Abstract
CD16A (FcγRIIIA) is an activating receptor mostly expressed on natural killer (NK) cells and monocytes/macrophages. It can mediate antibody-dependent cell-mediated cytotoxicity (ADCC) through low-affinity interaction with human immunoglobulin G (IgG) Fc. It can also mediate cell lysis if NK cells are guided by bispecific killer cells engagers (BiKEs). BiKEs showed some success in clinical trials of cancer and are promising candidate therapeutics. However, currently reported BiKEs are based on antibody fragments (scFvs) of relatively large size. The CD16A-specific antibodies are also typically from animal origin. Decreasing the BiKE size could result in enhanced penetration into solid tumor and normal tissues, and using fully human antibodies could decrease the likelihood of immunogenicity. Here we report the identification and characterization of two antibody domains, D6 and E11, isolated from a very large human VH antibody domain library displayed on phage. D6 and E11 bound CD16A with EC50 of 4 nM and 8 nM, respectively, but not other Fc gamma receptors (FcγRs) such as CD64 (FcγRI), CD32 (FcγRII) and CD16B (FcγRIIIB). They bound to both CD16A allotypes (158F,V) with equal affinity and competed with each other as well as with human IgG1 and the mouse anti-CD16A antibody 3G8. These and other results were used to build a molecular docking model predicting that D6 and E11 may bind to the CD16A membrane proximal D2 domain by interacting with its BC, C′E and EF loops. Importantly, cross-linked (bivalent) D6 and E11 induced secretion of IL-2 after binding to CD16A-expressing Jurkat T cells. The small size of these antibody domains combined with their high-affinity, specific, allotype-independent, activating interactions with CD16A could allow generation of novel highly effective BiKEs and other candidate protein therapeutics.
Keywords: Natural killer (NK) cells, CD16A, ADCC, Antibody domains, BiKE
1. Introduction
CD16 (FcγRIII) was first identified by using the monoclonal antibody (mAb) 3G8 which inhibited binding of IgG complexes to neutrophils (Fleit et al., 1982). Several groups found that in humans CD16 can be expressed in two isoforms (CD16A and CD16B) which are highly homologous but CD16A has a transmembrane domain and a cytoplasmic tail while CD16B is glycosylphosphatidylinositol (GPI)-anchored (Selvaraj et al., 1988; Lanier et al., 1989; Edberg et al., 1989; Ravetch and Perussia, 1989; Scallon et al., 1989; Ueda et al., 1989; Simmons and Seed, 1988). CD16A is associated with the immune-receptor tyrosine-based activation motifs (ITAM)-bearing proteins ζ and FcεRIγ and is expressed mainly on NK cells and macrophages as well as on some subsets of monocytes and T cells (van Sorge et al., 2003). Crosslinking of CD16 on NK cells could result in the association of the receptor complex with the p56lck tyrosine kinase and tyrosine phosphorylation of ζ and FcεRIγ followed by a number of events finally resulting in degranulation and cytokine production or in some cases apoptosis of NK cells. A major function of CD16A is to mediate antibody-dependent cell-mediated cytotoxicity (ADCC) through low-affinity interaction with human immunoglobulin G (IgG) Fc(Lanier, 2001). It can also mediate ADCC-independent cell lysis (Mandelboim et al., 1999).
The capability of NK cells to kill target cells specifically by using bispecific antibodies to both CD16 and target cells was demonstrated >30 years ago (Perez et al., 1985). However, difficulties in generating such antibodies and for other reasons it was not until relatively recently when such bispecific mAbs called Bispecific Killer cell Engagers (BiKEs) were successfully used in a clinical trial (Rothe et al., 2015).
A major component of a BiKE is the antibody that binds to CD6A. Most (but not all, e.g., (McCall et al., 1999)) previously reported antibodies (e.g., (Weiner et al., 1995)) are from animal origin. The animal antibodies can be humanized and some, e.g., from lamma (Behar et al., 2008), are similar in sequence to human antibodies; however, the probability for immunogenicity when administered in humans is still on average higher than that for fully human antibodies (Dimitrov, 2010). Here, we described two VH antibody domains (Ads) derived from a human library displayed on phage which bind to CD16A with high affinity, are highly specific and allotype independent. To our knowledge these are the first human Ads reported to bind CD16A. They could be used alone or as components of BiKEs and other fusion proteins for development of therapeutics.
2. Materials and methods
2.1. FcγRs, plasmids, antibodies and cells
Recombinant FcγRs ectodomains were purchased from Sino Biologic Inc. (North Wales, PA, USA). The pComb3X was kindly provided by Dennis Burton (Scripps Research Institute, La Jolla, CA, USA) and the pSecTag2 B was purchased from Invitrogen. IgG1 m336 and m912-mFc were produced in our group. The following antibodies were purchased: mouse anti-CD16A IgG1, 3G8 (Abcam, Cambridge, MA, USA); phycoerythrin (PE)-conjugate mouse anti-CD16A, PE conjugated mouse anti-FLAG (Miltenyi, Bergisch Gladbach, Germany); fluorescein isothiocyanate (FITC)-conjugated mouse anti-human CD64 (FcγRI) and CD32 (FcγRII) (Invitrogen); horseradish peroxidase (HRP) anti-M13 polyclonal (Pharmacia, Piscataway, NJ); HRP-conjugated mouse anti-FLAG tag, HRP conjugated goat anti-mouse IgG and HRP-conjugated goat anti-human IgG (Fc-specific) (Sigma-Aldrich). U937 cell was a gift from Anu Puri (National Cancer Institute, Frederick, MD). The following cell lines were purchased: Jurkat T (ATCC); 293 freestyle (Invitrogen) and Jurkat T over-expressing CD16A (Promega). Human blood was obtained from the NIH blood center.
2.2. Preparation of recombinant CD16A-mFc and its biotinylation
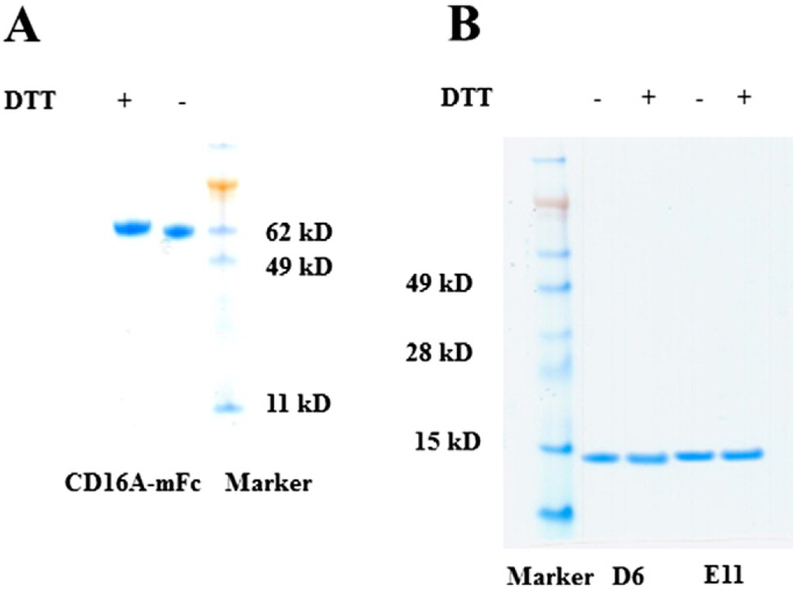
The CD16A gene was synthesized by Genescript (Piscataway, NJ). Its extracellular domain (ECD, Gly17-Gln208) was fused to monomeric Fc (mFc) which was generated in our group (Ying et al., 2012). The CD16A ECD was subcloned into modified pSecTag2 B vectors containing mFc by using the SfiI endonuclease restriction site. The recombinant CD16A-mFc was expressed in 293 freestyle cells by transient transfection, and purified by Protein A Sepharose 4 Fast Flow (GE Healthcare, Piscataway, NJ) resin according to the manufacturer’s protocols. The purity was checked by SDS-PAGE under both reducing and non-reducing conditions (Fig. 1A), and the homogeneity was confirmed by Superdex 200 chromatography (GE Healthcare). The protein was stored at −80 °C until use. We noticed that storage could change its properties but not to significant degree to affect conclusions. Biotinylation of CD16A-mFc was performed by EZ-Link Sulfo-NHS-Biotin reagent (Thermo Fisher) according to the manufacturer’s protocols.
Fig. 1.
SDS-PAGE of CD16A-mFc (A) used as the panning antigen, and the Ads, D6 and E11 (B) in presence or absence of reducing reagent, DTT. Under the reducing conditions, the samples were subjected to heating at 100°C for 10 min.
2.3. Panning, selection, expression and purification of binders
A large human antibody domain (Ad) phage library (size~1011) was constructed by grafting CDR loops from peripheral blood mononuclear cell (PBMC) cDNA of 40 healthy donors into human VH3–23 scaffold (T Ying and DS Dimitrov, unpublished). This library was panned against biotinylated CD16A-mFc in the presence of 10-fold excess of PD1-mFc as a competitive (depletion) protein. Amplified libraries of 1012 phage displayed Ads were incubated with 1, 0.5, 0.2, and 0.05 μg of CD16A-mFc during the first, second, third, and fourth rounds of panning, respectively. After four rounds of panning, the phage library was significantly enriched with CD16A binders as measured by polyclonal phage ELISA (ppELISA). Positive clones from the third and fourth rounds were selected by using supernatants from randomly picked clones which were incubated with immobilized antigens (semELISA) and measured. Then their DNA was sequenced. For protein preparation, these clones were transformed into HB2151 cells for expression, and proteins were purified by one-step Ni-NTA resin. Protein purity and homogeneity were analyzed by SDS-PAGE. Protein concentration was measured spectrophotometrically (NanoVue, GE Healthcare).
2.4. ELISA
96-well plates (Costar) were coated with CD16A-mFc protein at 50 ng/well in phosphate-buffered saline (PBS) overnight at 4 °C. Phage from each round of panning (ppELISA) or supernatants from randomly picked clones (semELISA) were incubated with immobilized antigens. Bound phage was detected with HRP-anti-M13-polyclonal Ab and bound proteins with HRP-conjugated mouse anti-FLAG mAb, which was also used for the determination of the binding EC50 to CD16A for the Ads. For evaluation of binding with other FcγRs, the commercial recombinant FcγRs were coated on plates and the bound Ads were detected by HRP-conjugated mouse anti-FLAG mAb. To determine whether the Ads obtained by panning against high-affinity CD16A-158V isoform could also bind to the low affinity CD16A-158F isoform, we performed ELISA using the two CD16A allotypes as coating antigens. For competition ELISA, 5 nM Ad was incubated with serially diluted competitors: E11-mFc; m336-IgG1-Fc and 3G8. For negative control, we used a mFc fusion protein, m912-mFc developed in our lab. After incubation with the coated antigen and extensive washing, the bound Ads were detected by HRP-conjugated mouse anti-FLAG tag Ab. The half-maximal binding (50% effective ELISA Concentration, EC50,) was estimated as the concentration at which OD is 50% of its maximal value.
2.5. Enrichment of NK cells from PBMC and flow cytometry (FACS)
PBMCs were isolated from peripheral blood of healthy donors by centrifugation on a Ficoll/Hypaque gradient (GE health). NK cells were enriched from human PBMC by using the NK cell isolation kit II in the negative selection modes (Miltenyi Biotec). Purified NK cells were detected by incubation with biotinylated anti-CD56 antibody (m909, developed by our group (Feng et al., 2016)) followed by streptavidin-FITC and PE conjugated mouse anti-human CD16 antibody. Ad binding to NK cells was measured by incubating 106 NK cells in 200 μL PBS containing 0.1% bovine serum albumin (BSA) (PBSA) with 10 nM of Ads for 30 min at room temperature. The cells were washed twice with 200 μL PBSA, followed by incubation with PE-conjugated-mouse anti-FLAG antibody (Miltenyi Biotec) for 30 min on ice. After washing, the cells were used for fluorescence-activated cell sorter (FACS) analysis. The second antibody alone was used as negative control, and the PE conjugated mouse anti-human CD16A antibody (Mouse IgMκ, clone VEP13) was used as a positive control. Jurkat T cells and phorbol myristate acetate (PMA) stimulated U937 cells, which express high levels of CD64 and CD32, but do not express CD16A were used as negative controls (Looney et al., 1986). For competition FACS, we incubated serially diluted competitors (3G8) with NK cells in the presence of 1 nM D6. After washing by PBSA, the bound D6 was detected by using PE-Anti-FLAG.
2.6. Molecular docking
Z-DOCK program (Chen et al., 2003) as part of the Accelrys Discovery studio (http://accelrys.com) was used to dock Ads onto CD16A. The three-dimensional structures for these Ads were modeled in Swiss-Model using homology modeling (Biasini et al., 2014). The structure of CD16A was extracted from PDB of 3WN5. The structure of CD16A-158F was generated by PyMOL mutagenesis wizard and energy optimized on Accelrys Discovery studio. Before docking, the structures of Ads and CD16A were processed as follows: water molecules were deleted and hydrogen atoms were added at pH 7.4 and an ionic strength of 0.145, and in a dielectric environment of 10; energy was minimized based on CHARMM (Sapay and Tieleman, 2011) with a cutoff of 0.9; loop regions were rebuilt according to SEQRES (Ordog et al., 2009) data; energy was used to judge the efficacy of the geometry optimization; and cysteine bridges in these proteins were defined as blocked regions. Then Z-DOCK performs a systematic search of a uniform sample of docked protein poses and uses an internal scoring algorithm to predict optimal interactions. 54,000 docked poses of sdAbs were initially produced, which were further filtered and re-ranked to obtain top 200 poses based on ZRank score using electrostatic and desolvation energy and non-deterministic FFT optimization. These 200 poses were then visually scrutinized. We finally chose the most reasonable pose as the model for the Ad-CD16A complex by combining considerations of general features of antibody-antigen interaction such as hydrogen bonds, salt bridges, hydrophobic packing and other interactions at the interface without any highly unusual features and steric clashes.
2.7. IL-2 secretion
Jurkat T-CD16A cells were used to evaluate the engagement of CD16A on the cell surface by Ads through monitoring of the IL-2 secretion. Jurkat T cells overexpressing CD16A were cultured in RPMI1640 medium (Invitrogen) containing selection reagents, G418 and hygromycin, and 10% FBS. Before incubation with Ads, Jurkat T-CD16A cells were activated overnight by PMA (50 ng/mL, Sigma). Then 106 cells were incubated with Ads (0.001 mg/mL) alone or with Ads/mouse anti-Flag mAb (1: 1000 dilution) mixture. The mouse anti-CD16A IgG1, 3G8, was used as a positive control. After incubation, cells were centrifuged for 10 min at 1200 ×g and the quantity of human IL-2 in the supernatant was measured by sandwich ELISA with the Duoset human IL-2 kit (Duoset Human IL-2, R&D Systems) according to the manufacturer’s protocol.
3. Results
3.1. Isolation and initial characterization of anti-CD16A antibody domains
To identify Ad binders to CD16A we first generated a recombinant CD16A fused to monomeric Fc (CD16A-mFc) (Fig. 1A) which was used as an antigen for competitive panning of a large human Ad library displayed on phage as described in the Materials and Methods. After four rounds of panning four dominant clones, D6, E11, C4 and G3 were identified from total of 200 clones tested. Two clones, D6 and E11, which exhibited the highest expression levels (yields > 50 mg/L) and binding, were selected for further characterizations. The purified domains exhibited single homogeneous bands on both reducing and non-reducing SDS-PAGE gels with molecular weights (MW) corresponding to the calculated ones of approximately 15 kDa (Fig. 1B). They were soluble and did not precipitate even at concentrations higher than 50 mg/mL. Therefore, they were characterized more extensively as described below.
3.2. High-affinity, specific and allotype-independent binding to recombinant CD16A
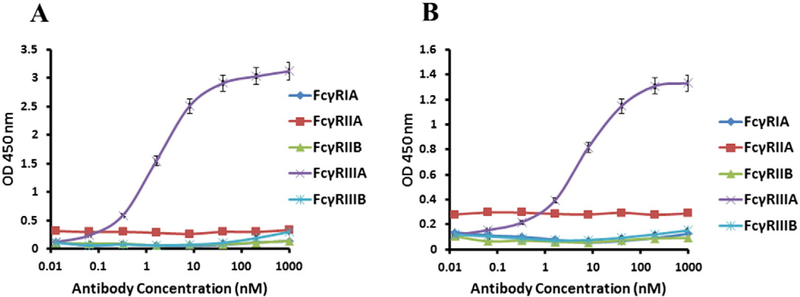
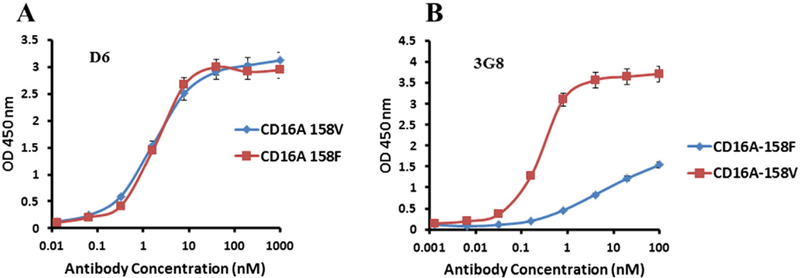
The affinity of D6 was higher than that of E11 as measured by ELISA (EC50 = 4 nM vs. 8 nM, respectively) (Fig. 2). To find out whether the binding is specific for CD16A we measured by ELISA also binding to other FcγRs including CD16B. The data showed that both D6 and E11 bound to CD16A but not to CD64A, CD32A, CD32B and CD16B (Fig. 2A, B). An important feature of CD16A is its polymorphism at position 158; the 158 V isoform has higher affinity to Fc than the 158F isoform (Koene et al., 1997). However, we found that D6 bound similarly to both CD16A isoforms as measured by ELISA; E11 behaved the same way (Fig. 3). These results show that both D6 and E11 had similar binding to CD16A 158V and F, in contrast to the mouse anti-CD16A antibody 3G8. Taken together, these data demonstrated a specific, high-affinity, allotype-independent binding of the new anti-CD16A Ads.
Fig. 2.
Binding affinity and specificity of D6 and E11 as measured by ELISA. FcγRs were coated on 96-well plates at a concentration of 1 μg/mL. Bound Ads were detected by HRP-conjugated mouse anti-FLAG antibody. Each measurement was repeated twice and the plotted data are means ± standard deviations. (A) D6 binding to different recombinant FcγRs. FcγRIIIA is also denoted as CD16A (B) E11 binding to different recombinant FcγRs.
Fig. 3.
Binding to CD16A allotypes by D6 (A) and the mouse IgG13G8 (B). CD16A containing 158V was produced as a monomeric Fc (mFc) fusion protein. CD16A carrying 158F isoform was prepared by site directed mutagenesis and expressed in 293 free style cells by the same procedure as CD16A-158V. Mouse anti-CD16A IgG1, 3G8, was purchased from Abcam. Each measurement was repeated twice and the plotted data are means ± standard deviations.
3.3. Specific binding of D6 and E11 to cell surface-associated CD16A
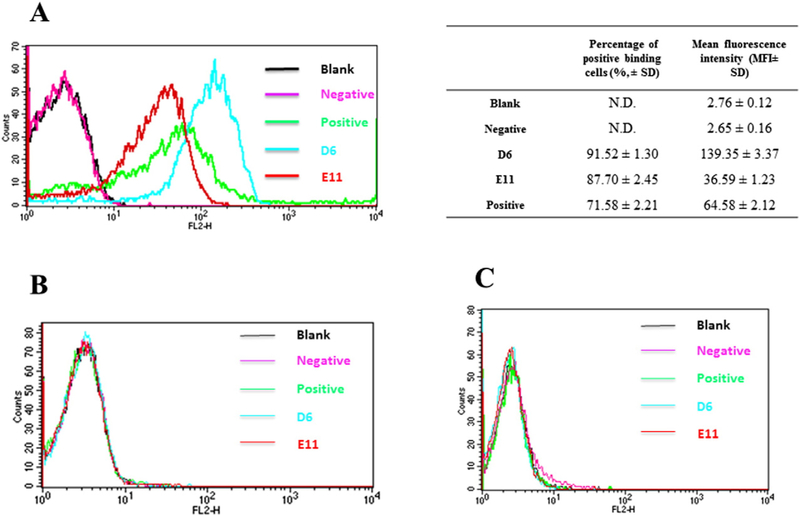
We next evaluated whether D6 and E11 binds specifically to CD16A expressing cells. Both D6 and E11 bound significantly to purified NK cells at a low concentration of 10 nM as measured by flow cytometry (Fig. 4A). D6 bound to NK cells more efficiently than E11 and a commercial mouse anti-CD16A antibody (Mouse IgMκ, clone VEP13). Neither of those antibodies bound to two CD16A negative cell lines, Jurkat T cells (Fig. 4B) and PMA stimulated U937 cells (Fig. 4C) which express FcγRI and FcγRII but not FcγRIII (Looney et al., 1986). These results suggest that D6 and E11 could be used as components of BiKEs to guide NK cells to target cells as well as research reagents.
Fig. 4.
Binding of D6 and E11 to human NK cells. Positive and negative controls refer to the binding of PE-conjugated mouse-anti-CD16A IgG1 and the PE-conjugated anti-FLAG antibody alone (in the absence of D6 or E11) binding to cells, respectively. Each measurement was repeated three times and the positive binding cells percentage and fluorescence data were listed as means ± standard deviations. (A) D6 and E11 binding to human NK cells expressing CD16A at low concentration of 10 nM. (B) D6 and E11 at the same concentrations binding to Jurkat T cells which do not express CD16A. (C) D6 and E11 binding to phorbol myristate acetate (PMA) activated U937 cells which express FcγRI and FcγRII but not FcγRIII.
3.4. Analysis of the D6 and E11 epitopes
We analyzed the epitopes of these Ads by competition ELISA and FACS. Competition ELISA results (Fig. 5A) showed the CD16A binding of D6 could be effectively competed by 3G8, and to much less extent competed by E11 and IgG1-Fc, which indicates that D6 may share similar epitopes with E11, and their epitopes overlap with those of IgG1-Fc and 3G8. To further confirm the competition between D6 and 3G8, we performed FACS to evaluate whether 3G8 could compete with D6 for binding to NK cells. Results showed 1 nM of D6 binding to NK cells could be significantly inhibited by 3G8 (Fig. 5B).
Fig. 5.
Epitopes analysis of D6 and E11 by competition ELISA and FACS. (A) D6 binding to CD16A could be competed by 3G8, E11 and IgG1-Fc. 5 nM D6 in the presence of the serially diluted competitors were incubated with the coated CD16A, and the bound Ads were detected by HRP-conjugated mouse anti-FLAG tag Ab. The negative competitor was a mFc fusion protein (m912-mFc) developed in our lab. (B) D6 binding to NK cells was competed by 3G8 in a concentration dependent manner. Serially diluted 3G8 were incubated with NK cells in the presence of 1 nM D6. After washing, the bound D6 were detected by PE-Anti-FLAG. (C) Comparison of D6 binding to CD16A in the absence or presence of human serum. After coating and blocking, D6 dissolved either in 3% MPBS or in complete human serum were incubated in the CD16A-coated plate for 2 h, followed by washing and detecting by HRP-conjugated mouse anti-FLAG tag Ab. Each measurement was repeated twice and the plotted data are means ± standard deviations.
The overlapping epitopes between D6 and IgG1-Fc raise concerns about the availability of D6 for targeting NK cells when used as intravenous therapy reagents due to the competition by the endogenous IgG1 in human serum. However, D6 could out-compete serum due to its high affinity. In human serum D6 began to bind to CD16A at about 3 nM and 50% binding was achieved at about 60 nM (Fig. 5C), i.e., 1 μg/mL, which is a relatively low concentration.
3.5. Computational modeling of the Ad-CD16A complex
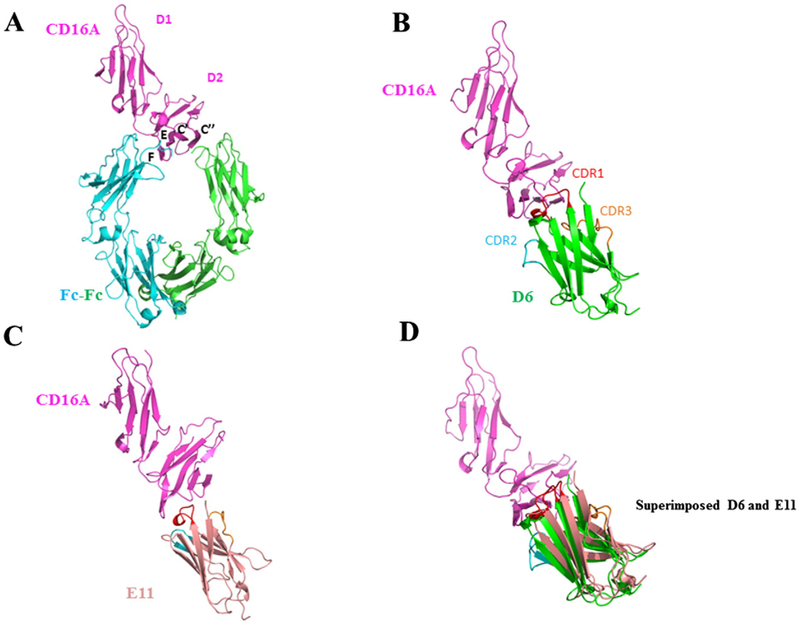
In an attempt to computationally map the epitopes of D6 and E11, we performed molecular docking calculations to elucidate structures of D6-CD16A and E11-CD16A complexes as described in Materials and Methods. Based on the competition ELISA results, during the docking process, we confined the interaction interface to the CDR loops of the Ads and the membrane proximal regions of the D2 domain of CD16A. The crystal structure of CD16A was previously solved (PDB ID: 3WN5) and kept fixed in the docking, the conformations of the eAbs were optimized at the CHARMM force field, and was searched against the CD16A D2 domain based on an energy minimization algorithm. The results showed that the D6 and E11 bound to the CD16A D2 domain in a similar manner as did Fc from IgG1 (Fig. 6). The CDR loops of D6 and E11 constituted large interfaces with the C’, C”, E, F strands of CD16A, and formed close contacts with EF, BC, C’C” loops of D2 domains. These binding models could reconcile the competition ELISA results showing that D6 and E11 could compete with Fc for binding to CD16A. Interestingly, the structures of D6 and E11 could be well superimposed at the interface regions of CD16A, which could explain their reciprocal competition with each other for CD16A binding.
Fig. 6.
Docking structures of CD16A complexed with D6 or E11. (A) Crystal structure of IgG1-Fc complex with CD16A (PDB ID: 3WN5). The interface β stands are denoted as C’, C”, E and F. CD16A is represented as magentas cartoon model, and Fc dimers with green and blue color. (B) The docking model of CD16A-D6 complex. CDR1, 2 and 3 of D6 are presented as red, blue and yellow loops. CD16A and D6 are depicted as magentas and green cartoon respectively. (C) The docking model of CD16A-E11 complex. E11 is represented as light pink cartoon. (D) The superimposed structures of D6 and E11.: 3WN5).
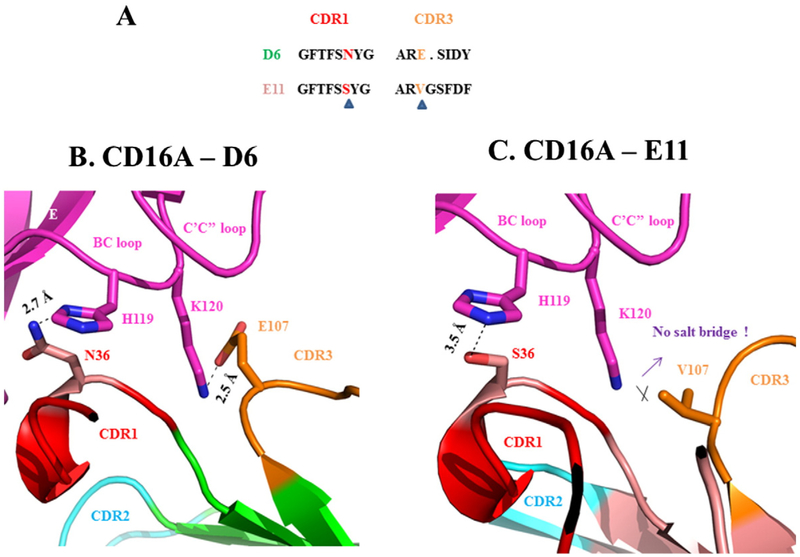
The structures of D6-CD16A and E11-CD16A could be also explored to explain why D6 has higher binding affinity to CD16A than E11. While the FR residues were almost the same for D6 and E11, their CDR loops were distinct. As the sequence alignments shown, Asn36 in CDR1 of D6 is at the same position as Ser in E11, and the Glu107 in CDR3 of D6 as Val in E11. These two different residues on CDR loops could significantly impact the antigen binding based on our docked complex structures. As Fig. 7 shows, while Asn36 formed strong hydrogen bond (~2.7 Å) to His119 from BC loop of CD16A, this corresponding hydrogen bond was severely compromised at E11 (~3.5 Å), probably since Ser is a poorer hydrogen bond donor than Asn. On the other hand, Glu107 formed strong salt bridge interaction with Lys120 from BC loop of CD16A (~2.5 Å), which is broken in E11 due to the inability of forming salt bridge for Val. Therefore, the favorable hydrogen bond and the extra salt bridge for D6 could explain its higher binding affinity to CD16A than E11.
Fig. 7.
Interactions likely to result in higher affinity of D6 than E11. (A) Sequences comparisons of CDR1 and CDR3 of D6 and E11. The key different residues are highlighted and indicated by triangle symbols. (B) Interaction interface between CD16A and D6. BC loop and C’C” loop from CD16A are depicted in magentas. The hydrogen bond between D6 N36 and CD16A H119, and the salt bridge of D6 E107 and CD16A K120 are present as stick models. (C) Interaction interface between CD16A and E11.
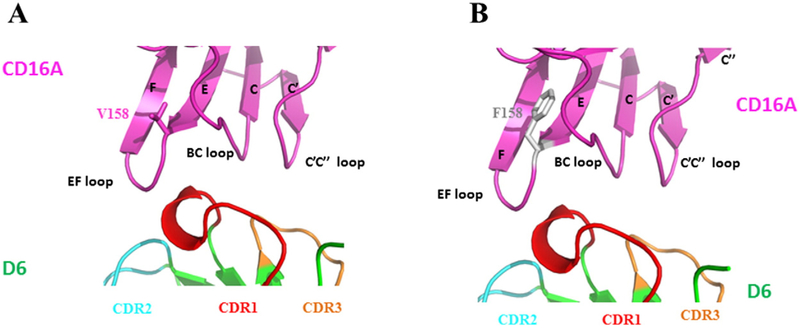
The calculated structures could also reconcile the same binding affinity of these Ads to the different CD16A allotypes. As shown in Fig. 8, although the residue 158 of CD16A locates at the interaction interface of D6 and CD16A, it is poised away from the directions of CDRs of D6, thus not involving the binding to CDR loop. Therefore, the 158F/V polymorphism of CD16A should not impact the binding of D6 and E11, as was also seen in the ELISA experiments.
Fig. 8.
Docking models of D6 complexed with both CD16A allotypes (158V, F) with highlighting the conformations of V158 (magentas stick) (A), and F158 (grey stick) (B) in EF loops. The key β strands of CD16A at the interaction interface are depicted as E, F, C’ and C” sheets. The CDR loops of D6 are represented as red, blue and yellow colors. The side chains of residue at position 158, either Val or Phe do not involve interaction with CDRs of D6.
3.6. Activation of T cells expressing CD16A by D6 and E11
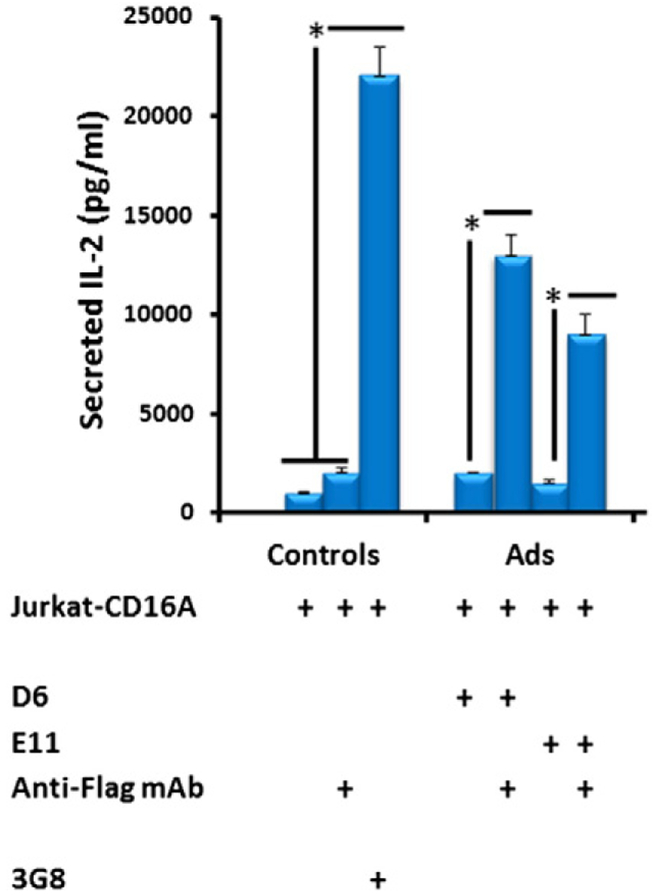
Finally, we assessed whether the binding of D6 and E11 to CD16A could activate cells expressing CD16A. Since the cell activation and subsequent release of cytokines requires cross-linking of CD16A, D6 and E11 were pre-incubated with the bivalent mouse anti-Flag antibody. The results showed that binding of the pre-incubated D6 or E11 to CD16+ Jurkat T cells leads to release of IL-2 which is consistent with the results obtained by using the bivalent 3G8 (Fig. 9). By contrast, the D6 and E11 or the anti-Flag IgG1 alone could not induce IL-2 secretion by the same cells. Interestingly, the higher affinity D6 induced more IL2 than E11 but less than 3G8 indicating that there could be correlation between affinity and cytokine release activity. These results demonstrate D6 and E11 could specifically engage CD16A on the effector cell surface, and activate signals eventually leading to ADCC.
Fig. 9.
IL-2 release by CD16A expressing Jurkat T cells after binding of anti-CD16A Ads (1 μg/mL) either alone or pre-incubated with mouse Anti-FLAG mAb. The Anti-FLAG mAb alone and 3G8 were set up as negative and positive controls respectively. IL-2 was detected by sandwich ELISA with the Duoset human IL-2 kit. Statistical tests were performed using GraphPad Prism version 5. Significant differences when comparing two groups were determined by Mann-Whitney U test. A p value <0.05 was considered significant. *p <0.05.
4. Discussion
A major result of this study is the identification of two novel human Ads that bind specifically and with high affinity to both CD16A allotypes. These Ads are fully human and may not exhibit immunogenicity if administered to humans. Importantly, the size of the domains is half the size of the scFvs which are typically used in BiKEs. This is important because the penetration in normal tissues and solid tumor could be significantly higher than an antibody fragment and fusion proteins based on it. For example, a previous study has shown that a full size antibody (IgG1) of MW about 150 kDa has an effective diffusion coefficient about 100-fold lower in solid tissues than molecules 10-fold smaller (Jain, 1990). Therefore, the effect of size could be significant. Previously, a lamma antibody domain that binds with high affinity to CD16A was identified (Behar et al., 2008) and used for generation of two BiKEs targeting carcinoembryonic antigen (CEA) (Rozan et al., 2013) and Her2 (Turini et al., 2014). These BiKEs were successful in CD16A mediated lysis of target cells thus demonstrating for the first time that antibody domains are functional as components of BiKEs. Therefore, one can expect that our newly identified antibody domains could be also used to generate functional BiKEs.
Similarly to the previously identified lamma domains the new Ads are of small (about 13 kDa) size, exhibit relatively high level of expression and bind to both allotypes of CD16A. The allotype-independent binding is important because the BiKEs based on these Ads are expected to be equally effective in humans with both allotypes. As shown before it appears that efficacy of some therapeutic antibodies which kill cancer cells by ADCC is dependent on the CD16A allotype with significantly lower efficacy for humans with the lower affinity allotype (158F) (Niwa et al., 2004). An analysis suggested that therapeutic mAbs usually have better performance when administered to CD16A 158V/V homozygous patients than to 158F/F ones (Cartron et al., 2002; Treon et al., 2005). The frequency of the low affinity allotype 158F (0.6) is higher than that of the high affinity one, 158V (0.4) (Koene et al., 1997). Thus, to effectively recruit NK cells in all patient population, it is important that anti-CD16A antibodies bind well to both allotypes which is the case with the new binders.
In contrast to the lamma domains the new Ads are fully human and do compete for binding to IgG1 Fc. As discussed in the Introduction the fully human nature of these domains decreases the likelihood of immunogenicity. However, only further experiments and ultimately human clinical trials will show whether they are immunogenic or not, and if immunogenic how immunogenic they are. Competition with Fc is typically considered as a property which is not desirable because it can lead to higher concentrations of the BiKE required for the same level of efficacy as non-competing BiKEs. However, if the CD16A binder is of sufficient affinity/avidity outcompeting IgGs could be of benefit in some cases. Outcompeting IgGs binding to CD16A on the NK cell surface could free those NK cells which otherwise are engaged with other targets including some autoimmune diseases (Schleinitz et al., 2010). This could increase the efficacy of BiKEs and simultaneously could have beneficial effect in cases when NK cells could promote disease by killing normal cells through ADCC. In such cases our binders alone could be used as therapeutic agents and inhibit ADCC if desirable. However, in some cases, e.g., some infections controlled by ADCC, the use of BiKEs based on our binders could be deleterious. One can also speculate that the simultaneous binding of IgG and BiKEs or other molecules based on non-competing CD16A binders could result in decreased efficacy or other unanticipated effects.
We also found that the affinities of the new binders are higher than that of previously reported llama anti-CD16A Ads. (10–100 nM) (Behar etal.,2008) and the human anti-CD16A scFv reported by A.M. McCall et al. (~20 nM) (McCall et al., 1999). Avidity effects can further increase the strength of binding of the Ads to cell surface associated CD16A. The high affinities could raise concerns about the binding specificity to CD16A. It is well established that FcγRs use similar epitopes to bind Fc (Jefferis and Lund, 2002). The non-specificity of anti-CD16A antibody could cause side effects. For example, severe pro-inflammatory reactions could be primed when FcγRI is non-specifically engaged. Besides, the effector function of immunological cells could be inhibited when the inhibitory receptor FcγRIIB is ligated. Furthermore, ADCC function would be significantly compromised when the decoy receptor FcγRIIIB, rather than the highly homologous CD16A, is bound by the ligands, since the FcγRIIIB + granulocytes could accelerate the clearance of bound ligands from the circulation. Our experiments provided strong evidence that the new binders are highly specific and did not bind to any of the FcγRs tested including CD16B.
We also developed computational models in attempt to try to elucidate the epitope of the two binders. The models help explain why binding is independent on the CD16A allotype and the competition with IgG Fc. However, remains a challenge to understand why they do not bind to CD16B. They can also serve to perform structure-based design of mutant libraries to further increase the affinity of the new binders.
The new Ads are not only with unique sequences but also with several unique features. They are the only reported fully human Ads binding to CD16A. They are highly specific, high-affinity, allotype independent binders which can outcompete human IgG Fc. They also possess good drugability properties including relatively high level of expression. Therefore, alone or in combination or as parts of larger biological molecules they could be useful in the development of novel therapeutics against cancer and other diseases.
Acknowledgements
We thank members of our group for help. This work was supported in part by the Intramural Research Program, National Cancer Institute, National Institutes of Health and by a Stand Up To Cancer-St. Baldrick’s Pediatric Dream Team translational research grant (SU2C-AACR-DT113). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
References
- Behar G, Siberil S, Groulet A, Chames P, Pugniere M, Boix C, Sautes-Fridman C, Teillaud JL, Baty D, 2008. Isolation and characterization of anti-FcgammaRIII (CD16) llama single-domain antibodies that activate natural killer cells. Protein Eng Des Sel 21 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T, 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42 (Web Server issue), W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H, 2002. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 99 (3), 754–758. [DOI] [PubMed] [Google Scholar]
- Chen R, Li L, Weng Z, 2003. ZDOCK: an initial-stage protein-docking algorithm. Proteins: Struct., Funct., Bioinf 52 (1), 80–87. [DOI] [PubMed] [Google Scholar]
- Dimitrov DS, 2010. Therapeutic antibodies, vaccines and antibodyomes. MAbs 2 (3), 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edberg JC, Redecha PB, Salmon JE, Kimberly RP, 1989. Human Fc gamma RIII (CD16). Isoforms with distinct allelic expression, extracellular domains, and membrane linkages on polymorphonuclear and natural killer cells. J. Immunol 143 (5), 1642–1649. [PubMed] [Google Scholar]
- Feng Y, Wang Y, Zhu Z, Li W, Sussman RT, Randall M, Bosse KR, Maris JM, Dimitrov DS, 2016. Differential killing of CD56-expressing cells by drug-conjugated human antibodies targeting membrane-distal and membrane-proximal non-overlapping epitopes. MAbs 8 (4), 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleit HB, Wright SD, Unkeless JC, 1982. Human neutrophil fc gamma receptor distribution and structure. Proc. Natl. Acad. Sci. U. S. A 79 (10), 3275–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, 1990. Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res. 50 (3 Suppl), 814s–819s. [PubMed] [Google Scholar]
- Jefferis R, Lund J, 2002. Interaction sites on human IgG-Fc for FcgammaR: current models. Immunol. Lett 82 (1–2), 57–65. [DOI] [PubMed] [Google Scholar]
- Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M, 1997. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood 90 (3), 1109–1114. [PubMed] [Google Scholar]
- Lanier LL, 2001. On guard-activating NK cell receptors. Nat. Immunol 2 (1), 23–27. [DOI] [PubMed] [Google Scholar]
- Lanier LL, Phillips JH, Testi R, 1989. Membrane anchoring and spontaneous release of CD16 (FcR III) by natural killer cells and granulocytes. Eur. J. Immunol 19 (4), 775–778. [DOI] [PubMed] [Google Scholar]
- Looney RJ, Abraham GN, Anderson CL, 1986. Human monocytes and U937 cells bear two distinct Fc receptors for IgG. J. Immunol 136 (5), 1641–1647. [PubMed] [Google Scholar]
- Mandelboim O, Malik P, Davis DM, Jo CH, Boyson JE, Strominger JL, 1999. Human CD16 as a lysis receptor mediating direct natural killer cell cytotoxicity. Proc. Natl. Acad. Sci. U. S. A 96 (10), 5640–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall AM, Adams GP, Amoroso AR, Nielsen UB, Zhang L, Horak E, Simmons H, Schier R, Marks JD, Weiner LM, 1999. Isolation and characterization of an anti-CD16 single-chain Fv fragment and construction of an anti-HER2/neu/anti-CD16 bispecific scFv that triggers CD16-dependent tumor cytolysis. Mol. Immunol 36 (7), 433–45. [DOI] [PubMed] [Google Scholar]
- Niwa R, Hatanaka S, Shoji-Hosaka E, Sakurada M, Kobayashi Y, Uehara A, Yokoi H, Nakamura K, Shitara K, 2004. Enhancement of the antibody-dependent cellular cytotoxicity of low-fucose IgG1 is independent of FcgammaRIIIa functional polymorphism. Clin. Cancer Res 10 (18 Pt 1), 6248–6255. [DOI] [PubMed] [Google Scholar]
- Ordog R, Szabadka Z, Grolmusz V, 2009. DECOMP: a PDB decomposition tool on the web. Bioinformation 3 (10), 413–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez P, Hoffman RW, Shaw S, Bluestone JA, Segal DM, 1985. Specific targeting of cytotoxic T cells by anti-T3 linked to anti-target cell antibody. Nature 316 (6026), 354–356. [DOI] [PubMed] [Google Scholar]
- Ravetch JV, Perussia B, 1989. Alternative membrane forms of fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J. Exp. Med 170 (2), 481–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe A, Sasse S, Topp MS, Eichenauer DA, Hummel H, Reiners KS, Dietlein M, Kuhnert G, Kessler J, Buerkle C, Ravic M, Knackmuss S, Marschner JP, Pogge von Strandmann E, Borchmann P, Engert A, 2015. A phase 1 study of the bispecific anti-CD30/CD16A antibody construct AFM13 in patients with relapsed or refractory Hodgkin lymphoma. Blood 125 (26), 4024–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozan C, Cornillon A, Petiard C, Chartier M, Behar G, Boix C, Kerfelec B, Robert B, Pelegrin A, Chames P, Teillaud JL, Baty D, 2013. Single-domain antibody-based and linker-free bispecific antibodies targeting FcgammaRIII induce potent antitumor activity without recruiting regulatory T cells. Mol. Cancer Ther 12 (8), 1481–1491. [DOI] [PubMed] [Google Scholar]
- Sapay N, Tieleman DP, 2011. Combination of the CHARMM27 force field with united-atom lipid force fields. J. Comput Chem 32 (7), 1400–1410. [DOI] [PubMed] [Google Scholar]
- Scallon BJ, Scigliano E, Freedman VH, Miedel MC, Pan YC, Unkeless JC, Kochan JP, 1989. A human immunoglobulin G receptor exists in both polypeptide-anchored and phosphatidylinositol-glycan-anchored forms. Proc. Natl. Acad. Sci. U. S. A 86 (13), 5079–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleinitz N, Vely F, Harle JR, Vivier E, 2010. Natural killer cells in human autoimmune diseases. Immunology 131 (4), 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj P, Rosse WF, Silber R, Springer TA, 1988. The major fc receptor in blood has a phosphatidylinositol anchor and is deficient in paroxysmal nocturnal haemoglobinuria. Nature 333 (6173), 565–567. [DOI] [PubMed] [Google Scholar]
- Simmons D, Seed B, 1988. The fc gamma receptor of natural killer cells is a phospholipid-linked membrane protein. Nature 333 (6173), 568–570. [DOI] [PubMed] [Google Scholar]
- Treon SP, Hansen M, Branagan AR, Verselis S, Emmanouilides C, Kimby E, Frankel SR, Touroutoglou N, Turnbull B, Anderson KC, Maloney DG, Fox EA, 2005. Polymorphisms in FcgammaRIIIA (CD16) receptor expression are associated with clinical response to rituximab in Waldenstrom’s macroglobulinemia. J. Clin. Oncol 23 (3), 474–481. [DOI] [PubMed] [Google Scholar]
- Turini M, Chames P, Bruhns P, Baty D, Kerfelec B, 2014. A FcgammaRIII-engaging bispecific antibody expands the range of HER2-expressing breast tumors eligible to antibody therapy. Oncotarget 5 (14), 5304–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda E, Kinoshita T, Nojima J, Inoue K, Kitani T, 1989. Different membrane anchors of fc gamma RIII (CD16) on K/NK-lymphocytes and neutrophils. Protein-vs lipid-anchor. J. Immunol 143 (4), 1274–1277. [PubMed] [Google Scholar]
- van Sorge NM, van der Pol WL, van de Winkel JG, 2003. FcgammaR polymorphisms: implications for function, disease susceptibility and immunotherapy. Tissue Antigens 61 (3), 189–202. [DOI] [PubMed] [Google Scholar]
- Weiner LM, Clark JI, Ring DB, Alpaugh RK, 1995. Clinical development of 2B1, a bispecific murine monoclonal antibody targeting c-erbB-2 and fc gamma RIII. J. Hematother 4 (5), 453–456. [DOI] [PubMed] [Google Scholar]
- Ying T, Chen W, Gong R, Feng Y, Dimitrov DS, 2012. Soluble monomeric IgG1 Fc. J. Biol. Chem 287 (23), 19399–19408. [DOI] [PMC free article] [PubMed] [Google Scholar]