Abstract
NORE1A (RASSF5) is a proapoptotic Ras effector that is frequently inactivated by promoter methylation in human tumors. It is structurally related to the RASSF1A tumor suppressor and is itself implicated as a tumor suppressor. In the presence of activated Ras, NORE1A is a potent inducer of apoptosis. However, when expressed at lower levels in the absence of activated Ras, NORE1A seems to promote cell cycle arrest rather than apoptosis. The mechanisms underlying NORE1A action are poorly understood. We have used microarray analysis of an inducible NORE1A system to screen for physiologic signaling targets of NORE1A action. Using this approach, we have identified several potential signaling pathways modulated by NORE1A. In particular, we identify the cyclin-dependent kinase inhibitor p21CIP1 as a target for NORE1A activation and show that it is a vital component of NORE1A-mediated growth inhibition. In primary human hepatocellular carcinomas (HCC), loss of NORE1A expression is frequent and correlates tightly with loss of p21CIP1 expression. NORE1A down-regulation in HCC also correlates with poor prognosis, enhanced proliferation, survival, and angiogenic tumor characteristics. Experimental inactivation of NORE1A results in the loss of p21CIP1 expression and promotes proliferation. The best characterized activator of p21CIP1 is the p53 master tumor suppressor. Further experiments showed that NORE1A activates p21CIP1 via promoting p53 nuclear localization. Thus, we define the molecular basis of NORE1A-mediated growth inhibition and implicate NORE1A as a potential component of the ill-defined connection between Ras and p53.
Introduction
NORE1A (novel Ras effector 1 or RASSF5) is a member of the RASSF family and is ~50% identical to the relatively well characterized RASSF1A tumor suppressor (1, 2). NORE1A is frequently down-regulated by promoter methylation in human tumors, and the NORE1A locus undergoes loss of heterozygosity (LOH) in some primary tumors (1, 3). Furthermore, a translocation involving NORE1A results in the manifestation of a hereditary human cancer syndrome (4). Exogenous expression of NORE1A can promote apoptosis (5, 6) or cell cycle arrest (7). Moreover, restoration of endogenous levels of NORE1A expression to a NORE1A-negative human tumor cell line blocks the tumorigenic phenotype (5). Thus, NORE1A seems to function as a human tumor suppressor, like the related RASSF1A (1, 2). However, the mechanism of action of NORE1A is poorly understood and the role of loss of NORE1A expression in tumor development has not been characterized.
NORE1A contains a Ras Association (RA) domain and was originally identified as a Ras binding protein in a two-hybrid screen (8). It directly binds the Ras oncoprotein in a GTP-dependent manner with an affinity comparable with that of other known Ras effectors (9). Moreover, NORE1A forms an endogenous complex with Ras in cells (6). Thus, NORE1A seems to be a bona fide Ras effector, which is also a tumor suppressor.
Activated forms of Ras, while being highly transforming, can also activate proapoptotic pathways (10). NORE1A has been identified as one of the proapoptotic effectors of Ras and may function in part by binding and activating the proapoptotic kinases MST1 and MST2 (6). Thus, loss of NORE1A function may enhance the transforming capacity of activated Ras by subverting its proapoptotic functions. This concept is supported by the observation that high levels of Ras activity correlate with low levels of NORE1A expression in hepatocellular carcinoma (HCC; ref. 11).
NORE1A promotes apoptosis when overexpressed or in the presence of activated Ras, but lower levels of NORE1A expression seem to promote G1 cell cycle arrest (7). The signaling pathways involved in NORE1A function, with the exception of the MST kinases (6), are completely unknown. In an attempt to determine the mechanisms of action of NORE1A, we performed a microarray analysis of kidney cells induced to express NORE1A at levels comparable with those seen in cells that retain endogenous NORE1A expression. Several alterations in gene expression that would be compatible with the action of a tumor suppressor were detected and validated by quantitative reverse transcription–PCR (qRT-PCR). One of the most interesting was the induction of the cyclin-dependent kinase (cdk) inhibitor p21CIP1. p21CIP1 has a complicated role in the regulation of multiple signaling pathways and can promote, apparently, contradictory biological activities (12). However, overexpression of p21CIP1 induces G1 arrest (13), like NORE1A. Moreover, p21CIP1 knockout mice are tumor prone and more sensitive to tumor induction by additional genetic lesions (14). Thus, p21CIP1 is a good candidate target for a novel NORE1A signaling pathway promoting G1 arrest.
Here, we show that NORE1A overexpression up-regulates p21CIP1 and that loss of NORE1A expression causes the reduction of p21CIP1 levels. We confirm that p21CIP1 is a key mediator of the growth inhibitory properties of NORE1A. Moreover, in primary liver tumors, loss of NORE1A expression correlated tightly with loss of p21CIP1 expression and an enhanced proliferative index. Experimentally, we found that stable knockdown of NORE1A enhanced proliferation and reduced contact inhibition of a nontransformed cell line.
Perhaps, the best-characterized activator of p21CIP1 is the master tumor suppressor p53 (15). Using dominant negatives and small interfering RNA (siRNA) against p53, we show that the activation of p21CIP1 by NORE1A is p53 dependent. Although NORE1A expression did not seem to affect the protein levels of p53, it did seem to activate the nuclear translocation of p53 by an unknown mechanism. Thus, we have identified multiple proteins implicated in tumor suppression that are activated by near-physiologic levels of NORE1A and identify NORE1A as a component of p53/p21CIP1 signaling pathways.
Materials and Methods
Plasmids.
pIND/SP1-NORE1A has been described previously (4), as have pCDNAFLAG and pZIP-NORE1A (5). HA-p53 wild-type and dominant-negative (16) expression plasmids were a generous gift of J. Isaacs (MUSC). p53 siRNA was from Applied Biosystems (ID 2714). The NORE1A short hairpin RNA (shRNA) sequences used were 367:GGCTGCTCAAGAAGTTCATGGTTGTGGAC and 971:GCGACGTGAGGAGCATCTTCGAGCAGCCG. Scram: CAGAAGATCGACAGCTACAACACGCGAGA cloned in the pRS vector (Origene) have been described previously (11).
Tissue culture, transfections, and treatment.
HEK-293ecr kidney (Invitrogen), HCT116 colonic (generous gift, C. Deng, NIH), A549 non–small cell lung cancer (NSCLC), WRL68 hepatoblast, and FOCUS, HuH2, HuH6, and HuH7 HCC (American Type Culture Collection) cell lines were grown in DMEM/10% fetal bovine serum. NIH 3T3 cells were grown as described previously (17). Transient transfections were performed using Lipofectamine 2000. NORE1A silencing experiments on FOCUS and WRL68 liver cell lines were performed, as previously reported (11), and results obtained at 24 and 36 h after transfection with siRNA were analyzed. For demethylating experiments, cell lines were plated and allowed 24-h growth before addition of 10 μmol/L 5-aza-cytidine (5-Aza-C; Sigma). All experiments were repeated at least thrice per each cell line.
Microarray analysis.
HEK-293ecr cells were stably transfected with vector or pIND/SP1 NORE1A, grown in triplicate, and induced with 5 μmol/L Ponasterone A for 48 h. Cells were lysed in Trizol, and extracted RNA was subjected to microarray analysis as described in Supplementary Materials and Methods. Differentially expressed genes between control versus NORE1A-expressing cells were identified with a P < 0.05 being considered statistically significant. qRT-PCR analysis was performed as described in the Supplementary Materials and Methods.
Tissue specimens, clinicopathologic data, methylation-specific PCR, microsatellite analysis, proliferation and apoptotic indices, and evaluation of microvessel density.
Western blots and immunoprecipitation.
The following antibodies were used: anti-p21CIP1 (Novus Biologicals); mouse monoclonal anti–cyclin A, anti–cyclin E, rabbit polyclonal anti-CDK2, p53-D0–1 (Santa Cruz Biotechnology); rabbit polyclonal anti-NORE1A (5); HA and FLAG antibodies (Sigma). Secondary antibodies were from Amersham. CDK2–cyclin A and CDK2–cyclin E immunocomplexes were assessed by immunoprecipitation with the anti-CDK2 antibody and probing the membranes with either the anti–cyclin A or anti–cyclin E antibody. Blots were developed using an enhanced chemiluminescence kit from Amersham. Nuclear and cytoplasmic extracts from HuH6 and HuH7 cell lines were prepared by using the NE-PER nuclear and cytoplasmic extraction kit (Pierce Biotechnology, Inc.). Antirabbit polyclonal antibodies against glyceraldehyde-3-phosphate dehydrogenase and histone H3 (Santa Cruz Biotechnology) were used to ascertain equal loading of cytoplasmic and nuclear pools, respectively.
Results
Microarray analysis of HEK-293ecr kidney cells stably induced for NORE1A implicates p21CIP1 as a downstream effector of NORE1A.
NORE1A can induce apoptosis (5, 6) and cell cycle arrest (7). The apoptotic properties of NORE1A are activated by Ras and may be mediated via the MST1 and MST2 kinases (5, 6). However, the mechanisms underlying the effects of NORE1A on the cell cycle remain completely unknown. To address this question, we performed microarray analysis of HEK-293ecr kidney cells induced to express NORE1A. The Invitrogen pIND/SPI/293ecr system was deliberately used to avoid artifacts due to excessive protein expression generated by transient transfection studies. The pIND/SPI plasmid system is induced by an insect hormone mimic, Ponasterone A, which has minimal effects on mammalian gene transcription.
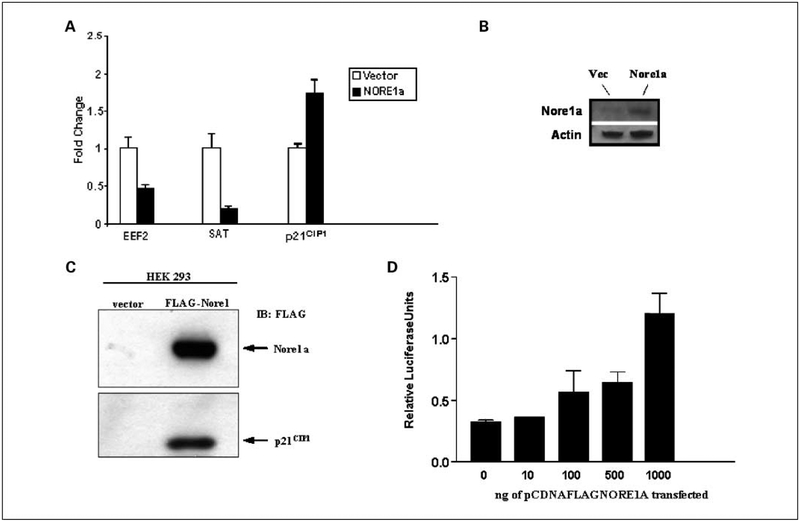
From these assays, we identified several genes that showed enhanced or reduced expression relative to the vector control (Table 1). Three of the targets that exhibited the greatest differential expression were subjected to qRT-PCR validation. EEF2 and SAT1 were confirmed as being down-regulated, whereas p21CIP1 was confirmed as being up-regulated (Fig. 1A). Induced expression of NORE1A was confirmed by Western blot analysis (Fig. 1B). NORE1A has previously been shown to induce G1 cell cycle arrest (7). As p21CIP1 has also been shown to be capable of this type of cell cycle arrest (13), we selected it for further study as a potential NORE1A signaling pathway component.
Table 1.
Summary of the most overt alterations in gene expression in 293ecr cells induced to express NORE1A compared with induced cells transfected with the empty vector
| Entrez gene ID | Gene | Description | Fold difference | Chromosomal location | Function |
|---|---|---|---|---|---|
| 1026 | CDKN1A | CDK inhibitor 1A (p21, Cip1) | 2.257365 | 6p21.2 | CDK inhibitor |
| 10981 | RAB32 | RAB32, member RAS oncogene family | 2.135154 | 6q24.3 | Participates in mitochondrial anchoring of PKA |
| 5906 | RAP1A | RAP1A, member of RAS oncogene family | 1.980178 | 1p13.3 | Interacts with RAS GAPs and RAF to counteract the mitogenic function of RAS |
| 5134 | PDCD2 | Programmed cell death 2 | 1.948819 | 6q27 | May play a role in cell death |
| 9689 | BZW1 | Basic leucine zipper and W2 domains 1 | 1.896422 | 2q33 | Enhances histone H4 gene transcription |
| 5116 | PCNT | Pericentrin | 1.883309 | 21q22.3 | Integral component of the pericentriolar material |
| 10950 | BTG3 | BTG family, member 3 | 1.881548 | 21q21.1-q21.2 | Involved in the negative regulation of cell proliferation |
| 5877 | RABIF | RAB interacting factor | 1.813152 | 1q32-q41 | Guanine nucleotide-releasing protein that acts on members of the SCE4/YPT1/RAB subfamily |
| 3301 | DNAJA1 | DnaJ (Hsp40) homologue, subfamily A, member 1 | 1.783573 | 9pl3-pl2 | May play a role in protein import into mitochondria |
| 9208 | LRRFIP1 | Leucine rich repeat (in FLII) interacting protein 1 | 1.714645 | 2q37.3 | Transcriptional repressor, may regulate expression of TNF, EGFR, and PDGFA |
| 1841 | DTYMK | Deoxythymidylate kinase (thymidylate kinase) | 1.698028 | 2q37.3 | Catalyzes the conversion of dTMP to dTDP |
| 4675 | NAP1L3 | Nucleosome assembly protein 1-like 3 | 1.676265 | Xq21.3-q22 | Involved in nucleosome assembly |
| 5707 | PSMD1 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 1.667157 | 2q37.1 | Regulatory subunit of the 26 proteasome, which is involved in the ATP-dependent degradation of ubiquitinated proteins |
| 5110 | PCMT1 | Protein L-isoaspartate (L-aspartate) O-methyltransferase | 1.627503 | 6q24-q25 | Catalyzes the methyl esterification of L-isoaspartyl and D-aspartyl residues |
| 7307 | U2AF1 | U2 small nRNA auxiliary factor 1 | 1.618806 | 21q22.3 | Plays a critical role in both constitutive and enhancer-dependent splicing |
| 2971 | GTF3A | General transcription factor IIIA | 1.615342 | 13q12.3-q13.1 | RNA polymerase III transcription factor inducing transcription of the 5S rRNA genes |
| 4192 | MDK | Midkine (neurite growth-promoting factor 2) | 1.605163 | 11p11.2 | Has heparin binding activity and growth-promoting activity |
| 23369 | PUM2 | Pumilio homologue 2 (Drosophila) | 0.756976 | 2p22-p21 | Sequence-specific RNA-binding protein that regulates translation and mRNA stability |
| 2195 | FAT1 | FAT tumor suppressor homologue 1 (Drosophila) | 0.749822 | 4q35 | May function as a cell-adhesion protein |
| 51690 | LSM7 | LSM7 homologue, U6 small nRNA associated (S. cerevisiae) | 0.74828 | 19p13.3 | Binds specifically to the 3’-terminal U-tract of U6 snRNA |
| 6209 | RPS15 | Ribosomal protein S15 | 0.733836 | 19p13.3 | component of the 40S ribosomal subunit |
| 4267 | CD99 | CD99 molecule | 0.708318 | Xp22.32; Yp11.3 | Involved in cell adhesion processes |
| 1153 | CIRBP | Cold inducible RNA binding protein | 0.700366 | 19p13.3 | May play an essential role in cold-induced suppression of cell proliferation |
| 10975 | UQCR | Ubiquinol-cytochrome c reductase, 6.4-kDa subunit | 0.663303 | 19p13.3 | May function as an iron-sulfur protein-binding factor |
| 1938 | EEF2 | Eukaryotic translation elongation factor 2 | 0.588235 | 19pter-q12 | Essential factor for protein synthesis |
| 6303 | SAT1 | Spermidine/spermine N1-acetyltransferase 1 | 0.427843 | Xp22.1 | Rate-limiting enzyme in the catabolic pathway of polyamine metabolism |
NOTE: Results are derived from three separate assays.
Figure 1.
A, qRT-PCR validation. qRT-PCR analysis was performed on the induced cells for EEF2, SAT1, and p21CIP1. Values for the vector were normalized to 1, and the fold change due to NORE1A was plotted. The results confirmed the microarray results. B, protein expression of NORE1A in the induced cells. C, NORE1A induces p21CIP1 protein expression. HEK-293 cells were transfected with pcDNAFLAG NORE1A or empty vector. After 24 h, the cells were lysed, and equal amounts of protein lysate were Western blotted for exogenous NORE1A (FLAG) and endogenous p21CIP1. D, increasing amounts of pcDNAFLAG NORE1A expression plasmid were transfected into NIH 3T3 cells with a p21-luciferase reporter and a Renilla-luciferase internal control plasmid, essentially as described previously (45).
NORE1A stimulates p21CIP1 protein expression.
To confirm that the microarray data reflected the effects of NORE1A on p21CIP1 protein expression, HEK-293ecr cells were transiently transfected with NORE1A and the levels of endogenous p21CIP1 protein measured after 24 hours. Western blotting revealed a dramatic increase in the levels of p21CIP1 protein in the NORE1A transfected cells (Fig. 1C). Further confirmation of the effects of NORE1A on p21CIP1 expression was obtained by performing luciferase assays with a p21-Luc reporter (18) and increasing amounts of NORE1A expression plasmid (Fig. 1D).
NORE1A inhibition reduces p21CIP1 expression.
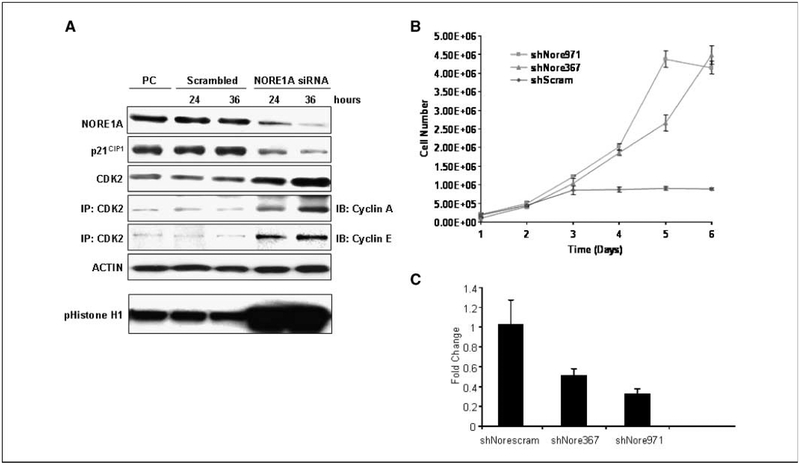
If NORE1A expression can enhance p21CIP1 levels, then inhibition of NORE1A should reduce the levels of p21CIP1 protein. The FOCUS (HCC) and WRL68 (hepatoblast) cell lines that retain NORE1A expression were transfected with siRNA against NORE1A, and the reduction of NORE1A protein expression was confirmed by Western blot analysis. The same protein samples were then analyzed for p21CIP1 expression (Fig. 2A). siRNA-mediated inhibition of NORE1A expression caused a corresponding decrease in the expression of p21CIP1. Furthermore, suppression of NORE1A resulted in upregulation of CDK2, whose activity is inhibited by p21CIP1. The CDK2 showed enhanced activation in the NORE1A knockdown cells, as measured by the presence of cyclin A and cyclin E in the complex, as well as by kinase activity toward histone H1 (Fig. 2A). These results support an enhanced G1-S transition after inactivation of NORE1A.
Figure 2.
A, inhibition of NORE1A expression leads to reduced p21CIP1 expression and increased G1-S transition markers in human liver cell lines. WRL68 and FOCUS cells were transfected with siRNA duplexes specific for human NORE1A. Results obtained from the FOCUS HCC cell line at 24 and 36 h after transfection with siRNA. Actin serves as the loading control. Equivalent results were obtained with WRL68 cells (data not shown). Experiments were repeated at least thrice per each cell line. PC, positive control (lysate from a human nontumor surrounding liver expressing NORE1A). The activity of CDK2 was measured by kinase assay of the immunoprecipitation (IP) on histone H1. B, down-regulation of NORE1A promotes modest enhanced growth and resistance to contact inhibition in NIH 3T3 cells C, down-regulation of NORE1A in the stable shRNA transfectants.
NORE1A inhibition leads to enhanced proliferation/loss of contact inhibition of NIH3T3 cells.
Several studies have examined the effects of exogenous expression of NORE1A on the suppression of cell growth/survival and the tumorigenic phenotype (1). However, the biological consequences of NORE1A loss of function have not been described experimentally. We used a nontransformed murine cell line (NIH 3T3) as a model. Two different shRNAs against murine NORE1A were used to generate stable NIH3T3 cell lines knocked down for the expression of endogenous NORE1A. A scrambled shRNA served as the control. The cells were examined for the effects on proliferation by growth curve analysis (Fig. 2B). In each case, the NORE1A shRNA-transfected cells showed modest growth enhancement but pronounced loss of contact inhibition. Confirmation of NORE1A knockdown was obtained by qRT-PCR (Fig. 2C).
Activation of p21CIP1 plays a vital role in NORE1A-mediated growth/survival inhibition.
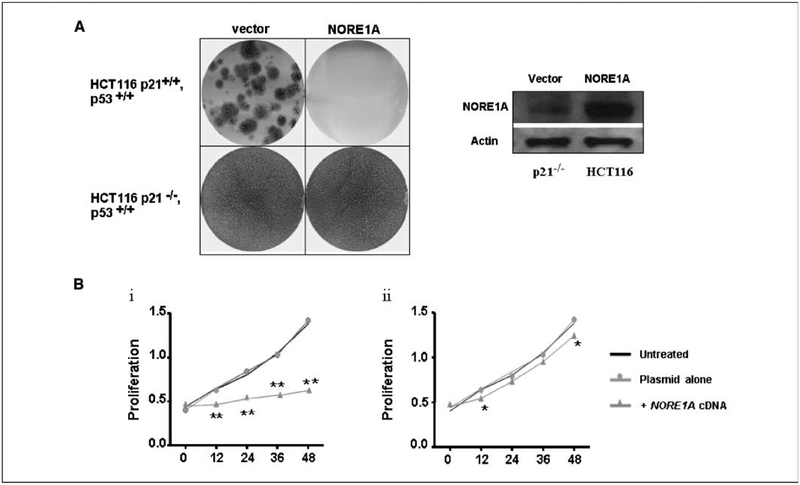
Having determined that NORE1A modulates p21CIP1, we sought to determine the importance of the p21CIP1 activation in NORE1A-mediated suppression of growth and survival. We transfected isogenic HCT116 cells that are (+) or (−) for p21CIP1 (19) while retaining wild-type p53 with pZIP-NORE1A and selected the cells in G418. Cells retaining p21CIP1 expression did not survive the transfection with NORE1A. The p21CIP1−/−cells showed multiple colony formation when transfected with NORE1A, although this was less than the cells transfected with vector (Fig. 3A). Confirmation that the p21CIP1−/−cells were expressing NORE1A protein is shown in the adjacent panel. We then transfected HuH6 HCC cells with a NORE1A expression plasmid in the absence (Fig. 3B, i) or presence (Fig. 3B, ii) of a p21CIP1 siRNA and measured the effects on proliferation. NORE1A inhibited proliferation, but this was significantly reduced in the presence of the p21CIP1 siRNA.
Figure 3.
NORE1A uses p21CIP1 to inhibit cell growth. A, a matched pair of HCT116 cells that were (+/−) for p21CIP1 were transfected with pZIP-NORE1A and selected in G418. Surviving colonies were stained with crystal violet (top). B, transfection of pCDNAF NORE1A led to marked growth restraint in the HuH6 cell line (i), which was lost when p21CIP1 was suppressed by siRNA (ii). Points, means of three experiments in triplicate. Tuckey-Kramer test, NORE1A-transfected versus control and plasmid alone; *, P < 0.05; **, P < 0.0001. Cell viability was determined by the WST-1 cell proliferation reagent (Roche Diagnostics).
Down-regulation of NORE1A expression correlates with poor prognosis and loss of p21CIP1 expression in HCC.
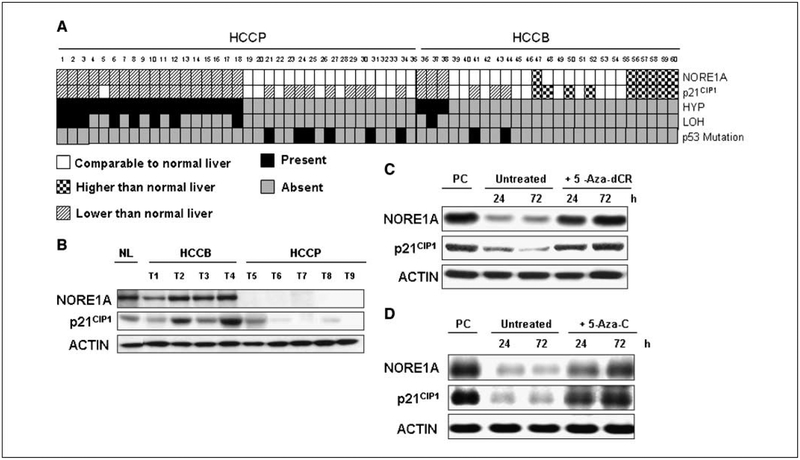
If NORE1A plays an important role in the regulation of basal p21CIP1 levels in vivo, then we might expect to see a correlation between the expression levels of NORE1A and p21CIP1 in primary tumors. We analyzed samples from a panel of HCC tumors that were classified as poor prognosis (HCCP; survival, <3 years after partial liver resection) or better prognosis (HCCB; survival, >3 years). Promoter hypermethylation of NORE1A was detected in 21 of 60 (35%) tumor samples (Fig. 4A), with no hypermethylation being found in normal and surrounding nonneoplastic livers (data not shown). Together with NORE1A promoter hypermethylation, 9 of 60 (15%) HCCs displayed LOH of the NORE1A locus. Importantly, 18 of 21 (85.7%) HCCs displaying promoter hypermethylation of NORE1A were characterized by a poor prognosis (HCCP), indicating that its down-regulation is a feature of HCC aggressiveness. Analysis of the HCCP panel for the relative levels of NORE1A and p21CIP1 protein expression showed promoter methylation of NORE1A correlated perfectly with reduced NORE1A expression (Fig. 4A). Furthermore, a significant, direct correlation was found between the levels of NORE1A and those of p21CIP1 in HCC by the Pearson’s correlation test (r2 = 0.6962, P < 0.001). These results were further confirmed by qRT-PCR in pooled samples that were +/− for NORE1A promoter methylation (Supplementary Fig. S1). In these experiments, reduction of NORE1A expression also correlated with increased EEF2 and SAT1 expression (Supplementary Fig. S1). Thus, the microarray results are again predictive of the situation in primary tumors. A representative Western blot of a panel of tumors that were NORE1A nonmethylated and of good prognosis (HCCB) compared with a panel of NORE1A-methylated tumors that were of poor prognosis (HCCP) is presented in Fig. 4B to illustrate the levels of NORE1A observed in HCCB versus HCCP.
Figure 4.
A, loss of NORE1A expression correlates with loss of p21CIP1 expression and inversely with p53 mutation in human HCC. HCC samples were examined for NORE1A promoter hypermethylation (HYP) and LOH, as well as for NORE1A and p21CIP1 expression and p53 mutation. Tumors 1 to 35 are HCCP (survival, <3 y). Tumors 36 to 60 are HCCB (survival, >3 y). Protein lysates were immunoblotted with NORE1A and p21CIP1 antibodies. In each case, Western blot analysis revealed that NORE1A methylation corresponded with reduced NORE1A expression compared with normal liver and, in every case but one, reduced p21CIP1 expression. NORE1A inactivation and p53 mutation were mutually exclusive. B, representative Western blot analysis is shown. Actin serves as a loading control. HCCB, HCC with better prognosis; HCCP, HCC with poorer prognosis; NL, normal livers. HuH2 human HCC (C) and A549 NSCLC (D) cell lines were treated with 5-Aza-C and analyzed by Western blot for the expression of NORE1A and p21CIP1. Actin serves as a loading control.
The tumor suppressor p53 is perhaps the best characterized regulator of p21CIP1 (15). If the NORE1A methylated tumors were all p53 mutant, then this might act as an alternative explanation of the reduction in p21CIP1 expression. Intriguingly, our analysis of the p53 mutation status of the tumors showed a total mutual exclusion of NORE1A promoter methylation/LOH and the mutation of p53 (Fig. 4A, bottom).
Correlation of NORE1A levels with clinicopathologic parameters of human HCC.
To further characterize the role of NORE1A in human HCC, we evaluated the proliferation and apoptotic indices and microvessel density in liver tumors with levels of NORE1A comparable or lower than normal livers, respectively. HCC with low levels of NORE1A displayed higher proliferation (35.02 ± 6.77 versus 21.87 ± 6.33; P = 1.92 ×10−6) and lower apoptotic index (1.03 ± 0.49 versus 2.24 ± 0.74; P = 8.19 ×10−8), as well as higher microvessel density (303.79 ± 58.14 versus129.65 ± 29.79; P = 3.03 ×10−15), when compared with HCC displaying NORE1A levels comparable with normal livers. This resulted in a significant shorter patient survival length for HCC with lower than normal NORE1A levels (11.26 ± 3.63 versus 51.60 ± 19.77; P = 3.93 ×10−6). Taken together, these data indicate that suppression of NORE1A is associated with increased proliferation, reduced apoptosis, and angiogenic properties of human liver tumors, leading to a rapid, adverse outcome (Supplementary Table S2).
Epigenetic therapy restores NORE1A signaling pathways.
NORE1A is down-regulated by aberrant promoter methylation, and this can be reversed by treating cells with the DNA methyltransferase inhibiting drug 5-Aza-C (5). To determine if such treatment not only restores NORE1A expression but allows reconstitution of NORE1A signaling pathways, we treated the HuH2 HCC (Fig. 4C) and the A549 NSCLC (Fig. 4D) cell lines that had suffered epigenetic inactivation of NORE1A (5)5 with 5-Aza-C. We then examined the cells by Western blot for NORE1A and p21CIP1 expression. 5-Aza-C not only increased NORE1A expression but also increased the expression of p21CIP1 in both cell lines (Fig. 4C and D).
NORE1A activates p21CIP1 via the p53 tumor suppressor.
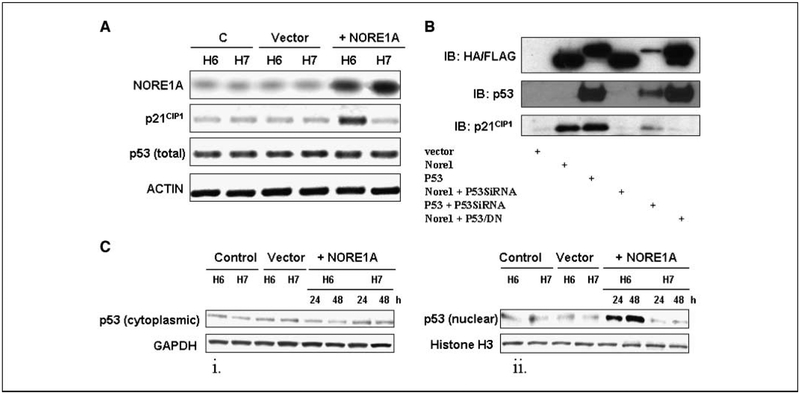
The best known activator of p21CIP1 is the tumor suppressor p53 (15). Activation of p21CIP1 by p53 has been shown to play a key role in the ability of p53 to induce cell cycle arrest (19). Consequently, we performed experiments to determine if NORE1A was acting via p53. First, we transfected two HCC cell lines, one wild type for p53 (HuH6) and one mutant for p53 (HuH7) with pCDNAFLAG NORE1A and analyzed the two cell lines for the expression of both NORE1A and p21CIP1 by Western blot. Figure 5A shows that p21CIP1 was only activated in the p53 wild-type cell line, despite the levels of transfected NORE1A expression being identical. Although the p53 mutant cell line was defective for p21CIP1 activation, we did not observe any apparent change in the levels of p53 protein (second to bottom panel). To confirm a role for p53 in the action of NORE1A, we transfected HEK-293ecr cells with NORE1A in the presence of a p53 dominant-negative (generous gift from J. Isaacs, MUSC) or p53 siRNA. Both inhibited the ability of NORE1A to stimulate p21CIP1 protein expression (Fig. 5B).
Figure 5.
NORE1A activates p21CIP1 protein expression via p53. A, HuH6 (p53 wild type; H6) and HuH7 (p53 mutant; H7) human HCC cell lines were transfected with pCDNAFLAG NORE1A, and the expression of NORE1A and p21CIP1 protein was determined by Western blot. NORE1A induced p21CIP1 only in the p53 wild-type cell line, whereas the total levels of p53 did not change in both cell lines. B, Hek-293 cells were transfected with NORE1A in the presence or absence of p53siRNA or a p53 dominant-negative construct. An HA-tagged p53 expression vector serves as a positive control. siRNA to p53 or the p53 dominant-negative inhibited NORE1A induced p21CIP1 expression. C, NORE1A induces nuclear translocation of wild-type p53. HuH6 (p53 wild type; H6) and HuH7 (p53 mutant; H7) HCC cell lines were transiently transfected with pCDNAFLAG NORE1A and fractionated into cytoplasmic (i) and nuclear (ii) fractions before Western blotting for p53. NORE1A had no effect on p53 levels in HuH7 cells, but it promoted progressive nuclear accumulation of p53 in HuH6 cells.
Examination of the levels of p53 in NORE1A transfected cells failed to show any obvious change (Fig. 5A). However, when we fractionated NORE1A-transfected HuH6 and HuH7cells, we found that the levels of endogenous p53 in the nucleus were increased (Fig. 5C, i and ii) in the HuH6 cells (containing wild-type p53) but not in the HuH7 cells (containing mutant p53). Nuclear translocation of p53 is indicative of activation (20). Thus, NORE1A seems to act via promoting the nuclear localization of p53.
Discussion
NORE1A was originally identified as a Ras effector protein and was subsequently shown to mediate some of the proapoptotic functions of Ras (5, 6). NORE1A has no apparent enzymatic activity and is hypothesized to function as a scaffolding molecule, as it has been shown to bind the MST kinases and recruit them to the plasma membrane in a complex with activated Ras (6). In the absence of activated Ras, NORE1A expression can promote G1 cell cycle arrest (7), but the mechanism of this action is not known. Thus, NORE1A can modulate apoptosis or cell cycle arrest, and the balance of these forces may be shifted toward apoptosis by the interaction with Ras.
Several lines of evidence suggest that in addition to serving as a Ras effector, NORE1A is also a tumor suppressor. Firstly, NORE1A expression is frequently impaired in a variety of tumors, particularly kidney, lung, and liver tumors (1). Secondly, restoration of NORE1A expression to normal levels in a NORE1A-negative tumor cell line blocks the ability of the cells to grow in soft agar (5). Thirdly, loss of NORE1A function is implicated in the development of a human hereditary cancer syndrome (4). Thus, NORE1A is strongly implicated as an important human tumor suppressor with a poorly understood mode of action.
To gain a better understanding of the function of NORE1A, we performed a microarray analysis to identify transcriptional alterations associated with the expression of NORE1A. In an attempt to maximize physiologic relevance, we used a kidney cell system stably transfected with an inducible form of NORE1A. Kidney tumors are the most frequently associated with NORE1A down-regulation (1), and the inducible system allowed us to control the expression levels of NORE1A, avoiding potential artifacts due to massive overexpression.
A series of transcriptional alterations due to NORE1A induction were observed. Three genes that showed some of the strongest changes in the microarray assay were subjected to quantification by qRT-PCR. These experiments confirmed that NORE1A expression induced decreases in EEF2 and SAT1 and an increase in p21CIP1 expression. Further analysis showed that in primary HCC, NORE1A expression correlated with p21CIP1 and inversely correlated with EEF2 and SAT1 expression (Supplementary Fig. S1).
EEF2 is a translation factor that mediates ribosomal translocation during peptide chain elongation and is activated by mitogenic stimuli (21). EEF2 is overexpressed in many tumors and seems to play an important role in rendering tumor cells resistant to the translation suppressing effects of hypoxia (22). Resistance to hypoxia is believed to play a critical role in the development of many tumors (23).
SAT1 is a spermidine kinase that plays a key role in the regulation of the intracellular levels of polyamines (24). Polyamines play an important role in neoplastic growth, and polyamine synthesis inhibitors are of interest as chemopreventive agents (25). Activated K-Ras, like NORE1A, inhibits SAT1 expression (25). Moreover, the related SAT2 has been implicated in the regulation of HIF-1α, a key component of the cellular response to hypoxia and an important component of the development of kidney cancer and HCC progression (26).
Previously, microarray analysis has been performed to determine the signaling profile of RASSF1A (27). RASSF1A is 50% identical to NORE1A, but the two proteins seem to promote quite different alterations in gene expression. SAT1 was the only target identified by both RASSF1A and NORE1A. This confirms that the functions of NORE1A and RASSF1A, while overlapping, are likely to be quite distinct.
Several other up-regulated targets identified in the array have also been associated with cell death and growth suppression. Among them, BTG3 is a putative tumor suppressor and target of p53 itself (28), and PDCD2 has been implicated in apoptosis (29) and proliferation control (30). Moreover, RAB32 down-regulation has been associated with colon cancer (31). Thus, NORE1A promotes a number of alterations in transcription that might be expected to repress transformation. Further work will be required to validate these targets and assess their role in NORE1A signaling.
The original purpose of initiating these experiments was to attempt to understand the basis of the nonapoptotic growth inhibition we had observed with relatively low levels of NORE1A expression. Consequently, the most interesting effect of NORE1A that we detected was the activation of transcription of the p21CIP1 cdk inhibitor.
Cell homeostasis is normally maintained by a complex, coordinated network of signaling pathways that balances proliferation, growth arrest, differentiation, and cell death. Errors in this balance can lead to the development of neoplasia. Correct regulation of the cell cycle is a key component of maintaining the homeostatic balance. Cell cycle regulation is mediated by cdks, and cdks are regulated, in part, by cdk inhibitors, such as p21CIP1 (32). p21CIP1 controls cell cycle progression through G1-S at several levels (33).
p21CIP1 has also been shown to induce resistance to apoptotic stimuli, such as DNA damage. The mechanism of this effect may be to delay the cell in G1 while DNA repair is affected. However, the role of p21CIP1 in the modulation of apoptosis is complex, and under some circumstances, p21CIP1 may exhibit proapoptotic functions (34, 35). p21CIP1 has also been identified as playing a key role in the induction of terminal differentiation and senescence (32, 36). Thus, p21CIP1 plays a complex and, at times, apparently contradictory role in modulating cellular behavior and survival. However, on the whole, loss of p21CIP1 expression seems to promote transformation. In primary human tumors, loss of p21CIP1 protein expression has been correlated with poor prognosis (37). Moreover, the inactivation of p21CIP1 in transgenic mice is, eventually, tumorigenic and enhances the transforming effects of other genetic lesions, including activated Ras (14, 38, 39). Therefore, p21CIP1 seems to serve as a tumor suppressor.
The ability of NORE1A to activate p21CIP1 explains the ability of NORE1A to induce G1 cell cycle arrest (7), as p21CIP1 blocks cell cycle at G1 by inhibiting cdk2 (32). Conversely, loss of NORE1A expression reduces p21CIP1 expression and enhances cdk2 activity. We also observed an increase in cdk2 protein levels, although this target was not identified in the microarray. Both effects could contribute to a large increase in kinase activity of the cdk2 kinase complex precipitated from the knockdown cells. The importance of p21CIP1 activation to NORE1A-mediated growth inhibition was confirmed by our observation that loss of p21CIP1 from the cell system impaired the growth/survival inhibitory effects of NORE1A.
NORE1A expression is often down-regulated in human tumors (1). Our siRNA experiments in cells lines suggested that cells require NORE1A for normal p21CIP1 expression. Examination of a panel of primary HCC showed that loss of NORE1A expression correlated closely with down-regulation of p21CIP1 expression. Preliminary data from our group show a similar pattern in a small collection of human renal, lung, and colorectal carcinomas (data not shown). These findings suggest a physiologic link between NORE1A and p21CIP1 in tumors. Loss of NORE1A and p21CIP1 expression also strongly correlated with poorer prognosis of HCC, and this may identify NORE1A as a good target for epigenetic therapy. Experimental restoration of NORE1A expression by demethylating drugs was successful in increasing p21CIP1 expression. This confirms the ability of these drugs to restore pathway function, not just NORE1A expression.
As the p53 tumor suppressor is the best characterized regulator p21CIP1 (40, 41), we examined the role of p53 in NORE1A mediated p21CIP1 activation. Dominant-negative and siRNA reagents showed that p53 was necessary for NORE1A to activate p21CIP1. Moreover, NORE1A could only activate p21CIP1 in a wild type p53-containing tumor cell line. p53 seems to be the most important tumor suppressor yet identified in human cancer with over 50% of all primary tumors showing defective p53 function (42). Thus, NORE1A participates in the modulation of what may be considered the major human tumor suppressor pathway. This conclusion is supported by the observation that, in primary human HCC s, the mutation of p53 and the inactivation of NORE1A are mutually exclusive.
The mechanism by which NORE1A can modulate p53 remains unknown. We have not detected NORE1A in complex with p53 nor have we observed changes in p53 protein levels due to NORE1A. However, we did observe an increase in the fraction of p53 present the nucleus in NORE1A-transfected cells. This suggests to us that NORE1A is promoting the nuclear localization of p53 via some posttranslational modification, such as phosphorylation or acetylation (43). We are currently examining which, if any, modifications p53 are affected by NORE1A.
Although NORE1A is frequently down-regulated in primary human tumors, the biological effects of inhibiting NORE1A function have not been determined. To identify the effects of NORE1A loss function on nontransformed p53 wild-type cells, we transfected NIH 3T3 cells with two different NORE1A shRNAs to make stable knockdown cell lines. The cells showed an enhanced rate of growth, but also a noticeable reduction in contact inhibition, and continued proliferate after the scrambled shRNA transfected cells had arrested. We have recently shown that defects in p53 function can lead to loss of contact inhibition in NIH 3T3 cells (44), so inactivation of NORE1A may have a similar effect via p53.
Thus, we identify a novel signaling pathway for NORE1A, connecting it to p53 and hence to the p21CIP1 cdk inhibitor. This may help explain the frequency with which NORE1A is inactivated in primary human tumors and why NORE1A and p53 inactivation is mutually exclusive in primary liver tumors. As NORE1A is a Ras effector, the data suggest that it could play a role in the well known but poorly understood connection between Ras and p53.
Supplementary Material
Acknowledgments
Grant support: NIH grant RR18733 (G.J. Clark) and National Cancer Institute intramural funds (G.J. Clark and M.J. Birrer).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
D.F. Calvisi, unpublished observation.
References
- 1.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci 2007;120:3163–72. [DOI] [PubMed] [Google Scholar]
- 2.Agathanggelou A, Cooper WN, Latif F. Role of the Rasassociation domain family 1 tumor suppressor gene in human cancers. Cancer Res 2005;65:3497–508. [DOI] [PubMed] [Google Scholar]
- 3.Steiner G, Cairns P, Polascik TJ, et al. High-density mapping of chromosomal arm 1q in renal collecting duct carcinoma: region of minimal deletion at 1q32.1–32.2. Cancer Res 1996;56:5044–6. [PubMed] [Google Scholar]
- 4.Chen J, Lui WO, Vos MD, et al. The t(1;3) breakpointspanning genes LSAMP and NORE1 are involved in clear cell renal cell carcinomas. Cancer Cell 2003;4:405–13. [DOI] [PubMed] [Google Scholar]
- 5.Vos MD, Martinez A, Ellis CA, Vallecorsa T, Clark GJ. The pro-apoptotic Ras effector Nore1 may serve as a Ras-regulated tumor suppressor in the lung. J Biol Chem 2003;278:21938–43. [DOI] [PubMed] [Google Scholar]
- 6.Khokhlatchev A, Rabizadeh S, Xavier R, et al. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol 2002;12:253–65. [DOI] [PubMed] [Google Scholar]
- 7.Aoyama Y, Avruch J, Zhang XF. Nore1 inhibits tumor cell growth independent of Ras or the MST1/2 kinases. Oncogene 2004;23:3426–33. [DOI] [PubMed] [Google Scholar]
- 8.Vavvas D, Li X, Avruch J, Zhang XF. Identification of Nore1 as a potential Ras effector. J Biol Chem 1998;273: 5439–42. [DOI] [PubMed] [Google Scholar]
- 9.Wohlgemuth S, Kiel C, Kramer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains: I. Biochemical analysis. J Mol Biol 2005;348:741–58. [DOI] [PubMed] [Google Scholar]
- 10.Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene 2003;22:8999–9006. [DOI] [PubMed] [Google Scholar]
- 11.Calvisi DF, Ladu S, Gorden A, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 2006;130:1117–28. [DOI] [PubMed] [Google Scholar]
- 12.Gartel AL, Tyner AL. The growth-regulatory role of p21 (WAF1/CIP1). Prog Mol Subcell Biol 1998;20:43–71. [DOI] [PubMed] [Google Scholar]
- 13.Gartel AL, Serfas MS, Tyner AL. p21-negative regulator of the cell cycle. Proc Soc Exp Biol Med 1996;213:138–49. [DOI] [PubMed] [Google Scholar]
- 14.Gartel AL. P21(WAF1/CIP1) may be a tumor suppressor after all. Cancer Biol Ther 2007;8:1171–2. [DOI] [PubMed] [Google Scholar]
- 15.El-Deiry WS, Tokino T, Velculescu A, et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75:817–25. [DOI] [PubMed] [Google Scholar]
- 16.Zacharatos PV, Gorgoulis VG, Kotsinas A, et al. Modulation of wild-type p53 activity by mutant p53 R273H depends on the p53 responsive element (p53RE). A comparative study between the p53REs of the MDM2, WAFI/Cip1 and Bax genes in the lung cancer environment. Anticancer Res 1999;1A:579–87. [PubMed] [Google Scholar]
- 17.Clark GJ, Cox AD, Graham SM, Der CJ. Biological assays for Ras transformation In: Balch W, Der CJ, editors. Methods in Enzymology. San Diego: Academic Press; 1995. Vol. 255. p. 395–412. [DOI] [PubMed] [Google Scholar]
- 18.Datto MB, Yu Y, Wang XF. Functional analysis of the transforming growth factor β responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem 1995;270: 28623–8. [DOI] [PubMed] [Google Scholar]
- 19.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res 1995;55:5187–90. [PubMed] [Google Scholar]
- 20.Beham A, Marin MC, Fernandez A, et al. Bcl-2 inhibits p53 nuclear import following DNA damage. Oncogene 1997;15:2767–72. [DOI] [PubMed] [Google Scholar]
- 21.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem 2004;279:12220–31. [DOI] [PubMed] [Google Scholar]
- 22.Connolly E, Braunstein S, Formenti S, Schneider RJ. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol Cell Biol 2006;10:3955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XF, O’Donoghue JA. Hypoxia in microscopic tumors. Cancer Lett 2008;264:172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pegg AE. Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator. Am J Physiol Endocrinol Metab 2008;294:995–1010. [DOI] [PubMed] [Google Scholar]
- 25.Gerner EW, Meyskens FL Jr. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer 2004; 10:781–92. [DOI] [PubMed] [Google Scholar]
- 26.Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest 2007;117:2713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agathanggelou A, Bieche I, Ahmed-Choudhury J, et al. Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma. Cancer Res 2003;63: 5344–51. [PMC free article] [PubMed] [Google Scholar]
- 28.Ou YH, Chung PH, Hsu FF, Sun TP, Chang WY, Shieh SY. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J 2007;26:3968–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron BW, Zeleznik-Le N, Baron MJ, et al. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of human B and T cell lymphomas. Proc Natl Acad Sci U S A 2007;104:7449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minakhina S, Druzhinina M, Steward R. Zfrp8, the Drosophila ortholog of PDCD2, functions in lymph gland development and controls cell proliferation. Development 2007;134:2387–96. [DOI] [PubMed] [Google Scholar]
- 31.Shibata D, Mori Y, Cai K, et al. RAB32 hypermethylation and microsatellite instability in gastric and endometrial adenocarcinomas. Int J Cancer 2006;119: 801–6. [DOI] [PubMed] [Google Scholar]
- 32.Weinberg WC, Denning MF. P21Waf1 control of epithelial cell cycle and cell fate. Crit Rev Oral Biol Med 2002;13:453–64. [DOI] [PubMed] [Google Scholar]
- 33.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999;13:1501–12. [DOI] [PubMed] [Google Scholar]
- 34.Gartel AL. The conflicting roles of the cdk inhibitor p21(CIP1/WAF1) in apoptosis. Leuk Res 2005;29:1237–8. [DOI] [PubMed] [Google Scholar]
- 35.Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther 2002; 1:639–49. [PubMed] [Google Scholar]
- 36.Dotto GP. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta 2000;1471:M43–56. [DOI] [PubMed] [Google Scholar]
- 37.Costa MJ, Hansen CL, Walls JE, Scudder SA. Immunohistochemical markers of cell cycle control applied to ovarian and primary peritoneal surface epithelial neoplasms: p21(WAF1/CIP1) predicts survival and good response to platinin-based chemotherapy. Hum Pathol 1999;30:640–7. [DOI] [PubMed] [Google Scholar]
- 38.Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci U S A 2006;103:19842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adnane J, Jackson RJ, Nicosia SV, Cantor AB, Pledger WJ, Sebti SM. Loss of p21WAF1/CIP1 accelerates Ras oncogenesis in a transgenic/knockout mammary cancer model. Oncogene 2000;19:5338–47. [DOI] [PubMed] [Google Scholar]
- 40.Kuribayashi K, El-Deiry WS. Regulation of programmed cell death by the p53 pathway. Adv Exp Med Biol 2008;615:201–21. [DOI] [PubMed] [Google Scholar]
- 41.El-Deiry WS. p21/p53, cellular growth control and genomic integrity. Curr Top Microbiol Immunol 1998; 227:121–37. [DOI] [PubMed] [Google Scholar]
- 42.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res 2000;60:6788–93. [PubMed] [Google Scholar]
- 43.Anderson CW, Appella E, Sakaguchi K. Posttranslational modifications involved in the DNA damage response. J Protein Chem 1998;17:527. [PubMed] [Google Scholar]
- 44.Donninger H, Binder A, Bohm L, Parker mitotic index (MI). Differential effects of novel tumour-derived p53 mutations on the transformation of NIH-3T3 cells. Biol Chem 2008;389:57–67. [DOI] [PubMed] [Google Scholar]
- 45.Ellis CA, Vos MD, Howell H, Vallecorsa T, Fults DW, Clark GJ. Rig is a novel Ras-related protein and potential neural tumor suppressor. Proc Natl Acad Sci U S A 2002; 99:9876–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.