits. Early results from the FOxTROT study indicate that neoadjuvant therapy decreases lymph node involvement and size of primary tumor(14). However, the application of neoadjuvant therapy is limited by our inability to accurately stage patients prior to surgery. Therefore, a serum test that may contribute to staging of colorectal cancer prior to surgery would prove to be an important advancement(14).
Increasingly, cancer serum biomarkers have become an attractive method for screening, prognostic, diagnostic, and recurrence monitoring in multiple diseases(15, 16). Recently FDA approval was granted for a Septin 9 DNA blood test for colorectal cancer(17, 18). However, like most biomarker tests, the Septin 9 test is only available for screening purposes. Moreover, the Septin 9 test does not have sensitivity and specificity (<50% sensitive for carcinoma) that is competitive with other available non-invasive screening tests(19). Here we present a study in which the sensitivity and specificity of blood serum protein biomarkers were investigated for colorectal cancer detection. Further, the initial trends for distinguishing localized colorectal cancer from regional lymph node positive disease are also described. The minimally invasive identification of cancer and lymph node status could substantially reduce morbidity and mortality from late stage diagnosis and complications arising from subsequent colorectal surgery.
MATERIALS AND METHODS
Study Design:
This prospective cross-sectional pilot study design was reviewed and approved by the Institutional Review Board (IRB) at the University of Wisconsin-Madison. Patients were prospectively enrolled in the study at the time of a routine screening colonoscopy or prior to colorectal surgery (for those patients undergoing a partial colectomy to remove colorectal cancer). For each enrolled patient, a 5mL blood draw was completed prior to the colonoscopy or colectomy procedure. Blood was processed into serum in accordance with guidelines set by the Early Detection Research Network(20). All serum was stored in 100μg aliquots in protein LoBind tubes (Eppendorf™) at −80°C until use and was thawed at room temperature. A targeted liquid chromatography tandem – mass spectrometry (LC-MS/MS) procedure was used to quantify biomarker analytes. Reference standards corresponding to target analytes were added to each patient sample for quantification purposes. For all target biomarkers, a relative chromatographic area ratio of the biomarker compared to its reference standard was calculated. The endpoint was a set of relative area ratios used for comparison across patient groups to evaluate biomarkers that could identify cancer compared to cancer-free cases as well as localized cancer (TNM stages 1&2) compared to regional cancer (TNM stage 3)(21, 22). The value of a panel of blood protein biomarkers was then assessed to identify regional lymph node involvement in non-metastatic cancer cases to provide a potentially new method for staging existing cancers.
Study Participants:
Participants were enrolled between 2014 and 2016 at the University of Wisconsin Hospital and Clinics (UWHC) after providing informed consent. Colonoscopy patients were ineligible to participate in the study if they had a history of IBD, and colectomy patients were ineligible if they had been previously diagnosed with metastatic (TNM stage 4) colorectal cancer or had received neoadjuvant treatment prior to the blood draw. We calculated that with 40 subjects per group we would have at least 80% power for identifying significance in the difference between the means of the cancer and non-cancer groups. We enrolled substantially more patients than the initially calculated 40 in cancer and non-cancer groups to take into account potential patient withdrawal and to achieve similar power for a completely polyp free control group. This study was not originally powered to identify differences between local and regional colorectal cancers. However, the staging analysis was included in this report due to the importance of the findings on potential minimally invasive staging, which currently cannot be performed with high sensitivity using other methods.
Selection of Candidate Biomarkers:
The candidate biomarkers considered arose from two studies previously performed in murine models at the University of Wisconsin-Madison(23, 24). The first study was conducted in the ApcMin/+ mouse to discover novel biomarkers in blood serum(25). Due to the disproportionate number of small intestinal polyps compared to colon in the ApcMin/+ mouse, a validation study was done in the more recently developed ApcPirc/+ rat model(26). The ApcPirc/+ rat has polyps with a substantially higher specificity to the colon that could be longitudinally monitored by colonoscopy for the purpose of identifying colon cancer-specific blood biomarkers. The ApcPirc/+ rat study validated differential expression of markers from the ApcMin/+ mouse study, verifying that the discovery study produced markers specific to colonic polyps. For the study presented herein, we selected 17 markers that were highly differentially expressed in tumor bearing versus control animals, had potential biological relevance to colorectal cancer, and may have been implicated as a blood biomarker in other published colorectal cancer studies.
Assay Methods:
Serum was depleted of the top seven most abundant proteins using antibody affinity chromatography (Agilent) according to manufacturer instructions. Remaining serum proteins were isolated using trichloroacetic acid precipitation, were resolubilized, and digested according to the referenced protocol(24). Peptide reference standards corresponding to target biomarkers were added to each patient sample. Reference standards contained one stable isotope labeled heavy amino acid in order to differentiate the endogenous peptide target from the reference standard (Appendix 1). LC-MS/MS was done according to the referenced targeted mass spectrometry procedure and also can be found replicated in our recent publication in Proceedings of the National Academy of Sciences of the United States of America (PNAS, “Conserved serum protein biomarkers associated with growing early colorectal adenomas”) examining colorectal adenoma biology (24). Resulting data was analyzed using Skyline open source software(27).
Statistical Analysis Methods:
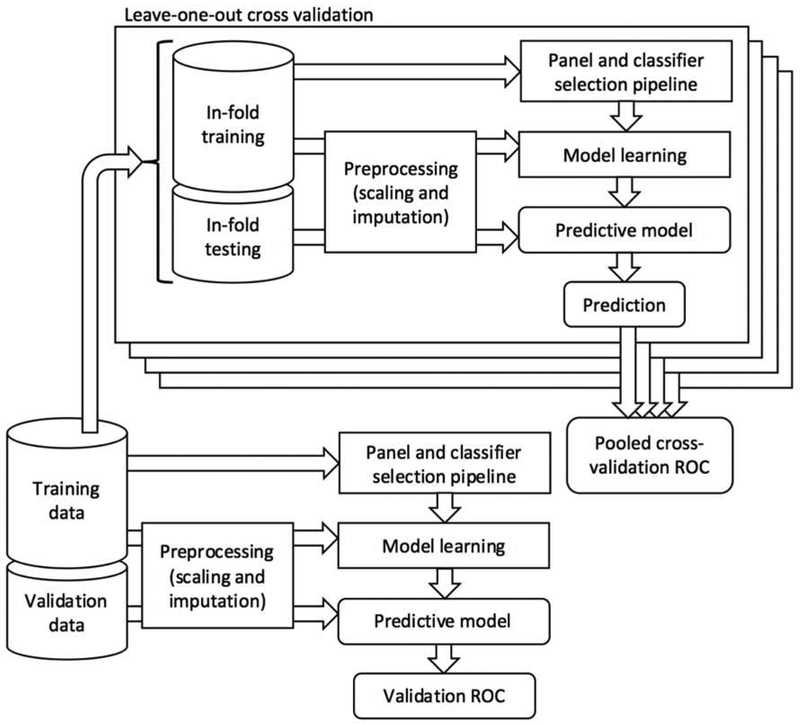
A comparison of single biomarkers between patient groups was done using a two-tailed Mann-Whitney test, with the false-discovery rate controlled at 0.05 using the Benjamini-Hochberg method. Using machine-learning methods, we sought to identify small panels of 2-5 biomarkers that could identify cancer compared to low-risk cancer-free cases and localized cancer compared to regional cancer. We cast the two screening tasks as classification tasks, and evaluated an array of machine-learning methods for each (Figure 1, Appendix 2). Specifically, we used logistic regression, support vector machines with linear and radial basis function (RBF) kernels, Gaussian naïve Bayes, decision trees, random forests, and extremely randomized trees. We used several feature selection methods for identifying candidate panels of biomarkers in a data-driven manner. Specifically, we evaluated L1-regularized logistic regression, L1-regularized linear support vector machines, and feature importance rankings for classification trees, random forests, and extremely randomized trees. We designed an analysis pipeline in which (1) a set of candidate panels is obtained using the feature selection methods, (2) every combination of panel and classification method is evaluated to produce an ROC curve, and (3) the combination that achieved maximum sensitivity above 80% specificity is selected.
Figure 1:
A schematic of the statistical analysis pipeline used. For each panel size of 2 up to 5 biomarkers, a pooled cross-validation ROC curve using the training data was produced by systematically selecting a classifier and panel using in-fold training data and testing on a prediction of the in-fold testing case. Validation was done on a separate set of data after selecting a classifier method and panel of markers using the training data. A validation ROC curve showing the performance of the resultant predictive model, trained on the training data and evaluated on the validation data was produced.
We evaluated our panel and classifier selection pipeline in terms of how well it generalized to data that was not used to build the models. We set aside almost half of the cases for a validation set and the remaining cases comprised a training set. The learning and validation cohorts were randomly assigned, though the percentage/number of cases assigned to the validation set was based on the discrete number of hyperparameters in our design (i.e. size/number of biomarker panels; 2-5 biomarkers). We used cross-validation within the training set to produce a receiver-operator characteristic (ROC) curve representative of the performance of the pipeline, and used training on the full training set and prediction on the validation set to produce an ROC curve demonstrating the performance of a specific selected panel and classification method for each of the screening tasks. Confidence intervals around the ROC curves were drawn using the referenced method(28). Additional method details can be found in Appendix 3. Raw data and code implementing the analysis is available at https://github.com/sverchkov/ivancic-panel-selection.
RESULTS
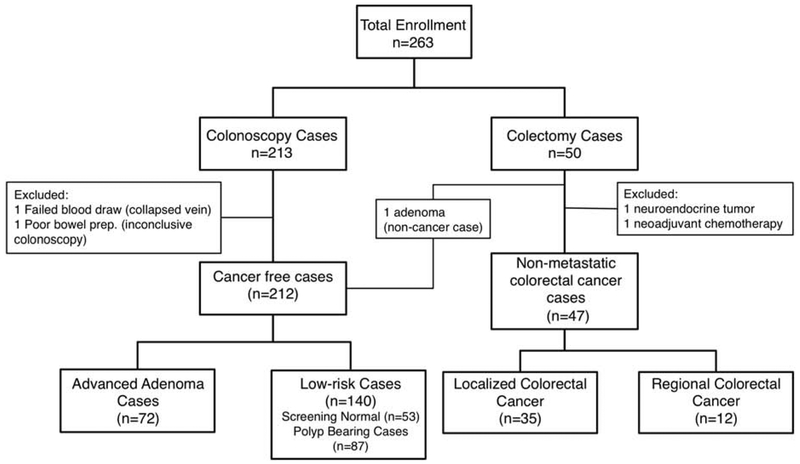
We enrolled 213 patients undergoing screening colonoscopy and 50 patients with non-metastatic colorectal cancer. Blood was collected prior to colonoscopy or surgery (Figure 2, Appendix 4). Patients in both groups underwent standard of practice bowel preparation procedures prior to the respective procedures. Two cases were discounted from the study due to error during blood draw and colonoscopy bowel preparation. Two surgery cases were removed because it was subsequently learned that one had received neoadjuvant treatment prior to the blood draw and the other had been diagnosed with a neuroendocrine tumor. Approximately equal numbers of males (n=130) and females (n=129) were enrolled in all of the patient groups. The median age at the time of blood draw for colonoscopy cases was 60 (range: 30-80) and the median age for colectomy cases was 62 (range: 37-88). A total of 35 localized cancer cases and 12 regional cancer cases were enrolled. We set aside 119 cases (109 cancer-free and 10 cancer cases as validation data, leaving the remaining 140 cases (103 cancer-free and 37 cancer cases) to comprise a training set.
Figure 2:
Overview of patient enrollment in this study. Blood was drawn prior to routine screening colonoscopy or prior to colectomy.
The individual biomarkers were assessed for statistical significance in detecting colorectal cancer from cancer-free cases. Over 70% of biomarkers showed significance by Mann-Whitney testing for distinguishing cancer cases compared to cancer-free cases (Table 1, Appendix 5). Multi-marker panels between 2 and 5 markers comparing cancer cases and cancer-free cases identified LRG1 (leucine rich alpha-2-glycoprotein 1) and EGFR (epidermal growth factor receptor) as members of all of the top marker panels in the classifier methods tested (Appendix 6). ITIH4 (inter-alpha-trypsin inhibitor heavy chain family member 4) was a member of the three-, four-, and five-marker panels.
Table 1:
A summary of Mann-Whitney P-values of all 17 tested markers. Significance was defined as a P-value less than or equal to 0.01532 after using Benjamini-Hochberg to control the false discovery rate at 0.05. Bolded values were considered significant.
| Protein Name | Protein Symbol |
Murine Studies Expression in Polyp Bearing vs. Polyp Free Control |
Human Expression in Cancer vs. Non- Cancer |
Cancer detection compared to non- cancer (p-value) |
|---|---|---|---|---|
| CD44 antigen | CD44 | Downregulated | Downregulated | <0.001 |
| Complement factor I | CFI | Upregulated | NS | 0.660 |
| Leucine-rich alpha-2-glycoprotein | LRG1 | Upregulated | Upregulated | <0.001 |
| Epidermal growth factor receptor | EGFR | Downregulated | Downregulated | <0.001 |
| Inter-alpha-trypsin inhibitor heavy chain H3 | ITIH3 | Upregulated | Upregulated | <0.001 |
| Coagulation factor V | F5 | Upregulated | Upregulated | 0.004 |
| Hemopexin | HPX | Upregulated | Upregulated | <0.001 |
| Vitamin D-binding protein | GC | Upregulated | NS | 0.170 |
| Inter-alpha-trypsin inhibitor, heavy chain 4 | ITIH4 | Upregulated | Upregulated | <0.001 |
| Serum amyloid P | APCS | Upregulated | NS | 0.864 |
| Fetuin B | FETUB | Upregulated | Downregulated | 0.003 |
| C-reactive protein | CRP | Upregulated | Upregulated | 0.001 |
| Sulfhydryl oxidase 1 | QSOX1 | Upregulated | Upregulated | 0.184 |
| Peptidase inhibitor 16 | PI16 | Upregulated | Downregulated | 0.001 |
| Cadherin-2 (N-Cadherin) | CDH2 | Upregulated | NS | 0.652 |
| Dipeptidyl peptidase 4 | DPP4 | Downregulated | Downregulated | <0.001 |
| Extracellular superoxide dismutase [Cu-Zn] | SOD3 | Downregulated | NS | <0.001 |
"NS" means not statistically significant
Bolded values were statistically significant by Mann-Whitney testing
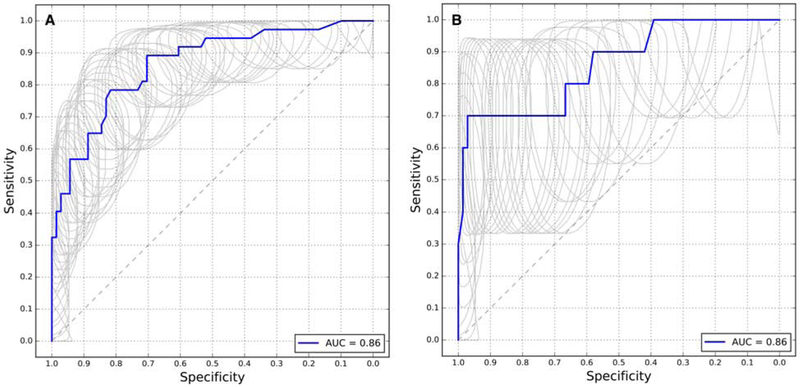
Our main focus in selecting an optimal protein biomarker panel for screening was to target panels with a higher specificity(29). Low specificity panels result in an increased frequency of follow-up colonoscopies due to excessive false positives. We concluded that to reduce the need for follow-up colonoscopies, the desired set of markers should have specificity above 80% in order to match current minimally invasive tests. Therefore, we targeted the highest sensitivity at more than 80% specificity within the training set cross-validated ROC curve in order to reduce false-positive cases for colorectal cancer screening. A five-marker panel consisting of LRG1, EGFR, ITIH4, HPX (hemopexin), and SOD3 (superoxide dismutase 3) identified using the random forest model performed the best according to this criterion, with 89% specificity at over 70% sensitivity in the validation set (Figure 3, Table 2).
Figure 3:
(A) A Pooled ROC curve produced from leave-one-out cross validation for panels of 5 biomarkers to distinguish non-metastatic cancer cases from low-risk cancer-free cases. (B) A ROC curve produced using the validation set to evaluate the selected 5-biomarker panel and method. The selected panel of biomarkers is HPX, ITIH4, LRG1, EGFR, and SOD3 with a random forest classifier model.
Table 2:
Table of counts showing how many times each panel and method were selected across leave-one-out cross-validation runs for panels of 5 biomarkers for the task of distinguishing cancer cases from low-risk cancer-free cases. Methods or panel candidates that were never selected as best within some fold are not shown. The cell corresponding to the method and panel combination that was selected when fitting to the entire training set (i.e. random forest and EGFR, HPX, ITIHR, LRG1, SOD3) is bolded
| Classification method | |||
|---|---|---|---|
| Panel | Extremely randomized trees | Random Forest | Total |
| DPP4, EGFR, ITIH3, ITIH4, LRG1 | 9 | 15 | 24 |
| EGFR, HPX, ITIH4, LRG1, SOD3 | 4 | 17 | 21 |
| DPP4, EGFR, ITIH4, LRG1, PI16 | 17 | 1 | 18 |
| CD44, EGFR, HPX, ITIH4, LRG1 | 11 | 4 | 15 |
| CD44, EGFR, ITIH4, LRG1, SOD3 | 12 | 3 | 15 |
| CD44, EGFR, ITIH3, ITIH4, LRG1 | 9 | 0 | 9 |
| EGFR, FETUB, ITIH4, LRG1, SOD3 | 1 | 2 | 3 |
| CFI, EGFR, ITIH4, LRG1, SOD3 | 2 | 0 | 2 |
| CD44, EGFR, ITIH4, LRG1, QSOX1 | 1 | 0 | 1 |
| Total | 66 | 42 | |
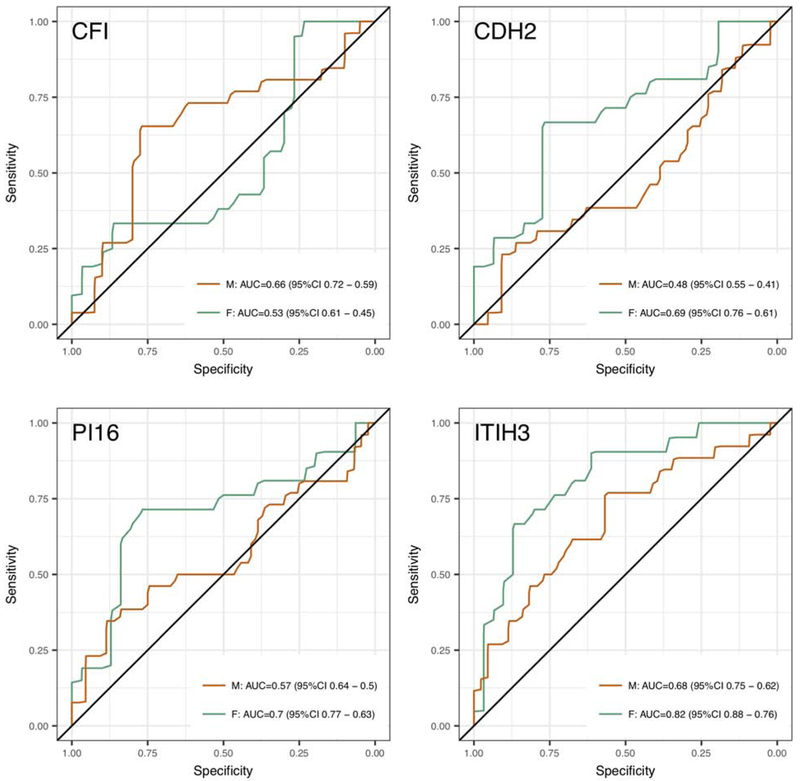
We considered that gender differences might affect protein expression. Therefore, we examined differences in cancer versus low-risk cases for males and females. CDH2 (cadherin2), CFI (complement factor 1), ITIH3 (inter-alpha-trypsin inhibitor heavy chain family member 3), and PI16 (peptidase inhibitor 16) showed potential differences in expression based on training set data as individual proteins (Figure 4). These proteins were within biomarker panels that were selected with high frequency in the leave-one-out cross validation analysis although they were not selected as part of the final panel (Table 2). We concluded that, in the current analysis, gender differences did not have adverse effects on identifying sensitive and specific panels for colorectal cancer detection. However, these differences may impact biomarker efficacy in a larger population study.
Figure 4:
Gender-related differences in sensitivity and specificity for predicting colorectal cancer compared to non-cancer cases using individual biomarkers. CFI, CDH2, ITIH3, and PI16 showed the greatest gender-related differences. For all of the markers shown, with the exception of CFI, females (F) showed higher AUCs than males (M) for the same marker.
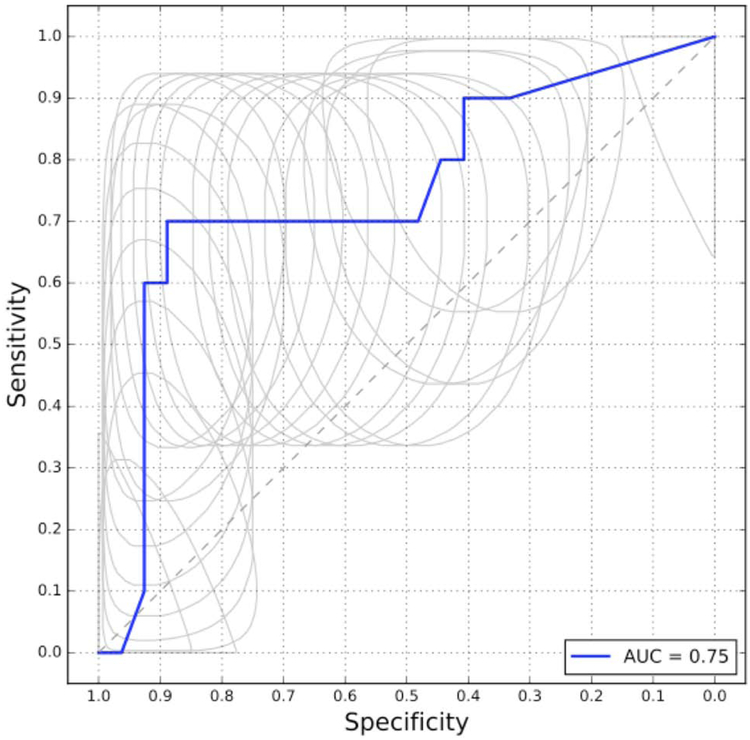
Finally, we examined how the biomarkers performed in discriminating cases with regional lymph node involvement from localized colorectal cancer cases. Cross-validation on the training set showed no predictive potential in 2-marker panels selected by our pipeline, 3-marker panels showed some ability to identify regional cancer, and the best performance was seen with 4-marker panels, with an AUC of 0.75 and 70% sensitivity at 89% specificity (Figure 5, Appendix 7). Five-marker panels did not show an increased performance over 4-marker panels. Using the entire training set, without cross-validation, the method and 4-marker panel selected by the pipeline was random forest with a panel consisting of GC (GC-vitamin D binding protein), CD44 (CD44 molecule), CRP (C-reactive protein), and ITIH3. These findings are of course limited by relatively small sample size.
Figure 5:
Pooled ROC curve produced from leave-one-out cross validation for panels of 4 biomarkers to distinguish non-metastatic cancer cases from low-risk cancer-free cases. The random forest method and a panel containing GC, CD44, CRP, and ITIH3 were identified using the entire training set without cross-validation.
DISCUSSION
The identification of non-invasive biomarkers for colorectal cancer detection with adequate sensitivity and specificity has been a significant challenge in the effort to increase patient compliance and subsequently reduce morbidity and mortality. New non-invasive tests are under scrutiny to meet a balance between sensitivity for true detection to ensure that patients receive proper care, and specificity to reduce the need for too many follow-up colonoscopies(30, 31). Our biomarker panel meets a critical specificity threshold of above 89% and 70% sensitivity nearly equaling the FIT stool-based test, which is considered a reliable alternative to colonoscopy, if done annually. The confidence intervals represented in the ROC curves show potential for increased sensitivity, although the large confidence interval range in the validation curve indicates potential for reduced sensitivity as well. A large screening trial where the biomarkers are put into clinical practice is necessary to determine the sensitivity and specificity more precisely across a large, average-risk population. Such a screening trial would likely require tens of thousands of patients in order to obtain enough cancer cases given that an average-risk routine screening population has a colorectal cancer prevalence of approximately 0.2%(18).
Given that a patient is approximately 25-50 times more likely to have an advanced adenoma than colorectal cancer, ideally, screening tests should also be able to identify advanced benign lesions with relatively high sensitivity and specificity(32, 33). In our study, we considered a comparison of advanced adenoma cases to a low-risk group adenoma. However, given our small sample size and assay variation factors, it is not surprising that we did not identify a biomarker panel with statistical significance in validation sets using any of the models learned on a training set. Further, it is likely that the concentration of serum biomarkers generated during adenomagenesis and carcinogenesis may differ to some degree, and this dynamic deserves further investigation. In fact, our group has investigated this phenomenon in a parallel adenomageneis study. Low sensitivity and specificity for detecting colorectal adenomas plagues the majority of screening tests, particularly non-invasive molecular tests. We predict that combining screening markers from multiple tests could solve this problem although potential increased costs could be prohibitive(19). Ideally, blood-based molecular diagnostics would be employed in conjunction with conventional endoscopic screening to perhaps bring to bear incredibly accurate and reliable screening of colorectal malignancies.
Colorectal surgical procedures remain a common cause of morbidity and mortality related to colorectal cancer. Although this study was not powered to look in depth at colorectal cancer staging markers, we did identify GC, CD44, CRP, and ITIH3 as having potential clinical significance. Other studies have found correlations with a few of these markers and colorectal cancer. Multiple studies have identified CRP as an indicator of pervasive inflammation associated with multiple diseases, including late-stage colorectal cancer(34-37). Work utilizing immunohistochemistry has shown a down-regulation of CD44 on lymph node positive colorectal cancer tissue(38). It is yet to be determined if CD44 expression can be linked to the presence of CRC. Further, low vitamin D levels have been implicated in poor colorectal cancer prognosis(39). However, the exact role of vitamin D binding protein (GC) concentration, freely circulating vitamin D, and bioavailability is unknown in this disease state(40). These factors are currently being studied in relation to colorectal cancer prognosis. Future studies examining cancer staging via protein biomarker measurements in blood serum should be done with sufficient power and in conjunction with a comparator method for identifying affected regional lymph nodes.
Our study displays a number of limitations that should be ameliorated with future projects focusing on protein-based molecular diagnostics for CRC. First, both the murine and rat models of intestinal adenomagenesis that laid the groundwork for this study do not exactly model the known step-wise pathogenesis of human large bowel malignancy. That being said, this study has proven that the differential expression of proteins released into serum of humans bearing colorectal cancer mimics to some degree these animal models. Further, our study did not examine the biomarker level differences between varying tumor differentiation, genetic basis of the tumors (microsatellite instability, BRAF/KRAS status, APC, etc), or tumor sidedness. Future work must account for these important clinicopathological descriptors of colorectal cancer as they have important prognostic and treatment consequences. There is also the issue of nonstandardized pathophysiologic timepoints for blood collections. We held to a protocol of standard of care bowel preparation, however given the pilot study nature of this project, we were not able to control other factors of blood draw timing (lead time to procedure, prior procedures, diet prior to prep, etc). There exists some chance that these factors could affect biomarker levels, however we feel that our panel is discerning enough, especially given the machine learning methods employed, to account for these variations. A final limitation of our study is a lack of real-world applicability, feasibility, or cost-benefit analyses. Unfortunately, the original design of the investigation herein did not include arrangements for these types of analyses. Our group plans to design and implement a study utilizing this technology to assess large-scale application, practical use, and patient attitudes among other real-world outcomes. We would hope that one day our process will improve adherence and increase access to cancer-detection methods among the general population.
CONCLUSIONS
In conclusion, we show that a blood protein panel consisting of LRG1, EGFR, ITIH4, HPX, and SOD3 has the potential to detect colorectal cancer with approximately 70% sensitivity at over 89% specificity in our population. When examining colorectal cancer stages, four of the proteins (GC, CD44, CRP, and ITIH3) could have the potential to distinguish cases with regional lymph node invasion from localized colorectal cancers. Future studies utilizing the protein biomarkers in clinical practice and increased numbers of patients will elucidate the utility of these protein panels in routine detection. Further work related to ours may one day reduce the disease burden of colorectal cancer via non-invasive high accuracy detection.
Supplementary Material
Acknowledgements:
Serge Aleshin-Guendel of the University of Wisconsin Department of Biostatistics and Medical Informatics was very involved with the day-to-day design of the machine learning methods found within and his assistance and expertise is much appreciated. Leigh W. Anson of the University of Wisconsin Biotechnology Center was instrumental in processing and organizing the serum samples as well as analyzing the peptides using the mass spectrometer.
Research Funding Support:
We acknowledge support from the Wisconsin Alumni Research Foundation (MMI, LWA, MRS); NIH T32 GM08349 (MMI); NIH T32 CA157322-03 (MMI); Advanced Opportunity Fellowship through SciMed Graduate Research Scholars at UW-Madison (MMI); NIH T15 LM007359 (YS); NIH U54 AI117924 (YS, SA-G, MC); NIH R01 CA169331-01 (PJP); NIH R01 CA144835-01 (PJP); NIH R01 CA155347-01 (PJP); NIH UL1TR000427 (MC); NIH R01 ES020900 (BM, GDK); The Institute for Clinical and Translational Research (ICTR) at UW-Madison (BM); University of Wisconsin School of Medicine and Public Health Shapiro Research Program (BM)
Footnotes
Conflicts of Interests:
The authors as a whole do not have any conflicts of interest to disclose, however Dr. Perry J. Pickhardt has unrelated financial relationships that we will list below for completeness and transparency. Dr. Pickhardt reports the following: Advisor to Bracco; Shareholder in SHINE and Elucent.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel RL, Miller KD, Jemal A Cancer Statistics, 2017. CA: a cancer journal for clinicians 2017:67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, et al. Colorectal cancer statistics, 2017. CA: a cancer journal for clinicians 2017:67:177–193. [DOI] [PubMed] [Google Scholar]
- 3.Knudsen AB, Zauber AG, Rutter CM, Naber SK, Doria-Rose VP, et al. Estimation of Benefits, Burden, and Harms of Colorectal Cancer Screening Strategies: Modeling Study for the US Preventive Services Task Force. JAMA : the journal of the American Medical Association 2016:315:2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inadomi JM, Vijan S, Janz NK, Fagerlin A, Thomas JP, et al. Adherence to colorectal cancer screening: a randomized clinical trial of competing strategies. Archives of internal medicine 2012:172:575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon K Colorectal cancer development and advances in screening. Clin Interv Aging 2016:11:967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharara AI, El Reda ZD, Harb AH, Abou Fadel CG, Sarkis FS, et al. The burden of bowel preparations in patients undergoing elective colonoscopy. United European Gastroenterol J 2016:4:314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Epling JW Jr., et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA : the journal of the American Medical Association 2016:315:2564–2575. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Levy BT, Daly JM, Bergus GR, Dunkelberg JC Comparison of patient preferences for fecal immunochemical test or colonoscopy using the analytic hierarchy process. BMC Health Serv Res 2015:15:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allison JE The best screening test for colorectal cancer is the one that gets done well. Gastrointestinal endoscopy 2010:71:342–345. [DOI] [PubMed] [Google Scholar]
- 10.Tevis SE, Kennedy GD Postoperative Complications: Looking Forward to a Safer Future. Clinics in colon and rectal surgery 2016:29:246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribeiro MS, Wallace MB Endoscopic Treatment of Early Cancer of the Colon. Gastroenterol Hepatol (N Y) 2015:11:445–452. [PMC free article] [PubMed] [Google Scholar]
- 12.Kijima S, Sasaki T, Nagata K, Utano K, Lefor AT, et al. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World journal of gastroenterology : WJG 2014:20:16964–16975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan YN, Li XF, Li JJ, Song YM, Jiang B, et al. The accuracy of computed tomography in the pretreatment staging of colorectal cancer. Hepatogastroenterology 2014:61:1207–1212. [PubMed] [Google Scholar]
- 14.Foxtrot Collaborative G Feasibility of preoperative chemotherapy for locally advanced, operable colon cancer: the pilot phase of a randomised controlled trial. The Lancet. Oncology 2012:13:1152–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America 2008:105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanash SM, Baik CS, Kallioniemi O Emerging molecular biomarkers--blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol 2011:8:142–150. [DOI] [PubMed] [Google Scholar]
- 17.Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC medicine 2011:9:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickhardt PJ Emerging stool-based and blood-based non-invasive DNA tests for colorectal cancer screening: the importance of cancer prevention in addition to cancer detection. Abdom Radiol (NY) 2016:41:1441–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song LL, Li YM Current noninvasive tests for colorectal cancer screening: An overview of colorectal cancer screening tests. World journal of gastrointestinal oncology 2016:8:793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, et al. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. Journal of proteome research 2009:8:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edge SB, American Joint Committee on Cancer AJCC cancer staging manual. New York: Springer, 2010. [DOI] [PubMed] [Google Scholar]
- 22.Ahnen D, Macrae FA Approach to the patient with colonic polyps. UpToDate. http://www.uptodate.com: Wolters Kluwer, 2016. [Google Scholar]
- 23.Ivancic MM, Irving AA, Jonakin KG, Dove WF, Sussman MR The concentrations of EGFR, LRG1, ITIH4, and F5 in serum correlate with the number of colonic adenomas in ApcPirc/+ rats. Cancer Prev Res (Phila) 2014:7:1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivancic MM, Huttlin EL, Chen X, Pleiman JK, Irving AA, et al. Candidate serum biomarkers for early intestinal cancer using 15N metabolic labeling and quantitative proteomics in the ApcMin/+ mouse. Journal of proteome research 2013:12:4152–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moser AR, Pitot HC, Dove WF A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 1990:247:322–324. [DOI] [PubMed] [Google Scholar]
- 26.Amos-Landgraf JM, Kwong LN, Kendziorski CM, Reichelderfer M, Torrealba J, et al. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proceedings of the National Academy of Sciences of the United States of America 2007:104:4036–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010:26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilbury JB, Van Eetvelt PW, Garibaldi JM, Curnow JS, Ifeachor EC Receiver operating characteristic analysis for intelligent medical systems--a new approach for finding confidence intervals. IEEE Trans Biomed Eng 2000:47:952–963. [DOI] [PubMed] [Google Scholar]
- 29.Pepe MS, Janes H, Li CI, Bossuyt PM, Feng Z, et al. Early-Phase Studies of Biomarkers: What Target Sensitivity and Specificity Values Might Confer Clinical Utility? Clinical chemistry 2016:62:737–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner S, Krause F, Rolny V, Strobl M, Morgenstern D, et al. Evaluation of a 5-Marker Blood Test for Colorectal Cancer Early Detection in a Colorectal Cancer Screening Setting. Clinical cancer research : an official journal of the American Association for Cancer Research 2016:22:1725–1733. [DOI] [PubMed] [Google Scholar]
- 31.Sovich JL, Sartor Z, Misra S Developments in Screening Tests and Strategies for Colorectal Cancer. Biomed Res Int 2015:2015:326728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regula J, Rupinski M, Kraszewska E, Polkowski M, Pachlewski J, et al. Colonoscopy in colorectal-cancer screening for detection of advanced neoplasia. The New England journal of medicine 2006:355:1863–1872. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman DA, Weiss DG, Bond JH, Ahnen DJ, Garewal H, et al. Use of colonoscopy to screen asymptomatic adults for colorectal cancer. Veterans Affairs Cooperative Study Group 380. The New England journal of medicine 2000:343:162–168. [DOI] [PubMed] [Google Scholar]
- 34.Tsilidis KK, Branchini C, Guallar E, Helzlsouer KJ, Erlinger TP, et al. C-reactive protein and colorectal cancer risk: a systematic review of prospective studies. International journal of cancer. Journal international du cancer 2008:123:1133–1140. [DOI] [PubMed] [Google Scholar]
- 35.Emerging Risk Factors C, Kaptoge S, Di Angelantonio E, Lowe G, Pepys MB, et al. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: an individual participant meta-analysis. Lancet 2010:375:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansar W, Ghosh S C-reactive protein and the biology of disease. Immunol Res 2013:56:131–142. [DOI] [PubMed] [Google Scholar]
- 37.Nozoe T, Matsumata T, Kitamura M, Sugimachi K Significance of preoperative elevation of serum C-reactive protein as an indicator for prognosis in colorectal cancer. Am J Surg 1998:176:335–338. [DOI] [PubMed] [Google Scholar]
- 38.Hong I, Hong SW, Chang YG, Lee WY, Lee B, et al. Expression of the Cancer Stem Cell Markers CD44 and CD133 in Colorectal Cancer: An Immunohistochemical Staining Analysis. Ann Coloproctol 2015:31:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira F, Larriba MJ, Munoz A Vitamin D and colon cancer. Endocr Relat Cancer 2012:19:R51–71. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Chen H, Zhao M, Peng P Prognostic value of circulating vitamin D binding protein, total, free and bioavailable 25-hydroxy vitamin D in patients with colorectal cancer. Oncotarget 2017:8:40214–40221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.